Integration of Next Generation Sequencing Data to Inform Survival Prediction of Patients with Spine Metastasis
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sequencing Analysis
2.3. Machine Learning Model
2.4. Statistical Analysis
3. Results
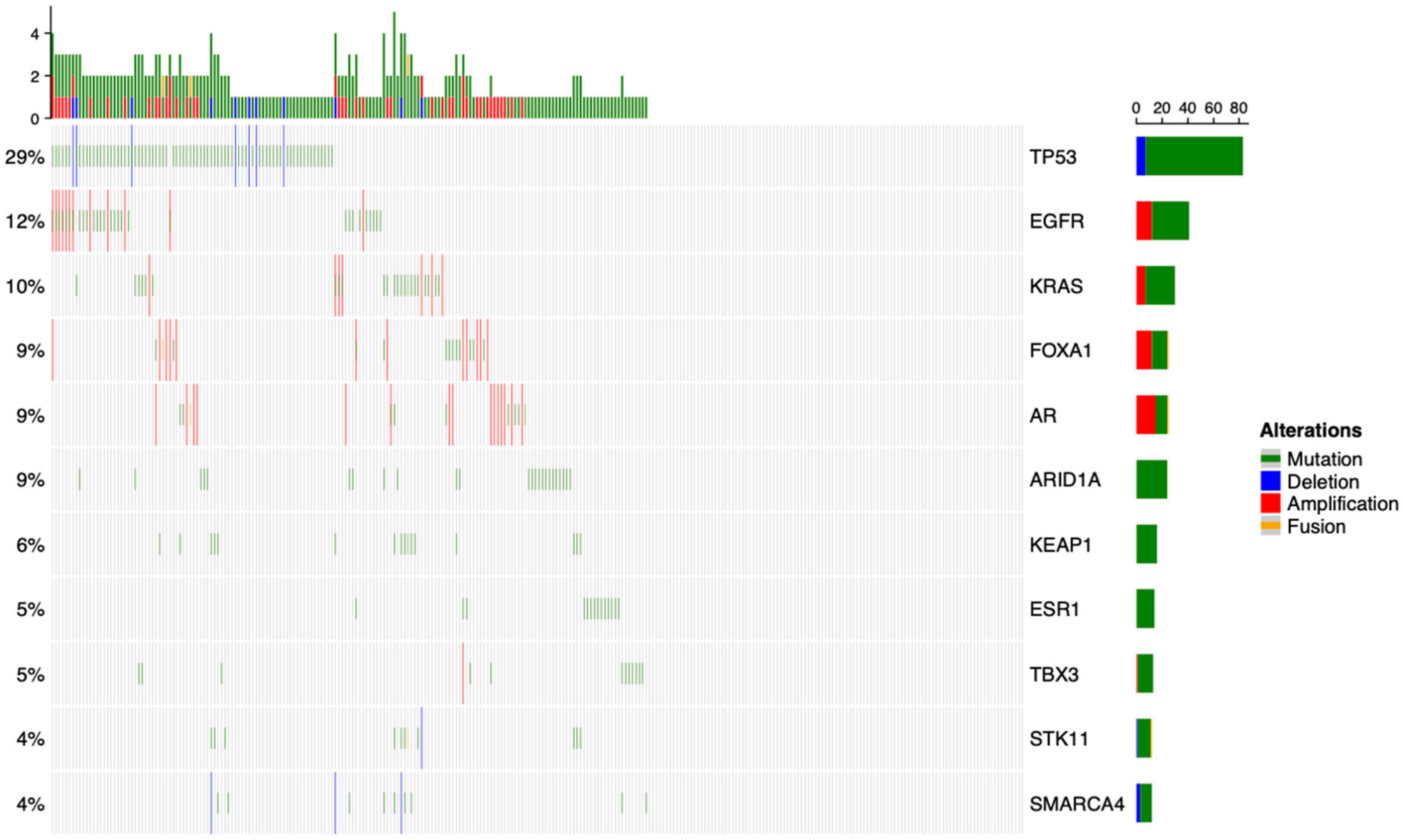
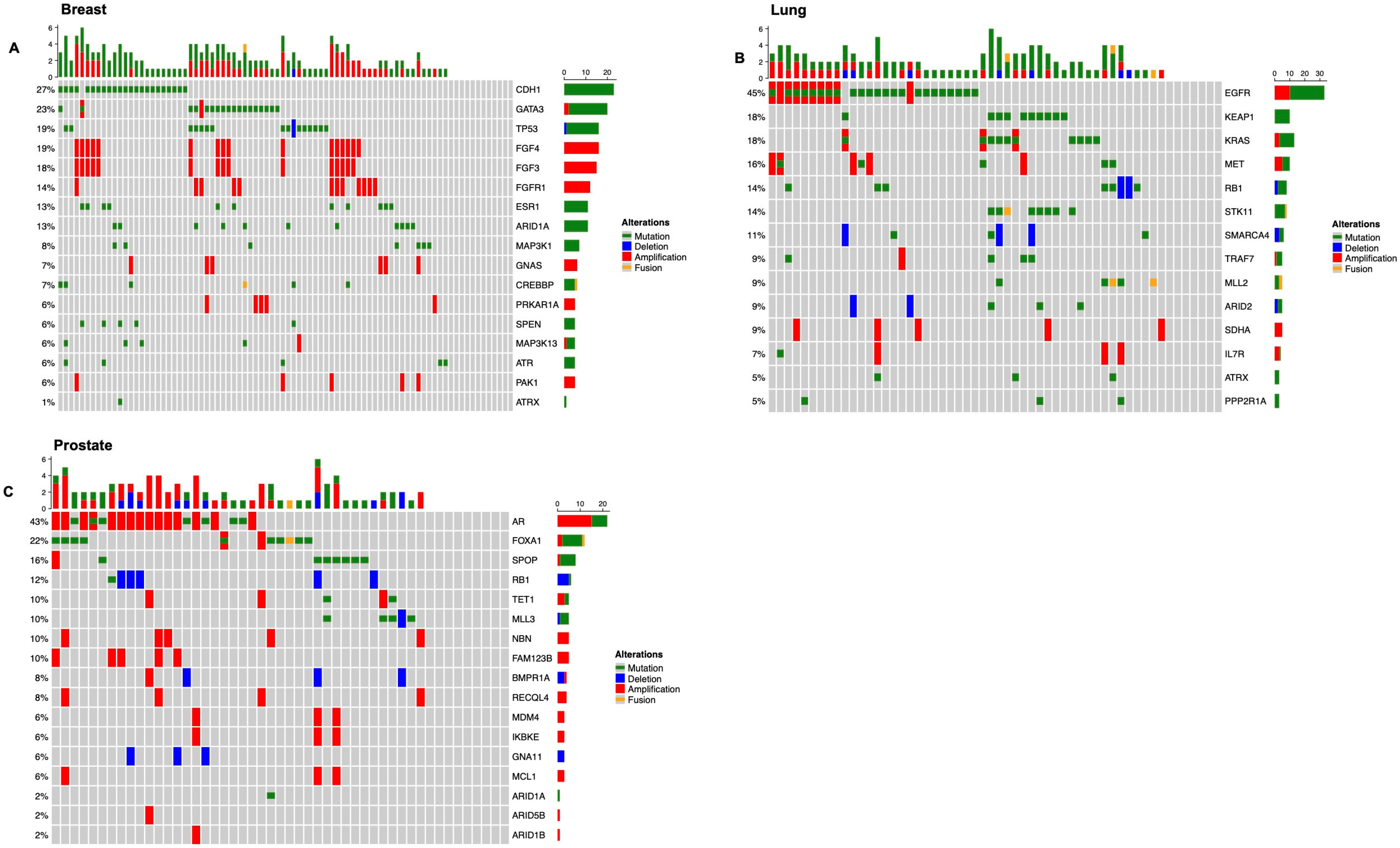
3.1. Overview of the Genomic Landscape for Spinal Metastases
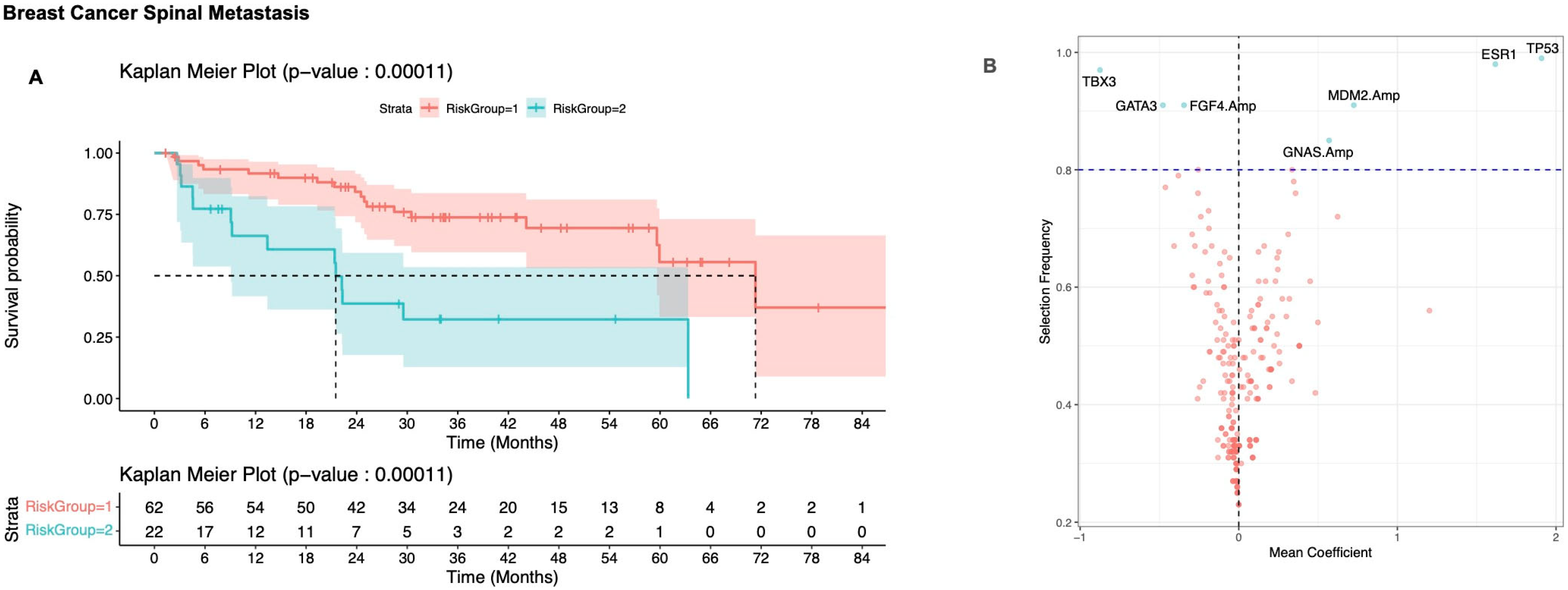
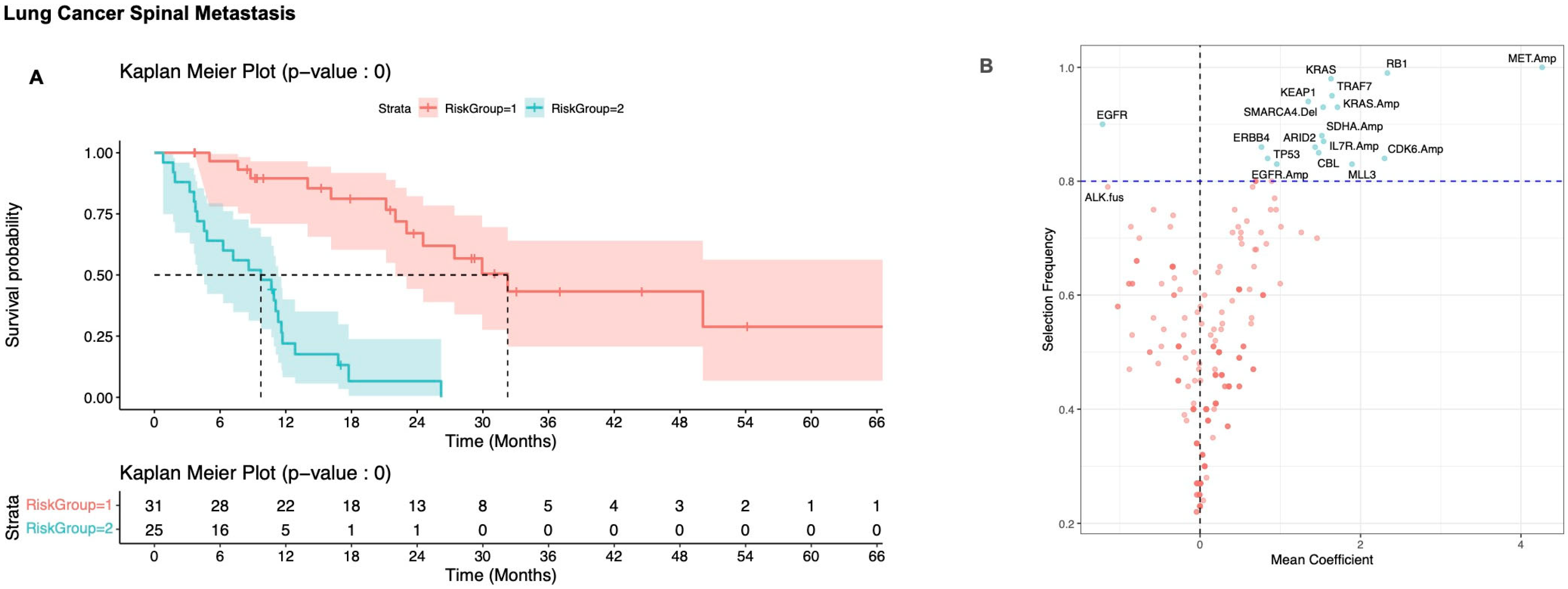
3.2. Defining Risk Subgroups
3.2.1. Breast Cancer
3.2.2. Non-Small Cell Lung Carcinoma
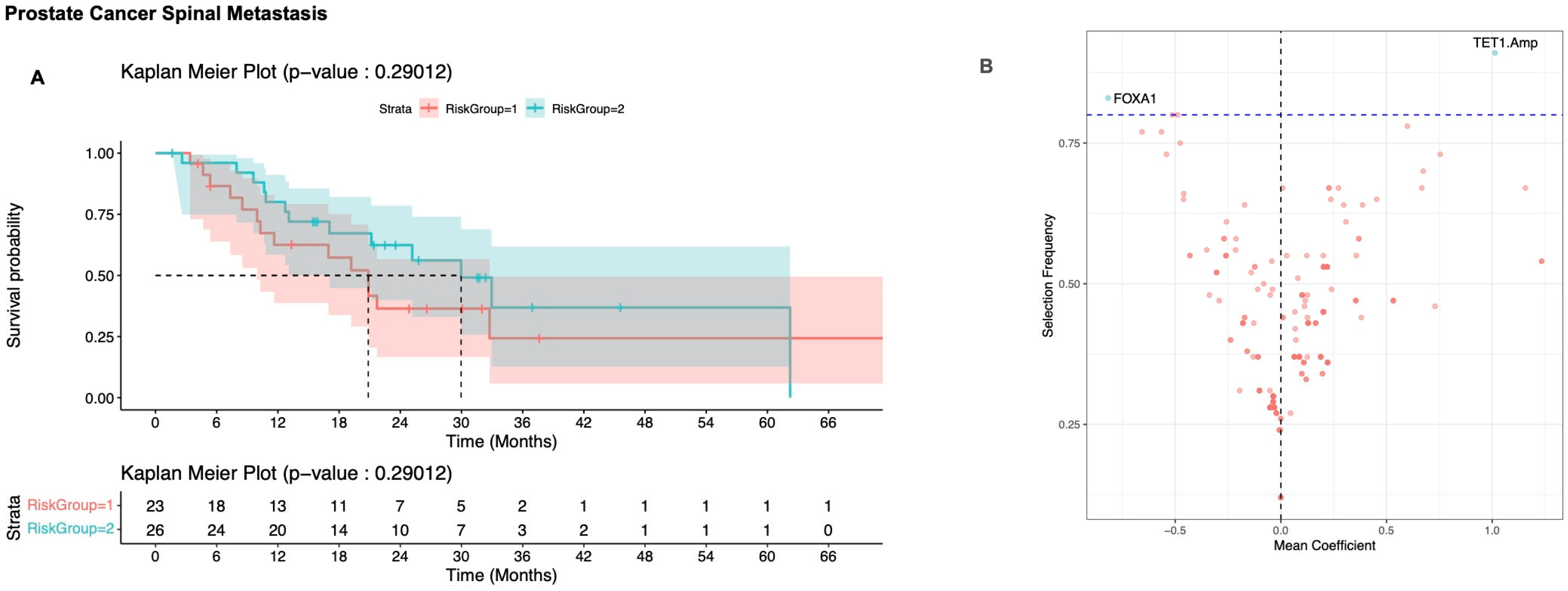
3.2.3. Prostate Cancer
4. Discussion
4.1. Key Findings and Significance
4.2. Clinical Implications
4.3. Histology-Specific Observations
4.4. Limitations
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| ML | Machine learning |
| MSK-IMPACT | Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets |
| NSCLC | Non-small cell lung cancer |
| OS | Overall survival |
References
- Barzilai, O.; Laufer, I.; Yamada, Y.; Higginson, D.S.; Schmitt, A.M.; Lis, E.; Bilsky, M.H. Integrating Evidence-Based Medicine for Treatment of Spinal Metastases Into a Decision Framework: Neurologic, Oncologic, Mechanicals Stability, and Systemic Disease. J. Clin. Oncol. 2017, 35, 2419–2427. [Google Scholar] [CrossRef]
- Spratt, D.E.; Beeler, W.H.; de Moraes, F.Y.; Rhines, L.D.; Gemmete, J.J.; Chaudhary, N.; Shultz, D.B.; Smith, S.R.; Berlin, A.; Dahele, M.; et al. An integrated multidisciplinary algorithm for the management of spinal metastases: An International Spine Oncology Consortium report. Lancet Oncol. 2017, 18, e720–e730. [Google Scholar] [CrossRef]
- Laufer, I.; Rubin, D.G.; Lis, E.; Cox, B.W.; Stubblefield, M.D.; Yamada, Y.; Bilsky, M.H. The NOMS framework: Approach to the treatment of spinal metastatic tumors. Oncologist 2013, 18, 744–751. [Google Scholar] [CrossRef]
