Reduction of NFX1-123 and HPV 16 E6 and E7 Decreased Telomerase and CENP-F in Cervical Cancer Cell Lines
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cells and Culture Conditions
2.2. Single-Cell RNA Library Preparation and Sequencing
2.3. Single-Cell Transcriptome Analysis
2.4. Gene Ontology Analysis of Differentially Expressed Genes Across Cell Clusters
2.5. RNA Isolation and Gene Expression Profiling by PCR
- INHBA-F: 5′-AAGTCGGGGAGAACGGGTATG-3′
- INHBA-R: 5′-TCTTCCTGGCTGTTCCTGAC-3′
- NEAT1-F: 5′-CCAGTTTTCCGAGAACCAAA-3′
- NEAT1-R: 5′-ATGCTGATCTGCTGCGTATG-3′
- RBM25-F: 5′-TGTCTTTTCCACCTCATTTGAATCG-3′
- RBM25 R: 5′-ATTGGTACAGGAATCATTGGGGT-3′
- PRRC2C-F: 5′-CCATCAGTAGCAAAAGTTCCC-3′
- PRRC2C-R: 5′-CTTCGCTCTTCCTCTTCACG-3′
- NEFM-F: 5′-TCAACGTCAAGATGGCTCTG-3′
- NEFM-R: 5′-GAGCTTCCACCTTGGGTTTC-3′
- DYNC1H1-F: 5′-GCCACCGTCAGTTTTGACAC-3′
- DYNC1H1-R: 5′-AAATTGCCTCCACCAAACGC-3′
- MKI67-F: 5′-CGTCCCAGTGGAAGAGTTGT-3′
- MKI67-R: 5′-CGACCCCGCTCCTTTTGATA-3′
- FTL-F: 5′-CAGCCTGGTCAATTTGTACCT-3′
- FLT-R: 5′-GCCAATTCG CGGAAGAAGTG-3′
- SAT1-F: 5′-GAGGCTTTGGCATAGGATCA-3′
- SAT1-R: 5′-TCCAACCCTCTTCACTGGAC-3′
- CENPF-F: 5′-CTCTCCCGTCAACAGCGTTC-3′,
- CENPF-R: 5′-GTTGTGCATATTCTTGGCTTGC-3′
- hTERT-F: 5′-CGAGCTGCTCAGGTCTTTCTTTTATG-3′
- hTERT-R: 5′-CCACGACGTAGTCCATGTTCACAATC-3′
- 36B4-F: 5′-TGCCAGTGTCTGTCTGCAGA-3′
- 36B4-R: 5′-ACAAAGGCAGATGGATCAGC-3′
2.6. Telomerase Activity Assay
2.7. Western Blot
2.8. TCGA Database Gene Expression Analysis in HPV-Associated and Non-HPV-Associated Cancers
2.9. DAVID Bioinformatic Analysis of Differentially Expressed Genes in CaSki Knock Out and Control Cells
3. Results
3.1. Three Cell Clusters Identified Using Single-Cell RNA Sequencing of NFX1-123 Knock Out CaSki Cells
3.2. Biological Processes Enriched in NFX1-123 KO Cell Clusters
3.3. Differentially Expressed Genes in NFX1-123 KO CaSki Cells
3.4. Gene Ontology Analysis of Decreased Genes in NFX1-123 KO CaSki Cells
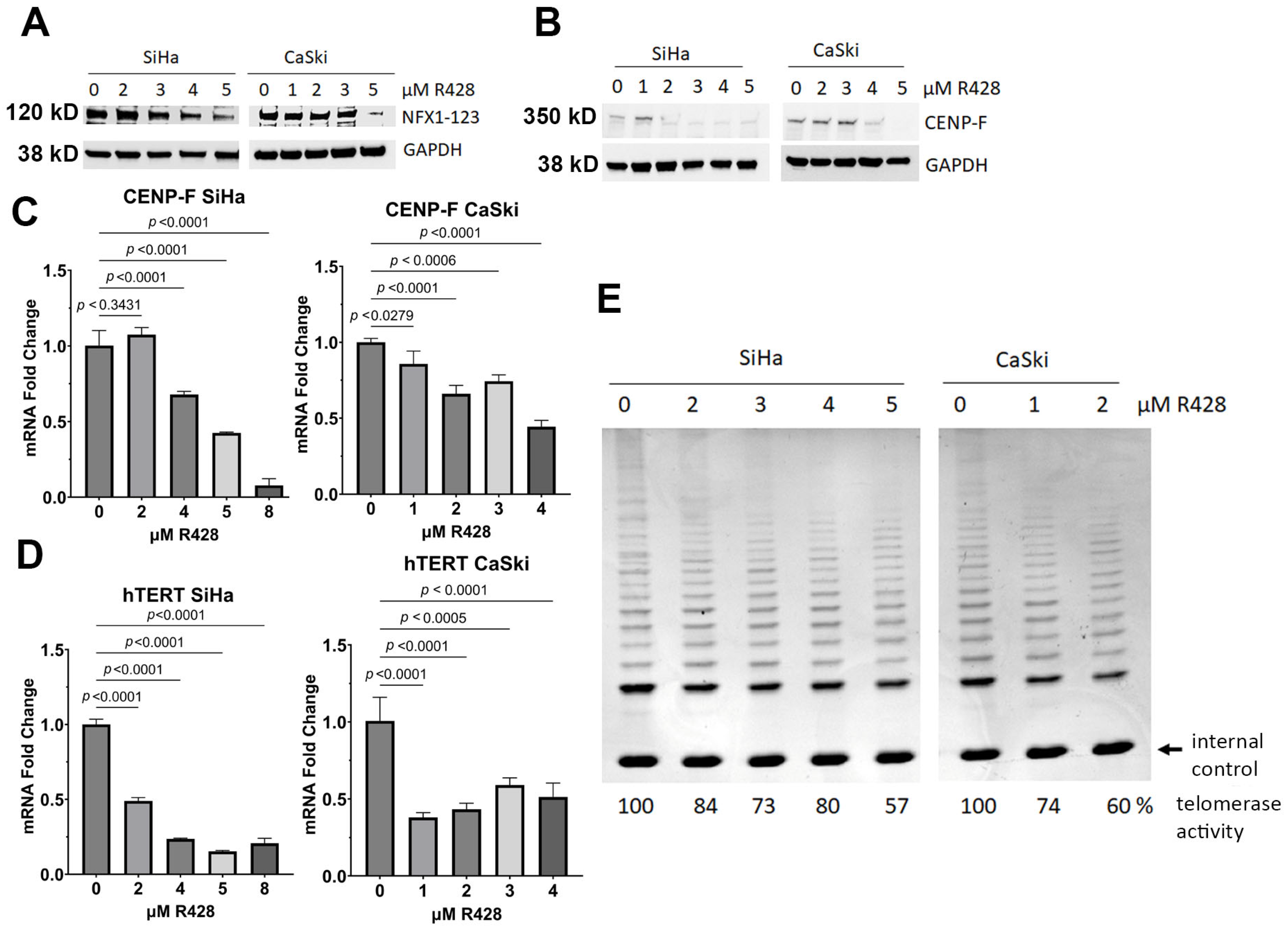
3.5. NFX1-123 Knock Out Decreased Telomerase Activity, hTERT, and CENP-F
3.6. Pharmacological Inhibition of NFX1-123 Decreased Telomerase Activity, hTERT, and CENP-F
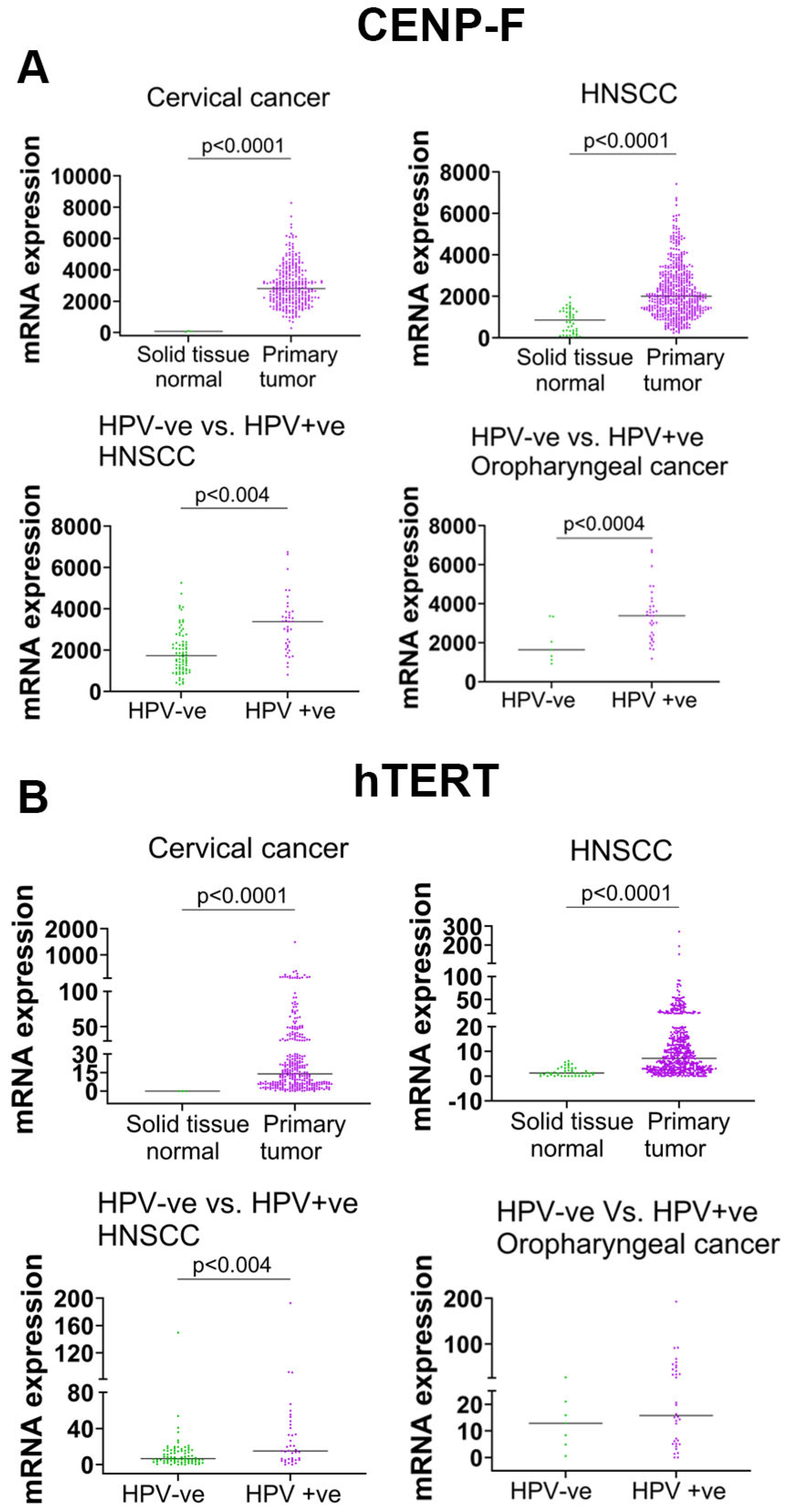
3.7. High-Risk HPV Increased hTERT and CENP-F mRNA in Primary Tumors of HPV-Associated Cancers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
| NFX1-123 | Nuclear transcription factor, X-box binding 1-123 |
| hTERT | Human telomerase reverse transcriptase |
| scRNAseq | Single-cell RNA sequencing |
| CENP-F | Centromere protein F |
References
- Chintala, S.; Dankoski, M.A.; Anbarasu, A.; Ramaiah, S.; Miryala, S.K.; Katzenellenbogen, R.A. NFX1-123: A potential therapeutic target in cervical cancer. J. Med. Virol. 2023, 95, e28856. [Google Scholar] [CrossRef] [PubMed]
