Mir-16 Decreases the Expression of VTI1B and SMPD1, Genes Involved in Membrane-Protein Trafficking in Melanoma
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Single Cell Expression and Tumor Cancer Genome Atlas (TCGA) Data Analysis
2.2. Generation of Melanoma Cell Lines and Transfection
2.3. Cloning
2.4. RNA Extraction and Analysis
2.5. Luciferase Reporter Assays
2.6. Western Blot Analysis
2.7. Statistical Analysis
3. Results
3.1. Immune Checkpoint mRNAs Are Co-Expressed in the Melanoma Microenvironment
3.2. Several Co-Expressed Checkpoint Genes Contain Putative Binding Sites for miRNAs from the miR-15/16 Family
3.3. miR-16 Over-Expression Leads to Reduced Expression of CD40 and CD80 mRNAs
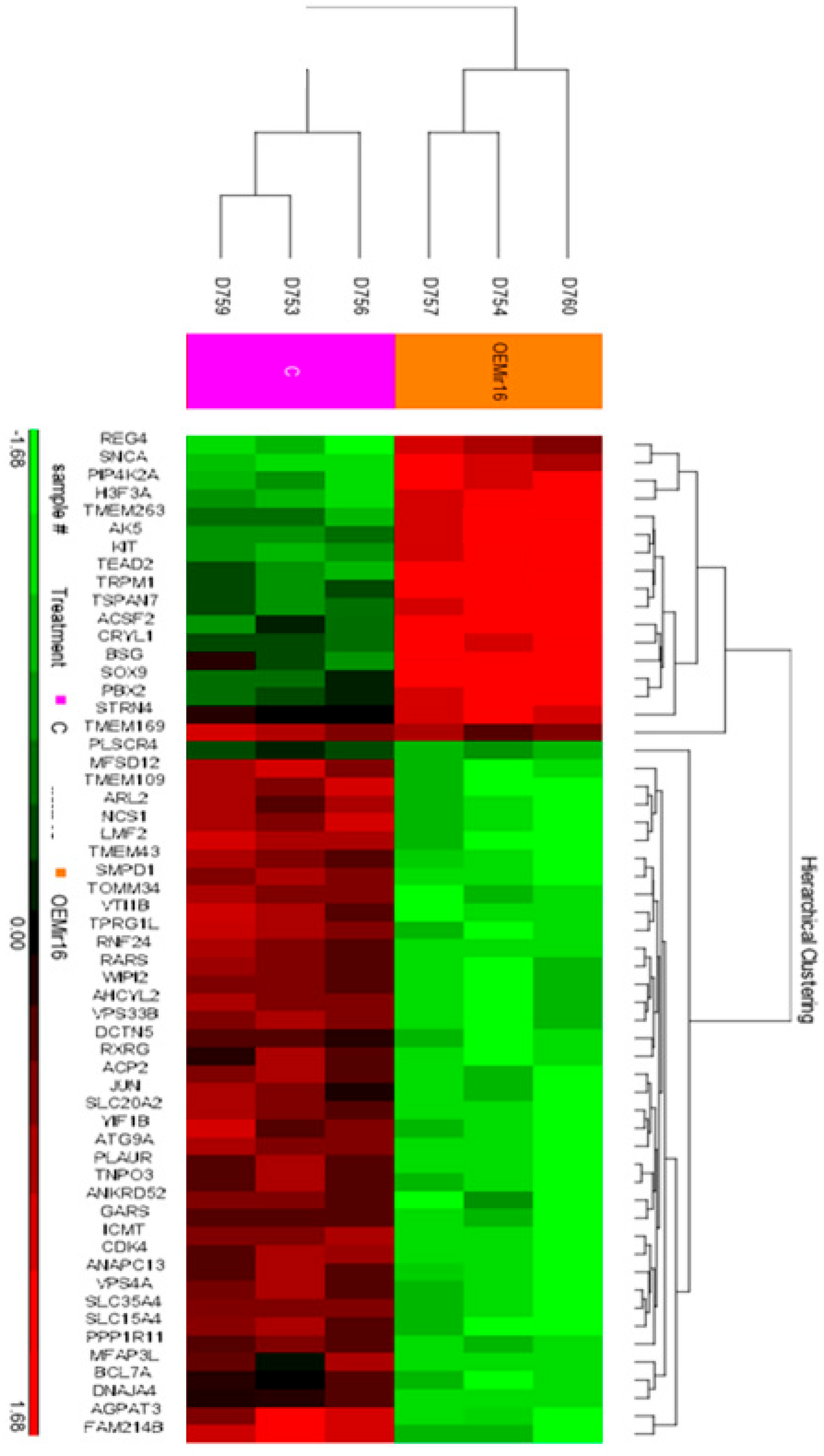
3.4. Over-Expression of miR-16 Leads to a Significant Decrease in the Expression of mRNAs Involved in Intracellular Vesicle-Mediated Transport and Protein Trafficking
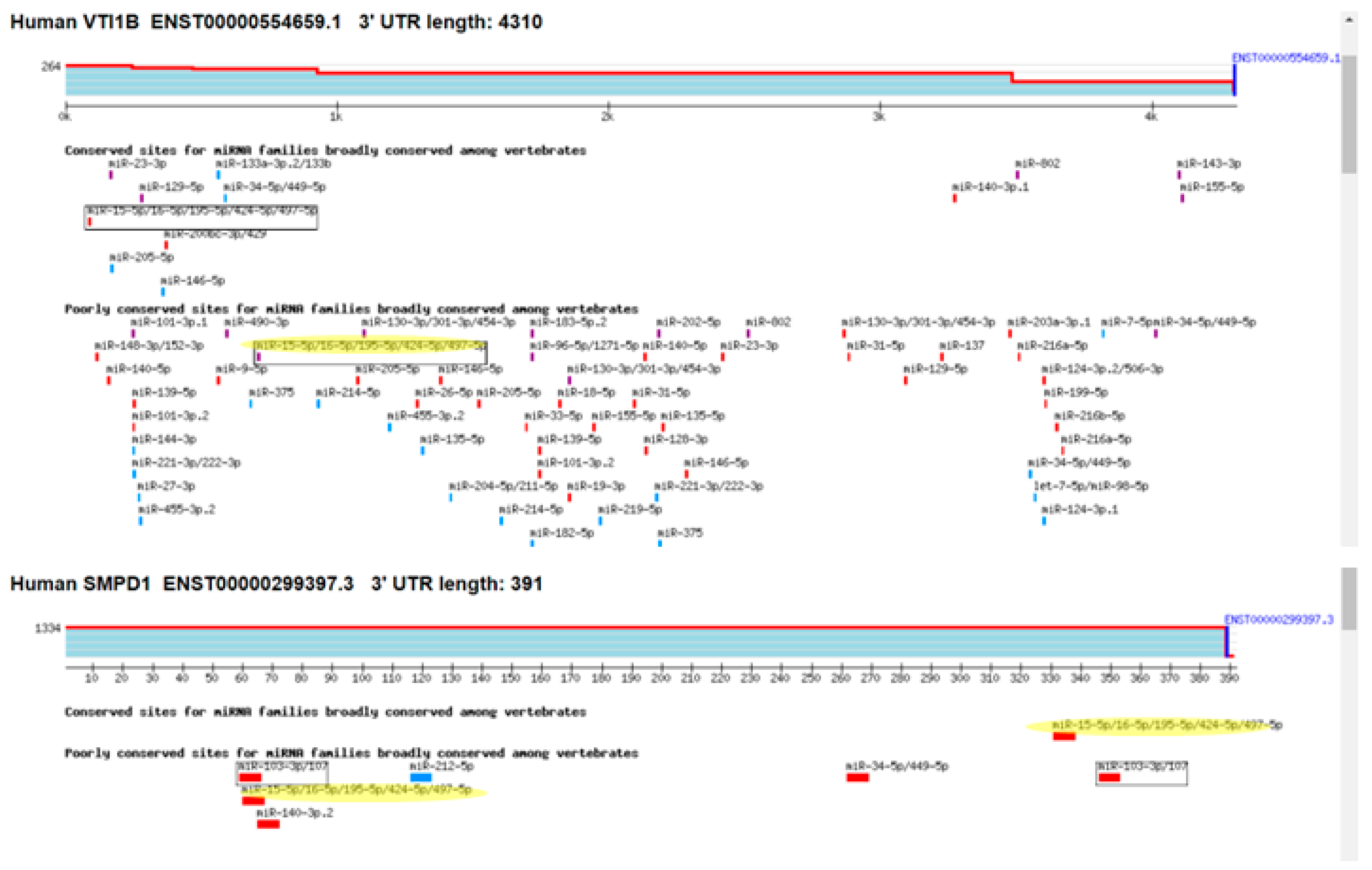
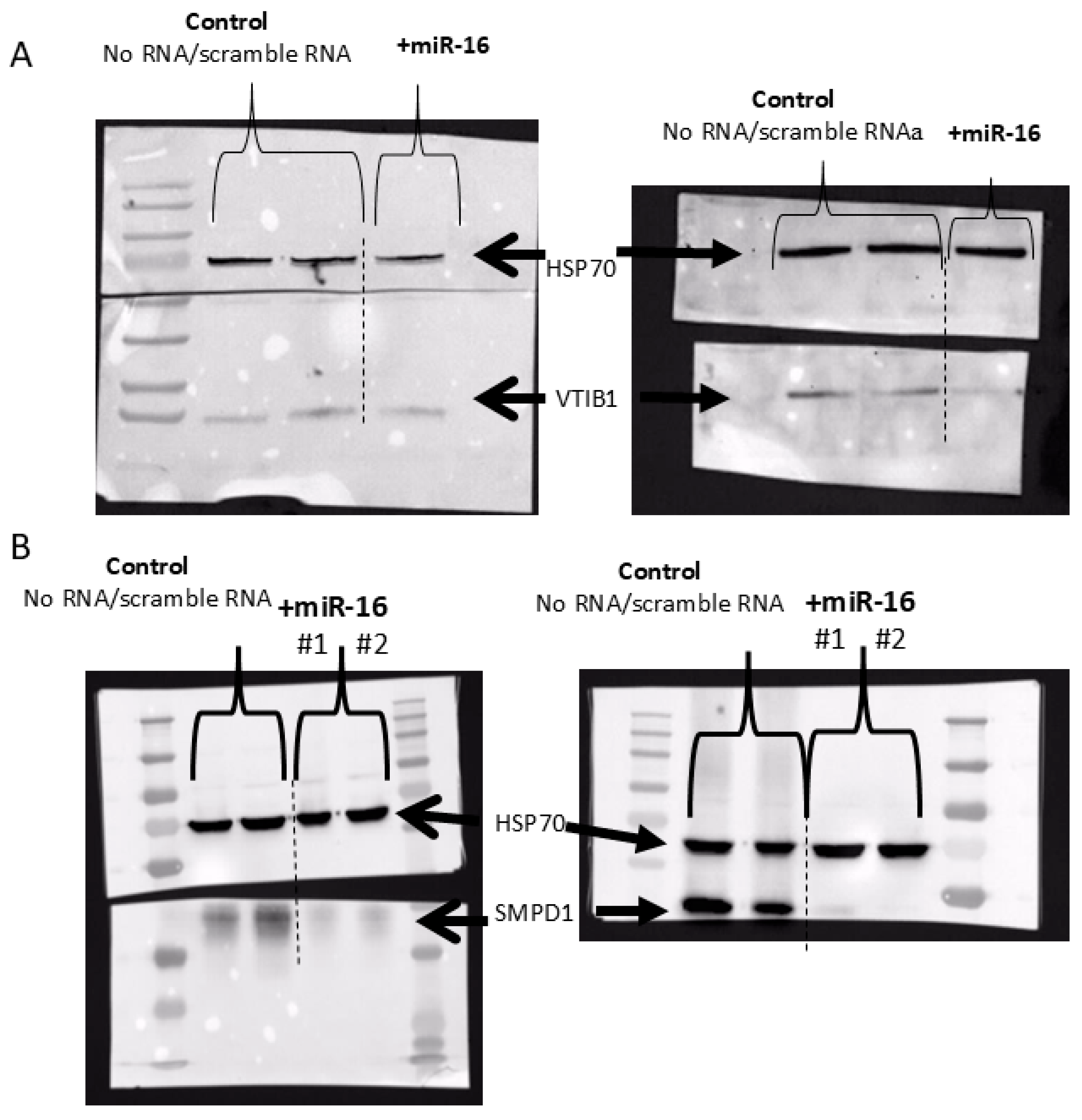
3.5. miR-16 Directly Targets the 3′UTR of SMPD1 and VTI1B and Decreases Their Protein Expression
4. Discussion
4.1. Direct Effects of miR-16 on Checkpoint Proteins
4.2. Direct Effects of miR-16 on VTI1B and SMPD1
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| FDR | False Discovery Rate |
| IS | Immunological Synapse |
| MHC | Major Histocompatibility Complex |
| miRNA | micro-RNA |
| TCGA | Tumor Cancer Genome Atlas |
| TCR | T-Cell Receptor |
References
- Saginala, K.; Barsouk, A.; Aluru, J.S.; Rawla, P.; Barsouk, A. Epidemiology of Melanoma. Med. Sci. 2021, 9, 63. [Google Scholar] [CrossRef] [PubMed]
- Lauss, M.; Phung, B.; Borch, T.H.; Harbst, K.; Kaminska, K.; Ebbesson, A.; Hedenfalk, I.; Yuan, J.; Nielsen, K.; Ingvar, C.; et al. Molecular patterns of resistance to immune checkpoint blockade in melanoma. Nat. Commun. 2024, 15, 3075. [Google Scholar] [CrossRef] [PubMed]