- Barzilai, O.; Fisher, C.G.; Bilsky, M.H. State of the Art Treatment of Spinal Metastatic Disease. Neurosurgery 2018, 82, 757–769. [Google Scholar] [CrossRef] [PubMed]
- Rothrock, R.J.; Barzilai, O.; Reiner, A.S.; Lis, E.; Schmitt, A.M.; Higginson, D.S.; Yamada, Y.; Bilsky, M.H.; Laufer, I. Survival Trends After Surgery for Spinal Metastatic Tumors: 20-Year Cancer Center Experience. Neurosurgery 2021, 88, 402–412. [Google Scholar] [CrossRef] [PubMed]
- Leithner, A.; Radl, R.; Gruber, G.; Hochegger, M.; Leithner, K.; Welkerling, H.; Rehak, P.; Windhager, R. Predictive value of seven preoperative prognostic scoring systems for spinal metastases. Eur. Spine J. 2008, 17, 1488–1495. [Google Scholar] [CrossRef] [PubMed]
- Tokuhashi, Y.; Ajiro, Y.; Umezawa, N. Outcome of treatment for spinal metastases using scoring system for preoperative evaluation of prognosis. Spine 2009, 34, 69–73. [Google Scholar] [CrossRef]
- Zubair, T.; Bandyopadhyay, D. Small Molecule EGFR Inhibitors as Anti-Cancer Agents: Discovery, Mechanisms of Action, and Opportunities. Int. J. Mol. Sci. 2023, 24, 2651. [Google Scholar] [CrossRef]
- Klein, M.E.; Kovatcheva, M.; Davis, L.E.; Tap, W.D.; Koff, A. CDK4/6 Inhibitors: The Mechanism of Action May Not Be as Simple as Once Thought. Cancer Cell 2018, 34, 9–20. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The repertoire of mutational signatures in human cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Robinson, D.R.; Wu, Y.M.; Lonigro, R.J.; Vats, P.; Cobain, E.; Everett, J.; Cao, X.; Rabban, E.; Kumar-Sinha, C.; Raymond, V.; et al. Integrative clinical genomics of metastatic cancer. Nature 2017, 548, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Zehir, A.; Benayed, R.; Shah, R.H.; Syed, A.; Middha, S.; Kim, H.R.; Srinivasan, P.; Gao, J.; Chakravarty, D.; Devlin, S.M.; et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat. Med. 2017, 23, 703–713. [Google Scholar] [CrossRef]
