Curcumin Induces Homologous Recombination Deficiency by BRCA2 Degradation in Breast Cancer and Normal Cells
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. PDX Tissue Handling
2.3. Cell and Tissue Treatment
2.4. Irradiation
2.5. Immunostaining
2.6. Imaging Acquisition and Analysis
2.7. Western Blotting
2.8. Clonogenic Survival Assay
2.9. Statistics
3. Results
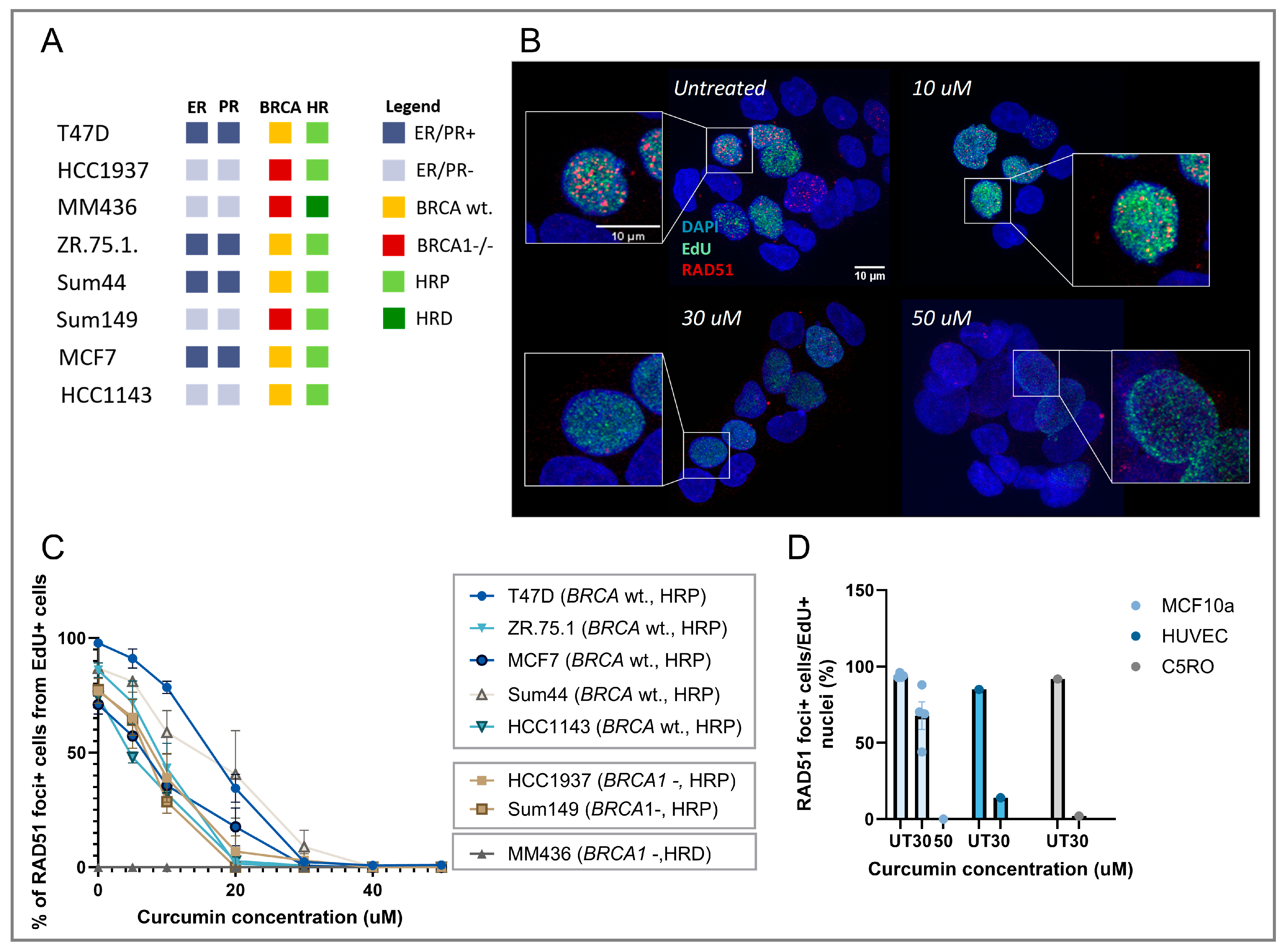
3.1. Curcumin Treatment Inhibits RAD51 IRIF Formation in Replicating Cells
