Performance and Prognostic Relevance of Lymph Node Assessment by One-Step Nucleic Acid Amplification Assay in Rectal Cancer: A Multicenter Study
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Eligibility of Patients
2.2. Lymph Node Processing and Examination
2.3. One-Step Nucleic Acid Amplification Assay (OSNA)
2.4. CK19 Immunohistochemistry
2.5. Estimation of Sample Size
2.6. Statistical Analysis
3. Results
3.1. Patient Characteristics
3.2. Diagnostic Performance of OSNA Versus H&E
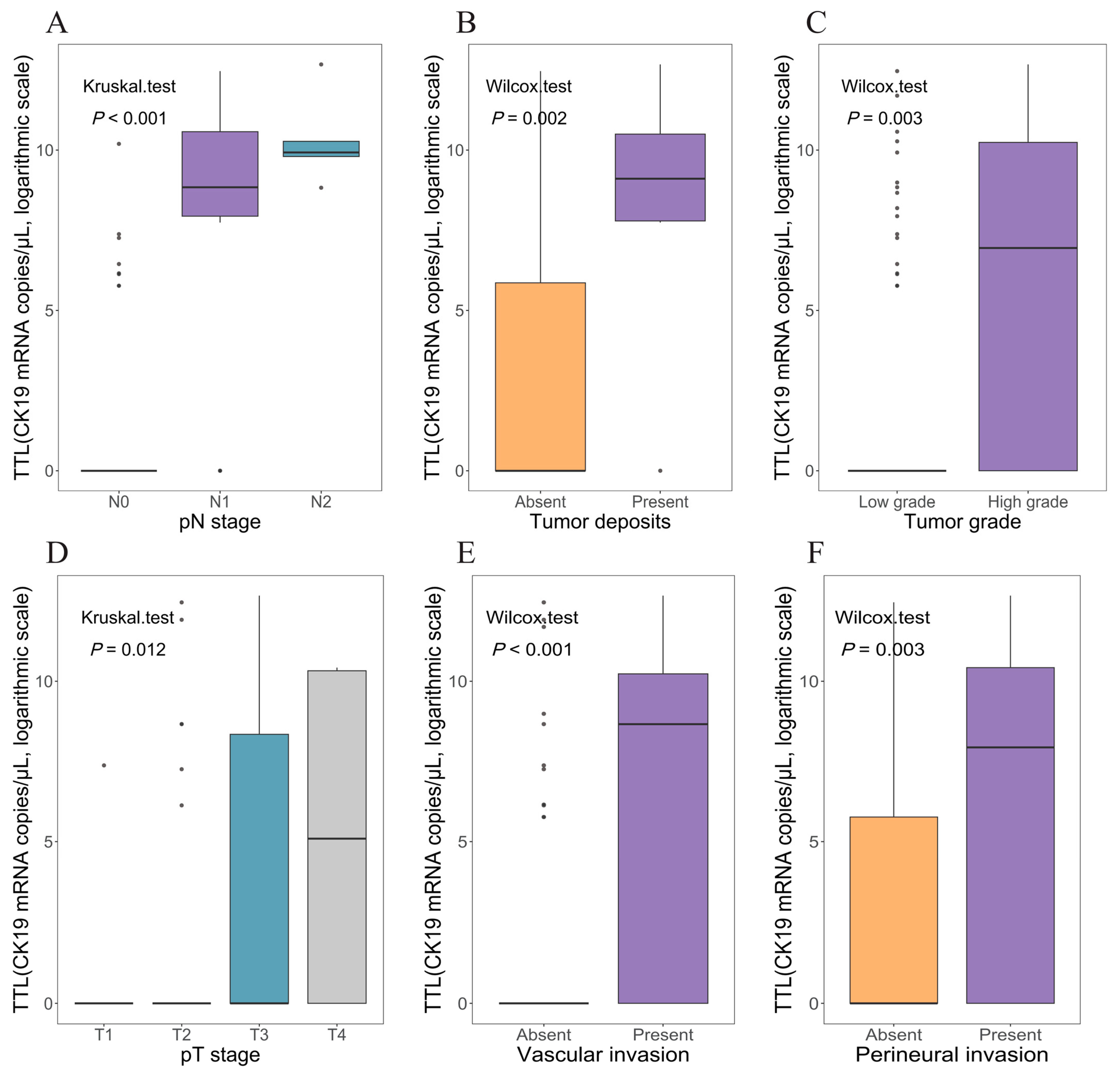
3.3. Association Between TTL and Clinicopathological Characteristics
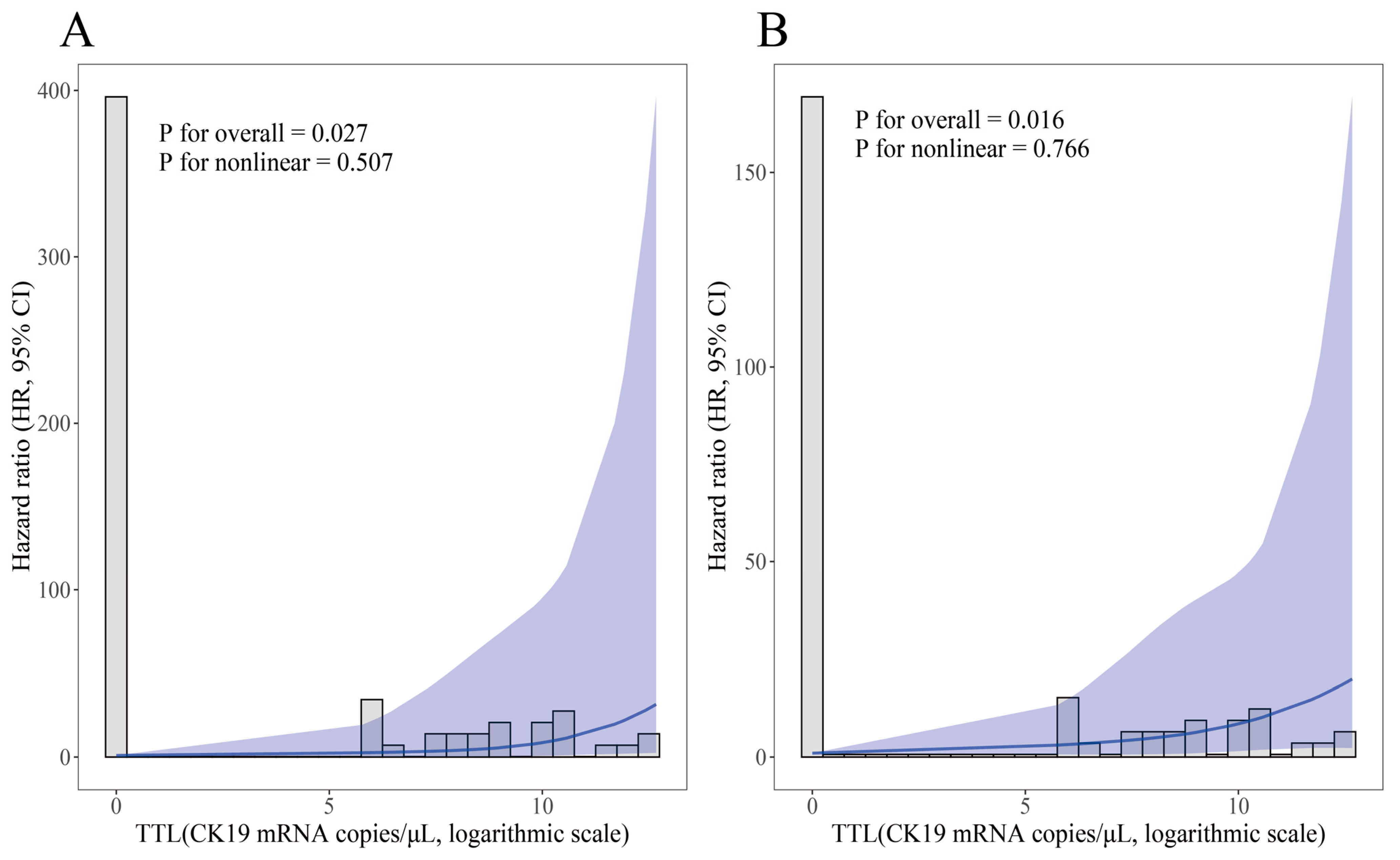
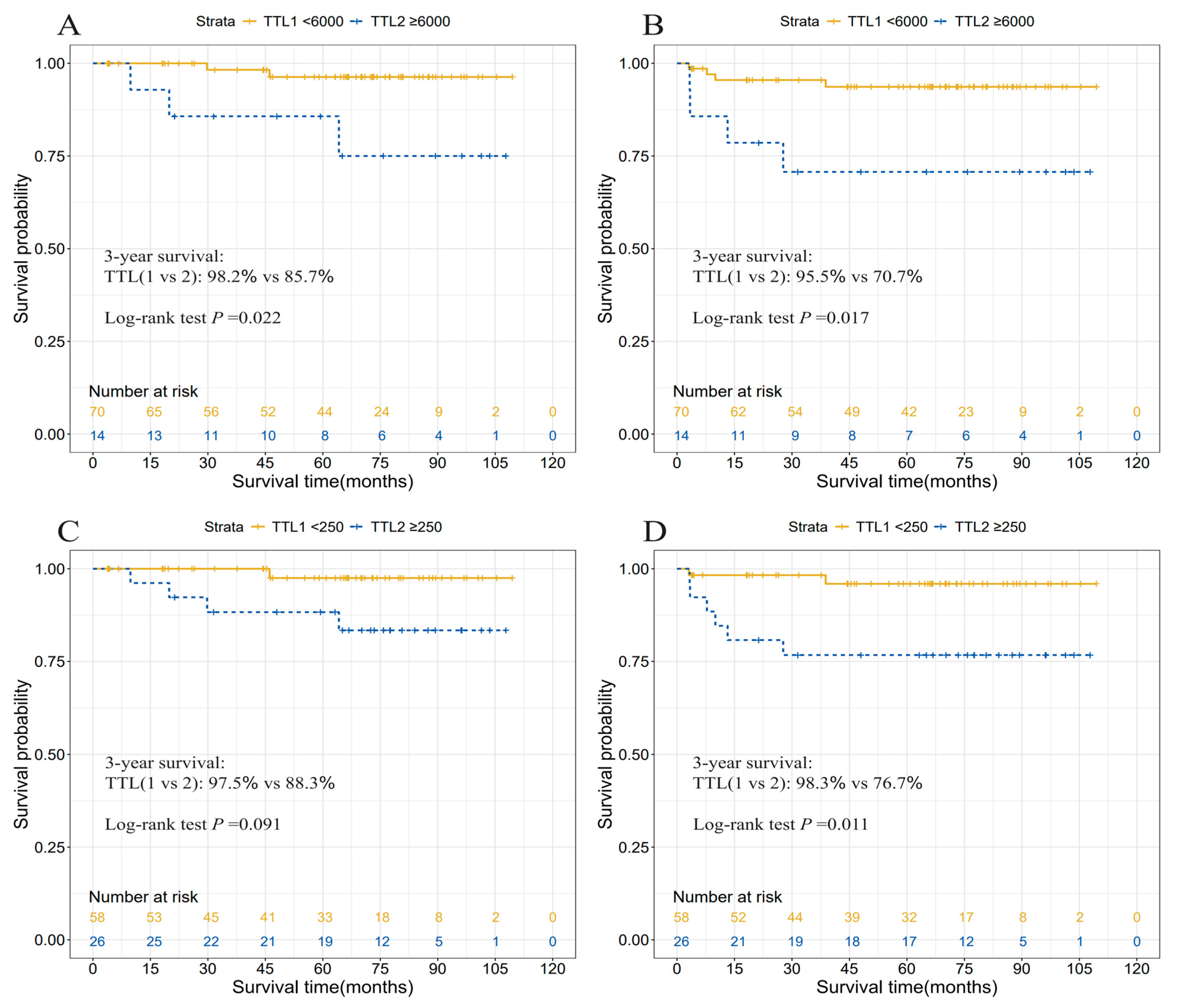
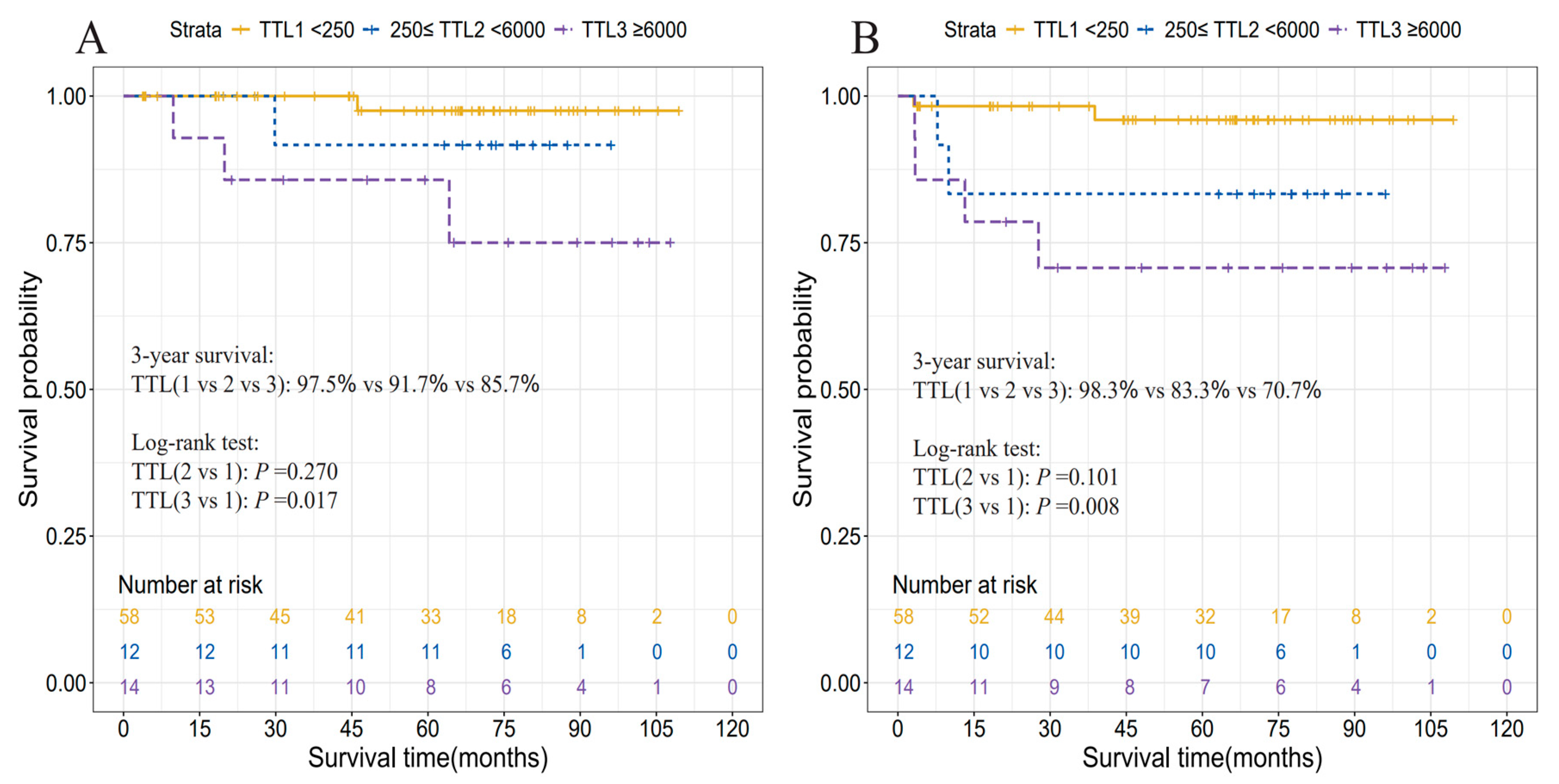
3.4. Prognostic Relevance of TTL
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Stoffel, E.M.; Murphy, C.C. Epidemiology and Mechanisms of the Increasing Incidence of Colon and Rectal Cancers in Young Adults. Gastroenterology 2020, 158, 341–353. [Google Scholar] [CrossRef]
- Rawla, P.; Sunkara, T.; Barsouk, A. Epidemiology of Colorectal Cancer: Incidence, Mortality, Survival, and Risk Factors. Prz. Gastroenterol. 2019, 14, 89–103. [Google Scholar] [CrossRef]
- Losurdo, P.; Giacca, M.; Biloslavo, A.; Fracon, S.; Sereni, E.; Giudici, F.; Generali, D.; de Manzini, N. Colorectal Cancer-Screening Program Improves Both Short- and Long-Term Outcomes: A Single-Center Experience in Trieste. Updates Surg. 2020, 72, 89–96. [Google Scholar] [CrossRef]
- Jiang, H.; Zhang, P.; Gu, K.; Gong, Y.; Peng, P.; Shi, Y.; Ai, D.; Chen, W.; Fu, C. Cost-Effectiveness Analysis of a Community-Based Colorectal Cancer Screening Program in Shanghai, China. Front. Public Health 2022, 10, 986728. [Google Scholar] [CrossRef] [PubMed]
- Emile, S.H.; Barsom, S.H.; Wexner, S.D. An Updated Review of the Methods, Guidelines of, and Controversies on Screening for Colorectal Cancer. Am. J. Surg. 2022, 224, 339–347. [Google Scholar] [CrossRef] [PubMed]
- Cubiella, J.; Lorenzo, M.; Baiocchi, F.; Tejido, C.; Conde, A.; Sande-Meijide, M.; Castro, M. Impact of a Colorectal Cancer Screening Program Implantation on Delays and Prognosis of Non-Screening Detected Colorectal Cancer. World J. Gastroenterol. 2021, 27, 6689–6700. [Google Scholar] [CrossRef]
- Kim, H.J.; Choi, G. Clinical Implications of Lymph Node Metastasis in Colorectal Cancer: Current Status and Future Perspectives. Ann. Coloproctol. 2019, 35, 109–117. [Google Scholar] [CrossRef]
- Iddings, D.; Ahmad, A.; Elashoff, D.; Bilchik, A. The Prognostic Effect of Micrometastases in Previously Staged Lymph Node Negative (N0) Colorectal Carcinoma: A Meta-Analysis. Ann. Surg. Oncol. 2006, 13, 1386–1392. [Google Scholar] [CrossRef]
- Sloothaak, D.A.M.; Sahami, S.; van der Zaag-Loonen, H.J.; van der Zaag, E.S.; Tanis, P.J.; Bemelman, W.A.; Buskens, C.J. The Prognostic Value of Micrometastases and Isolated Tumour Cells in Histologically Negative Lymph Nodes of Patients with Colorectal Cancer: A Systematic Review and Meta-Analysis. Eur. J. Surg. Oncol. 2014, 40, 263–269. [Google Scholar] [CrossRef]
- Rahbari, N.N.; Bork, U.; Motschall, E.; Thorlund, K.; Büchler, M.W.; Koch, M.; Weitz, J. Molecular Detection of Tumor Cells in Regional Lymph Nodes Is Associated with Disease Recurrence and Poor Survival in Node-Negative Colorectal Cancer: A Systematic Review and Meta-Analysis. J. Clin. Oncol. 2012, 30, 60–70. [Google Scholar] [CrossRef] [PubMed]