- Rezazadeh-Gavgani, E.; Majidazar, R.; Lotfinejad, P.; Kazemi, T.; Shamekh, A. Immune Checkpoint Molecules: A Review on Pathways and Immunotherapy Implications. Immun. Inflamm. Dis. 2025, 13, e70196. [Google Scholar] [CrossRef]
- Bredel, D.; Tihic, E.; Mouraud, S.; Danlos, F.-X.; Susini, S.; Aglave, M.; Mohamed-Djalim, C.; Rouanne, M.; Halse, H.; Bigorgne, A.; et al. Immune checkpoints are predominantly co-expressed by clonally expanded CD4+FoxP3+ intratumoral T-cells in primary human cancers. J. Exp. Clin. Cancer Res. 2023, 42, 333. [Google Scholar] [CrossRef]
- Nirschl, C.J.; Drake, C.G. Molecular pathways: Coexpression of immune checkpoint molecules: Signaling pathways and implications for cancer immunotherapy. Clin. Cancer Res. 2013, 19, 4917–4924. [Google Scholar] [CrossRef] [PubMed]
- Thomas, W.D.; Smith, M.J.; Si, Z.; Hersey, P. Expression of the co-stimulatory molecule CD40 on melanoma cells. Int. J. Cancer 1996, 68, 795–801. [Google Scholar] [CrossRef]
- Sviatoha, V.; Rundgren, A.; Tani, E.; Hansson, J.; Kleina, R.; Skoog, L. Expression of CD40, CD44, bcl-2 antigens and rate of cell proliferation on fine needle aspirates from metastatic melanoma. Cytopathology 2002, 13, 11–21. [Google Scholar] [CrossRef]
- Kalbasi, A.; Fonsatti, E.; Natali, P.G.; Altomonte, M.; Bertocci, E.; Cutaia, O.; Calabrò, L.; Chiou, M.; Tap, W.; Chmielowski, B.; et al. CD40 expression by human melanocytic lesions and melanoma cell lines and direct CD40 targeting with the therapeutic anti-CD40 antibody CP-870,893. J. Immunother. 2010, 33, 810–816. [Google Scholar] [CrossRef]
- Obeid, J.M.; Erdag, G.; Smolkin, M.E.; Deacon, D.H.; Patterson, J.W.; Chen, L.; Bullock, T.N.; Slingluff, C.L. PD-L1, PD-L2 and PD-1 expression in metastatic melanoma: Correlation with tumor-infiltrating immune cells and clinical outcome. Oncoimmunology 2016, 5, e1235107. [Google Scholar] [CrossRef]