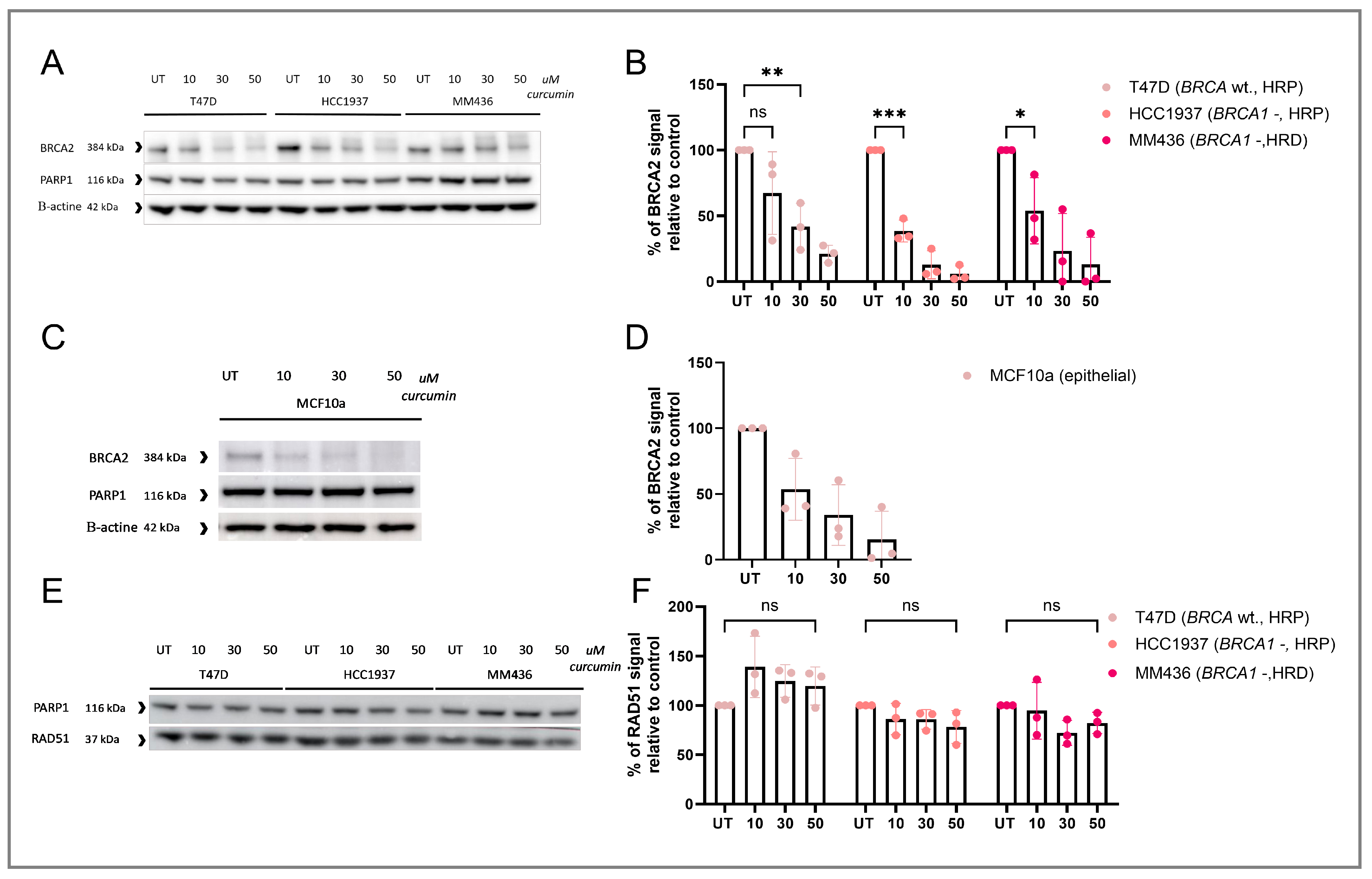
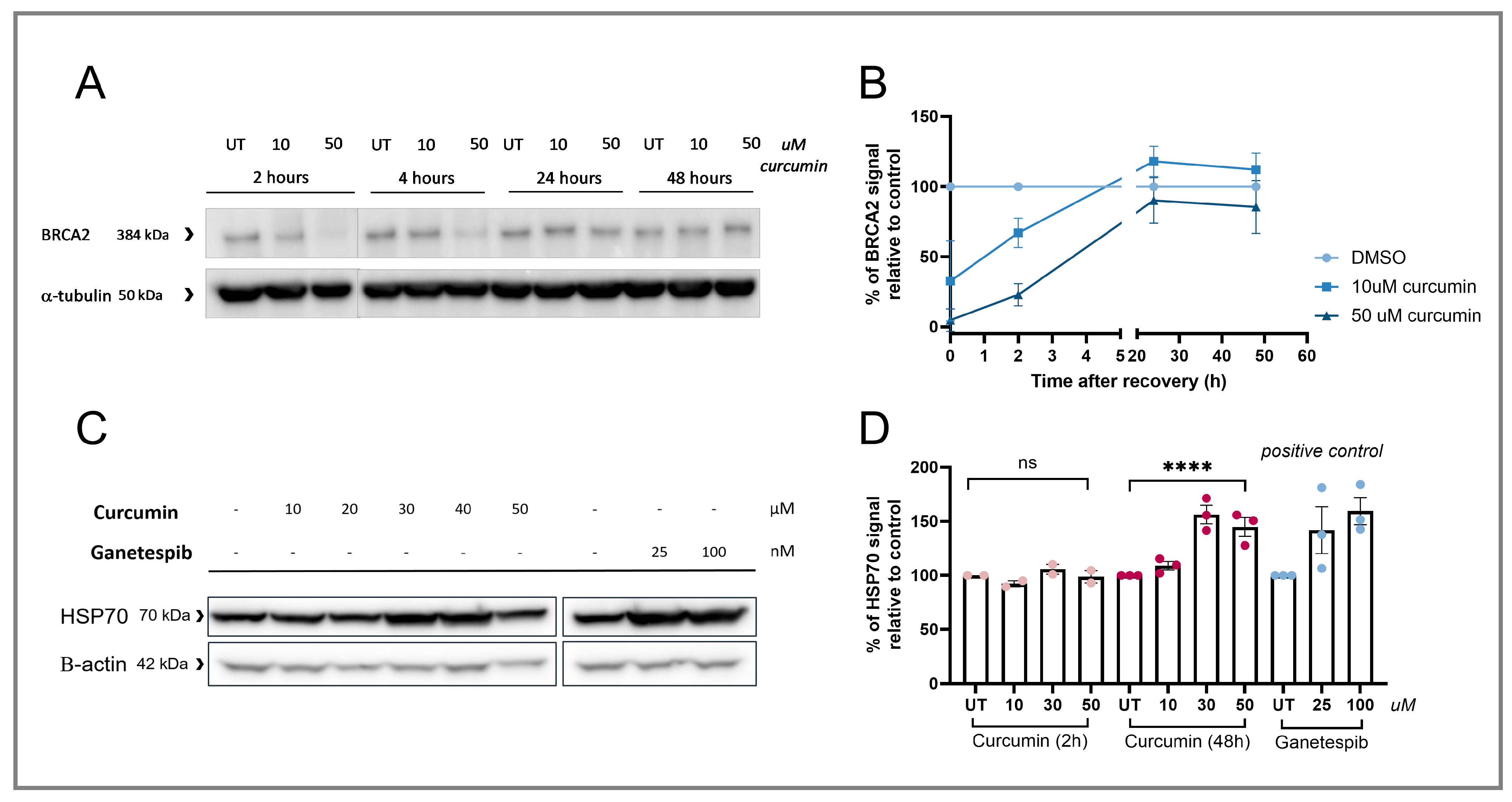
3.2. RAD51 IRIF Loss Is Caused by Curcumin-Induced BRCA2 Degradation
3.3. Curcumin Treatment Specifically Induces HRD and Sensitizes HRP Cell Lines to PARPi Therapy
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| BC | Breast Cancer |
| BRCA1/2 | Breast cancer 1/2 protein |
| DSB | Double stranded break |
| EdU | 5-Ethynyl-2′-doxyuridine |
| HR | Homologous Recombination |
| HRD | Homologous Recombination deficiency |
| HRP | Homologous Recombination proficiency |
| HSP90 | Heat shock protein 90 |
| OTC | Over-the-counter |
| PARPi | Poly (ADP-ribose) polymerase (PARP)-inhibitors |
| PDX | Patient derived Xenografts |
| RAD51 IRIF | RAD51 Ionizing radiation induced foci |
References
- WHO. WHO Launches New Roadmap on Breast Cancer. 2023. Available online: https://www.who.int/news/item/03-02-2023-who-launches-new-roadmap-on-breast-cancer (accessed on 19 November 2024).