- Märkl, B.; Herbst, C.; Cacchi, C.; Schaller, T.; Krammer, I.; Schenkirsch, G.; Probst, A.; Spatz, H. Prognostic Significance of Histologically Detected Lymph Node Micrometastases of Sizes between 0.2 and 2 mm in Colorectal Cancer. Int. J. Color. Dis. 2013, 28, 977–983. [Google Scholar] [CrossRef]
- Hitchcock, C.L.; Sampsel, J.; Young, D.C.; Martin, E.W.; Arnold, M.W. Limitations with Light Microscopy in the Detection of Colorectal Cancer Cells. Dis. Colon Rectum 1999, 42, 1046–1052. [Google Scholar] [CrossRef]
- De Robles, M.S.; O’Neill, R.S.; Mourad, A.P.; Winn, R.; Putnis, S.; Kang, S. Survival in Stage IIB/C Compared to Stage IIIA Rectal Cancer: An Australian Experience Affirming That Size Does Matter. ANZ J. Surg. 2021, 91, 1866–1873. [Google Scholar] [CrossRef] [PubMed]
- Mo, S.; Dai, W.; Xiang, W.; Huang, B.; Li, Y.; Feng, Y.; Li, Q.; Cai, G. Survival Contradiction Between Stage IIA and Stage IIIA Rectal Cancer: A Retrospective Study. J. Cancer 2018, 9, 1466–1475. [Google Scholar] [CrossRef]
- Vedire, Y.R.; Mukherjee, S.; Dondapati, S.; Yendamuri, S. Association Between Visceral Obesity, Metformin Use, and Recurrence Risk in Early-Stage Colorectal Cancer. Sci. Rep. 2023, 13, 8401. [Google Scholar] [CrossRef]
- Nakagami, Y.; Hazama, S.; Suzuki, N.; Yoshida, S.; Tomochika, S.; Matsui, H.; Shindo, Y.; Tokumitsu, Y.; Matsukuma, S.; Watanabe, Y.; et al. CD4 and FOXP3 as Predictive Markers for the Recurrence of T3/T4a Stage II Colorectal Cancer: Applying a Novel Discrete Bayes Decision Rule. BMC Cancer 2022, 22, 1071. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H. Micrometastasis in Lymph Nodes of Colorectal Cancer. Ann. Gastroenterol. Surg. 2022, 6, 466–473. [Google Scholar] [CrossRef]
- Argilés, G.; Tabernero, J.; Labianca, R.; Hochhauser, D.; Salazar, R.; Iveson, T.; Laurent-Puig, P.; Quirke, P.; Yoshino, T.; Taieb, J.; et al. Localised Colon Cancer: ESMO Clinical Practice Guidelines for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2020, 31, 1291–1305. [Google Scholar] [CrossRef]
- Noura, S.; Yamamoto, H.; Miyake, Y.; Kim, B.n.; Takayama, O.; Seshimo, I.; Ikenaga, M.; Ikeda, M.; Sekimoto, M.; Matsuura, N.; et al. Immunohistochemical Assessment of Localization and Frequency of Micrometastases in Lymph Nodes of Colorectal Cancer. Clin. Cancer Res. 2002, 8, 759–767. [Google Scholar]
- You, X.; Wang, Y.; Wu, J.; Liu, Q.; Chen, D.; Tang, D.; Wang, D. Aberrant Cytokeratin 20 mRNA Expression in Peripheral Blood and Lymph Nodes Indicates Micrometastasis and Poor Prognosis in Patients with Gastric Carcinoma. Technol. Cancer Res. Treat. 2019, 18, 1533033819832856. [Google Scholar] [CrossRef] [PubMed]
- Manzotti, M.; Dell’Orto, P.; Maisonneuve, P.; Zurrida, S.; Mazzarol, G.; Viale, G. Reverse Transcription-Polymerase Chain Reaction Assay for Multiple mRNA Markers in the Detection of Breast Cancer Metastases in Sentinel Lymph Nodes. Int. J. Cancer 2001, 95, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, H.; Sekimoto, M.; Oya, M.; Yamamoto, N.; Konishi, F.; Sasaki, J.; Yamada, S.; Taniyama, K.; Tominaga, H.; Tsujimoto, M.; et al. OSNA-Based Novel Molecular Testing for Lymph Node Metastases in Colorectal Cancer Patients: Results from a Multicenter Clinical Performance Study in Japan. Ann. Surg. Oncol. 2011, 18, 1891–1898. [Google Scholar] [CrossRef]