- McCarthy, D.O.; Glowacki, N.; Schell, K.; Emler, C.A.; Albertini, M.R. Antigenicity of human melanoma cells transfected to express the B7-1 co-stimulatory molecule (CD80) varies with the level of B7-1 expression. Cancer Immunol. Immunother. 2000, 49, 85–93. [Google Scholar] [CrossRef]
- Cortez, M.A.; Ivan, C.; Zhou, P.; Wu, X.; Ivan, M.; Calin, G.A. microRNAs in cancer: From bench to bedside. Adv. Cancer Res. 2010, 108, 113–157. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Guo, W.; Li, S.; Dai, W.; Zhang, N.; Zhao, T.; Wang, H.; Ma, J.; Yi, X.; Ge, R.; et al. Serum miR-16: A Potential Biomarker for Predicting Melanoma Prognosis. J. Investig. Dermatol. 2016, 136, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.-X.; Guo, H.-B.; Liu, S.-Y.; Feng, H.-P.; Shi, J. ETS1 promoted cell growth, metastasis and epithelial-mesenchymal transition process in melanoma by regulating miR-16-mediated SOX4 expression. Melanoma Res. 2021, 31, 298–308. [Google Scholar] [CrossRef]
- Ghafouri-Fard, S.; Khoshbakht, T.; Hussen, B.M.; Abdullah, S.T.; Taheri, M.; Samadian, M. A review on the role of mir-16-5p in the carcinogenesis. Cancer Cell Int. 2022, 22, 342. [Google Scholar] [CrossRef] [PubMed]
- Xu, S.; Tao, Z.; Hai, B.; Liang, H.; Shi, Y.; Wang, T.; Song, W.; Chen, Y.; OuYang, J.; Chen, J.; et al. miR-424(322) reverses chemoresistance via T-cell immune response activation by blocking the PD-L1 immune checkpoint. Nat. Commun. 2016, 7, 11406. [Google Scholar] [CrossRef]
- Tirosh, I.; Izar, B.; Prakadan, S.M.; Wadsworth, M.H.; Treacy, D.; Trombetta, J.J.; Rotem, A.; Rodman, C.; Lian, C.; Murphy, G.; et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science 2016, 352, 189–196. [Google Scholar] [CrossRef]
