Simple Summary
The NOD2 protein plays a crucial role in regulating intestinal inflammation. The dysregulation of NOD2, often due to genetic variations (polymorphisms), has been implicated in chronic gut inflammation and, consequently, increased colorectal cancer (CRC) risk. However, prior research on the association between NOD2 polymorphisms and CRC susceptibility has yielded inconsistent results. This meta-analysis aimed to synthesize the existing evidence to provide a more robust assessment of this association. Our findings indicate that two specific NOD2 polymorphisms, rs2066845 and rs2066847, are significantly associated with an elevated risk of CRC. These insights may contribute to the identification of individuals predisposed to CRC, thereby facilitating early detection and potentially guiding personalized preventive strategies in clinical practice.
Abstract
Background: Nucleotide-binding oligomerization domain-containing protein 2, encoded by NOD2, can trigger chronic gut inflammation that leads to colorectal cancer (CRC). However, studies that have investigated the association of NOD2 polymorphisms and CRC susceptibility have produced inconsistent findings. To clarify this relationship, a meta-analysis was conducted to integrate data from previous studies to achieve a more precise evaluation of the risk association. Methods: PubMed, Scopus, and Web of Science databases were systematically searched to identify relevant studies on the association of NOD2 polymorphisms with CRC risk. Genetic risk association was quantitatively assessed under five genetic models: homozygous, heterozygous, dominant, recessive, and allele. Thirteen studies, comprising 5,013 cases and 4,463 controls, were included in this study. Four NOD2 polymorphisms were investigated in these studies, namely rs2066842, rs2066844, rs2066845, and rs2066847. Results: Of these, only rs2066845 and rs2066847 were found to be significantly associated with increased CRC risk (rs2066845, heterozygous OR = 1.544, 95% CI = 1.014–2.349, P = 0.043; dominant OR = 1.561, 95% CI = 1.035–2.354, P = 0.034; allele OR = 1.572, 95% CI = 1.040–2.375, P = 0.032; rs2066847, heterozygous OR = 1.321, 95% CI = 1.060–1.647, P = 0.013; dominant OR = 1.402, 95% CI = 1.147–1.713, P = 0.001; allele OR = 1.345, 95% CI = 1.088–1.663, P = 0.006). Conclusions: In conclusion, the NOD2 rs2066845 and rs2066847 polymorphisms are associated with an increased risk of CRC and may potentially serve as predisposition biomarkers for the cancer.
1. Introduction
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers and a leading cause of cancer-related deaths worldwide [1]. Although dietary and environmental factors have been well-established as major risk factors in the development of CRC, genetic factors, particularly genetic polymorphisms, have been shown to be an equally important element in determining individual susceptibility to the disease [2]. Therefore, identification of genetic polymorphisms associated with CRC may facilitate the early detection of at-risk individuals, which will allow preventive strategies to be taken before symptoms appear [3].
It has been known for some time that CRC may be preceded by inflammatory bowel disease (IBD), which includes Crohn’s disease (CD) and ulcerative colitis (UC) [4]. Therefore, genes associated with IBD pathogenesis are ideal candidates for biomarker-based studies on CRC. One such gene is nucleotide-binding oligomerization domain 2 (NOD2) that is located on chromosome 16q12 [5]. NOD2 is one of the most important members of the caspase activation and recruitment domain subfamily capable of recognizing the conserved muramyl dipeptide (MDP) structure present in virtually all bacterial types [6,7]. The gene encodes an intracellular protein that belongs to the Nod-like receptors (NLRs), which contain a C-terminal sensor domain, a central nucleotide-binding oligomerization domain, and an N-terminal effector domain [8].
The NOD2 protein is directly involved in the regulation of the immune response through the activation of nuclear factor-kappa B (NF-κB) via the RIP2/IKK pathway [9]. In response to the presence of MDP, NOD2 interacts with RIP2 kinase to activate NF-κB and mitogen-activated protein kinase, leading to the transcription of proinflammatory mediators [10]. Mutations in NOD2 result in increased NF-κB activity, a phenomenon observed in various human malignancies such as colorectal, thyroid, breast, and lung cancers [11,12]. NOD2 also routinely stimulates host defense when it detects elevated levels of MDP following partial degradation of bacterial peptidoglycan [13]. In addition, the protein is actively involved in the recycling and degradation of the bacterial cell components through autophagy [2]. As the role of the gut microbiome in the development of CRC becomes increasingly clear, the ability of NOD2 to modulate bacterial growth suggests involvement of the protein in carcinogenesis.
Polymorphisms in NOD2 may affect the functionality of its protein product and thus the risk for various diseases. There are four major NOD2 single nucleotide polymorphisms that have been extensively studied, namely rs2066842 (conventionally known as Pro268Ser), rs2066844 (conventionally known as Arg702Trp), rs2066845 (conventionally known as Gly908Arg), and rs2066847 (conventionally known as 3020insC/Leu1007fsX1008). These polymorphisms are located in the coding region of NOD2 and cause a change in the amino acid sequence of the protein product, which subsequently affects its expression and normal function [8]. For this reason, these polymorphisms have been found to be associated with a higher risk of CD [14,15,16]. In CRC, however, the association between NOD2 polymorphisms and the risk of developing cancer remains controversial. For example, although [5,17,18,19] reported that there was no apparent association between the four polymorphisms and the risk of CRC, a few other studies demonstrated a significant association between the polymorphisms and CRC risk [11,20,21,22]. These discrepancies could be attributed to the different genetic and environmental backgrounds of the study subjects in different studies. To address these inconsistencies, a meta-analysis was conducted by [8] to combine the results of studies published before July 1, 2013. Nevertheless, numerous newer studies have been published more recently, and the inclusion of these more recent studies could potentially lead to a different conclusion. Thus, in this work, we performed an updated meta-analysis to investigate the association between NOD2 polymorphisms and CRC risk.
2. Materials and Methods
2.1. Literature Search Strategy and Study Selection
This systematic review was conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) 2020 guidelines. A thorough literature search was conducted on the Web of Science, PubMed, and Scopus databases on 28 May 2025. No language limitations were applied during the search. The following terms were used: “NOD2” AND “polymorphism” AND “colorectal cancer”. Two investigators independently screened the studies for inclusion in the meta-analysis, based on the following eligibility criteria: (1) assessing the associations between NOD2 rs2066842, rs2066844, rs2066845, and rs2066847 polymorphisms and CRC risk; (2) case–control study; and (3) providing sufficient data for the odds ratio (OR) and 95% confidence interval (CI) calculation. Non-original studies (including review, editorials, and letters to the editor) were excluded. Furthermore, the reference lists of the included studies were also screened for additional potentially relevant articles. If two or more publications were available for the same population, the one containing either the highest number of samples or the latest dataset was selected. No prospective registration was carried out for this review.
2.2. Data Extraction and Quality Assessment
For each eligible study, the following information was extracted independently by the two investigators in an Excel sheet: name of the first author, publication year, country, ethnicity, the genotype, and allele frequencies of the four NOD2 polymorphisms, and deviation from the Hardy–Weinberg equilibrium (HWE). The ethnicity of the study populations was categorized as Caucasian, African, Asian, and others. The quality of the included studies was evaluated based on the Modified Newcastle-Ottawa Scale for Case-Control Studies of Genetic Association [23] independently by the two investigators. Studies with a rating of 5 stars or above were considered as having a high methodological quality.
2.3. Statistical Analysis
The pooled OR and the 95% CI were calculated to evaluate the associations of NOD2 rs2066842, rs2066844, rs2066845, and rs2066847 polymorphisms with CRC risk. Meta-analysis was performed only when data from at least three studies were available. For all calculations, wild-type genotype and/or allele were used as the reference group. The significance of the overall OR was determined using the Z-test. Cochran’s Chi-squared-based Q test and I2 test were used to assess the presence of statistical heterogeneity among the studies. If significant heterogeneity was present (as indicated by an I2 value ≥ 50% and P < 0.10, the random-effects model (the DerSimonian–Laird method) was used to calculate the pooled OR. Meanwhile, a fixed-effects model (the Mantel–Haenszel method) was used to calculate the pooled OR when there was no apparent between-study heterogeneity. The stability of the test results was determined using sensitivity analysis by excluding one study at a time. Subgroup analysis by ethnicity was also performed. In addition, funnel plots, Egger’s test, and Begg’s test were applied to examine the presence of publication bias. All statistical analyses were performed using Stata version 18.0 software (StataCorp, College Station, TX, USA), by assuming P < 0.05 as statistically significant, unless otherwise stated.
3. Results
3.1. Characteristics of Studies
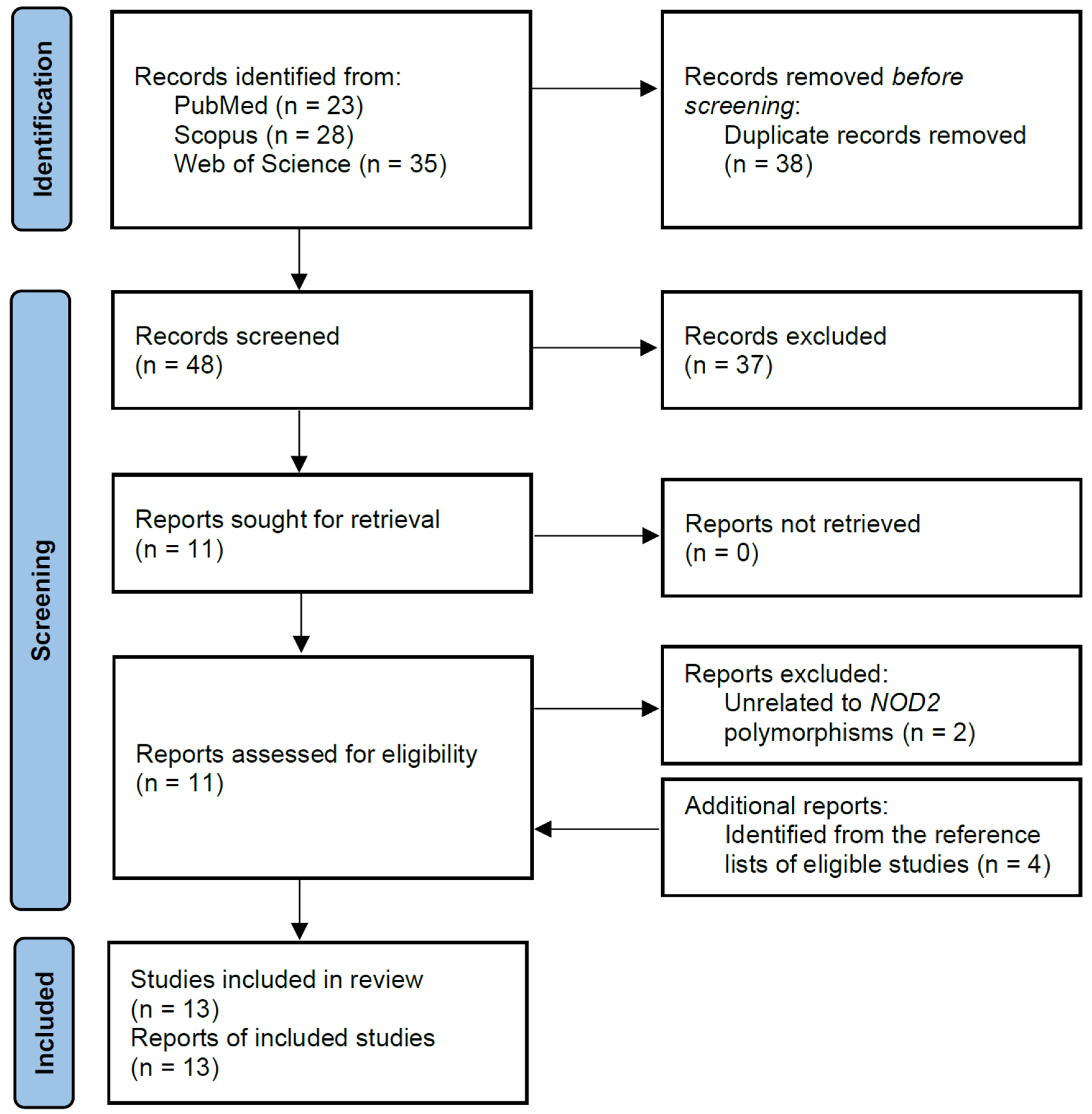
Using the search strategy, a total of 86 records were found in the PubMed, Scopus, and Web of Science databases. Of these, 38 duplicated records were removed, leaving 48 articles that were screened based on titles and abstracts. After reviewing the titles and abstracts, 11 articles were determined as being potentially relevant and were further assessed for eligibility. Based on the eligibility criteria, two articles were subsequently removed as they did not contain information on the polymorphisms of interest. After the screening process, four additional articles were identified from the reference lists of eligible studies and were included in this meta-analysis, resulting in a total of 13 studies comprising 9476 subjects (5013 cases and 4463 controls). The flow chart of the study selection process is summarized in Figure 1, and the characteristics of the included articles are shown in Table 1.