- Arnold, M.; Morgan, E.; Rumgay, H.; Mafra, A.; Singh, D.; Laversanne, M.; Vignat, J.; Gralow, J.R.; Cardoso, F.; Siesling, S. Current and future burden of breast cancer: Global statistics for 2020 and 2040. Breast 2022, 66, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Damery, S.; Gratus, C.; Grieve, R.; Warmington, S.; Jones, J.; Routledge, P.; Greenfield, S.; Dowswell, G.; Sherriff, J.; Wilson, S. The use of herbal medicines by people with cancer: A cross-sectional survey. Br. J. Cancer 2011, 104, 927–933. [Google Scholar] [CrossRef] [PubMed]
- Strimpakos, A.S.; Sharma, R.A. Curcumin: Preventive and therapeutic properties in laboratory studies and clinical trials. Antioxid. Redox Signal. 2008, 10, 511–546. [Google Scholar] [CrossRef] [PubMed]
- He, Z.-Y.; Shi, C.-B.; Wen, H.; Li, F.-L.; Wang, B.-L.; Wang, J. Upregulation of p53 expression in patients with colorectal cancer by administration of curcumin. Cancer Investig. 2011, 29, 208–213. [Google Scholar] [CrossRef]
- Dhillon, N.; Aggarwal, B.B.; Newman, R.A.; Wolff, R.A.; Kunnumakkara, A.B.; Abbruzzese, J.L.; Ng, C.S.; Badmaev, V.; Kurzrock, R. Phase II trial of curcumin in patients with advanced pancreatic cancer. Clin. Cancer Res. 2008, 14, 4491–4499. [Google Scholar] [CrossRef]
- Ghasemi, F.; Shafiee, M.; Banikazemi, Z.; Pourhanifeh, M.H.; Khanbabaei, H.; Shamshirian, A.; Moghadam, S.A.; ArefNezhad, R.; Sahebkar, A.; Avan, A. Curcumin inhibits NF-kB and Wnt/β-catenin pathways in cervical cancer cells. Pathol. Res. Pract. 2019, 215, 152556. [Google Scholar] [CrossRef]
- Nagaraju, G.P.; Aliya, S.; Zafar, S.F.; Basha, R.; Diaz, R.; El-Rayes, B.F. The impact of curcumin on breast cancer. Integr. Biol. 2012, 4, 996–1007. [Google Scholar] [CrossRef]
- Pillai, G.R.; Srivastava, A.S.; Hassanein, T.I.; Chauhan, D.P.; Carrier, E. Induction of apoptosis in human lung cancer cells by curcumin. Cancer Lett. 2004, 208, 163–170. [Google Scholar] [CrossRef]
- Yu, J.; Peng, Y.; Wu, L.-C.; Xie, Z.; Deng, Y.; Hughes, T.; He, S.; Mo, X.; Chiu, M.; Wang, Q.-E. Curcumin down-regulates DNA methyltransferase 1 and plays an anti-leukemic role in acute myeloid leukemia. PLoS ONE 2013, 8, e55934. [Google Scholar] [CrossRef]
- Prasad, S.; Tyagi, A.K.; Aggarwal, B.B. Recent developments in delivery, bioavailability, absorption and metabolism of curcumin: The golden pigment from golden spice. Cancer Res. Treat. Off. J. Korean Cancer Assoc. 2014, 46, 2–18. [Google Scholar] [CrossRef]
- Greil, R.; Greil-Ressler, S.; Weiss, L.; Schönlieb, C.; Magnes, T.; Radl, B.; Bolger, G.T.; Vcelar, B.; Sordillo, P.P. A phase 1 dose-escalation study on the safety, tolerability and activity of liposomal curcumin (Lipocurc™) in patients with locally advanced or metastatic cancer. Cancer Chemother. Pharmacol. 2018, 82, 695–706. [Google Scholar] [CrossRef]
- Sharma, R.A.; Euden, S.A.; Platton, S.L.; Cooke, D.N.; Shafayat, A.; Hewitt, H.R.; Marczylo, T.H.; Morgan, B.; Hemingway, D.; Plummer, S.M. Phase I clinical trial of oral curcumin: Biomarkers of systemic activity and compliance. Clin. Cancer Res. 2004, 10, 6847–6854. [Google Scholar] [CrossRef]