- Cheng, D.T.; Mitchell, T.N.; Zehir, A.; Shah, R.H.; Benayed, R.; Syed, A.; Chandramohan, R.; Liu, Z.Y.; Won, H.H.; Scott, S.N.; et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. J. Mol. Diagn. 2015, 17, 251–264. [Google Scholar] [CrossRef] [PubMed]
- Shen, R.; Martin, A.; Ni, A.; Hellmann, M.; Arbour, K.C.; Jordan, E.; Arora, A.; Ptashkin, R.; Zehir, A.; Kris, M.G.; et al. Harnessing Clinical Sequencing Data for Survival Stratification of Patients with Metastatic Lung Adenocarcinomas. JCO Precis Oncol. 2019, 3. [Google Scholar] [CrossRef]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res. 2023, 83, 3861–3867. [Google Scholar] [CrossRef]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef]
- Brown, S.; Lavery, J.A.; Shen, R.; Martin, A.S.; Kehl, K.L.; Sweeney, S.M.; Lepisto, E.M.; Rizvi, H.; McCarthy, C.G.; Schultz, N.; et al. Implications of Selection Bias Due to Delayed Study Entry in Clinical Genomic Studies. JAMA Oncol. 2022, 8, 287–291. [Google Scholar] [CrossRef]
- Chakravarty, D.; Gao, J.; Phillips, S.M.; Kundra, R.; Zhang, H.; Wang, J.; Rudolph, J.E.; Yaeger, R.; Soumerai, T.; Nissan, M.H.; et al. OncoKB: A Precision Oncology Knowledge Base. JCO Precis Oncol. 2017, 2017. [Google Scholar] [CrossRef]
- Cheng, D.T.; Prasad, M.; Chekaluk, Y.; Benayed, R.; Sadowska, J.; Zehir, A.; Syed, A.; Wang, Y.E.; Somar, J.; Li, Y.; et al. Comprehensive detection of germline variants by MSK-IMPACT, a clinical diagnostic platform for solid tumor molecular oncology and concurrent cancer predisposition testing. BMC Med. Genom. 2017, 10, 33. [Google Scholar] [CrossRef]
- Barzilai, O.; Martin, A.; Reiner, A.S.; Laufer, I.; Schmitt, A.; Bilsky, M.H. Clinical reliability of genomic data obtained from spinal metastatic tumor samples. Neuro Oncol. 2022, 24, 1090–1100. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; Kawahara, N.; Kobayashi, T.; Yoshida, A.; Murakami, H.; Akamaru, T. Surgical strategy for spinal metastases. Spine 2001, 26, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Tokuhashi, Y.; Matsuzaki, H.; Oda, H.; Oshima, M.; Ryu, J. A revised scoring system for preoperative evaluation of metastatic spine tumor prognosis. Spine 2005, 30, 2186–2191. [Google Scholar] [CrossRef] [PubMed]
- Bauer, H.C.; Wedin, R. Survival after surgery for spinal and extremity metastases. Prognostication in 241 patients. Acta Orthop. Scand. 1995, 66, 143–146. [Google Scholar] [CrossRef]
- Paulino Pereira, N.R.; McLaughlin, L.; Janssen, S.J.; van Dijk, C.N.; Bramer, J.A.M.; Laufer, I.; Bilsky, M.H.; Schwab, J.H. The SORG nomogram accurately predicts 3- and 12-months survival for operable spine metastatic disease: External validation. J. Surg. Oncol. 2017, 115, 1019–1027. [Google Scholar] [CrossRef]
- Conti, A.; Acker, G.; Kluge, A.; Loebel, F.; Kreimeier, A.; Budach, V.; Vajkoczy, P.; Ghetti, I.; Germano, A.F.; Senger, C. Decision Making in Patients with Metastatic Spine. The Role of Minimally Invasive Treatment Modalities. Front. Oncol. 2019, 9, 915. [Google Scholar] [CrossRef]
- Majeed, H.; Kumar, S.; Bommireddy, R.; Klezl, Z.; Calthorpe, D. Accuracy of prognostic scores in decision making and predicting outcomes in metastatic spine disease. Ann. R. Coll. Surg. Engl. 2012, 94, 28–33. [Google Scholar] [CrossRef]
- Shiovitz, S.; Korde, L.A. Genetics of breast cancer: A topic in evolution. Ann. Oncol. 2015, 26, 1291–1299. [Google Scholar] [CrossRef]
- Chandarlapaty, S.; Chen, D.; He, W.; Sung, P.; Samoila, A.; You, D.; Bhatt, T.; Patel, P.; Voi, M.; Gnant, M.; et al. Prevalence of ESR1 Mutations in Cell-Free DNA and Outcomes in Metastatic Breast Cancer: A Secondary Analysis of the BOLERO-2 Clinical Trial. JAMA Oncol. 2016, 2, 1310–1315. [Google Scholar] [CrossRef]