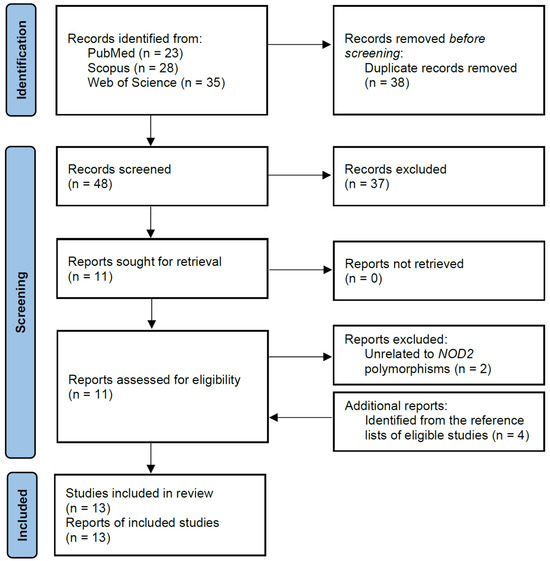
Figure 1.
Flow chart of selection of eligible studies.

Table 1.
Main characteristics of included studies.
For rs2066842, there were three studies comprising a total of 314 cases and 513 controls that were eligible for meta-analysis. In addition, eight studies with 2718 cases and 2310 controls were included for investigating the association between the rs2066844 polymorphism and CRC risk. On the other hand, 7 studies with 2616 cases and 2045 controls were included for rs2066845, and 11 studies involving 3945 cases and 3690 controls were included for the rs2066847 polymorphism. These studies were conducted in Denmark, Northern Germany, Hungary, Poland, Romania, Malaysia, Tunisia, Finland, Greece, and New Zealand. The genotype distributions in the controls of all studies were consistent with the HWE, with the exception of Szeliga et al. [22] (for all the four polymorphisms studied), Mockelmann et al. [20] (for rs2066847), and Lau et al. [12] (for rs2066844, rs2066845, and rs2066847). All studies had high methodological quality as assessed using the Newcastle-Ottawa Scale (Table 2).

Table 2.
Newcastle-Ottawa Scale for assessing the quality of the included studies.
3.2. Quantitative Data Synthesis
The pooled association of the NOD2 rs2066844, rs2066845, and rs2066847 polymorphisms with the risk of CRC is shown in Table 3, Table 4 and Table 5. Meta-analysis was not performed for rs2066842, as well as the homozygous and recessive models of the three aforementioned polymorphisms, because the number of included studies was too small to be analyzed after excluding studies with zero-count cells, i.e., studies where no events occurred in either the case or control group. Subgroup analysis by study quality was also not performed as all studies had high methodological quality. In addition, subgroup analysis by ethnicity was performed only for the Caucasian and “other ethnicity” subgroups, but not for the Asian subgroup, as there was only one study involving Asians.