- Cheng, A.-L.; Hsu, C.-H.; Lin, J.-K.; Hsu, M.-M.; Ho, Y.-F.; Shen, T.-S.; Ko, J.-Y.; Lin, J.-T.; Lin, B.-R.; Wu, M.-S.; et al. Phase I clinical trial of curcumin, a chemopreventive agent, in patients with high-risk or pre-malignant lesions. Anticancer Res. 2001, 21, e2900. [Google Scholar]
- Ogiwara, H.; Ui, A.; Shiotani, B.; Zou, L.; Yasui, A.; Kohno, T. Curcumin suppresses multiple DNA damage response pathways and has potency as a sensitizer to PARP inhibitor. Carcinogenesis 2013, 34, 2486–2497. [Google Scholar] [CrossRef] [PubMed]
- Levine, A.J. Spontaneous and inherited TP53 genetic alterations. Oncogene 2021, 40, 5975–5983. [Google Scholar] [CrossRef] [PubMed]
- Heijink, A.M.; Stok, C.; Porubsky, D.; Manolika, E.M.; de Kanter, J.K.; Kok, Y.P.; Everts, M.; de Boer, H.R.; Audrey, A.; Bakker, F.J. Sister chromatid exchanges induced by perturbed replication can form independently of BRCA1, BRCA2 and RAD51. Nature Commun. 2022, 13, 6722. [Google Scholar] [CrossRef]
- Mateo, J.; Moreno, V.; Gupta, A.; Kaye, S.B.; Dean, E.; Middleton, M.R.; Friedlander, M.; Gourley, C.; Plummer, R.; Rustin, G. An adaptive study to determine the optimal dose of the tablet formulation of the PARP inhibitor olaparib. Target. Oncol. 2016, 11, 401–415. [Google Scholar] [CrossRef]
- Krawczyk, P.M.; Eppink, B.; Essers, J.; Stap, J.; Rodermond, H.; Odijk, H.; Zelensky, A.; van Bree, C.; Stalpers, L.J.; Buist, M.R. Mild hyperthermia inhibits homologous recombination, induces BRCA2 degradation, and sensitizes cancer cells to poly (ADP-ribose) polymerase-1 inhibition. Proc. Natl. Acad. Sci. USA 2011, 108, 9851–9856. [Google Scholar] [CrossRef]
- Tan, S.L.W.; Chadha, S.; Liu, Y.; Gabasova, E.; Perera, D.; Ahmed, K.; Constantinou, S.; Renaudin, X.; Lee, M.; Aebersold, R. A class of environmental and endogenous toxins induces BRCA2 haploinsufficiency and genome instability. Cell 2017, 169, 1105–1118.e15. [Google Scholar] [CrossRef]
- Neri, P.; Ren, L.; Gratton, K.; Stebner, E.; Johnson, J.; Klimowicz, A.; Duggan, P.; Tassone, P.; Mansoor, A.; Stewart, D.A. Bortezomib-induced “BRCAness” sensitizes multiple myeloma cells to PARP inhibitors. Blood J. Am. Soc. Hematol. 2011, 118, 6368–6379. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, Y.; Uray, I.P.; Shen, J.; Wang, L.; Peng, X.; Brown, P.H.; Tu, W.; Peng, G. Natural product β-thujaplicin inhibits homologous recombination repair and sensitizes cancer cells to radiation therapy. DNA Repair 2017, 60, 89–101. [Google Scholar] [CrossRef] [PubMed]
- Zelensky, A.N.; Sanchez, H.; Ristic, D.; Vidic, I.; van Rossum-Fikkert, S.E.; Essers, J.; Wyman, C.; Kanaar, R. Caffeine suppresses homologous recombination through interference with RAD51-mediated joint molecule formation. Nucleic Acids Res. 2013, 41, 6475–6489. [Google Scholar] [CrossRef]
- Naipal, K.A.T.; Verkaik, N.S.; Ameziane, N.; van Deurzen, C.H.M.; Ter Brugge, P.; Meijers, M.; Sieuwerts, A.M.; Martens, J.W.; O’Connor, M.J.; Vrieling, H. Functional ex vivo assay to select homologous recombination–deficient breast tumors for PARP inhibitor treatment. Clin. Cancer Res. 2014, 20, 4816–4826. [Google Scholar] [CrossRef] [PubMed]
- Debnath, J.; Muthuswamy, S.K.; Brugge, J.S. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods 2003, 30, 256–268. [Google Scholar] [CrossRef]