- Yamamoto, H.; Tomita, N.; Inomata, M.; Furuhata, T.; Miyake, Y.; Noura, S.; Kato, T.; Murata, K.; Hayashi, S.; Igarashi, S.; et al. OSNA-Assisted Molecular Staging in Colorectal Cancer: A Prospective Multicenter Trial in Japan. Ann. Surg. Oncol. 2016, 23, 391–396. [Google Scholar] [CrossRef]
- Diaz-Mercedes, S.; Archilla, I.; Camps, J.; De Lacy, A.; Gorostiaga, I.; Momblan, D.; Ibarzabal, A.; Maurel, J.; Chic, N.; Bombí, J.A.; et al. Budget Impact Analysis of Molecular Lymph Node Staging Versus Conventional Histopathology Staging in Colorectal Carcinoma. Appl. Health Econ. Health Policy 2019, 17, 655–667. [Google Scholar] [CrossRef]
- Archilla, I.; Díaz-Mercedes, S.; Aguirre, J.J.; Tarragona, J.; Machado, I.; Rodrigo, M.T.; Lopez-Prades, S.; Gorostiaga, I.; Landolfi, S.; Alén, B.O.; et al. Lymph Node Tumor Burden Correlates With Tumor Budding and Poorly Differentiated Clusters: A New Prognostic Factor in Colorectal Carcinoma? Clin. Transl. Gastroenterol. 2021, 12, e00303. [Google Scholar] [CrossRef]
- Itabashi, M.; Yamamoto, H.; Tomita, N.; Inomata, M.; Murata, K.; Hayashi, S.; Miyake, Y.; Igarashi, S.; Kato, T.; Noura, S.; et al. Lymph Node Positivity in One-Step Nucleic Acid Amplification Is a Prognostic Factor for Postoperative Cancer Recurrence in Patients with Stage II Colorectal Cancer: A Prospective, Multicenter Study. Ann. Surg. Oncol. 2020, 27, 1077–1083. [Google Scholar] [CrossRef] [PubMed]
- Paschke, S.; Jafarov, S.; Staib, L.; Kreuser, E.-D.; Maulbecker-Armstrong, C.; Roitman, M.; Holm, T.; Harris, C.C.; Link, K.-H.; Kornmann, M. Are Colon and Rectal Cancer Two Different Tumor Entities? A Proposal to Abandon the Term Colorectal Cancer. Int. J. Mol. Sci. 2018, 19, 2577. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Gong, Z.; Ye, H.; Zhao, X.; Li, J.; Zhang, X.; Li, S.; Zhu, W.; Wang, M.; et al. Distinguishing Rectal Cancer from Colon Cancer Based on the Support Vector Machine Method and RNA-Sequencing Data. Curr. Med. Sci. 2021, 41, 368–374. [Google Scholar] [CrossRef]
- Wu, X.; Yang, Q.; Leng, L.; Yang, P.; Zhu, Z. Altered Metabolic Profiles in Colon and Rectal Cancer. Sci. Rep. 2025, 15, 11310. [Google Scholar] [CrossRef]
- Markowski, A.R.; Błachnio-Zabielska, A.U.; Pogodzińska, K.; Markowska, A.J.; Zabielski, P. Diverse Sphingolipid Profiles in Rectal and Colon Cancer. Int. J. Mol. Sci. 2023, 24, 10867. [Google Scholar] [CrossRef]
- Tamas, K.; Walenkamp, A.M.E.; de Vries, E.G.E.; van Vugt, M.A.T.M.; Beets-Tan, R.G.; van Etten, B.; de Groot, D.J.A.; Hospers, G.A.P. Rectal and Colon Cancer: Not Just a Different Anatomic Site. Cancer Treat. Rev. 2015, 41, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Kornmann, M.; Staib, L.; Wiegel, T.; Kron, M.; Henne-Bruns, D.; Link, K.-H.; Formentini, A.; Study Group Oncology of Gastrointestinal Tumors (FOGT). Long-Term Results of 2 Adjuvant Trials Reveal Differences in Chemosensitivity and the Pattern of Metastases Between Colon Cancer and Rectal Cancer. Clin. Color. Cancer 2013, 12, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Staib, L.; Link, K.H.; Blatz, A.; Beger, H.G. Surgery of Colorectal Cancer: Surgical Morbidity and Five- and Ten-Year Results in 2400 Patients—Monoinstitutional Experience. World J. Surg. 2002, 26, 59–66. [Google Scholar] [CrossRef]