Table 3.
Association between NOD2 rs2066844 polymorphism and colorectal cancer risk.

Table 4.
Association between NOD2 rs2066845 polymorphism and colorectal cancer risk.

Table 5.
Association between NOD2 rs2066847 polymorphism and colorectal cancer risk.
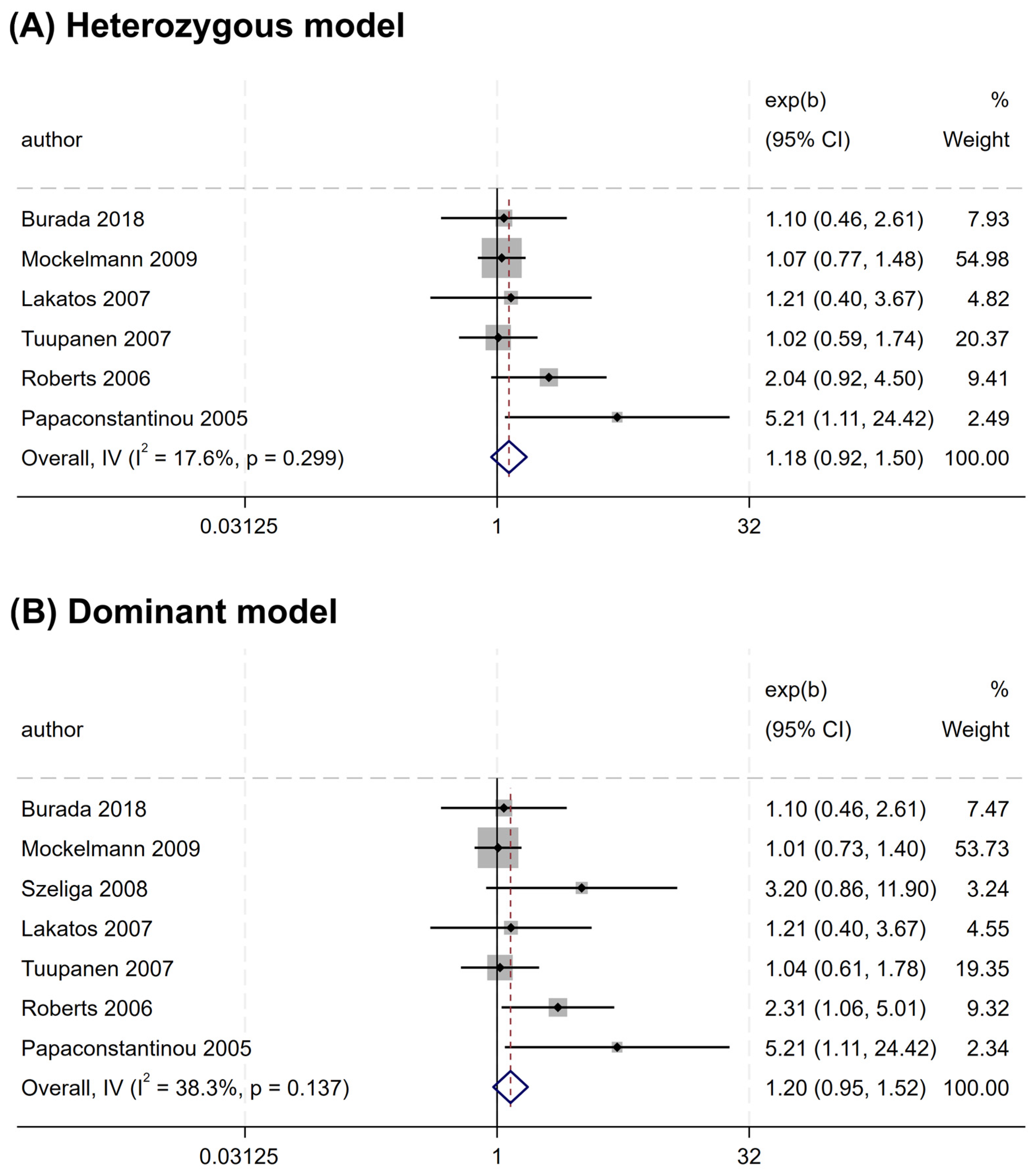
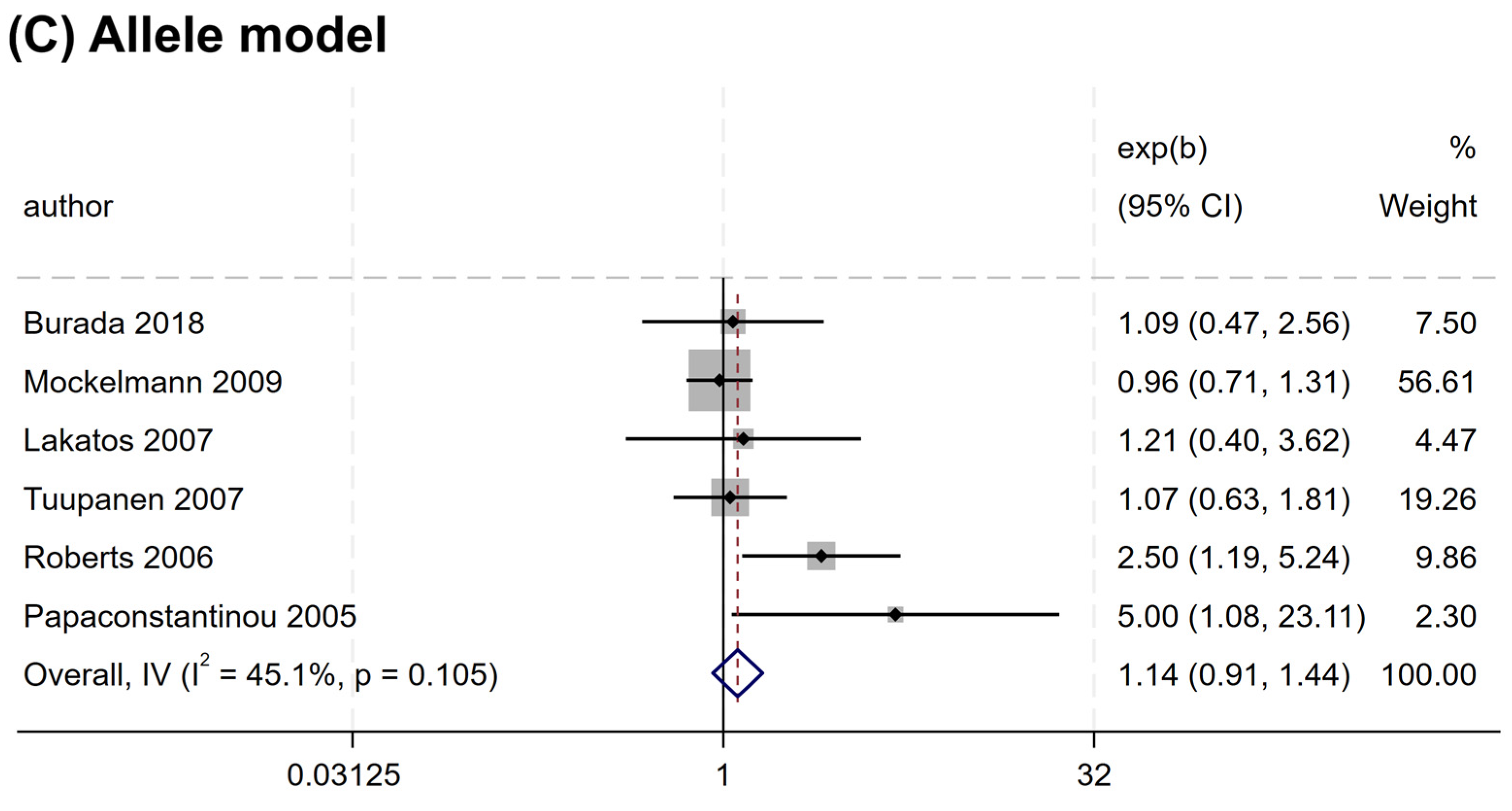
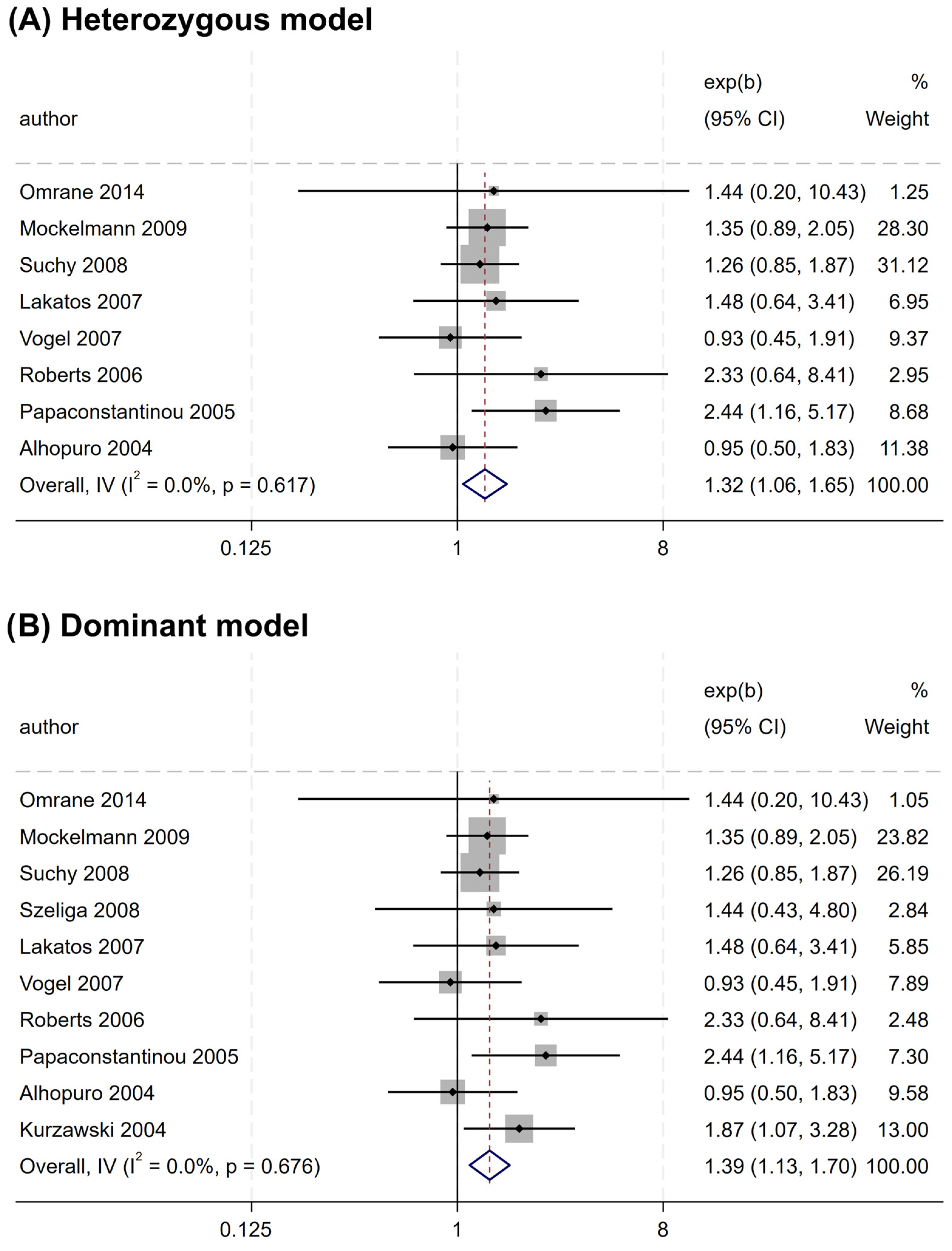
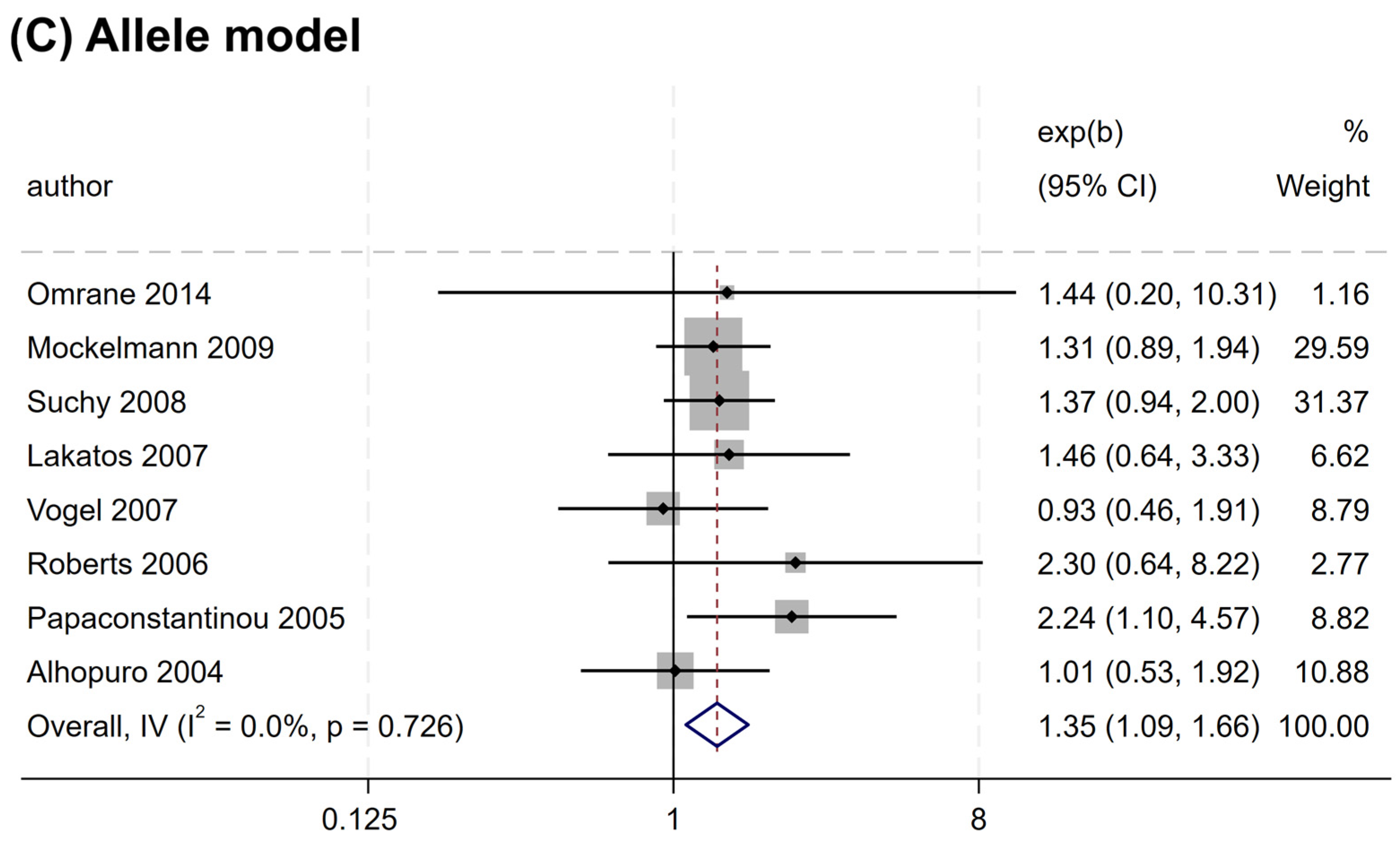
Overall, no significant association was found between the rs2066844 polymorphism and the risk of CRC under all genetic models analyzed (heterozygous model, OR = 1.176, 95% CI = 0.922–1.501, P = 0.191; dominant model, OR = 1.253, 95% CI = 0.989–1.589, P = 0.062; allele model, OR = 1.243, 95% CI = 0.983–1.571, P = 0.069) (Table 3 and Figure 2). In the subgroup analysis based on ethnicity, rs2066844 was also not significantly associated with the risk of CRC in either the Caucasian or “other ethnicity” subgroups (P > 0.05).
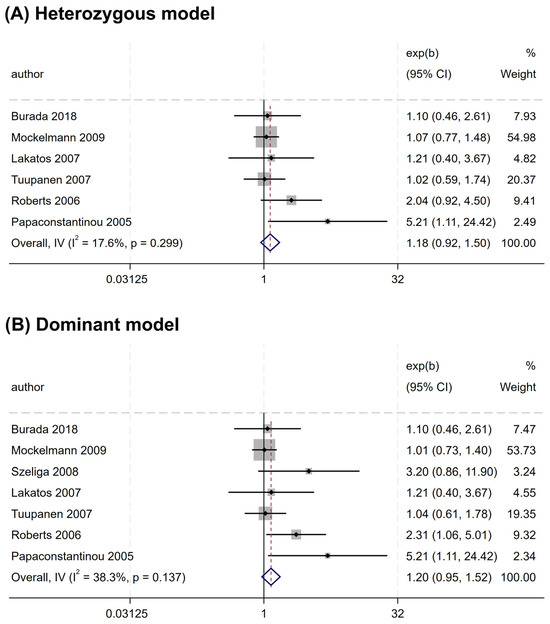
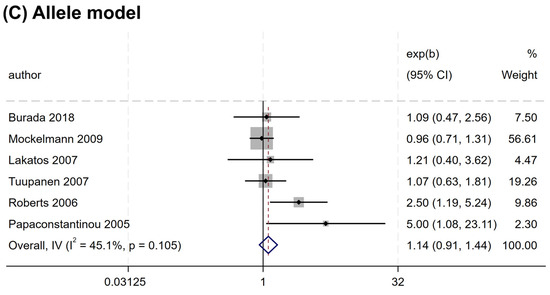
Figure 2.
Forest plots of association between NOD2 rs2066844 polymorphism and colorectal cancer risk. (A) Heterozygous model; (B) Dominant model; (C) Allele model.
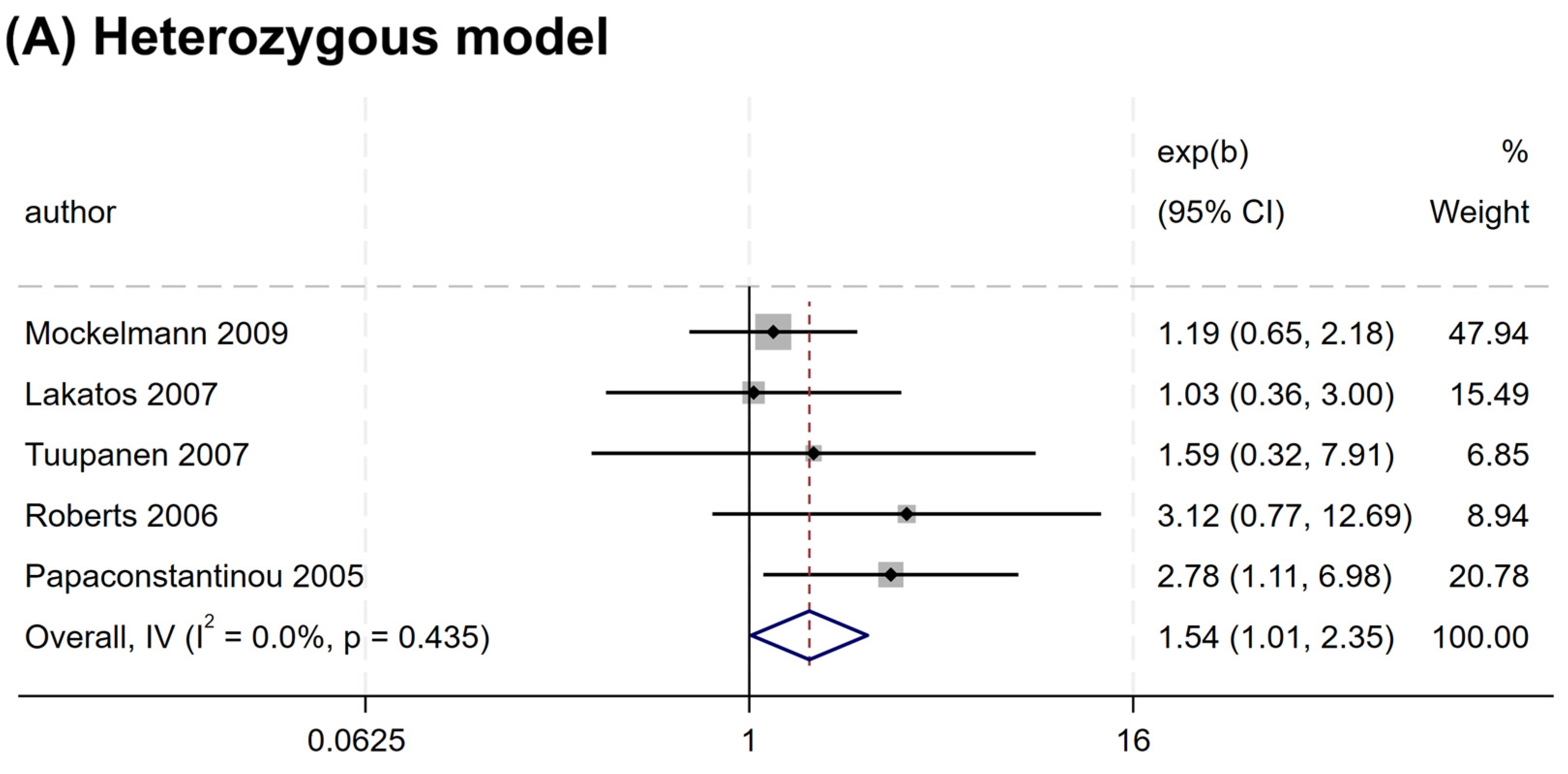
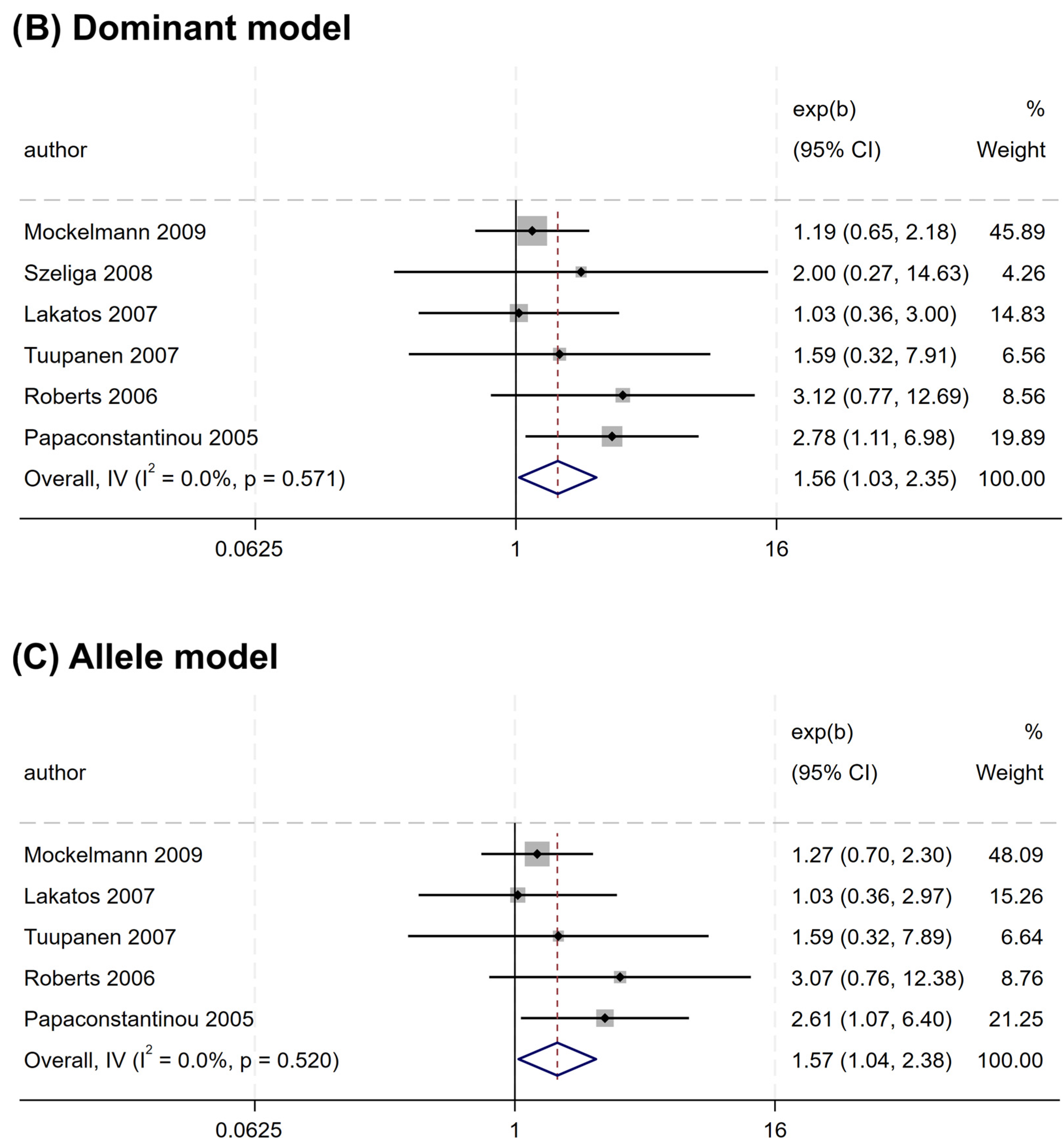
Nevertheless, a significant association was observed for rs2066845 and rs2066847. An increased risk association was noted for rs2066845 (heterozygous model, OR = 1.544, 95% CI = 1.014–2.349, P = 0.043; dominant model, OR = 1.561, 95% CI = 1.035–2.354, P = 0.034; allele model, OR = 1.572, 95% CI = 1.040–2.375, P = 0.032) (Table 4 and Figure 3). Despite this, none of the subgroups showed a significant association (P > 0.05).