- Chintala, S.; Levan, J.; Robinson, K.; Quist, K.; Katzenellenbogen, R.A. Genes Regulated by HPV 16 E6 and High Expression of NFX1-123 in Cervical Cancers. Onco Targets Ther. 2020, 13, 6143–6156. [Google Scholar] [CrossRef]
- Chintala, S.; Quist, K.M.; Gonzalez-DeWhitt, P.A.; Katzenellenbogen, R.A. High expression of NFX1-123 in HPV positive head and neck squamous cell carcinomas. Head Neck 2022, 44, 177–188. [Google Scholar] [CrossRef] [PubMed]
- Chintala, S.; Katzenellenbogen, R.A. NFX1, Its Isoforms and Roles in Biology, Disease and Cancer. Biology 2021, 10, 279. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, R.A.; Vliet-Gregg, P.; Xu, M.; Galloway, D.A. NFX1-123 increases hTERT expression and telomerase activity posttranscriptionally in human papillomavirus type 16 E6 keratinocytes. J. Virol. 2009, 83, 6446–6456. [Google Scholar] [CrossRef]
- Katzenellenbogen, R.A.; Vliet-Gregg, P.; Xu, M.; Galloway, D.A. Cytoplasmic poly (A) binding proteins regulate telomerase activity and cell growth in human papillomavirus type 16 E6-expressing keratinocytes. J. Virol. 2010, 84, 12934–12944. [Google Scholar] [CrossRef]
- Vliet-Gregg, P.A.; Hamilton, J.R.; Katzenellenbogen, R.A. NFX1-123 and human papillomavirus 16E6 increase Notch expression in keratinocytes. J. Virol. 2013, 87, 13741–13750. [Google Scholar] [CrossRef]
- Levan, J.; Vliet-Gregg, P.A.; Robinson, K.L.; Matsumoto, L.R.; Katzenellenbogen, R.A. HPV type 16 E6 and NFX1-123 augment JNK signaling to mediate keratinocyte differentiation and L1 expression. Virology 2019, 531, 171–182. [Google Scholar] [CrossRef]
- Levan, J.; Vliet-Gregg, P.A.; Robinson, K.L.; Katzenellenbogen, R.A. Human papillomavirus type 16 E6 and NFX1-123 mislocalize immune signaling proteins and downregulate immune gene expression in keratinocytes. PLoS ONE 2017, 12, e0187514. [Google Scholar] [CrossRef]
- Chen, X.; Lu, D.; Gao, J.; Zhu, H.; Zhou, Y.; Gao, D.; Zhou, H. Identification of a USP9X Substrate NFX1-123 by SILAC-based Quantitative Proteomics. J. Proteome Res. 2019, 18, 2654–2665. [Google Scholar] [CrossRef]
- Li, S.; Ikeuchi, K.; Kato, M.; Buschauer, R.; Sugiyama, T.; Adachi, S.; Kusano, H.; Natsume, T.; Berninghausen, O.; Matsuo, Y.; et al. Sensing of individual stalled 80S ribosomes by Fap1 for nonfunctional rRNA turnover. Mol. Cell 2022, 82, 3424–3437.e3428. [Google Scholar] [CrossRef] [PubMed]
- Tornesello, M.L.; Cerasuolo, A.; Starita, N.; Tornesello, A.L.; Bonelli, P.; Tuccillo, F.M.; Buonaguro, L.; Isaguliants, M.G.; Buonaguro, F.M. The Molecular Interplay between Human Oncoviruses and Telomerase in Cancer Development. Cancers 2022, 14, 5257. [Google Scholar] [CrossRef] [PubMed]