- Meijer, T.G.; Martens, J.W.M.; Prager-van der Smissen, W.J.C.; Verkaik, N.S.; Beaufort, C.M.; van Herk, S.; Robert-Finestra, T.; Hoogenboezem, R.M.; Ruigrok-Ritstier, K.; Paul, M.W. Functional Homologous Recombination (HR) Screening Shows the Majority of BRCA1/2-Mutant Breast and Ovarian Cancer Cell Lines Are HR-Proficient. Cancers 2024, 16, 741. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, E.; Vincent-Salomon, A.; Auger, N.; Degeorges, A.; Assayag, F.; De Cremoux, P.; De Plater, L.; Guyader, C.; De Pinieux, G.; Judde, J.-G. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 2007, 13, 3989–3998. [Google Scholar] [CrossRef]
- Coussy, F.; de Koning, L.; Lavigne, M.; Bernard, V.; Ouine, B.; Boulai, A.; El Botty, R.; Dahmani, A.; Montaudon, E.; Assayag, F. A large collection of integrated genomically characterized patient-derived xenografts highlighting the heterogeneity of triple-negative breast cancer. Int. J. Cancer 2019, 145, 1902–1912. [Google Scholar] [CrossRef]
- Naipal, K.A.T.; Verkaik, N.S.; Sánchez, H.; van Deurzen, C.H.M.; den Bakker, M.A.; Hoeijmakers, J.H.J.; Kanaar, R.; Vreeswijk, M.P.G.; Jager, A.; van Gent, D.C. Tumor slice culture system to assess drug response of primary breast cancer. BMC Cancer 2016, 16, 78. [Google Scholar] [CrossRef]
- Guney Eskiler, G.; Sahin, E.; Deveci Ozkan, A.; Cilingir Kaya, O.T.; Kaleli, S. Curcumin induces DNA damage by mediating homologous recombination mechanism in triple negative breast cancer. Nutr. Cancer 2020, 72, 1057–1066. [Google Scholar] [CrossRef]
- Essers, J.; Houtsmuller, A.B.; van Veelen, L.; Paulusma, C.; Nigg, A.L.; Pastink, A.; Vermeulen, W.; Hoeijmakers, J.H.J.; Kanaar, R. Nuclear dynamics of RAD52 group homologous recombination proteins in response to DNA damage. EMBO J. 2002, 21, 2030–2037. [Google Scholar] [CrossRef]
- Van Den Tempel, N.; Zelensky, A.N.; Odijk, H.; Laffeber, C.; Schmidt, C.K.; Brandsma, I.; Demmers, J.; Krawczyk, P.M.; Kanaar, R. On the mechanism of hyperthermia-induced BRCA2 protein degradation. Cancers 2019, 11, 97. [Google Scholar] [CrossRef]
- Van Der Burg, M.; Ijspeert, H.; Verkaik, N.S.; Turul, T.; Wiegant, W.W.; Morotomi-Yano, K.; Mari, P.-O.; Tezcan, I.; Chen, D.J.; Zdzienicka, M.Z. A DNA-PKcs mutation in a radiosensitive T–B–SCID patient inhibits Artemis activation and nonhomologous end-joining. J. Clin. Investig. 2009, 119, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Stael, S.; Miller, L.P.; Fernandez-Fernandez, A.D.; Van Breusegem, F. Detection of damage-activated metacaspase activity by western blot in plants. In Plant Proteases and Plant Cell Death: Methods and Protocols; Humana: New York, NY, USA, 2022; pp. 127–137. [Google Scholar]
- Pellegrini, L.; Yu, D.S.; Lo, T.; Anand, S.; Lee, M.; Blundell, T.L.; Venkitaraman, A.R. Insights into DNA recombination from the structure of a RAD51–BRCA2 complex. Nature 2002, 420, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Davies, E.; Hiscox, S. New therapeutic approaches in breast cancer. Maturitas 2011, 68, 121–128. [Google Scholar] [CrossRef] [PubMed]
- Kudryavtsev, V.A.; Khokhlova, A.V.; Mosina, V.A.; Selivanova, E.I.; Kabakov, A.E. Induction of Hsp70 in tumor cells treated with inhibitors of the Hsp90 activity: A predictive marker and promising target for radiosensitization. PLoS ONE 2017, 12, e0173640. [Google Scholar] [CrossRef]