- Toy, W.; Shen, Y.; Won, H.; Green, B.; Sakr, R.A.; Will, M.; Li, Z.; Gala, K.; Fanning, S.; King, T.A.; et al. ESR1 ligand-binding domain mutations in hormone-resistant breast cancer. Nat. Genet. 2013, 45, 1439–1445. [Google Scholar] [CrossRef]
- Toy, W.; Weir, H.; Razavi, P.; Lawson, M.; Goeppert, A.U.; Mazzola, A.M.; Smith, A.; Wilson, J.; Morrow, C.; Wong, W.L.; et al. Activating ESR1 Mutations Differentially Affect the Efficacy of ER Antagonists. Cancer Discov. 2017, 7, 277–287. [Google Scholar] [CrossRef] [PubMed]
- Zundelevich, A.; Dadiani, M.; Kahana-Edwin, S.; Itay, A.; Sella, T.; Gadot, M.; Cesarkas, K.; Farage-Barhom, S.; Saar, E.G.; Eyal, E.; et al. ESR1 mutations are frequent in newly diagnosed metastatic and loco-regional recurrence of endocrine-treated breast cancer and carry worse prognosis. Breast Cancer Res. 2020, 22, 16. [Google Scholar] [CrossRef]
- Brett, J.O.; Spring, L.M.; Bardia, A.; Wander, S.A. ESR1 mutation as an emerging clinical biomarker in metastatic hormone receptor-positive breast cancer. Breast Cancer Res. 2021, 23, 85. [Google Scholar] [CrossRef]
- Yang, P.; Du, C.W.; Kwan, M.; Liang, S.X.; Zhang, G.J. The impact of p53 in predicting clinical outcome of breast cancer patients with visceral metastasis. Sci. Rep. 2013, 3, 2246. [Google Scholar] [CrossRef] [PubMed]
- Ungerleider, N.A.; Rao, S.G.; Shahbandi, A.; Yee, D.; Niu, T.; Frey, W.D.; Jackson, J.G. Breast cancer survival predicted by TP53 mutation status differs markedly depending on treatment. Breast Cancer Res. 2018, 20, 115. [Google Scholar] [CrossRef]
- Yuan, M.; Huang, L.L.; Chen, J.H.; Wu, J.; Xu, Q. The emerging treatment landscape of targeted therapy in non-small-cell lung cancer. Signal Transduct. Target. Ther. 2019, 4, 61. [Google Scholar] [CrossRef]
- Choi, B.D.; Shankar, G.M.; Sivaganesan, A.; Van Beaver, L.A.; Oh, K.; Shin, J.H. Implication of Biomarker Mutations for Predicting Survival in Patients With Metastatic Lung Cancer to the Spine. Spine 2018, 43, E1274–E1280. [Google Scholar] [CrossRef] [PubMed]