- Branca, M.; Giorgi, C.; Ciotti, M.; Santini, D.; Di Bonito, L.; Costa, S.; Benedetto, A.; Bonifacio, D.; Di Bonito, P.; Paba, P.; et al. Upregulation of telomerase (hTERT) is related to the grade of cervical intraepithelial neoplasia, but is not an independent predictor of high-risk human papillomavirus, virus persistence, or disease outcome in cervical cancer. Diagn. Cytopathol. 2006, 34, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Katzenellenbogen, R.A. Activation of telomerase by HPVs. Virus Res. 2017, 231, 50–55. [Google Scholar] [CrossRef]
- Liu, X.; Dakic, A.; Zhang, Y.; Dai, Y.; Chen, R.; Schlegel, R. HPV E6 protein interacts physically and functionally with the cellular telomerase complex. Proc. Natl. Acad. Sci. USA 2009, 106, 18780–18785. [Google Scholar] [CrossRef]
- Panczyszyn, A.; Boniewska-Bernacka, E.; Glab, G. Telomeres and Telomerase During Human Papillomavirus-Induced Carcinogenesis. Mol. Diagn. Ther. 2018, 22, 421–430. [Google Scholar] [CrossRef]
- Gewin, L.; Myers, H.; Kiyono, T.; Galloway, D.A. Identification of a novel telomerase repressor that interacts with the human papillomavirus type-16 E6/E6-AP complex. Genes. Dev. 2004, 18, 2269–2282. [Google Scholar] [CrossRef]
- James, M.A.; Lee, J.H.; Klingelhutz, A.J. HPV16-E6 associated hTERT promoter acetylation is E6AP dependent, increased in later passage cells and enhanced by loss of p300. Int. J. Cancer 2006, 119, 1878–1885. [Google Scholar] [CrossRef]
- Kelley, M.L.; Keiger, K.E.; Lee, C.J.; Huibregtse, J.M. The global transcriptional effects of the human papillomavirus E6 protein in cervical carcinoma cell lines are mediated by the E6AP ubiquitin ligase. J. Virol. 2005, 79, 3737–3747. [Google Scholar] [CrossRef]
- Liu, X.; Yuan, H.; Fu, B.; Disbrow, G.L.; Apolinario, T.; Tomaic, V.; Kelley, M.L.; Baker, C.C.; Huibregtse, J.; Schlegel, R. The E6AP ubiquitin ligase is required for transactivation of the hTERT promoter by the human papillomavirus E6 oncoprotein. J. Biol. Chem. 2005, 280, 10807–10816. [Google Scholar] [CrossRef]
- Xu, M.; Luo, W.; Elzi, D.J.; Grandori, C.; Galloway, D.A. NFX1 interacts with mSin3A/histone deacetylase to repress hTERT transcription in keratinocytes. Mol. Cell Biol. 2008, 28, 4819–4828. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, L.C.; da Silva, I.D.; Correa, J.C.; Ribalta, J.C. Survivin and telomerase expression in the uterine cervix of women with human papillomavirus-induced lesions. Int. J. Gynecol. Cancer 2011, 21, 15–21. [Google Scholar] [CrossRef]
- Reddy, V.G.; Khanna, N.; Jain, S.K.; Das, B.C.; Singh, N. Telomerase-A molecular marker for cervical cancer screening. Int. J. Gynecol. Cancer 2001, 11, 100–106. [Google Scholar] [CrossRef]