- van de Kamp, G.; Heemskerk, T.; Kanaar, R.; Essers, J. DNA double strand break repair pathways in response to different types of ionizing radiation. Front. Genet. 2021, 12, 738230. [Google Scholar] [CrossRef]
- Fan, Y.-J.; Zhou, Y.-X.; Zhang, L.-R.; Lin, Q.-F.; Gao, P.-Z.; Cai, F.; Zhu, L.-P.; Liu, B.; Xu, J.-H. C1206, a novel curcumin derivative, potently inhibits Hsp90 and human chronic myeloid leukemia cells in vitro. Acta Pharmacol. Sin. 2018, 39, 649–658. [Google Scholar] [CrossRef]
- Giommarelli, C.; Zuco, V.; Favini, E.; Pisano, C.; Dal Piaz, F.; De Tommasi, N.; Zunino, F. The enhancement of antiproliferative and proapoptotic activity of HDAC inhibitors by curcumin is mediated by Hsp90 inhibition. Cell. Mol. Life Sci. 2010, 67, 995–1004. [Google Scholar] [CrossRef]
- Ying, W.; Du, Z.; Sun, L.; Foley, K.P.; Proia, D.A.; Blackman, R.K.; Zhou, D.; Inoue, T.; Tatsuta, N.; Sang, J. Ganetespib, a unique triazolone-containing Hsp90 inhibitor, exhibits potent antitumor activity and a superior safety profile for cancer therapy. Mol. Cancer Ther. 2012, 11, 475–484. [Google Scholar] [CrossRef]
- Jiang, J.; Lu, Y.; Li, Z.; Li, L.; Niu, D.; Xu, W.; Liu, J.; Fu, L.; Zhou, Z.; Gu, Y. Ganetespib overcomes resistance to PARP inhibitors in breast cancer by targeting core proteins in the DNA repair machinery. Investig. New Drugs 2017, 35, 251–259. [Google Scholar] [CrossRef]
- Baell, J.; Walters, M.A. Chemistry: Chemical con artists foil drug discovery. Nature 2014, 513, 481–483. [Google Scholar] [CrossRef] [PubMed]
- Nelson, K.M.; Dahlin, J.L.; Bisson, J.; Graham, J.; Pauli, G.F.; Walters, M.A. The essential medicinal chemistry of curcumin: Miniperspective. J. Med. Chem. 2017, 60, 1620–1637. [Google Scholar]
- Shi, M.; Cai, Q.; Yao, L.; Mao, Y.; Ming, Y.; Ouyang, G. Antiproliferation and apoptosis induced by curcumin in human ovarian cancer cells. Cell Biol. Int. 2006, 30, 221–226. [Google Scholar] [PubMed]
- Hu, S.; Xu, Y.; Meng, L.; Huang, L.; Sun, H. Curcumin inhibits proliferation and promotes apoptosis of breast cancer cells. Exp. Ther. Med. 2018, 16, 1266–1272. [Google Scholar] [PubMed]
- Senft, C.; Polacin, M.; Priester, M.; Seifert, V.; Kögel, D.; Weissenberger, J. The nontoxic natural compound Curcumin exerts anti-proliferative, anti-migratory, and anti-invasive properties against malignant gliomas. BMC Cancer 2010, 10, 491. [Google Scholar] [CrossRef]
- Yallapu, M.M.; Maher, D.M.; Sundram, V.; Bell, M.C.; Jaggi, M.; Chauhan, S.C. Curcumin induces chemo/radio-sensitization in ovarian cancer cells and curcumin nanoparticles inhibit ovarian cancer cell growth. J. Ovarian Res. 2010, 3, 11. [Google Scholar]
- Chen, P.; Li, J.; Jiang, H.-G.; Lan, T.; Chen, Y.-C. Curcumin reverses cisplatin resistance in cisplatin-resistant lung cancer cells by inhibiting FA/BRCA pathway. Tumor Biol. 2015, 36, 3591–3599. [Google Scholar]
- Zhao, Q.; Guan, J.; Qin, Y.; Ren, P.; Zhang, Z.; Lv, J.; Sun, S.; Zhang, C.; Mao, W. Curcumin sensitizes lymphoma cells to DNA damage agents through regulating Rad51-dependent homologous recombination. Biomed. Pharmacother. 2018, 97, 115–119. [Google Scholar] [CrossRef]