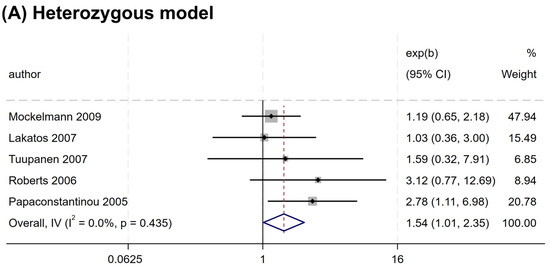
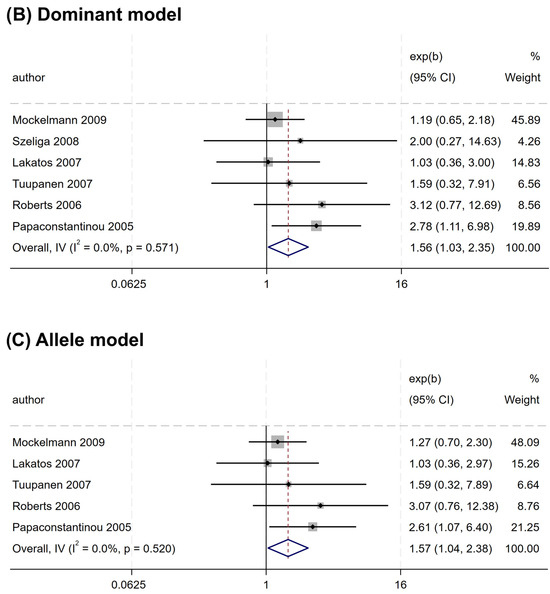
Figure 3.
Forest plots of association between NOD2 rs2066845 polymorphism and colorectal cancer risk. (A) Heterozygous model; (B) Dominant model; (C) Allele model.
Similarly, rs2066847 was also associated with an increased CRC risk (heterozygous model, OR = 1.321, 95% CI = 1.060–1.647, P = 0.013; dominant model, OR = 1.402, 95% CI = 1.147–1.713, P = 0.001; allele model, OR = 1.345, 95% CI = 1.088–1.663, P = 0.006) (Table 5 and Figure 4), and subgroup analysis revealed that the association was present only in subjects of “other ethnicity” (heterozygous model, OR = 1.343, 95% CI = 1.047–1.722, P = 0.020; dominant model, OR = 1.434, 95% CI = 1.149–1.788, P = 0.001; allele model, OR = 1.364, 95% CI = 1.076–1.729, P = 0.010), whereas no significant association was observed in the Caucasians (P > 0.05).
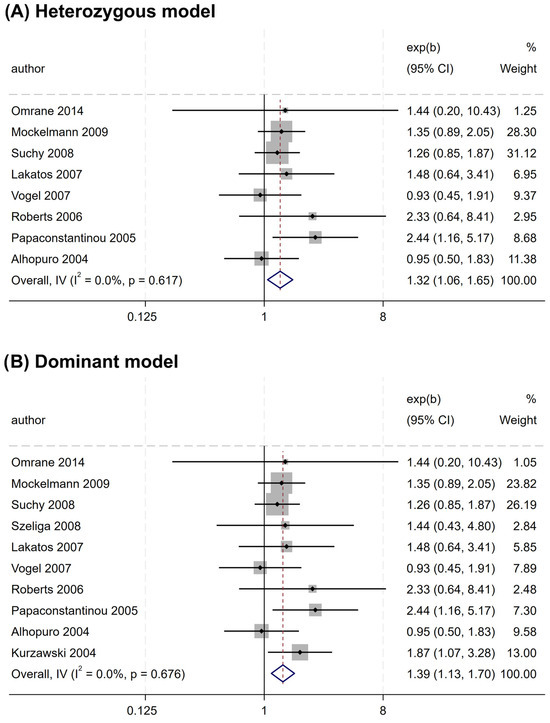
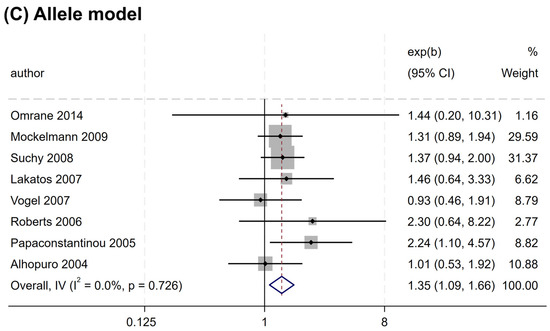
Figure 4.
Forest plots illustrating the relationship between the NOD2 rs2066847 variant and the risk of CRC. (A) Heterozygous model; (B) Dominant model; (C) Allele model.
3.3. Sensitivity Analysis
Sensitivity analysis revealed that the omission of a few studies changed the association of rs2066844 with CRC from non-significant to significant under the dominant and allele models (Supplementary Information online). Likewise, but in the opposite direction, the omission of a few studies removed the significance of the CRC risk association conferred by rs2066845 (under all genetic models) and rs2066847 (under the heterozygous model). Nevertheless, this change was not surprising, given that the lower bound of the 95% CI was very near to the cutoff of 1.000.
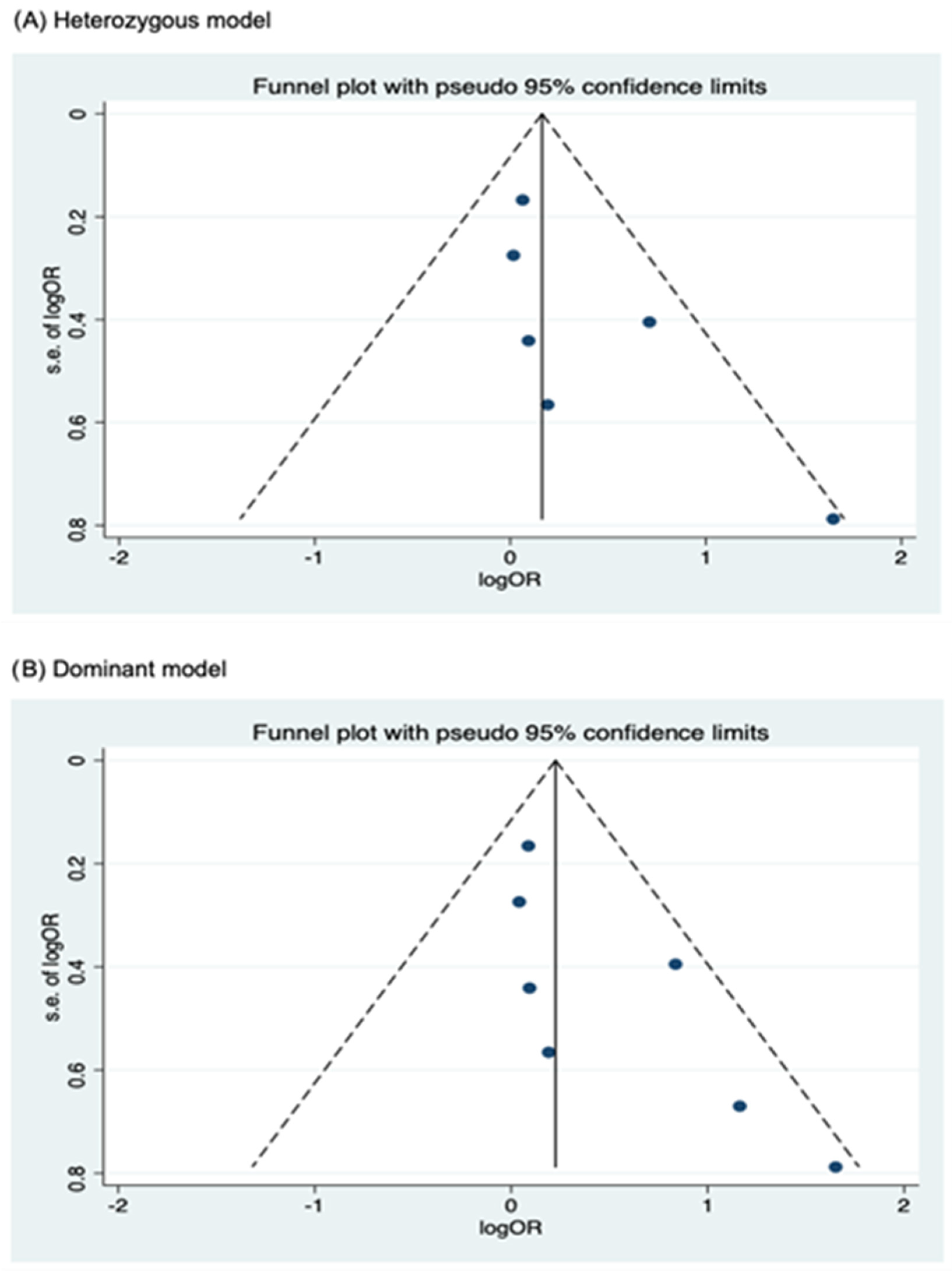
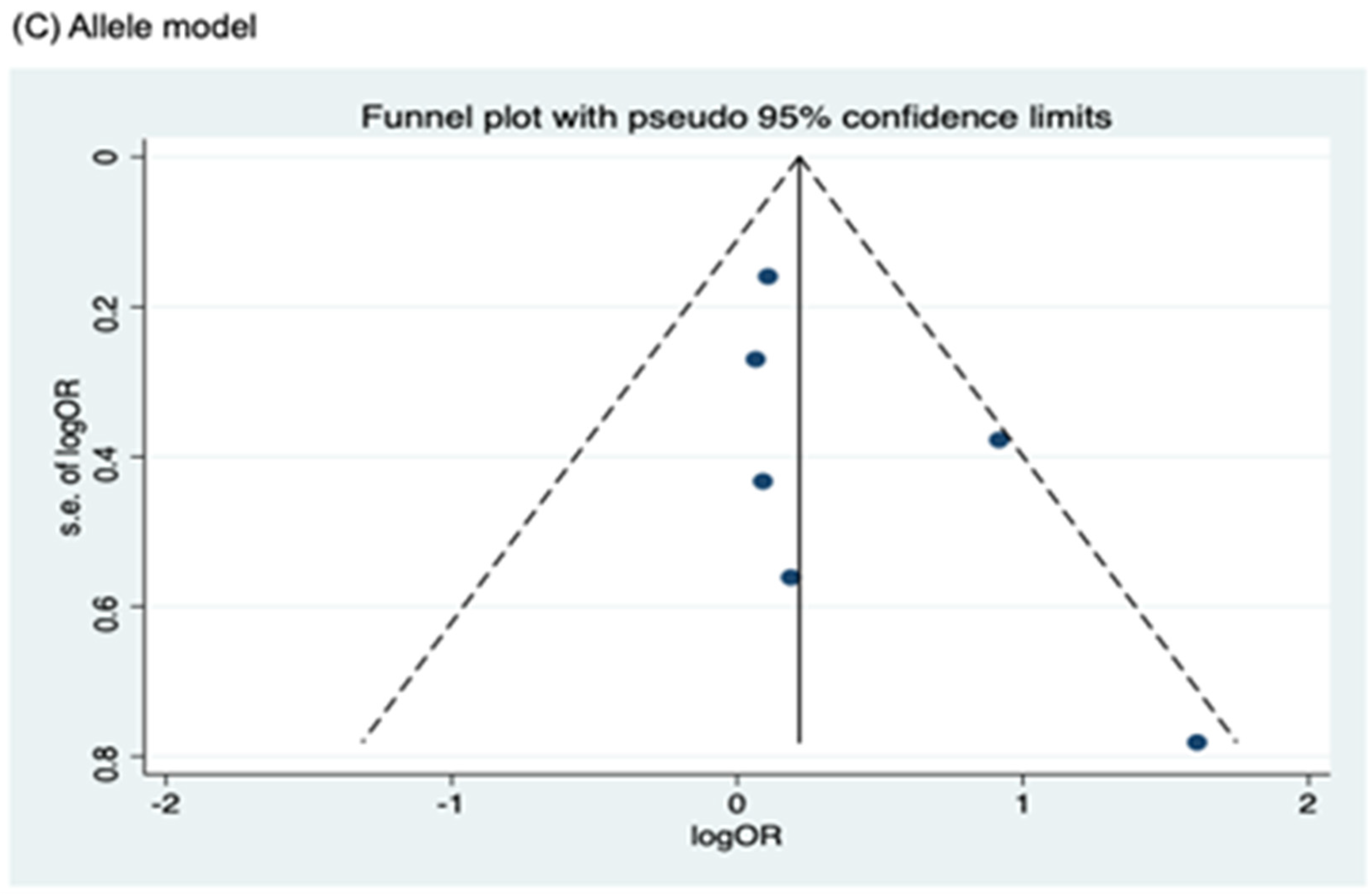
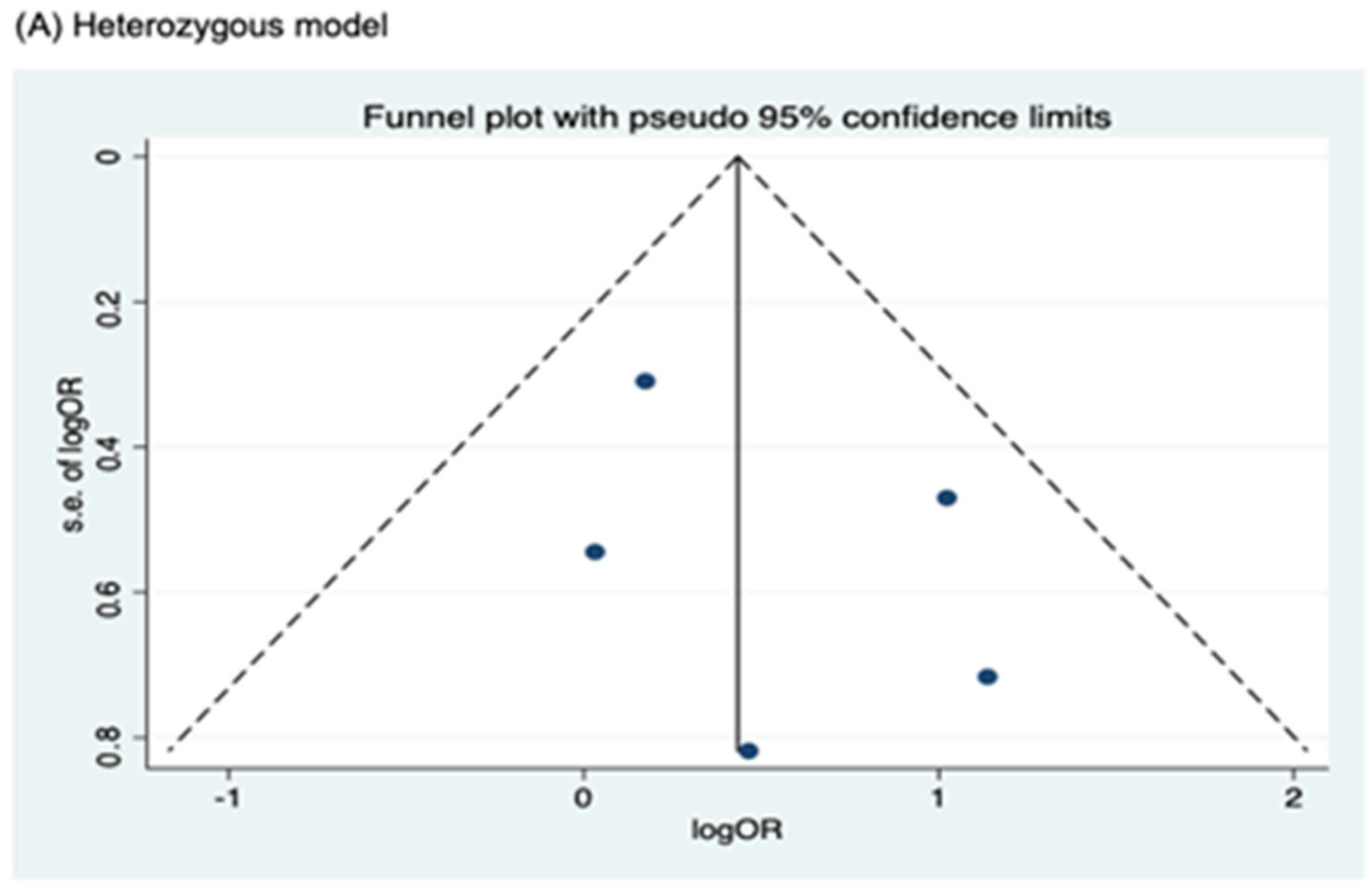
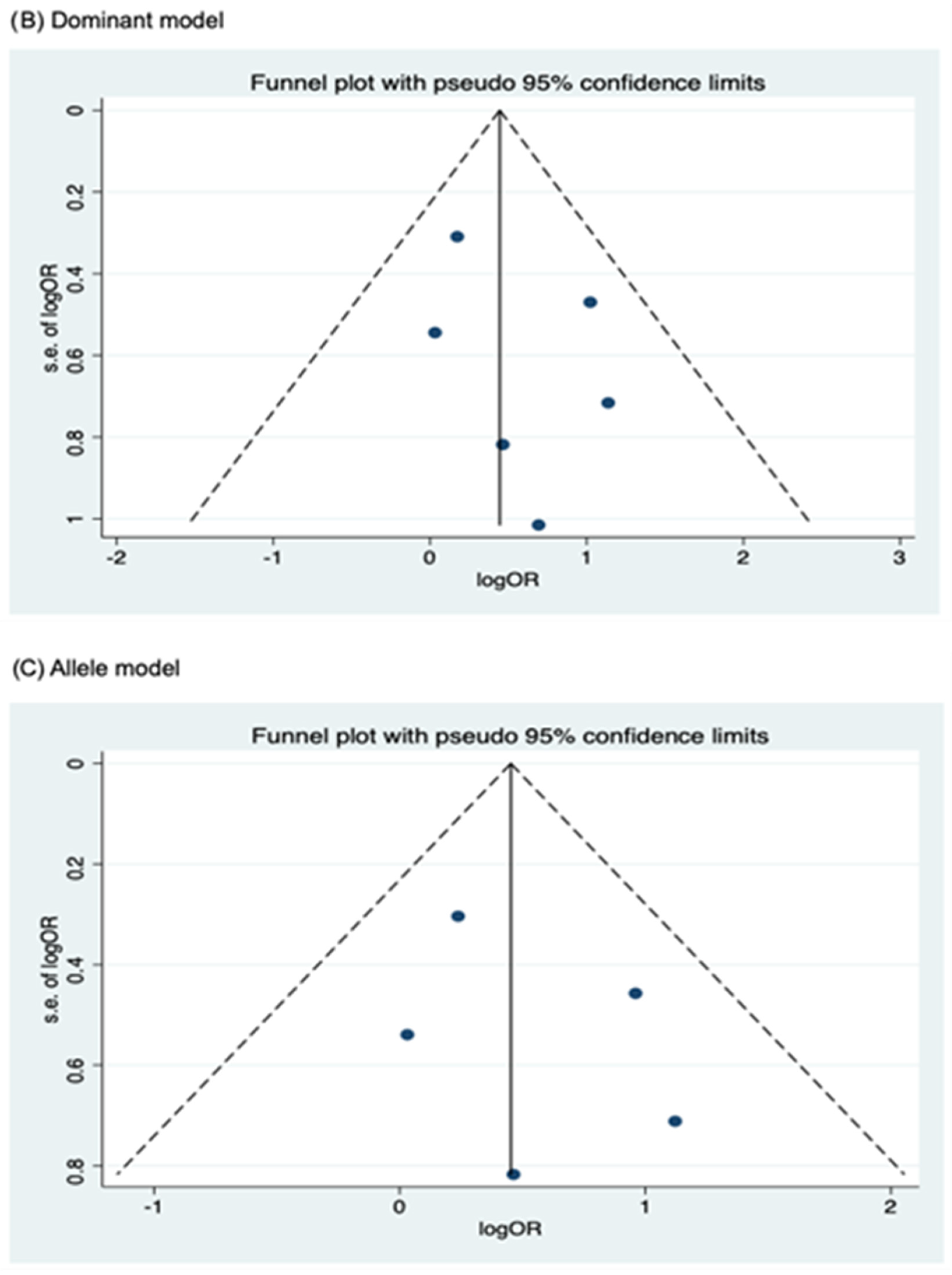
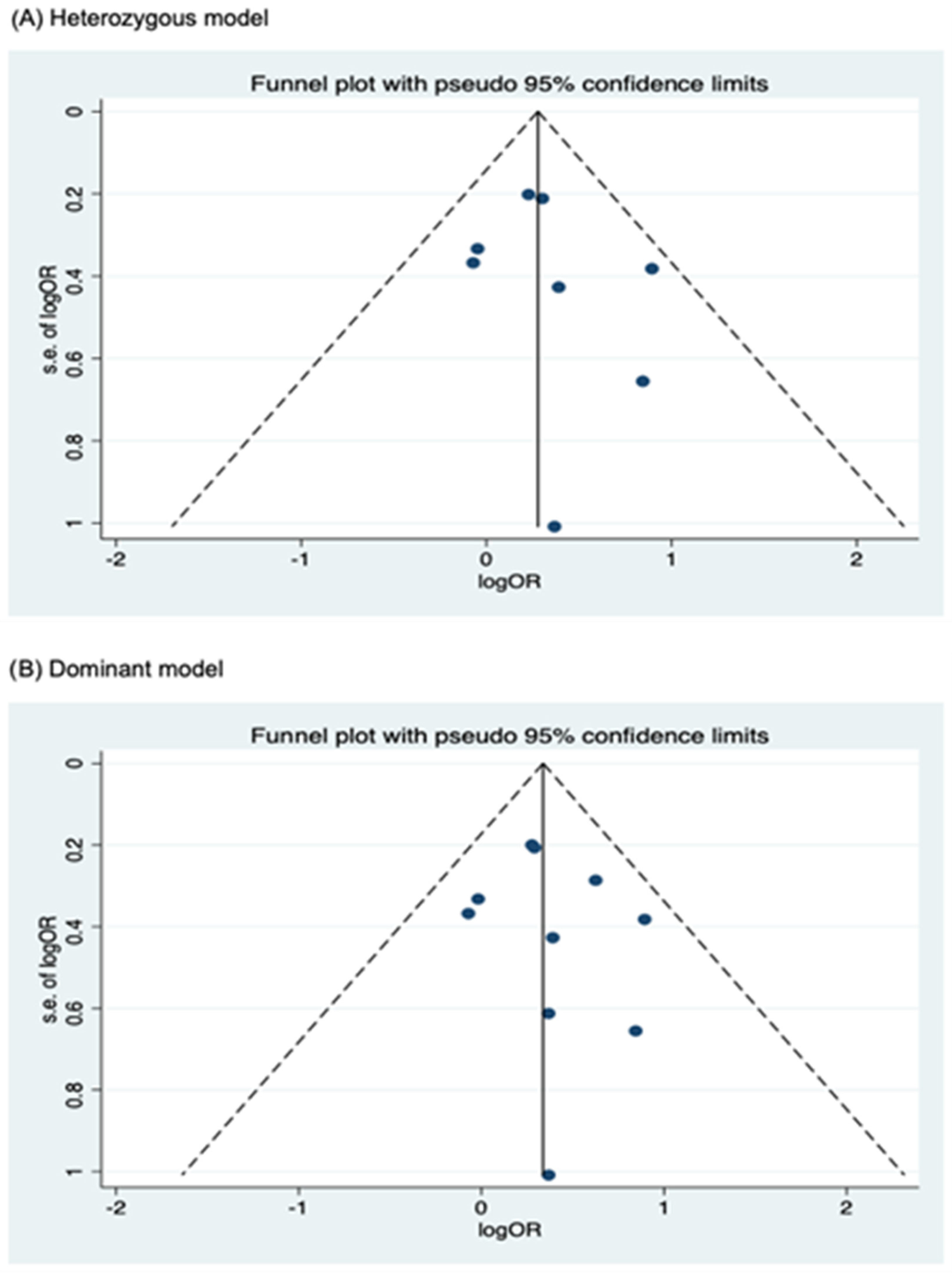