- Boscolo-Rizzo, P.; Da Mosto, M.C.; Rampazzo, E.; Giunco, S.; Del Mistro, A.; Menegaldo, A.; Baboci, L.; Mantovani, M.; Tirelli, G.; De Rossi, A. Telomeres and telomerase in head and neck squamous cell carcinoma: From pathogenesis to clinical implications. Cancer Metastasis Rev. 2016, 35, 457–474. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Rampazzo, E.; Perissinotto, E.; Piano, M.A.; Giunco, S.; Baboci, L.; Spinato, G.; Spinato, R.; Tirelli, G.; Da Mosto, M.C.; et al. Telomere shortening in mucosa surrounding the tumor: Biosensor of field cancerization and prognostic marker of mucosal failure in head and neck squamous cell carcinoma. Oral Oncol. 2015, 51, 500–507. [Google Scholar] [CrossRef] [PubMed]
- Boscolo-Rizzo, P.; Rampazzo, E.; Polesel, J.; Giunco, S.; Menegaldo, A.; Mantovani, M.; Stellin, M.; Bandolin, L.; Spinato, G.; Del Mistro, A.; et al. Predictive and prognostic significance of telomerase levels/telomere length in tissues and peripheral blood in head and neck squamous cell carcinoma. Sci. Rep. 2019, 9, 17572. [Google Scholar] [CrossRef]
- Fustero-Torre, C.; Jimenez-Santos, M.J.; Garcia-Martin, S.; Carretero-Puche, C.; Garcia-Jimeno, L.; Ivanchuk, V.; Di Domenico, T.; Gomez-Lopez, G.; Al-Shahrour, F. Beyondcell: Targeting cancer therapeutic heterogeneity in single-cell RNA-seq data. Genome Med. 2021, 13, 187. [Google Scholar] [CrossRef]
- Smit, M.M.; Feller, K.J.; You, L.; Storteboom, J.; Begce, Y.; Beerens, C.; Chien, M.P. Spatially Annotated Single Cell Sequencing for Unraveling Intratumor Heterogeneity. Front. Bioeng. Biotechnol. 2022, 10, 829509. [Google Scholar] [CrossRef]
- O’Brien, S.L.; Fagan, A.; Fox, E.J.; Millikan, R.C.; Culhane, A.C.; Brennan, D.J.; McCann, A.H.; Hegarty, S.; Moyna, S.; Duffy, M.J.; et al. CENP-F expression is associated with poor prognosis and chromosomal instability in patients with primary breast cancer. Int. J. Cancer 2007, 120, 1434–1443. [Google Scholar] [CrossRef]
- Yu, B.; Chen, L.; Zhang, W.; Li, Y.; Zhang, Y.; Gao, Y.; Teng, X.; Zou, L.; Wang, Q.; Jia, H.; et al. TOP2A and CENPF are synergistic master regulators activated in cervical cancer. BMC Med. Genom. 2020, 13, 145. [Google Scholar] [CrossRef]
- Gierahn, T.M.; Wadsworth, M.H., 2nd; Hughes, T.K.; Bryson, B.D.; Butler, A.; Satija, R.; Fortune, S.; Love, J.C.; Shalek, A.K. Seq-Well: Portable, low-cost RNA sequencing of single cells at high throughput. Nat. Methods 2017, 14, 395–398. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; McInnes, L.; Healy, J.; Dutertre, C.A.; Kwok, I.W.H.; Ng, L.G.; Ginhoux, F.; Newell, E.W. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol. 2018, 37, 38–44. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Katzenellenbogen, R.A.; Egelkrout, E.M.; Vliet-Gregg, P.; Gewin, L.C.; Gafken, P.R.; Galloway, D.A. NFX1-123 and poly (A) binding proteins synergistically augment activation of telomerase in human papillomavirus type 16 E6-expressing cells. J. Virol. 2007, 81, 3786–3796. [Google Scholar] [CrossRef]