- Araghi, M.; Arnold, M.; Rutherford, M.J.; Guren, M.G.; Cabasag, C.J.; Bardot, A.; Ferlay, J.; Tervonen, H.; Shack, L.; Woods, R.R.; et al. Colon and Rectal Cancer Survival in Seven High-Income Countries 2010–2014: Variation by Age and Stage at Diagnosis (the ICBP SURVMARK-2 Project). Gut 2021, 70, 114–126. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.-C.; Lee, Y.-L.; Chuang, J.-P.; Lee, J.-C. Differences in Survival between Colon and Rectal Cancer from SEER Data. PLoS ONE 2013, 8, e78709. [Google Scholar] [CrossRef]
- Buchwald, P.; Hall, C.; Davidson, C.; Dixon, L.; Dobbs, B.; Robinson, B.; Frizelle, F. Improved Survival for Rectal Cancer Compared to Colon Cancer: The Four Cohort Study. ANZ J. Surg. 2018, 88, E114–E117. [Google Scholar] [CrossRef]
- Fischer, J.; Hellmich, G.; Jackisch, T.; Puffer, E.; Zimmer, J.; Bleyl, D.; Kittner, T.; Witzigmann, H.; Stelzner, S. Outcome for Stage II and III Rectal and Colon Cancer Equally Good after Treatment Improvement over Three Decades. Int. J. Color. Dis. 2015, 30, 797–806. [Google Scholar] [CrossRef]
- von Elm, E.; Altman, D.G.; Egger, M.; Pocock, S.J.; Gøtzsche, P.C.; Vandenbroucke, J.P.; STROBE Initiative. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) Statement: Guidelines for Reporting Observational Studies. PLoS Med. 2007, 4, e296. [Google Scholar] [CrossRef]
- Rakislova, N.; Montironi, C.; Aldecoa, I.; Fernandez, E.; Bombi, J.A.; Jimeno, M.; Balaguer, F.; Pellise, M.; Castells, A.; Cuatrecasas, M. Lymph Node Pooling: A Feasible and Efficient Method of Lymph Node Molecular Staging in Colorectal Carcinoma. J. Transl. Med. 2017, 15, 14. [Google Scholar] [CrossRef]
- Saez De Gordoa, K.; Rodrigo-Calvo, M.T.; Archilla, I.; Lopez-Prades, S.; Diaz, A.; Tarragona, J.; Machado, I.; Ruiz Martín, J.; Zaffalon, D.; Daca-Alvarez, M.; et al. Lymph Node Molecular Analysis with OSNA Enables the Identification of pT1 CRC Patients at Risk of Recurrence: A Multicentre Study. Cancers 2023, 15, 5481. [Google Scholar] [CrossRef]
- Aldecoa, I.; Atares, B.; Tarragona, J.; Bernet, L.; Sardon, J.D.; Pereda, T.; Villar, C.; Mendez, M.C.; Gonzalez-Obeso, E.; Elorriaga, K.; et al. Molecularly Determined Total Tumour Load in Lymph Nodes of Stage I–II Colon Cancer Patients Correlates with High-Risk Factors. A Multicentre Prospective Study. Virchows Arch. 2016, 469, 385–394. [Google Scholar] [CrossRef]
- Tranoulis, A.; Georgiou, D.; Yap, J.; Attard-Montalto, S.; Twigg, J.; Elattar, A.; Singh, K.; Balega, J.; Kehoe, S. The Evolving Role of One-Step Nucleic Acid Amplification (OSNA) for the Intra-Operative Detection of Lymph Node Metastases: A Diagnostic Accuracy Meta-Analysis. Eur. J. Surg. Oncol. 2021, 47, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Wang, X.; Jiang, L.; Chen, X.; Bao, X.; Chen, X. The Diagnostic Value of One Step Nucleic Acid Amplification (OSNA) in Differentiating Lymph Node Metastasis of Tumors: A Systematic Review and Meta-Analysis. Int. J. Surg. 2018, 56, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Firth, D. Bias Reduction of Maximum Likelihood Estimates. Biometrika 1993, 80, 27–38. [Google Scholar] [CrossRef]