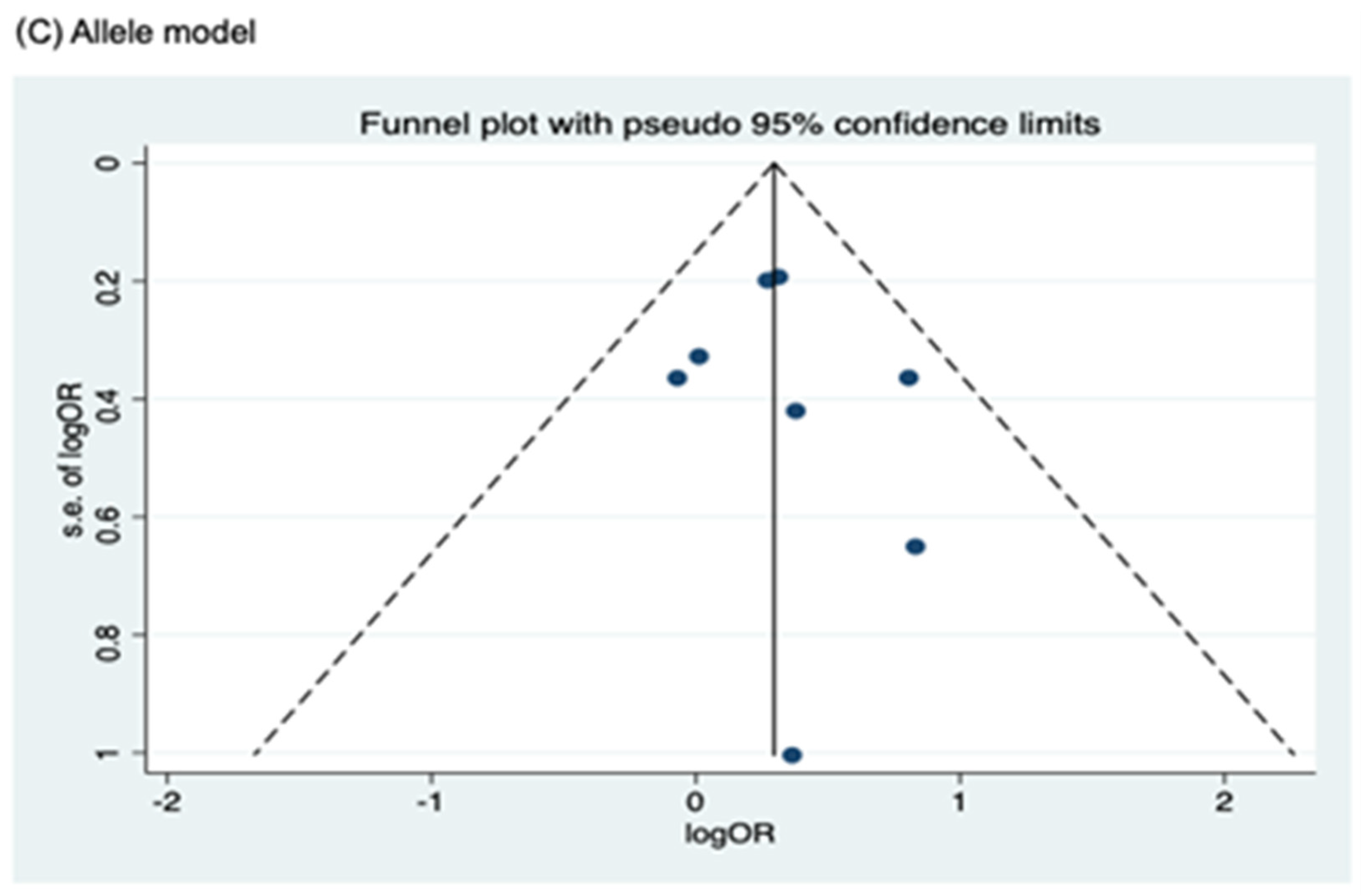
3.4. Publication Bias
Funnel plots for assessing publication bias for rs2066844, rs2066845, and rs2066847 are shown in Figure 5, Figure 6, and Figure 7, respectively. Formal assessments with the Begg’s test revealed significant publication bias in the heterozygous and dominant models for rs2066844 (heterozygous model, P = 0.039; dominant model, P = 0.024), although no significant bias was detected with the Egger’s test (heterozygous model, P = 0.118; dominant model, P = 0.057). The discrepancy between Begg’s and Egger’s tests was not unexpected, as both tests have different sensitivity and statistical power, especially when the number of included studies is small. Performing both tests, especially when combined with visual inspection of the funnel plot, provides a more robust assessment, as each captures different aspects of potential publication bias and helps to validate findings through complementary statistical approaches. Nevertheless, a “trim and fill” analysis was performed for the two genetic models. In each model, one potentially missing study was identified and correction for the missing study did not significantly alter the results (heterozygous model, P = 0.310; dominant model, P = 0.110).
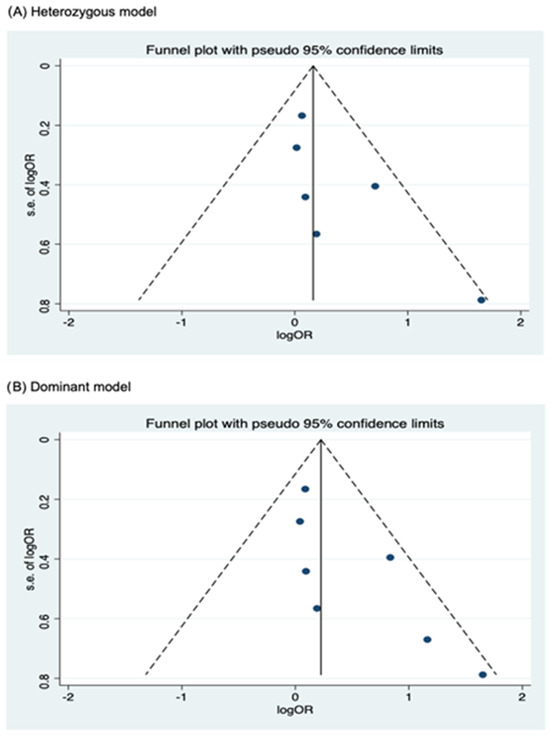
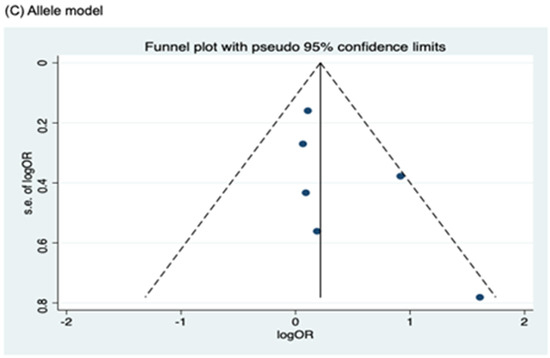
Figure 5.
Funnel plot analysis to detect publication bias for NOD2 rs2066844 polymorphism and colorectal cancer risk. (A) Heterozygous model; (B) Dominant model; (C) Allele model.
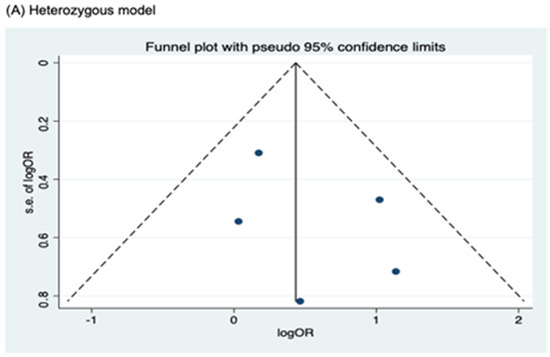
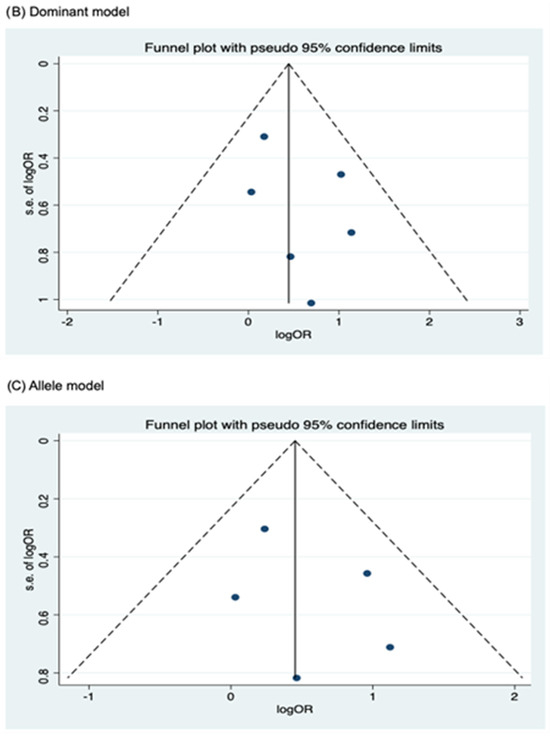
Figure 6.
Funnel plot analysis to detect publication bias for NOD2 rs2066845 polymorphism and colorectal cancer risk. (A) Heterozygous model; (B) Dominant model; (C) Allele model.
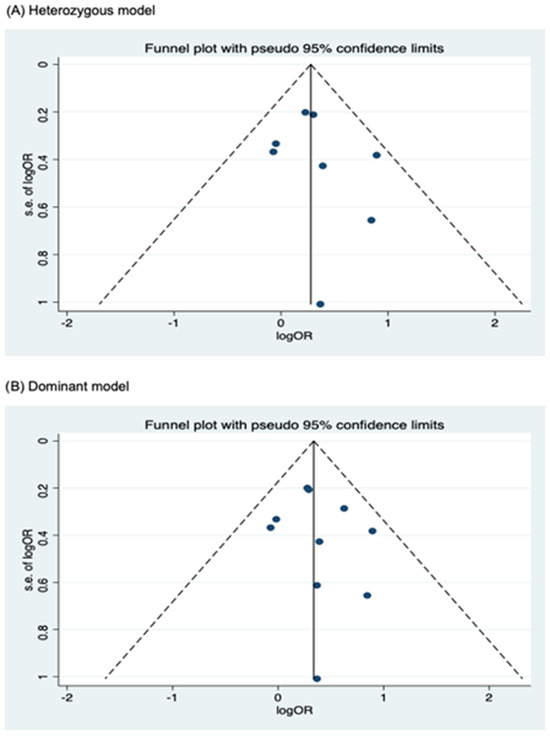
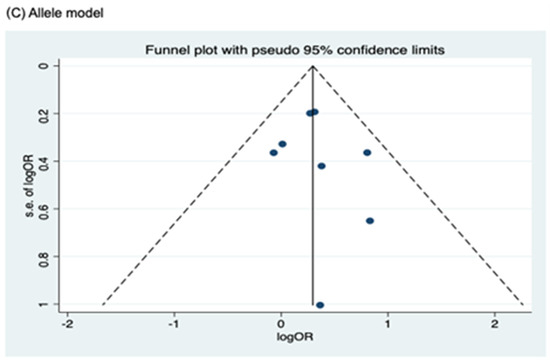
Figure 7.
Funnel plot analysis to detect publication bias for NOD2 rs2066847. (A) Heterozygous model; (B) Dominant model; (C) Allele model.