- Hayflick, L.; Moorhead, P.S. The serial cultivation of human diploid cell strains. Exp. Cell Res. 1961, 25, 585–621. [Google Scholar] [CrossRef]
- Tang, X.H.; Zhao, T.N.; Guo, L.; Liu, X.Y.; Zhang, W.N.; Zhang, P. Cell-Cycle-related Protein Centromere Protein F Deficiency Inhibits Cervical Cancer Cell Growth by Inducing Ferroptosis Via Nrf2 Inactivation. Cell Biochem. Biophys. 2024, 82, 997–1006. [Google Scholar] [CrossRef]
- Carlson, S.M.; Soulette, C.M.; Yang, Z.; Elias, J.E.; Brooks, A.N.; Gozani, O. RBM25 is a global splicing factor promoting inclusion of alternatively spliced exons and is itself regulated by lysine mono-methylation. J. Biol. Chem. 2017, 292, 13381–13390. [Google Scholar] [CrossRef]
- Ge, Y.; Schuster, M.B.; Pundhir, S.; Rapin, N.; Bagger, F.O.; Sidiropoulos, N.; Hashem, N.; Porse, B.T. The splicing factor RBM25 controls MYC activity in acute myeloid leukemia. Nat. Commun. 2019, 10, 172. [Google Scholar] [CrossRef]
- Bohlen, J.; Roiuk, M.; Neff, M.; Teleman, A.A. PRRC2 proteins impact translation initiation by promoting leaky scanning. Nucleic Acids Res. 2023, 51, 3391–3409. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Xu, J.; Chen, R. Silencing proline-rich coiled-coil 2C inhibit the proliferation and metastasis of liver cancer cells. J. Gastrointest. Oncol. 2023, 14, 287–301. [Google Scholar] [CrossRef] [PubMed]
- Palaniappan, A.; Ramar, K.; Ramalingam, S. Computational Identification of Novel Stage-Specific Biomarkers in Colorectal Cancer Progression. PLoS ONE 2016, 11, e0156665. [Google Scholar] [CrossRef] [PubMed]
- Pan, H.; Chai, W.; Liu, X.; Yu, T.; Sun, L.; Yan, M. DYNC1H1 regulates NSCLC cell growth and metastasis by IFN-gamma-JAK-STAT signaling and is associated with an aberrant immune response. Exp. Cell Res. 2021, 409, 112897. [Google Scholar] [CrossRef]
- Xu, X.S.; Ma, Y.S.; Dai, R.H.; Zhang, H.L.; Yang, Q.X.; Fan, Q.Y.; Liu, X.Y.; Liu, J.B.; Feng, W.W.; Meng, H.; et al. Identification of novel genomic hotspots and tumor-relevant genes via comprehensive analysis of HPV integration in Chinese patients of cervical cancer. Am. J. Cancer Res. 2024, 14, 4665–4682. [Google Scholar] [CrossRef]
- Schneider, M.A.; Spoden, G.A.; Florin, L.; Lambert, C. Identification of the dynein light chains required for human papillomavirus infection. Cell Microbiol. 2011, 13, 32–46. [Google Scholar] [CrossRef]
- Ke, S.; Wang, C.; Su, Z.; Lin, S.; Wu, G. Integrated Analysis Reveals Critical Ferroptosis Regulators and FTL Contribute to Cancer Progression in Hepatocellular Carcinoma. Front. Genet. 2022, 13, 897683. [Google Scholar] [CrossRef]
- Li, A.; Li, Y.; Li, X.; Tang, C.; Yang, Y.; Li, N.; Jin, Y. Ferritin light chain as a potential biomarker for the prognosis of liver hepatocellular carcinoma. Heliyon 2024, 10, e36040. [Google Scholar] [CrossRef]