- Engelman, J.A.; Zejnullahu, K.; Mitsudomi, T.; Song, Y.; Hyland, C.; Park, J.O.; Lindeman, N.; Gale, C.M.; Zhao, X.; Christensen, J.; et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science 2007, 316, 1039–1043. [Google Scholar] [CrossRef]
- Finocchiaro, G.; Toschi, L.; Gianoncelli, L.; Baretti, M.; Santoro, A. Prognostic and predictive value of MET deregulation in non-small cell lung cancer. Ann. Transl. Med. 2015, 3, 83. [Google Scholar] [CrossRef]
- Baldacci, S.; Kherrouche, Z.; Cockenpot, V.; Stoven, L.; Copin, M.C.; Werkmeister, E.; Marchand, N.; Kyheng, M.; Tulasne, D.; Cortot, A.B. MET amplification increases the metastatic spread of EGFR-mutated NSCLC. Lung Cancer 2018, 125, 57–67. [Google Scholar] [CrossRef]
- Wu, Y.L.; Cheng, Y.; Zhou, J.; Lu, S.; Zhang, Y.; Zhao, J.; Kim, D.W.; Soo, R.A.; Kim, S.W.; Pan, H.; et al. Tepotinib plus gefitinib in patients with EGFR-mutant non-small-cell lung cancer with MET overexpression or MET amplification and acquired resistance to previous EGFR inhibitor (INSIGHT study): An open-label, phase 1b/2, multicentre, randomised trial. Lancet Respir. Med. 2020, 8, 1132–1143. [Google Scholar] [CrossRef] [PubMed]
- Zotti, T.; Scudiero, I.; Vito, P.; Stilo, R. The Emerging Role of TRAF7 in Tumor Development. J. Cell Physiol. 2017, 232, 1233–1238. [Google Scholar] [CrossRef] [PubMed]
- Jaramillo, M.C.; Zhang, D.D. The emerging role of the Nrf2-Keap1 signaling pathway in cancer. Genes Dev. 2013, 27, 2179–2191. [Google Scholar] [CrossRef] [PubMed]
- Cuadrado, A.; Rojo, A.I.; Wells, G.; Hayes, J.D.; Cousin, S.P.; Rumsey, W.L.; Attucks, O.C.; Franklin, S.; Levonen, A.L.; Kensler, T.W.; et al. Therapeutic targeting of the NRF2 and KEAP1 partnership in chronic diseases. Nat. Rev. Drug Discov. 2019, 18, 295–317. [Google Scholar] [CrossRef]