- Zehavi, L.; Schayek, H.; Jacob-Hirsch, J.; Sidi, Y.; Leibowitz-Amit, R.; Avni, D. MiR-377 targets E2F3 and alters the NF-kB signaling pathway through MAP3K7 in malignant melanoma. Mol. Cancer 2015, 14, 68. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- McGeary, S.E.; Lin, K.S.; Shi, C.Y.; Pham, T.M.; Bisaria, N.; Kelley, G.M.; Bartel, D.P. The biochemical basis of microRNA targeting efficacy. Science 2019, 366, eaav1741. [Google Scholar] [CrossRef]
- Smolle, M.A.; Calin, H.N.; Pichler, M.; Calin, G.A. Noncoding RNAs and immune checkpoints—Clinical implications as cancer therapeutics. FEBS J. 2017, 284, 1952–1966. [Google Scholar] [CrossRef] [PubMed]
- Zehavi, L.; Avraham, R.; Barzilai, A.; Bar-Ilan, D.; Navon, R.; Sidi, Y.; Avni, D.; Leibowitz-Amit, R. Silencing of a large microRNA cluster on human chromosome 14q32 in melanoma: Biological effects of mir-376a and mir-376c on insulin growth factor 1 receptor. Mol. Cancer 2012, 11, 44. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Thomas, P.D. PANTHER in 2013: Modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013, 41, D377–D386. [Google Scholar] [CrossRef]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef]
- Lee, A.; Papangeli, I.; Park, Y.; Jeong, H.-N.; Choi, J.; Kang, H.; Jo, H.-N.; Kim, J.; Chun, H.J. A PPARγ-dependent miR-424/503-CD40 axis regulates inflammation mediated angiogenesis. Sci. Rep. 2017, 7, 2528. [Google Scholar] [CrossRef]
- Grassmé, H.; Jendrossek, V.; Bock, J.; Riehle, A.; Gulbins, E. Ceramide-rich membrane rafts mediate CD40 clustering. J. Immunol. 2002, 168, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Grassme, H.; Jekle, A.; Riehle, A.; Schwarz, H.; Berger, J.; Sandhoff, K.; Kolesnick, R.; Gulbins, E. CD95 signaling via ceramide-rich membrane rafts. J. Biol. Chem. 2001, 276, 20589–20596. [Google Scholar] [CrossRef]