- Park, K.S.; Kim, H.; Kim, N.G.; Cho, S.Y.; Choi, K.H.; Seong, J.K.; Paik, Y.K. Proteomic analysis and molecular characterization of tissue ferritin light chain in hepatocellular carcinoma. Hepatology 2002, 35, 1459–1466. [Google Scholar] [CrossRef]
- Zhang, L.; Chen, Z.; Xu, A. FTL: A novel predictor in gastric cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 7865–7872. [Google Scholar]
- Siegel, E.M.; Patel, N.; Lu, B.; Lee, J.H.; Nyitray, A.G.; Huang, X.; Villa, L.L.; Franco, E.L.; Giuliano, A.R. Circulating biomarkers of iron storage and clearance of incident human papillomavirus infection. Cancer Epidemiol. Biomarkers Prev. 2012, 21, 859–865. [Google Scholar] [CrossRef] [PubMed]
- Rattner, J.B.; Rao, A.; Fritzler, M.J.; Valencia, D.W.; Yen, T.J. CENP-F is a ca 400 kDa kinetochore protein that exhibits a cell-cycle dependent localization. Cell Motil. Cytoskeleton 1993, 26, 214–226. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Sun, X. Correlation of INHBA Overexpression with Pathological Features, Antitumor Immune Response and Clinical Prognosis in Cervical Cancer. Medicina 2023, 59, 495. [Google Scholar] [CrossRef]
- Zhao, K.; Yi, Y.; Ma, Z.; Zhang, W. INHBA is a Prognostic Biomarker and Correlated With Immune Cell Infiltration in Cervical Cancer. Front. Genet. 2021, 12, 705512. [Google Scholar] [CrossRef] [PubMed]
- Friede, R.L.; Samorajski, T. Axon caliber related to neurofilaments and microtubules in sciatic nerve fibers of rats and mice. Anat. Rec. 1970, 167, 379–387. [Google Scholar] [CrossRef]
- Hoffman, P.N.; Griffin, J.W.; Price, D.L. Control of axonal caliber by neurofilament transport. J. Cell Biol. 1984, 99, 705–714. [Google Scholar] [CrossRef]
- Emi, M.; Fujiwara, Y.; Nakajima, T.; Tsuchiya, E.; Tsuda, H.; Hirohashi, S.; Maeda, Y.; Tsuruta, K.; Miyaki, M.; Nakamura, Y. Frequent loss of heterozygosity for loci on chromosome 8p in hepatocellular carcinoma, colorectal cancer, and lung cancer. Cancer Res. 1992, 52, 5368–5372. [Google Scholar]
- Vogelstein, B.; Fearon, E.R.; Kern, S.E.; Hamilton, S.R.; Preisinger, A.C.; Nakamura, Y.; White, R. Allelotype of colorectal carcinomas. Science 1989, 244, 207–211. [Google Scholar] [CrossRef]
- Yaremko, M.L.; Kutza, C.; Lyzak, J.; Mick, R.; Recant, W.M.; Westbrook, C.A. Loss of heterozygosity from the short arm of chromosome 8 is associated with invasive behavior in breast cancer. Genes. Chromosomes Cancer 1996, 16, 189–195. [Google Scholar] [CrossRef]
- Alholle, A.; Brini, A.T.; Gharanei, S.; Vaiyapuri, S.; Arrigoni, E.; Dallol, A.; Gentle, D.; Kishida, T.; Hiruma, T.; Avigad, S.; et al. Functional epigenetic approach identifies frequently methylated genes in Ewing sarcoma. Epigenetics 2013, 8, 1198–1204. [Google Scholar] [CrossRef]