| Characteristic | Breast, N = 84 1 | Lung, N = 56 1 | Prostate, N = 49 1 | Other, N = 93 1 | p-Value |
|---|---|---|---|---|---|
| Age | 57 (50, 66) | 67 (58, 72) | 70 (63, 74) | 60 (50, 68) | <0.001 2 |
| Surgery or Biopsy | <0.001 3 | ||||
| Biopsy | 70 (83%) | 30 (54%) | 42 (86%) | 38 (41%) | |
| Surgery | 14 (17%) | 26 (46%) | 7 (14%) | 55 (59%) | |
| Spinal Level | 0.003 4 | ||||
| Lumbar | 25 (30%) | 16 (29%) | 16 (33%) | 31 (33%) | |
| Other | 2 (2.4%) | 5 (8.9%) | 0 (0%) | 15 (16%) | |
| Sacral | 16 (19%) | 4 (7.1%) | 13 (27%) | 9 (9.7%) | |
| Thoracic | 41 (49%) | 31 (55%) | 20 (41%) | 38 (41%) |
| Characteristic | N | HR 1 | 95% CI 1 | p-Value | q-Value 2 | Mutation Frequency |
|---|---|---|---|---|---|---|
| KEAP1 | 282 | 3.95 | 2.24, 6.98 | <0.001 * | <0.001 ** | 0.06 |
| TP53 | 282 | 1.80 | 1.26, 2.56 | 0.001 * | 0.015 ** | 0.27 |
| KRAS | 282 | 1.87 | 1.11, 3.16 | 0.019 * | 0.14 | 0.08 |
| CDH1 | 282 | 0.43 | 0.21, 0.88 | 0.021 * | 0.14 | 0.09 |
| GATA3 | 282 | 0.50 | 0.23, 1.07 | 0.076 | 0.4 | 0.07 |
| PIK3CA | 282 | 0.71 | 0.46, 1.11 | 0.14 | 0.5 | 0.18 |
| AR.Amp | 282 | 1.60 | 0.84, 3.05 | 0.2 | 0.5 | 0.05 |
| NF1 | 282 | 1.54 | 0.81, 2.94 | 0.2 | 0.5 | 0.06 |
| APC | 282 | 0.59 | 0.26, 1.33 | 0.2 | 0.5 | 0.06 |
| MLL3 | 282 | 0.67 | 0.36, 1.24 | 0.2 | 0.5 | 0.09 |
| Characteristic | No Spine Met (%), N = 5658 1 | Spine Met (%), N = 84 1 | p-Value 2 | q-Value 3 |
|---|---|---|---|---|
| Breast | ||||
| TP53 | 2314 (41) | 15 (18) | <0.001 * | <0.001 ** |
| CDH1 | 789 (14) | 23 (27) | 0.001 * | 0.016 ** |
| ARID1A | 320 (5.7) | 11 (13) | 0.008 * | 0.067 |
| MYC.Amp | 640 (11) | 3 (3.6) | 0.022 * | 0.13 |
| KMT2C | 456 (8.1) | 12 (14) | 0.045 * | 0.2 |
| TBX3 | 306 (5.4) | 9 (11) | 0.048 * | 0.2 |
| Lung Adenocarcinoma | ||||
| CDKN2A.Del | 635 (9.7) | 15 (27) | <0.001 * | 0.008 ** |
| CDKN2AP14ARF.Del | 623 (9.5) | 14 (25) | <0.001 * | 0.009 ** |
| CDKN2AP16INK4A.Del | 635 (9.7) | 14 (25) | <0.001 * | 0.009 ** |