- Ogundimu, E.O.; Altman, D.G.; Collins, G.S. Adequate Sample Size for Developing Prediction Models Is Not Simply Related to Events per Variable. J. Clin. Epidemiol. 2016, 76, 175–182. [Google Scholar] [CrossRef]
- Sychowski, G.; Romanowicz, H.; Smolarz, B. Application of the OSNA Technique (One-Step Nucleic Acid Amplification Test) in Breast Cancer. Int. J. Mol. Sci. 2025, 26, 656. [Google Scholar] [CrossRef] [PubMed]
- Llompart-Coll, M.M.; Domínguez-Garijo, P.; Manyalich-Blasi, M.; Domènech-Gómez, G.; Perales-Galan, I.; Saavedra-Pérez, D.; Rodrigo, M.T.; Vidal-Sicart, S.; Pera-Roman, M.; Vidal-Pérez, O. Systematic Review and Meta-Analysis of Diagnostic Accuracy of One-Step Nucleic Acid Amplification for Lymph Node Metastases of Papillary Thyroid Carcinoma. Langenbecks Arch. Surg. 2025, 410, 184. [Google Scholar] [CrossRef]
- Märkl, B.; Grosser, B.; Bauer, K.; Vlasenko, D.; Schenkirsch, G.; Probst, A.; Kriening, B. Ultrastaging Using Ex Vivo Sentinel Lymph Node Mapping and One-Step Nucleic Acid Amplification (OSNA) in Gastric Cancer: Experiences of a European Center. Cancers 2021, 13, 2683. [Google Scholar] [CrossRef]
- Diaz-Mercedes, S.; Archilla, I.; Lahoz, S.; Rodrigo-Calvo, M.T.; Lopez-Prades, S.; Tarragona, J.; Landolfi, S.; Concha, A.; Machado, I.; Maurel, J.; et al. Cytology Smears: An Enhanced Alternative Method for Colorectal Cancer pN Stage—A Multicentre Study. Cancers 2022, 14, 6072. [Google Scholar] [CrossRef]
- Rodrigo-Calvo, M.T.; Saez De Gordoa, K.; Lopez-Prades, S.; Archilla, I.; Diaz, A.; Berrios, M.; Camps, J.; Musulen, E.; Cuatrecasas, M. Tumour Cell Seeding to Lymph Nodes from In Situ Colorectal Cancer. Cancers 2023, 15, 842. [Google Scholar] [CrossRef] [PubMed]
- Weixler, B.; Teixeira da Cunha, S.; Warschkow, R.; Demartines, N.; Güller, U.; Zettl, A.; Vahrmeijer, A.; van de Velde, C.J.H.; Viehl, C.T.; Zuber, M. Molecular Lymph Node Staging with One-Step Nucleic Acid Amplification and its Prognostic Value for Patients with Colon Cancer: The First Follow-up Study. World J. Surg. 2021, 45, 1526–1536. [Google Scholar] [CrossRef] [PubMed]
- Marhic, A.; Tremblay, J.-F.; Kaci, R.; André, T.; Eveno, C.; Pocard, M. Molecular Analysis of Sentinel Lymph Node in Colon Carcinomas by One-Step Nucleic Acid Amplification (OSNA) Reduces Time to Adjuvant Chemotherapy Interval. Dig. Liver Dis. 2017, 49, 924–928. [Google Scholar] [CrossRef]
- How, P.; Shihab, O.; Tekkis, P.; Brown, G.; Quirke, P.; Heald, R.; Moran, B. A Systematic Review of Cancer Related Patient Outcomes after Anterior Resection and Abdominoperineal Excision for Rectal Cancer in the Total Mesorectal Excision Era. Surg. Oncol. 2011, 20, e149–e155. [Google Scholar] [CrossRef] [PubMed]
- Swedish Rectal Cancer Trial; Cedermark, B.; Dahlberg, M.; Glimelius, B.; Påhlman, L.; Rutqvist, L.E.; Wilking, N. Improved Survival with Preoperative Radiotherapy in Resectable Rectal Cancer. N. Engl. J. Med. 1997, 336, 980–987. [Google Scholar] [CrossRef]
- Letaief, F.; Nasri, M.; Ayadi, M.; Meddeb, K.; Mokrani, A.; Yahyaoui, Y.; Chraiet, N.; Raies, H.; Mezlini, A. Potential Predictive Factors for Pathologic Complete Response after the Neoadjuvant Treatment of Rectal Adenocarcinoma: A Single Center Experience. Cancer Biol. Med. 2017, 14, 327–334. [Google Scholar] [CrossRef][Green Version]