Meanwhile, no publication bias was observed for the allele model of rs2066844 (Begg’s test, P = 0.091; Egger’s test, P = 0.179), and for all genetic models of rs2066845 (heterozygous model, Begg’s test, P = 0.624, Egger’s test, P = 0.430; dominant model, Begg’s test, P = 0.573, Egger’s test, P = 0.371; allele model, Begg’s test, P = 0.624, Egger’s test, P = 0.484), as well as rs2066847 (heterozygous model, Begg’s test, P = 0.322, Egger’s test, P = 0.503; dominant model, Begg’s test, P = 0.531, Egger’s test, P = 0.542; allele model, Begg’s test, P = 0.805, Egger’s test, P = 0.601).
4. Discussion
Polymorphisms in NOD2 have been associated with the risk of many cancers, including lymphoma, CRC, gastric cancer, breast cancer, ovarian cancer, lung cancer, and laryngeal cancer [9]. Although a number of genetic association studies have been conducted to investigate the association between NOD2 polymorphisms and CRC risk, there is still no clear consensus on their role, as many studies have reported conflicting results. Therefore, in the present study, we performed a meta-analysis of 13 independent case–control studies that included 5,013 cases and 4,463 controls to evaluate the association of NOD2 polymorphisms with CRC risk. The main finding of the present meta-analysis was that both rs2066845 and rs2066847 were associated with an increased risk of CRC under heterozygous, dominant, and allele genetic models. Interestingly, subgroup analysis by ethnicity revealed that rs2066847 was not associated with increased CRC risk in Caucasians but was significant in participants of other ethnicities. On the contrary, for the rs2066844 polymorphism, we found no significant association with CRC risk. Analysis of the rs2066842 polymorphism was not possible because the allele frequency of this polymorphism was very low, resulting in zero-count cells that did not allow calculation of the pooled data.