- Xiong, X.; Lee, C.-F.; Li, W.; Yu, J.; Zhu, L.; Kim, Y.; Zhang, H.; Sun, H. Acid sphingomyelinase regulates the localization and trafficking of palmitoylated proteins. Biol. Open 2019, 8, bio040311. [Google Scholar] [CrossRef]
- Zhu, L.; Xiong, X.; Kim, Y.; Okada, N.; Lu, F.; Zhang, H.; Sun, H. Acid sphingomyelinase is required for cell surface presentation of Met receptor tyrosine kinase in cancer cells. J. Cell Sci. 2016, 129, 4238–4251. [Google Scholar] [CrossRef]
- Bollinger, C.R.; Teichgräber, V.; Gulbins, E. Ceramide-enriched membrane domains. Biochim. Biophys. Acta 2005, 1746, 284–294. [Google Scholar] [CrossRef] [PubMed]
- Varshney, P.; Yadav, V.; Saini, N. Lipid rafts in immune signalling: Current progress and future perspective. Immunology 2016, 149, 13–24. [Google Scholar] [CrossRef] [PubMed]
- Emperador-Melero, J.; Toonen, R.F.; Verhage, M. Vti Proteins: Beyond Endolysosomal Trafficking. Neuroscience 2019, 420, 32–40. [Google Scholar] [CrossRef]
- Wade, N.; Bryant, N.J.; Connolly, L.M.; Simpson, R.J.; Luzio, J.P.; Piper, R.C.; James, D.E. Syntaxin 7 complexes with mouse Vps10p tail interactor 1b, syntaxin 6, vesicle-associated membrane protein (VAMP)8, and VAMP7 in b16 melanoma cells. J. Biol. Chem. 2001, 276, 19820–19827. [Google Scholar] [CrossRef] [PubMed]
- Music, A.; Tejeda-González, B.; Cunha, D.M.; Fischer von Mollard, G.; Hernández-Pérez, S.; Mattila, P.K. The SNARE protein Vti1b is recruited to the sites of BCR activation but is redundant for antigen internalisation, processing and presentation. Front. Cell Dev. Biol. 2022, 10, 987148. [Google Scholar] [CrossRef]
- You, L.; Mao, L.; Wei, J.; Jin, S.; Yang, C.; Liu, H.; Zhu, L.; Qian, W. The crosstalk between autophagic and endo-/exosomal pathways in antigen processing for MHC presentation in anticancer T cell immune responses. J. Hematol. Oncol. 2017, 10, 165. [Google Scholar] [CrossRef]