- Dubrowinskaja, N.; Gebauer, K.; Peters, I.; Hennenlotter, J.; Abbas, M.; Scherer, R.; Tezval, H.; Merseburger, A.S.; Stenzl, A.; Grunwald, V.; et al. Neurofilament Heavy polypeptide CpG island methylation associates with prognosis of renal cell carcinoma and prediction of antivascular endothelial growth factor therapy response. Cancer Med. 2014, 3, 300–309. [Google Scholar] [CrossRef]
- Kim, M.S.; Chang, X.; LeBron, C.; Nagpal, J.K.; Lee, J.; Huang, Y.; Yamashita, K.; Trink, B.; Ratovitski, E.A.; Sidransky, D. Neurofilament heavy polypeptide regulates the Akt-beta-catenin pathway in human esophageal squamous cell carcinoma. PLoS ONE 2010, 5, e9003. [Google Scholar] [CrossRef]
- Revill, K.; Wang, T.; Lachenmayer, A.; Kojima, K.; Harrington, A.; Li, J.; Hoshida, Y.; Llovet, J.M.; Powers, S. Genome-wide methylation analysis and epigenetic unmasking identify tumor suppressor genes in hepatocellular carcinoma. Gastroenterology 2013, 145, 1424–1435.e25. [Google Scholar] [CrossRef] [PubMed]
- Calmon, M.F.; Jeschke, J.; Zhang, W.; Dhir, M.; Siebenkas, C.; Herrera, A.; Tsai, H.C.; O’Hagan, H.M.; Pappou, E.P.; Hooker, C.M.; et al. Epigenetic silencing of neurofilament genes promotes an aggressive phenotype in breast cancer. Epigenetics 2015, 10, 622–632. [Google Scholar] [CrossRef] [PubMed]
- Li, D.; Zhao, W.; Zhang, X.; Lv, H.; Li, C.; Sun, L. NEFM DNA methylation correlates with immune infiltration and survival in breast cancer. Clin. Epigenetics 2021, 13, 112. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zou, Q.; Chen, Q.; Wang, S.; Du, Q.; Mai, Q.; Wang, X.; Lin, X.; Du, L.; Yao, S.; et al. Methylation-related differentially expressed genes as potential prognostic biomarkers for cervical cancer. Heliyon 2024, 10, e36240. [Google Scholar] [CrossRef]
- Bae, D.H.; Lane, D.J.R.; Jansson, P.J.; Richardson, D.R. The old and new biochemistry of polyamines. Biochim. Biophys. Acta Gen. Subj. 2018, 1862, 2053–2068. [Google Scholar] [CrossRef]
- Mou, Y.; Zhang, L.; Liu, Z.; Song, X. Abundant expression of ferroptosis-related SAT1 is related to unfavorable outcome and immune cell infiltration in low-grade glioma. BMC Cancer 2022, 22, 215. [Google Scholar] [CrossRef]
- Pegg, A.E.; Casero, R.A., Jr. Current status of the polyamine research field. Methods Mol. Biol. 2011, 720, 3–35. [Google Scholar] [CrossRef]
- Brett-Morris, A.; Wright, B.M.; Seo, Y.; Pasupuleti, V.; Zhang, J.; Lu, J.; Spina, R.; Bar, E.E.; Gujrati, M.; Schur, R.; et al. The polyamine catabolic enzyme SAT1 modulates tumorigenesis and radiation response in GBM. Cancer Res. 2014, 74, 6925–6934. [Google Scholar] [CrossRef]