- Kasi, A.; Abbasi, S.; Handa, S.; Al-Rajabi, R.; Saeed, A.; Baranda, J.; Sun, W. Total Neoadjuvant Therapy vs Standard Therapy in Locally Advanced Rectal Cancer: A Systematic Review and Meta-Analysis. JAMA Netw. Open 2020, 3, e2030097. [Google Scholar] [CrossRef]
- Brouwer, N.P.M.; Bos, A.C.R.K.; Lemmens, V.E.P.P.; Tanis, P.J.; Hugen, N.; Nagtegaal, I.D.; de Wilt, J.H.W.; Verhoeven, R.H.A. An Overview of 25 Years of Incidence, Treatment and Outcome of Colorectal Cancer Patients. Int. J. Cancer 2018, 143, 2758–2766. [Google Scholar] [CrossRef]
- Duraes, L.C.; Steele, S.R.; Valente, M.A.; Lavryk, O.A.; Connelly, T.M.; Kessler, H. Right Colon, Left Colon, and Rectal Cancer Have Different Oncologic and Quality of Life Outcomes. Int. J. Color. Dis. 2022, 37, 939–948. [Google Scholar] [CrossRef]
- Boustani, J.; Caubet, M.; Bosset, J.-F. Adjuvant Chemotherapy in Rectal Cancer after Chemoradiotherapy. Clin. Oncol. R. Coll. Radiol. 2016, 28, 140–145. [Google Scholar] [CrossRef]
- Tamburini, E.; Tassinari, D.; Ramundo, M.; De Stefano, A.; Viola, M.G.; Romano, C.; Elia, M.T.; Zanaletti, N.; Rudnas, B.; Casadei-Gardini, A.; et al. Adjuvant Chemotherapy after Neoadjuvant Chemo-Radiotherapy and Surgery in Locally Advanced Rectal Cancer. A Systematic Review of Literature with a Meta-Analysis of Randomized Clinical Trials. Crit. Rev. Oncol. Hematol. 2022, 172, 103627. [Google Scholar] [CrossRef]
- Glimelius, B. Adjuvant Chemotherapy in Rectal Cancer: State of the Art and Future Perspectives. Curr. Opin. Oncol. 2020, 32, 377–383. [Google Scholar] [CrossRef] [PubMed]
- Bregni, G.; Akin Telli, T.; Camera, S.; Deleporte, A.; Moretti, L.; Bali, A.M.; Liberale, G.; Holbrechts, S.; Hendlisz, A.; Sclafani, F. Adjuvant Chemotherapy for Rectal Cancer: Current Evidence and Recommendations for Clinical Practice. Cancer Treat. Rev. 2020, 83, 101948. [Google Scholar] [CrossRef]
- Quasar Collaborative Group. Adjuvant Chemotherapy versus Observation in Patients with Colorectal Cancer: A Randomised Study. Lancet 2007, 370, 2020–2029. [Google Scholar] [CrossRef] [PubMed]
- Song, J.H.; Lee, J.H.; Kim, S.H.; Um, J.W.; Korean Clinical Practice Guideline for Colon, Rectal Cancer Committee. Oxaliplatin-Based Adjuvant Chemotherapy Rather than Fluorouracil-Based Chemotherapy in Rectal Cancer Is More Efficient to Decrease Distant Metastasis and Increase Survival after Preoperative Chemoradiotherapy and Surgery: A Meta-Analysis. Int. J. Color. Dis. 2022, 37, 649–656. [Google Scholar] [CrossRef]
- Ma, B.; Ren, Y.; Chen, Y.; Lian, B.; Jiang, P.; Li, Y.; Shang, Y.; Meng, Q. Is Adjuvant Chemotherapy Necessary for Locally Advanced Rectal Cancer Patients with Pathological Complete Response after Neoadjuvant Chemoradiotherapy and Radical Surgery? A Systematic Review and Meta-Analysis. Int. J. Color. Dis. 2019, 34, 113–121. [Google Scholar] [CrossRef] [PubMed]
- Japanese Study Group for Postoperative Follow-up of Colorectal Cancer; Ahiko, Y.; Shida, D.; Kudose, Y.; Nakamura, Y.; Moritani, K.; Yamauchi, S.; Sugihara, K.; Kanemitsu, Y. Recurrence Hazard of Rectal Cancer Compared with Colon Cancer by Adjuvant Chemotherapy Status: A Nationwide Study in Japan. J. Gastroenterol. 2021, 56, 371–381. [Google Scholar] [CrossRef]