- Zou, J.; Zhu, L.; Jiang, X.; Wang, Y.; Wang, Y.; Wang, X.; Chen, B. Curcumin increases breast cancer cell sensitivity to cisplatin by decreasing FEN1 expression. Oncotarget 2018, 9, 11268–11278. [Google Scholar]
- Syng-Ai, C.; Kumari, A.L.; Khar, A. Effect of curcumin on normal and tumor cells: Role of glutathione and bcl-2. Mol. Cancer Ther. 2004, 3, 1101–1108. [Google Scholar] [CrossRef]
- Hussein, H.A.; Khaphi, F.L. The apoptotic activity of Curcumin against Oral cancer cells without affecting Normal cells in comparison to paclitaxel activity. Appl. Biochem. Biotechnol. 2023, 195, 5019–5033. [Google Scholar] [CrossRef]
- Bolger, G.T.; Licollari, A.; Bagshaw, R.; Tan, A.; Greil, R.; Vcelar, B.; Majeed, M.; Sordillo, P. Intense uptake of liposomal curcumin by multiple myeloma cell lines: Comparison to normal lymphocytes, red blood cells and chronic lymphocytic leukemia cells. Anticancer Res. 2019, 39, 1161–1168. [Google Scholar] [CrossRef] [PubMed]
- Heger, M.; van Golen, R.F.; Broekgaarden, M.; Michel, M.C. The molecular basis for the pharmacokinetics and pharmacodynamics of curcumin and its metabolites in relation to cancer. Pharmacol. Rev. 2014, 66, 222–307. [Google Scholar] [PubMed]
- Anand, P.; Kunnumakkara, A.B.; Newman, R.A.; Aggarwal, B.B. Bioavailability of curcumin: Problems and promises. Mol. Pharm. 2007, 4, 807–818. [Google Scholar] [CrossRef] [PubMed]
- Hussaarts, K.G.A.M.; Hurkmans, D.P.; Oomen-de Hoop, E.; van Harten, L.J.; Berghuis, S.; van Alphen, R.J.; Spierings, L.E.A.; van Rossum-Schornagel, Q.C.; Vastbinder, M.B.; van Schaik, R.H.N. Impact of curcumin (with or without piperine) on the pharmacokinetics of tamoxifen. Cancers 2019, 11, 403. [Google Scholar] [CrossRef]
- Gera, M.; Sharma, N.; Ghosh, M.; Huynh, D.L.; Lee, S.J.; Min, T.; Kwon, T.; Jeong, D.K. Nanoformulations of curcumin: An emerging paradigm for improved remedial application. Oncotarget 2017, 8, 66680–66698. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Komar, Z.M.; Ladan, M.M.; Verkaik, N.S.; Dahmani, A.; Montaudon, E.; Marangoni, E.; Kanaar, R.; Nonnekens, J.; Houtsmuller, A.B.; Jager, A.; et al. Curcumin Induces Homologous Recombination Deficiency by BRCA2 Degradation in Breast Cancer and Normal Cells. Cancers 2025, 17, 2109. https://doi.org/10.3390/cancers17132109
Komar ZM, Ladan MM, Verkaik NS, Dahmani A, Montaudon E, Marangoni E, Kanaar R, Nonnekens J, Houtsmuller AB, Jager A, et al. Curcumin Induces Homologous Recombination Deficiency by BRCA2 Degradation in Breast Cancer and Normal Cells. Cancers. 2025; 17(13):2109. https://doi.org/10.3390/cancers17132109
Chicago/Turabian StyleKomar, Zofia M., Marjolijn M. Ladan, Nicole S. Verkaik, Ahmed Dahmani, Elodie Montaudon, Elisabetta Marangoni, Roland Kanaar, Julie Nonnekens, Adriaan B. Houtsmuller, Agnes Jager, and et al. 2025. "Curcumin Induces Homologous Recombination Deficiency by BRCA2 Degradation in Breast Cancer and Normal Cells" Cancers 17, no. 13: 2109. https://doi.org/10.3390/cancers17132109
APA StyleKomar, Z. M., Ladan, M. M., Verkaik, N. S., Dahmani, A., Montaudon, E., Marangoni, E., Kanaar, R., Nonnekens, J., Houtsmuller, A. B., Jager, A., & Gent, D. C. v. (2025). Curcumin Induces Homologous Recombination Deficiency by BRCA2 Degradation in Breast Cancer and Normal Cells. Cancers, 17(13), 2109. https://doi.org/10.3390/cancers17132109