| CDKN2B.Del | 602 (9.2) | 13 (23) | 0.001 * | 0.012 ** |
| EGFR.Amp | 392 (6.0) | 10 (18) | 0.002 * | 0.012 ** |
| EGFR | 1624 (25) | 23 (41) | 0.008 * | 0.043 ** |
| TERT.Amp | 418 (6.4) | 9 (16) | 0.009 * | 0.043 ** |
| Prostate | ||||
| AR.Amp | 320 (13) | 15 (31) | 0.002 * | 0.034 ** |
| APC | 202 (8.3) | 9 (18) | 0.032 * | 0.2 |
| MYC.Amp | 159 (6.6) | 7 (14) | 0.042 * | 0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Giantini-Larsen, A.; Ramos, A.D.; Martin, A.; Panageas, K.S.; Kostrzewa, C.E.; Abou-Mrad, Z.; Schmitt, A.; Bromberg, J.F.; Safonov, A.; Rudin, C.M.; et al. Integration of Next Generation Sequencing Data to Inform Survival Prediction of Patients with Spine Metastasis. Cancers 2025, 17, 2218. https://doi.org/10.3390/cancers17132218
Giantini-Larsen A, Ramos AD, Martin A, Panageas KS, Kostrzewa CE, Abou-Mrad Z, Schmitt A, Bromberg JF, Safonov A, Rudin CM, et al. Integration of Next Generation Sequencing Data to Inform Survival Prediction of Patients with Spine Metastasis. Cancers. 2025; 17(13):2218. https://doi.org/10.3390/cancers17132218
Chicago/Turabian StyleGiantini-Larsen, Alexandra, Alexander D. Ramos, Axel Martin, Katherine S. Panageas, Caroline E. Kostrzewa, Zaki Abou-Mrad, Adam Schmitt, Jacqueline F. Bromberg, Anton Safonov, Charles M. Rudin, and et al. 2025. "Integration of Next Generation Sequencing Data to Inform Survival Prediction of Patients with Spine Metastasis" Cancers 17, no. 13: 2218. https://doi.org/10.3390/cancers17132218
APA StyleGiantini-Larsen, A., Ramos, A. D., Martin, A., Panageas, K. S., Kostrzewa, C. E., Abou-Mrad, Z., Schmitt, A., Bromberg, J. F., Safonov, A., Rudin, C. M., Newman, W. C., Bilsky, M. H., & Barzilai, O. (2025). Integration of Next Generation Sequencing Data to Inform Survival Prediction of Patients with Spine Metastasis. Cancers, 17(13), 2218. https://doi.org/10.3390/cancers17132218