The rs2066842 polymorphism involves an amino acid change from proline to serine, but it has been speculated to have no adverse effects on the protein function [29]. For this reason, previous studies conducted in the German and New Zealand Caucasian populations have reported that the rs2066842 polymorphism was not associated with the risk of gastric cancer and CRC, respectively [19,30]. It was also found that the polymorphism was not able to alter gene function when assessed alone [31]. Moreover, unlike the other three NOD2 polymorphisms, this polymorphism was located neither in between nor within the key protein domains of NOD2 and therefore could not directly trigger NF-κB activation in response to bacterial lipopolysaccharide and peptidoglycan [32]. Nevertheless, the rs2066842 polymorphism has been described to have a protective effect against CD in the Arab population of Kuwait and against tuberculosis in the African American population [33,34]. In another study by Szeliga et al. [22], it was found that the T allele of this polymorphism may be associated with a higher risk of rectal cancer in the Polish population. In these cases, it is postulated that the polymorphism was in linkage disequilibrium (LD) with other causal variants that caused the disease, resulting in the presence of disease associations in some populations studied [35].
In addition, although previous studies on gastric carcinoma [36], gastric lymphoma [37], glioblastoma [38], and CRC [19,21] have all shown significant associations with the rs2066844 polymorphism, these results were not reflected in this meta-analysis. We found no significant association between this polymorphism and the risk of CRC, which is consistent with studies reported on other diseases, including malignant melanoma and gastrointestinal diseases [39,40,41]. In contrast to the location of the rs2066845 and rs2066847 polymorphisms, which are located in the leucine-rich repeat (LRR) region and contribute to a loss-of-function phenotype, rs2066844 is positioned between the LRR and nucleotide-binding domains [42]. Given this position, rs2066844 is most likely innocuous and may not affect responses to MDP and/or downstream signaling pathways, explaining our observation of the lack of significant association [43]. Nevertheless, there is a possibility that the polymorphism has a minor effect or may co-occur with other polymorphisms to alter NOD2 protein function, explaining why a significant association was observed in several cancers [44].
In addition, we demonstrated that the NOD2 rs2066845 polymorphism was significantly associated with CRC risk under all genetic models analyzed. This result contrasted with that observed in Germany, Hungary, Portugal, and Finland [17,18,20,45]. Nonetheless, there are also studies showing significant associations between the rs2066845 polymorphism and the risk of CRC [21] and other diseases, such as pangastritis [46], CD [47], pulmonary non-tuberculous mycobacterial infections [48], and sarcoidosis [49]. These significant associations could be explained by the location of the polymorphism in the LRR domain, which mediates the protein–ligand interaction of NOD2. In fact, the positive association of this polymorphism was reaffirmed by a functionality assessment, which showed that the polymorphism may have deleterious effects on the function of the receptor [50]. Moreover, it was bioinformatically predicted that rs2066845 may cause impairment to the protein structure [50], further justifying our observation that the polymorphism was significantly associated with CRC risk.
Finally, rs2066847 is perhaps the most studied polymorphism among all NOD2 polymorphisms. This polymorphism involves a cytosine insertion that results in a premature stop codon and thus LRR domain truncation [44]. Consequently, the variant allele is incapable of stimulating an appropriate response to activate the NF-κB, as it can only recognize lipopolysaccharide instead of MDP [51]. All the four polymorphisms are well known for their association with Crohn’s disease, which is characterized by defective innate immune responses and dysregulated intestinal inflammation. As mentioned above, these polymorphisms can impair the ability of NOD2 to recognize MDP, leading to compromised activation of the NF-κB signaling pathway. This defect disrupts epithelial barrier function and impairs bacterial clearance, contributing to persistent intestinal inflammation. Chronic inflammation, as seen in Crohn’s disease, is a known risk factor for colorectal cancer, and this inflammatory microenvironment may provide a biological basis for the observed association between NOD2 polymorphisms and increased CRC risk.
In our study, we observed a significant association of the rs2066847 polymorphism with CRC risk. This result is consistent with the findings reported by several other studies on different types of solid tumors such as breast, lung, and gastric cancers, as well as non-Hodgkin’s lymphoma [39,52,53,54]. In addition, [11] concluded in their study of 12 different cancers that the lifetime risk of cancer increases by around 25% to 35% in the presence of the rs2066847 polymorphism. It has also been suggested that rs2066847, unlike the rs2066844 and rs2066845 polymorphisms, causes greater life-threatening disease progression due to its frameshift mutation in the LRR region, which plays an important role in immunological modulation [55]. However, there are also studies showing that this polymorphism is not associated with the risk of multiple myeloma and lung cancer in the Turkish population [44,56]. Thus, with all the controversial published findings, it is reasonable to assume that every key aspect, including geographic variability, source of control, prevalence of polymorphisms in specific populations, genotyping methods, differences in sample size, ethnicity, environmental and genetic factors, complex gene–gene or gene–environment interactions, and even mere chance, could play an important role in determining how these polymorphisms affect the development of disease risk [2,13,57].
Several limitations to this meta-analysis should be noted. First, the effects of gene–environment interactions could not be effectively assessed due to the limited number of studies that reported on this aspect. Second, for the rs2066842 polymorphism, only a small number of studies was included in the analysis and its allele frequencies were very low. This resulted in cells with null values, so quantitative data synthesis was not possible. Third, ethnicity was also not proportionally distributed in the included studies, as the majority of individuals analyzed for rs2066847 belonged to the “other ethnicity” subgroup, and only one of the included studies was conducted in Asia. Ethnic variation can influence the results of genetic association studies in different ways. For example, linkage disequilibrium patterns may be different among ethnic groups, meaning that a polymorphism may be in LD with a causal variant in one population but not in another. In addition, the effect size or direction of the association may be influenced by gene–environment and gene–gene interactions; thus, different ethnicities, which may have distinct genetic backgrounds, environmental exposures, lifestyle factors, and microbiome compositions, could exhibit variable risk profiles for the same polymorphism. To address the influence of ethnic variation in genetic association, future studies should include diverse ethnic groups, especially the underrepresented populations, in order to ensure a more comprehensive and generalizable assessment. Another limitation of this paper is that although we significantly improved the statistical power of the study, the sample size may still be too small to reliably assess the risk association. Finally, the certainty of the evidence was not formally assessed using a structured framework such as GRADE. Despite these limitations, our meta-analysis had several strengths. For instance, the quality of the included studies in this meta-analysis was considered high and met our inclusion criteria as presented in the Newcastle-Ottawa Scale. In addition, we did not find any publication bias in this meta-analysis, especially for the two polymorphisms that showed an association with the risk of CRC (rs2066845 and rs2066847). This means that the pooled results were unbiased.
5. Conclusions
The results of this meta-analysis showed that NOD2 rs2066845 and rs2066847, but not NOD2 rs2066842 and rs2066844, are associated with CRC risk. Interestingly, when stratified by ethnicity, the association of rs2066847 proved significant only in participants of “other ethnicities”, but not in Caucasians. However, the “other ethnicities” subgroup was itself a mixture of many different ancestries, and the small sample size did not allow us to further subdivide the subgroup into more specific ancestries. Therefore, future large-scale studies in different ethnicities are needed to obtain a convincing result on the influence of NOD2 polymorphisms on CRC risk. Nevertheless, the current results showed that the rs2066845 and rs2066847 polymorphisms can potentially serve as predisposition biomarkers for CRC, although further validation work is needed.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/cancers17121999/s1, Figure S1: Sensitivity analysis of NOD2 rs2066844 and colorectal cancer risk.
Author Contributions
Conceptualization, S.C.T.; methodology, M.A.K.S. and S.C.T.; software, M.A.K.S. and S.C.T.; validation, S.C.T.; formal analysis, M.A.K.S. and S.C.T.; investigation, M.A.K.S.; resources, S.C.T.; data curation, S.C.T., M.A.K.S. and H.S.; writing—original draft preparation, M.A.K.S.; writing—review and editing, M.A.I., R.J. and S.C.T.; visualization, S.C.T., H.S. and R.J.; supervision, S.C.T.; project administration, S.C.T.; funding acquisition, S.C.T. and M.A.I. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fundamental Research Grant Scheme of the Ministry of Higher Education, Malaysia (grant number: FRGS/1/2019/SKK08/UKM/02/9) and the APC was funded by the University of Wolverhampton, UK.
Data Availability Statement
Data is contained within the article or Supplementary Material. The review protocol is available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Ferlay, J.; Ervik, M.; Lam, F.; Colombet, M.; Mery, L.; Piñeros, M.; Znaor, A.; Soerjomataram, I.; Bray, F. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2023, 73, 377–406. [Google Scholar]
- Zinkeng, A.; Taylor, F.L.; Cheong, S.H.; Song, H.; Merchant, J.L. Early Onset Colorectal Cancer: Molecular Underpinnings Accelerating Occurrence. Cell Mol. Gastroenterol. Hepatol. 2025, 19, 101425. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.C. Low penetrance genetic polymorphisms as potential biomarkers for colorectal cancer predisposition. J. Gene Med. 2018, 20, e3010. [Google Scholar] [CrossRef]
- Papadakos, S.P.; Georgiadou, C.; Argyrou, A.; Michailidou, E.; Thanos, C.; Vogli, S.; Siakavellas, S.I.; Manolakopoulos, S.; Theocharis, S. Inflammatory Bowel Disease (IBD)-Associated Colorectal Cancer (CRC): Is cGAS-STING Pathway Targeting the Key to Chemoprevention? Int. J. Mol. Sci. 2025, 26, 4979. [Google Scholar] [CrossRef] [PubMed]
- Burada, F.; Mirea, C.S.; Cucu, G.; Vilcea, I.D.; Cimpoeru, A.; Ioana, M. The Association between Nod2 R702w Polymorphism and Susceptibility to Colorectal Cancer in Romanian Patients. Curr. Health Sci. J. 2018, 44, 135–139. [Google Scholar]
- Li, Z.; Shang, D. NOD1 and NOD2: Essential Monitoring Partners in the Innate Immune System. Curr. Issues Mol. Biol. 2024, 46, 9463–9479. [Google Scholar] [CrossRef]
- Kong, L.; Cao, Y.; He, Y.; Zhang, Y. Role and Molecular Mechanism of NOD2 in Chronic Non-Communicable Diseases. J. Mol. Med. 2024, 102, 787–799. [Google Scholar] [CrossRef]
- Omaru, N.; Watanabe, T.; Kamata, K.; Minaga, K.; Kudo, M. Activation of NOD1 and NOD2 in the Development of Liver Injury and Cancer. Front. Immunol. 2022, 13, 1004439. [Google Scholar] [CrossRef]