- Marano, L.; Verre, L.; Carbone, L.; Poto, G.E.; Fusario, D.; Venezia, D.F.; Calomino, N.; Kaźmierczak-Siedlecka, K.; Polom, K.; Marrelli, D.; et al. Current Trends in Volume and Surgical Outcomes in Gastric Cancer. J. Clin. Med. 2023, 12, 2708. [Google Scholar] [CrossRef]
- Güller, U.; Zettl, A.; Worni, M.; Langer, I.; Cabalzar-Wondberg, D.; Viehl, C.T.; Demartines, N.; Zuber, M. Molecular Investigation of Lymph Nodes in Colon Cancer Patients Using One-Step Nucleic Acid Amplification (OSNA): A New Road to Better Staging? Cancer 2012, 118, 6039–6045. [Google Scholar] [CrossRef]
- Yamamoto, H.; Murata, K.; Fukunaga, M.; Ohnishi, T.; Noura, S.; Miyake, Y.; Kato, T.; Ohtsuka, M.; Nakamura, Y.; Takemasa, I.; et al. Micrometastasis Volume in Lymph Nodes Determines Disease Recurrence Rate of Stage II Colorectal Cancer: A Prospective Multicenter Trial. Clin. Cancer Res. 2016, 22, 3201–3208. [Google Scholar] [CrossRef]
- O’Connor, E.S.; Greenblatt, D.Y.; LoConte, N.K.; Gangnon, R.E.; Liou, J.-I.; Heise, C.P.; Smith, M.A. Adjuvant Chemotherapy for Stage II Colon Cancer with Poor Prognostic Features. J. Clin. Oncol. 2011, 29, 3381–3388. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | RC Patients (n = 97) | CC Patients (n = 574) | p-Value |
|---|---|---|---|
| Sex (male), n (%) | 58 (59.8) | 344 (59.9) | 0.980 |
| Age (years), median (IQR) | 67.0 (59.0, 78.0) | 71.0 (62.0, 80.0) | 0.083 |
| Range (years) | 41–90 | 24–94 | |
| Tumor size (cm), median (IQR) | 3.2 (2.1, 4.0) | 3.1 (2.0, 4.5) | 0.779 |
| Range (cm) | 0.3–7.1 | 0.2–13 | |
| Tumor grade, n (%) | 0.004 | ||
| Low (G1–G2) | 77 (79.4) | 371 (64.6) | |
| High (G3–G4) | 20 (20.6) | 203 (35.4) | |
| Tumor budding, n (%) † | 0.475 | ||
| Bd1 | 68 (73.9) | 372 (69.5) | |
| Bd2 | 14 (15.2) | 108 (20.2) | |
| Bd3 | 10 (10.9) | 55 (10.3) | |
| Perineural invasion, n (%) † | 0.326 | ||
| Absent | 82 (84.5) | 503 (88.1) | |
| Present | 15 (15.5) | 68 (11.9) | |
| Vascular invasion, n (%) | 0.855 | ||
| Absent | 71 (73.2) | 415 (72.3) | |
| Present | 26 (26.8) | 159 (27.7) | |
| Tumor deposits, n (%) † | 0.738 | ||
| Absent | 90 (92.8) | 534 (93.7) | |
| Present | 7 (7.2) | 36 (6.3) | |
| Total LNs, median (IQR) | 19.0 (15.0, 24.0) | 19.0 (15.0, 25.0) | 0.719 |
| Fresh LNs, median (IQR) | 16.0 (12.0, 21.0) | 16.0 (11.0, 20.8) | 0.607 |
| Post-fixation LNs, median (IQR) | 1.0 (0, 6.0) | 2.0 (0, 5.0) | 0.292 |
| pT stage, n (%) | 0.084 | ||
| T1 | 18 (18.6) | 103 (17.9) | |
| T2 | 27 (27.8) | 117 (20.4) | |
| T3 | 46 (47.4) | 277 (48.3) | |
| T4 | 6 (6.2) | 77 (13.4) | |
| pN stage, n (%) | 0.673 | ||
| N0 | 72 (74.2) | 430 (74.9) | |
| N1 | 19 (19.6) | 102 (17.8) | |
| N2 | 6 (6.2) | 42 (7.3) | |
| pStage, n (%) | 0.452 | ||
| I | 40 (41.2) | 199 (34.7) | |
| II | 32 (33.0) | 231 (40.2) | |
| III | 25 (25.8) | 144 (25.1) | |
| TTL, median (IQR) | 0 (0, 2300) | 0 (0, 1350) | 0.993 |
| Range | 0–318,000 | 0–1,724,500 | |
| Mean | 15,873.40 | 20,608.80 | |
| OSNA positive a, n (%) | 33 (34.0) | 205 (35.7) | 0.747 |
| OSNA clinically relevant b, n (%) | 19 (19.6) | 87 (15.2) | 0.268 |
| Adjuvant chemotherapy, n (%) † | 0.561 | ||
| No | 75 (77.3) | 416 (74.6) | |
| Yes | 22 (22.7) | 142 (25.4) | |
| 3-year OS (IQR, %) † | 92.3 (86.5, 98.4) | 89.9 (87.0, 92.9) | 0.364 |
| 3-year DFS (IQR, %) † | 89.0 (82.4, 96.1) | 82.6 (79.0, 86.2) | 0.184 |