- Yao, H.; Lan, J.; Li, C.; Shi, H.; Brosseau, J.-P.; Wang, H.; Lu, H.; Fang, C.; Zhang, Y.; Liang, L.; et al. Inhibiting PD-L1 palmitoylation enhances T-cell immune responses against tumours. Nat. Biomed. Eng. 2019, 3, 306–317. [Google Scholar] [CrossRef]




| Name | Sequence: (5′ to 3′) |
|---|---|
| psi_smpd1F | CCCGGGAATTCGTTTGGCCCCAGGGCCCACATTTG |
| psi_smpd1R | GGCCGCTCTAGGTTTTGGAGTCCAAGTCTCTTATC |
| psi_vti1bF | CCCGGGAATTCGTTTACTTCTATAGGGAAGGGTTT |
| psi_vti1bR | GGCCGCTCTAGGTTTGTGGCATACATTTTGCCAAC |
| psi_CD80F | CCCGGGAATTCGTTTCTAACTCTGGTGCTCTTTCC |
| psi_CD80R | GGCCGCTCTAGGTTTTGGTGTTAGACCTCTCTGCC |
| psi_CD40F | CCCGGGAATTCGTTTGGAACCCCAGGAGATCAATTTTCC |
| psi_CD40R | GGCCGCTCTAGGTTTACCCTTCTTCCCCACCCCAGACG |
| miR-16F | AGCTTGTCAGCAGTGCCTTAGCAGCACGTAAATATTGGCGTTAAGATTCTAAAATTATCT CCAGTATTAACTGTGCTGCTGAAGTAAGGTTGACG |
| miR-16R | AATTCGTCAACCTTACTTCAGCAGCACAGTTAATACTGGAGATAATTTTAGAATCTTAACG CCAATATTTACGTGCTGCTAAGGCACTGCTGACA |
| PVRL2 | TNSFSF18 | TNSFF4 | ICOSLG | CD274 | CD40 | LGALS9 | CD80 | PDCD1LG2 | |
|---|---|---|---|---|---|---|---|---|---|
| 0.24 | 0.47 | 0.51 | 0.61 | 0.73 | 0.74 | 0.79 | 0.90 | 0.90 | CD86 |
| 0.17 | 0.49 | 0.49 | 0.52 | 0.83 | 0.67 | 0.73 | 0.89 | PDCD1LG2 | |
| 0.14 | 0.45 | 0.47 | 0.53 | 0.78 | 0.70 | 0.72 | CD80 | ||
| 0.27 | 0.26 | 0.34 | 0.54 | 0.57 | 0.60 | LGALS9 | |||
| 0.25 | 0.32 | 0.44 | 0.56 | 0.55 | CD40 | ||||
| 0.05 | 0.40 | 0.47 | 0.38 | CD274 | |||||
| 0.22 | 0.21 | 0.36 | ICOSLG | ||||||
| 0.01 | 0.31 | TNSFF4 | |||||||
| 0.03 | TNSFSF18 |
| Gene Symbol | Fold Change— Over-Express miR-16 vs. Control | p-Value | Bioinformatics Possible Target of miR-16/15 |
|---|---|---|---|
| VPS33B | −1.36448 | 0.0375063 | √ |
| WIPI2 | −1.36762 | 0.0375063 | √ |
| MFAP3L | −1.37428 | 0.0654535 | √ |
| PPP1R11 | −1.39183 | 0.0624488 | √ |
| TOMM34 | −1.39962 | 0.0442763 | √ |
| TNPO3 | −1.41463 | 0.0579612 | |