- Thakur, V.S.; Aguila, B.; Brett-Morris, A.; Creighton, C.J.; Welford, S.M. Spermidine/spermine N1-acetyltransferase 1 is a gene-specific transcriptional regulator that drives brain tumor aggressiveness. Oncogene 2019, 38, 6794–6800. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.; Zhu, L.; Luo, Y.; Tang, Y.; Tan, Q.; Zou, Y.; Chen, K.; Deng, X.; Tang, H.; Li, H.; et al. Autophagy Deficiency Induced by SAT1 Potentiates Tumor Progression in Triple-Negative Breast Cancer. Adv. Sci. 2024, 11, e2309903. [Google Scholar] [CrossRef]
- Tang, S.; Chen, L. The recent advancements of ferroptosis of gynecological cancer. Cancer Cell Int. 2024, 24, 351. [Google Scholar] [CrossRef] [PubMed]
- Pennisi, G.; Bruzzaniti, P.; Burattini, B.; Piaser Guerrato, G.; Della Pepa, G.M.; Sturiale, C.L.; Lapolla, P.; Familiari, P.; La Pira, B.; D’Andrea, G.; et al. Advancements in Telomerase-Targeted Therapies for Glioblastoma: A Systematic Review. Int. J. Mol. Sci. 2024, 25, 8700. [Google Scholar] [CrossRef] [PubMed]
- Chiappori, A.A.; Kolevska, T.; Spigel, D.R.; Hager, S.; Rarick, M.; Gadgeel, S.; Blais, N.; Von Pawel, J.; Hart, L.; Reck, M.; et al. A randomized phase II study of the telomerase inhibitor imetelstat as maintenance therapy for advanced non-small-cell lung cancer. Ann. Oncol. 2015, 26, 354–362. [Google Scholar] [CrossRef]
- Bruedigam, C.; Porter, A.H.; Song, A.; Vroeg In de Wei, G.; Stoll, T.; Straube, J.; Cooper, L.; Cheng, G.; Kahl, V.F.S.; Sobinoff, A.P.; et al. Imetelstat-mediated alterations in fatty acid metabolism to induce ferroptosis as a therapeutic strategy for acute myeloid leukemia. Nat. Cancer 2024, 5, 47–65. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chintala, S.; Dankoski, M.A.; Maina, T.K.; Oduor, C.I.; Quist, K.M.; Bailey, J.A.; Katzenellenbogen, R.A. Reduction of NFX1-123 and HPV 16 E6 and E7 Decreased Telomerase and CENP-F in Cervical Cancer Cell Lines. Cancers 2025, 17, 2044. https://doi.org/10.3390/cancers17122044
Chintala S, Dankoski MA, Maina TK, Oduor CI, Quist KM, Bailey JA, Katzenellenbogen RA. Reduction of NFX1-123 and HPV 16 E6 and E7 Decreased Telomerase and CENP-F in Cervical Cancer Cell Lines. Cancers. 2025; 17(12):2044. https://doi.org/10.3390/cancers17122044
Chicago/Turabian StyleChintala, Sreenivasulu, Maura A. Dankoski, Titus K. Maina, Cliff I. Oduor, Kevin M. Quist, Jeffrey A. Bailey, and Rachel A. Katzenellenbogen. 2025. "Reduction of NFX1-123 and HPV 16 E6 and E7 Decreased Telomerase and CENP-F in Cervical Cancer Cell Lines" Cancers 17, no. 12: 2044. https://doi.org/10.3390/cancers17122044
APA StyleChintala, S., Dankoski, M. A., Maina, T. K., Oduor, C. I., Quist, K. M., Bailey, J. A., & Katzenellenbogen, R. A. (2025). Reduction of NFX1-123 and HPV 16 E6 and E7 Decreased Telomerase and CENP-F in Cervical Cancer Cell Lines. Cancers, 17(12), 2044. https://doi.org/10.3390/cancers17122044