| 3-year RFS (IQR, %) † | 93.0 (87.7, 98.6) | 86.9 (83.8, 90.2) | 0.197 |
| H&E, pN Stage | n | Concordance (95% CI) | Sensitivity (95% CI) | Specificity (95% CI) | PPV (95% CI) | NPV (95% CI) | Kappa Index | McNemar’s p-Value | |||
|---|---|---|---|---|---|---|---|---|---|---|---|
| N0 | N1a~2b | ||||||||||
| OSNA | − | 61 | 2 † | 96 | 0.865 (0.780, 0.926) | 0.917 (0.730, 0.990) | 0.847 (0.743, 0.921) | 0.667 (0.511, 0.946) | 0.968 (0.882, 0.985) | 0.679 (0.517, 0.841) | 0.013 |
| + | 11 | 22 | |||||||||
| Nodal Status Assessed by OSNA/H&E | No. of Patients (%) | CSS | RFS | ||
|---|---|---|---|---|---|
| HR (95% CI) | p-Value | HR (95% CI) | p-Value | ||
| TTL (logarithmic scale), continuous | 84 (100) | 1.27 (1.02, 1.56) | 0.010 | 1.24 (1.06, 1.45) | 0.003 |
| OSNA positive a | |||||
| No | 58 (69.0) | ref | ref | ||
| Yes | 26 (31.0) | 5.72 (0.76, 43.35) | 0.091 | 5.86 (1.25, 27.52) | 0.011 |
| OSNA clinically relevant b | |||||
| No | 70 (83.3) | ref | ref | ||
| Yes | 14 (16.7) | 6.90 (1.17, 40.85) | 0.022 | 5.34 (1.33, 21.39) | 0.017 |
| TTL subgroups, copies/μL | |||||
| [0, 250) | 58 (69.0) | ref | ref | ||
| [250, 6000) | 12 (14.3) | 3.73 (0.28, 49.42) | 0.270 | 4.63 (0.68, 31.28) | 0.101 |
| [6000, ~) | 14 (16.7) | 9.07 (1.02, 80.26) | 0.017 | 7.90 (1.46, 42.63) | 0.008 |
| pN stage | |||||
| N0 | 63 (75.0) | ref | ref | ||
| N1/2 | 21 (25.0) | 8.66 (1.14, 65.60) | 0.012 | 8.48 (1.80, 39.89) | 0.002 |
| pN stage | |||||
| N0 | 63 (75.0) | ref | ref | ||
| N1 | 16 (19.0) | 5.97 (0.59, 60.08) | 0.080 | 5.56 (0.95, 32.59) | 0.051 |
| N2 | 5 (6.0) | 24.72 (2.45, 249.61) | 0.003 | 26.58 (4.44, 158.99) | <0.001 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Q.; Lopez-Prades, S.; Saez de Gordoa, K.; Rodrigo-Calvo, M.; Garcia, M.; Ruiz Martin, J.; Romo, A.; Pinilla, I.; Tarragona, J.; Alen, B.O.; et al. Performance and Prognostic Relevance of Lymph Node Assessment by One-Step Nucleic Acid Amplification Assay in Rectal Cancer: A Multicenter Study. Cancers 2025, 17, 2141. https://doi.org/10.3390/cancers17132141
Liu Q, Lopez-Prades S, Saez de Gordoa K, Rodrigo-Calvo M, Garcia M, Ruiz Martin J, Romo A, Pinilla I, Tarragona J, Alen BO, et al. Performance and Prognostic Relevance of Lymph Node Assessment by One-Step Nucleic Acid Amplification Assay in Rectal Cancer: A Multicenter Study. Cancers. 2025; 17(13):2141. https://doi.org/10.3390/cancers17132141
Chicago/Turabian StyleLiu, Qing, Sandra Lopez-Prades, Karmele Saez de Gordoa, Maite Rodrigo-Calvo, Mireia Garcia, Juan Ruiz Martin, Angel Romo, Ignacio Pinilla, Jordi Tarragona, Begoña Otero Alen, and et al. 2025. "Performance and Prognostic Relevance of Lymph Node Assessment by One-Step Nucleic Acid Amplification Assay in Rectal Cancer: A Multicenter Study" Cancers 17, no. 13: 2141. https://doi.org/10.3390/cancers17132141
APA StyleLiu, Q., Lopez-Prades, S., Saez de Gordoa, K., Rodrigo-Calvo, M., Garcia, M., Ruiz Martin, J., Romo, A., Pinilla, I., Tarragona, J., Alen, B. O., Camps, J., Archilla, I., & Cuatrecasas, M. (2025). Performance and Prognostic Relevance of Lymph Node Assessment by One-Step Nucleic Acid Amplification Assay in Rectal Cancer: A Multicenter Study. Cancers, 17(13), 2141. https://doi.org/10.3390/cancers17132141