| CDK4 | −1.43391 | 0.0375063 | √ |
| RNF24 | −1.43433 | 0.0375063 | √ |
| SLC15A4 | −1.4514 | 0.0381192 | √ |
| TPRG1L | −1.47329 | 0.0375063 | √ |
| ACP2 | −1.50057 | 0.0375063 | √ |
| TMEM109 | −1.51314 | 0.0375063 | √ |
| VPS4A | −1.5148 | 0.0442763 | √ |
| AHCYL2 | −1.55187 | 0.0375063 | √ |
| VTI1B | −1.56299 | 0.0375063 | √ |
| AGPAT3 | −1.56375 | 0.0375063 | √ |
| MFSD12 | −1.57871 | 0.0375063 | |
| ICMT | −1.59596 | 0.0375063 | √ |
| SLC35A4 | −1.65548 | 0.0546149 | √ |
| SLC20A2 | −1.72137 | 0.0375063 | √ |
| ANAPC13 | −1.74267 | 0.0466223 | √ |
| LMF2 | −1.77151 | 0.0375063 | √ |
| YIF1B | −1.8387 | 0.0375063 | √ |
| ATG9A | −1.95334 | 0.0375063 | √ |
| RARS | −1.99813 | 0.0375063 | |
| ARL2 | −2.10958 | 0.0381192 | √ |
| ANKRD52 | −2.16298 | 0.0577935 | |
| SMPD1 | −2.45929 | 0.0375063 | √ |
| Analysis Type: | PANTHER Overrepresentation Test (Released 11 July 2019) | ||||||
| Annotation Version and Release Date: | GO Ontology Database; Released 3 July 2019 | ||||||
| Analyzed List: | Client Text Box Input (Homo sapiens) | ||||||
| Reference List: | Homo sapiens (all genes in database) | ||||||
| Test Type: | FISHER | ||||||
| Correction: | FDR | ||||||
| GO cellular component complete | Homo sapiens—REFLIST (20,996) | −41 | (expected) | (over/under) | (-fold enrichment) | (raw p-value) | (FDR) |
| Late endosome membrane (GO:0031902) | 136 | 4 | 0.27 | + | 15.06 | 1.57 × 10−4 | 4.00 × 10−2 |
| Membrane-bounded organelle (GO:0043227) | 12,535 | 36 | 24.48 | + | 1.47 | 1.61 × 10−4 | 3.28 × 10−2 |
| Cytoplasmic part (GO:0044444) | 9790 | 31 | 19.12 | + | 1.62 | 2.25 × 10−4 | 4.17 × 10−2 |
| Endomembrane system (GO:0012505) | 4494 | 22 | 8.78 | + | 2.51 | 6.18 × 10−6 | 4.20 × 10−3 |
| Bounding membrane of organelle (GO:0098588) | 2091 | 15 | 4.08 | + | 3.67 | 4.90 × 10−6 | 4.99 × 10−3 |
| Organelle membrane (GO:0031090) | 3525 | 20 | 6.88 | + | 2.91 | 2.31 × 10−6 | 4.70 × 10−3 |
| Whole membrane (GO:0098805) | 1683 | 12 | 3.29 | + | 3.65 | 6.25 × 10−5 | 2.55 × 10−2 |
| Vacuolar membrane (GO:0005774) | 417 | 6 | 0.81 | + | 7.37 | 1.58 × 10−4 | 3.58 × 10−2 |
| Vacuolar part (GO:0044437) | 575 | 7 | 1.12 | + | 6.23 | 1.19 × 10−4 | 3.46 × 10−2 |
| Vacuole (GO:0005773) | 789 | 9 | 1.54 | + | 5.84 | 1.83 × 10−5 | 9.31 × 10−3 |
| Intracellular membrane-bounded organelle (GO:0043231) | 11,046 | 33 | 21.57 | + | 1.53 | 2.62 × 10−4 | 4.44 × 10−2 |
| Lysosome (GO:0005764) | 684 | 7 | 1.34 | + | 5.24 | 3.40 × 10−4 | 5.33 × 10−2 |
| Lytic vacuole (GO:0000323) | 685 | 7 | 1.34 | + | 5.23 | 3.43 × 10−4 | 4.99 × 10−2 |
| Secretory vesicle (GO:0099503) | 995 | 9 | 1.94 | + | 4.63 | 1.09 × 10−4 | 3.70 × 10−2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Layani, A.; Meningher, T.; Sidi, Y.; Avni, D.; Leibowitz, R. Mir-16 Decreases the Expression of VTI1B and SMPD1, Genes Involved in Membrane-Protein Trafficking in Melanoma. Cancers 2025, 17, 2197. https://doi.org/10.3390/cancers17132197
Layani A, Meningher T, Sidi Y, Avni D, Leibowitz R. Mir-16 Decreases the Expression of VTI1B and SMPD1, Genes Involved in Membrane-Protein Trafficking in Melanoma. Cancers. 2025; 17(13):2197. https://doi.org/10.3390/cancers17132197
Chicago/Turabian StyleLayani, Adi, Tal Meningher, Yechezkel Sidi, Dror Avni, and Raya Leibowitz. 2025. "Mir-16 Decreases the Expression of VTI1B and SMPD1, Genes Involved in Membrane-Protein Trafficking in Melanoma" Cancers 17, no. 13: 2197. https://doi.org/10.3390/cancers17132197
APA StyleLayani, A., Meningher, T., Sidi, Y., Avni, D., & Leibowitz, R. (2025). Mir-16 Decreases the Expression of VTI1B and SMPD1, Genes Involved in Membrane-Protein Trafficking in Melanoma. Cancers, 17(13), 2197. https://doi.org/10.3390/cancers17132197