- Mecca, M.; Picerno, S.; Cortellino, S. The Killer’s Web: Interconnection between Inflammation, Epigenetics and Nutrition in Cancer. Int. J. Mol. Sci. 2024, 25, 2750. [Google Scholar] [CrossRef]
- Hoffmann, P.; Lamerz, D.; Hill, P.; Kirchner, M.; Gauss, A. Gene Polymorphisms of NOD2, IL23R, PTPN2 and ATG16L1 in Patients with Crohn’s Disease: On the Way to Personalized Medicine? Genes 2021, 12, 866. [Google Scholar] [CrossRef]
- Lubinski, J.; Huzarski, T.; Kurzawski, G.; Suchy, J.; Masojc, B.; Mierzejewski, M.; Cybulski, C.; Gronwald, J.; Dębniak, T.; Drosik, A.; et al. The 3020insC Allele of NOD2 Predisposes to Cancers of Multiple Organs. Hered. Cancer Clin. Pract. 2005, 3, 59–63. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lau, T.; Roslani, A.; Lian, L.H.; Lee, P.S.; Hilmi, I.; Goh, K.L.; Vadiveloo, M. NOD2/CARD15 variants in Malaysian patients with sporadic colorectal cancer. Genet. Mol. Res. 2014, 13, 7079–7085. [Google Scholar] [CrossRef]
- Ferrand, A.; al Nabhani, Z.; Tapias, N.S.; Mas, E.; Hugot, J.P.; Barreau, F. NOD2 Expression in Intestinal Epithelial Cells Protects Toward the Development of Inflammation and Associated Carcinogenesis. Cell Mol. Gastroenterol. Hepatol. 2019, 7, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Horowitz, J.E.; Warner, N.; Staples, J.; Crowley, E.; Gosalia, N.; Murchie, R.; Cohen, A.; Mack, D.; Leis, J.A.; Critch, J.; et al. Mutation spectrum of NOD2 reveals recessive inheritance as a main driver of Early Onset Crohn’s Disease. Sci. Rep. 2021, 11, 5595. [Google Scholar] [CrossRef]
- Park, S.C.; Jeen, Y.T. Genetic Studies of Inflammatory Bowel Disease-Focusing on Asian Patients. Cells 2019, 8, 404. [Google Scholar] [CrossRef]
- Turpin, W.; Bedrani, L.; Espin-Garcia, O.; Xu, W.; Silverberg, M.S.; Smith, M.I.; Somasekharan, S.; Guttman, D.S.; Paterson, A.D. Associations of NOD2 polymorphisms with Erysipelotrichaceae in stool of in healthy first degree relatives of Crohn’s disease subjects. BMC Med. Genet. 2020, 21, 204. [Google Scholar] [CrossRef]
- Tuupanen, S.; Alhopuro, P.; Mecklin, J.P.; Järvinen, H.; Aaltonen, L.A. No evidence for association of NOD2 R702W and G908R with colorectal cancer. Int. J. Cancer 2007, 121, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Lakatos, P.L.; Hitre, E.; Szalay, F.; Zinober, K.; Fuszek, P.; Lakatos, L.; Varga, M.; Balog, A.; Kiss, L.S.; Lakatos, B. Common NOD2/CARD15 variants are not associated with susceptibility or the clinicopathologic characteristics of sporadic colorectal cancer in Hungarian patients. BMC Cancer 2007, 7, 54. [Google Scholar] [CrossRef]
- Roberts, R.L.; Gearry, R.B.; Allington, M.D.E.; Morrin, H.R.; Robinson, B.A.; Frizelle, F.A. Caspase Recruitment Domain-Containing Protein 15 Mutations in Patients with Colorectal Cancer. Cancer Res. 2006, 66, 2532–2535. [Google Scholar] [CrossRef]
- Mockelmann, N.; von Schonfels, W.; Buch, S.; von Kampen, O.; Sipos, B.; Egberts, J.H.; König, A.O.; Held, H.; Hampe, J.; Schreiber, S.; et al. Investigation of innate immunity genes CARD4, CARD8 and CARD15 as germline susceptibility factors for colorectal cancer. BMC Gastroenterol. 2009, 9, 79. [Google Scholar] [CrossRef]
- Papaconstantinou, I.; Theodoropoulos, G.; Gazouli, M.; Panoussopoulos, D.; Mantzaris, G.J.; Felekouras, E.; Klonaris, C.; Katsaragakis, S.; Lazaris, A.; Patsouris, E.; et al. Association between mutations in the CARD15/NOD2 gene and colorectal cancer in a Greek population. Int. J. Cancer 2005, 114, 433–435. [Google Scholar] [CrossRef]
- Szeliga, J.; Sondka, Z.; Jackowski, M.; Jarkiewicz-Tretyn, J.; Tretyn, A.; Malenczyk, M. NOD2/CARD15 polymorphism in patients with rectal cancer. Med. Sci. Monit. 2008, 14, CR480–CR484. [Google Scholar] [PubMed]
- Yang, H.; Yang, P.; Liu, H.; Lin, J. The Association of rs4753426 Polymorphism in the Melatonin Receptor 1B (MTNR1B) Gene and Susceptibility to Adolescent Idiopathic Scoliosis: A Systematic Review and Meta-analysis. Pain Physician 2015, 18, 419–431. [Google Scholar] [CrossRef]
- Alhopuro, P.; Ahvenainen, T.; Mecklin, J.P.; Juhola, M.; Järvinen, H.J.; Karhu, A.; Ollikainen, M.; Lehtonen, H.; Aaltonen, L.A. NOD2 3020insC Alone Is Not Sufficient for Colorectal Cancer Predisposition. Cancer Res. 2004, 64, 7245–7247. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kurzawski, G.; Suchy, J.; Kładny, J.; Grabowska, E.; Mierzejewski, M.; Jakubowska, A.; Cybulski, C.; Gronwald, J.; Dębniak, T.; Lubinski, J. The NOD2 3020insC Mutation and the Risk of Colorectal Cancer. Cancer Res. 2004, 64, 1604–1606. [Google Scholar] [CrossRef] [PubMed]
- Omrane, I.; Mezlini, A.; Baroudi, O.; Stambouli, N.; Bougatef, K.; Ayari, H.; Laouani, A.; Ben Ahmed, M. 3020insC NOD2/CARD15 polymorphism associated with treatment of colorectal cancer. Med. Oncol. 2014, 31, 954. [Google Scholar] [CrossRef]
- Suchy, J.; Kłujszo-Grabowska, E.; Kładny, J.; Cybulski, C.; Wokołorczyk, D.; Szymańska-Pasternak, J.; Mierzejewski, M.; Kurzawski, G.; Lubinski, J. Inflammatory response gene polymorphisms and their relationship with colorectal cancer risk. BMC Cancer 2008, 8, 112. [Google Scholar] [CrossRef]
- Vogel, U.; Christensen, J.; Dybdahl, M.; Friis, S.; Hansen, R.D.; Wallin, H.; Jensen, M.; Olsen, A.; Overvad, K.; Tjonneland, A.; et al. Prospective study of interaction between alcohol, NSAID use and polymorphisms in genes involved in the inflammatory response in relation to risk of colorectal cancer. Mutat. Res. Fundam. Mol. Mech. Mutagen. 2007, 624, 88–100. [Google Scholar] [CrossRef]
- Mentzer, A.; Nayee, S.; Omar, Y.; Hullah, E.; Taylor, K.; Goel, R.; Lomer, M.; Powell, M.; Prasad, N.; Borrelli, V.; et al. Genetic Association Analysis Reveals Differences in the Contribution of NOD2 Variants to the Clinical Phenotypes of Orofacial Granulomatosis. Inflamm. Bowel Dis. 2016, 22, 1552–1558. [Google Scholar] [CrossRef]
- Wex, T.; Ebert, M.P.; Siegfried, K.; Dierkes, J.; Schuttler, K.; Rocken, C.; Malfertheiner, P. Gene polymorphisms of the NOD-2/CARD-15 gene and the risk of gastric cancer in Germany. Anticancer Res. 2008, 28, 757–762. [Google Scholar]
- Cubillos-Angulo, J.M.; Fernandes, C.D.; Araújo, D.N.; Carmo, C.A.; Arriaga, M.B.; Andrade, B.B. The influence of single nucleotide polymorphisms of NOD2 or CD14 on the risk of Mycobacterium tuberculosis diseases: A systematic review. Syst. Rev. 2021, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Hedegaard, C.J.; Enevold, C.; Sellebjerg, F.; Bendtzen, K.; Nielsen, C.H.; Sechi, L.A. Variation in NOD2 Augments Th2-and Th17 Responses to Myelin Basic Protein in Multiple Sclerosis. PLoS ONE 2011, 6, e20253. [Google Scholar] [CrossRef]
- Siddique, I.; Mustafa, A.; Khan, I.; Ziyab, A.; Altarrah, M.; Sulaiman, R.; Al-Tarakmah, R.; Owayed, S.; Akbar, H.; Abdulwahab, A.; et al. Detection of mutations in NOD2/CARD15 gene in Arab patients with Crohn’s disease: A case-control study. Saudi J. Gastroenterol. 2021, 27, 240–248. [Google Scholar] [CrossRef]
- Austin, C.M.; Ma, X.; Graviss, E.A. Common Nonsynonymous Polymorphisms in the NOD2 Gene Are Associated with Resistance or Susceptibility to Tuberculosis Disease in African Americans. J. Infect. Dis. 2008, 197, 1713–1716. [Google Scholar] [CrossRef]
- Lesage, S.; Zouali, H.; Cézard, J.P.; Almer, S.; Tysk, C.; O’morain, C.; Gassull, M.; Travis, S.; Annese, V.; Cottone, M.; et al. CARD15/NOD2 Mutational Analysis and Genotype-Phenotype Correlation in 612 Patients with Inflammatory Bowel Disease. Am. J. Hum. Genet. 2002, 70, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Angeletti, S.; Galluzzo, S.; Santini, D.; Ruzzo, A.; Vincenzi, B.; Ferraro, E.; Nasti, G.; Coppola, R.; Dicuonzo, G.; Delogu, G. NOD2/CARD15 polymorphisms impair innate immunity and increase susceptibility to gastric cancer in an Italian population. Hum. Immunol. 2009, 70, 729–732. [Google Scholar] [CrossRef] [PubMed]
- Rosenstiel, P.; Hellmig, S.; Hampe, J.; Ott, S.; Till, A.; Fischbach, W.; Raedler, A.; Fölsch, U.R.; Schreiber, S. Influence of polymorphisms in the NOD1/CARD4 and NOD2/CARD15 genes on the clinical outcome of Helicobacter pylori infection. Cell Microbiol. 2006, 8, 1188–1198. [Google Scholar] [CrossRef]
- Bueno-Martínez, E.; Lara-Almunia, M.; Rodríguez-Arias, C.; Otero-Rodríguez, A.; Garfias-Arjona, S.; González-Sarmiento, R. Polymorphisms in autophagy genes are genetic susceptibility factors in glioblastoma development. BMC Cancer 2022, 22, 146. [Google Scholar] [CrossRef]
- Freire, P.; Figueiredo, P.; Cardoso, R.; Donato, M.M.; Sá, A.; Portela, F.; Amaro, P.; Ferreira, M.J.; Dias, J.A. Card15 mutations and gastric cancer in a Portuguese population. Scand. J. Gastroenterol. 2013, 48, 1188–1197. [Google Scholar] [CrossRef]
- Dȩbniak, T.; Kurzawski, G.; Huzarski, T.; Byrski, T.; Gronwald, J.; Dȩbniak, B.; Górski, B.; Lubinski, J. NOD2 variants and the risk of malignant melanoma. Eur. J. Cancer Prev. 2005, 14, 143–146. [Google Scholar] [CrossRef]
- Yazdanyar, S.; Nordestgaard, B.G. NOD2/CARD15 genotype, cardiovascular disease and cancer in 43 600 individuals from the general population. J. Intern. Med. 2010, 268, 162–170. [Google Scholar] [CrossRef] [PubMed]
- Yao, Q. Nucleotide-binding oligomerization domain containing 2: Structure, function, and diseases. Semin. Arthritis Rheum. 2013, 43, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, T.; Hovingh, E.S.; Foerster, E.G.; Abdel-Nour, M.; Philpott, D.J.; Girardin, S.E. NOD1 and NOD2 in inflammation, immunity and disease. Arch. Biochem. Biophys. 2019, 670, 69–81. [Google Scholar] [CrossRef]
- Zmorzynski, S.; Popek-Marciniec, S.; Styk, W.; Wojcierowska-Litwin, M.; Korszen-Pilecka, I.; Szudy-Szczyrek, A.; Szuba, A.; Dryka, M.; Kruk, M.; Maj, J. The Impact of the NOD2/CARD15 Variant (3020insC) and PSMA6 Polymorphism (-8C>G) on the Development and Outcome of Multiple Myeloma. BioMed Res. Int. 2020, 2020, 7629456. [Google Scholar] [CrossRef] [PubMed]
- Freire, P.; Portela, F.; Donato, M.M.; Figueiredo, P.; Ferreira, M.; Amaro, P.; Dias, J.A. CARD15 mutations and colorectal cancer in a South European country. Int. J. Colorectal Dis. 2010, 25, 1211–1219. [Google Scholar] [CrossRef][Green Version]
- Rigoli, D.L.; Rigoli, L.; di Bella, C.; Fedele, F.; Procopio, V.; Amorini, M.; Currò, M.; Ientile, R.; Venza, M.; Di Bella, C. TLR4 and NOD2/CARD15 Genetic Polymorphisms and their Possible Role in Gastric Carcinogenesis. Anticancer Res. 2010, 30, 513–518. [Google Scholar]
- Abdelnaby, H.; Ndiaye, N.C.; D’amico, F.; Fouad, A.M.; Hassan, S.; Elshafey, A.; Al-Tarakmah, R.; Al-Qallaf, B.; Akbar, H.; Abdulwahab, A.; et al. NOD2/CARD15 polymorphisms (P268S, IVS8 +158, G908R, L1007fs, R702W) among Kuwaiti patients with Crohn’s disease: A case-control study. Saudi J. Gastroenterol. 2021, 27, 249–256. [Google Scholar] [CrossRef]
- Gonzalez-Mancera, M.S.; Forghani, I.; Mirsaeidi, M. Missense (p.Glu778Lys) and (p.Gly908Arg) variants of NOD2 gene are associated with recurrent pulmonary non-tuberculous mycobacterial infections. Scand. J. Immunol. 2020, 92, e12935. [Google Scholar] [CrossRef]
- Besnard, V.; Calender, A.; Bouvry, D.; Pacheco, Y.; Chapelon-Abric, C.; Jeny, F.; Valeyre, D.; Cadranel, J. G908R NOD2 variant in a family with sarcoidosis. Respir. Res. 2018, 19, 44. [Google Scholar] [CrossRef]
- Alipoor, S.D.; Mirsaeidi, M. Inborn Errors in the LRR Domain of Nod2 and Their Potential Consequences on the Function of the Receptor. Cells 2021, 10, 2031. [Google Scholar] [CrossRef]
- Jaskula, E.; Lange, A.; Kyrcz-Krzemien, S.; Markiewicz, M.; Dzierzak-Mietla, M.; Jedrzejczak, W.W.; Szewczyk, M. NOD2/CARD15 single nucleotide polymorphism 13 (3020insC) is associated with risk of sepsis and single nucleotide polymorphism 8 (2104C>T) with herpes viruses reactivation in patients after allogeneic hematopoietic stem cell transplantation. Biol. Blood Marrow Transplant. 2014, 20, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Huzarski, T.; Lener, M.; Domagała, W.; Gronwald, J.; Byrski, T.; Kurzawski, G.; Suchy, J.; Cybulski, C.; Dębniak, T.; Górski, B.; et al. The 3020insC allele of NOD2 predisposes to early-onset breast cancer. Breast Cancer Res. Treat. 2005, 89, 91–93. [Google Scholar] [CrossRef] [PubMed]
- Lener, M.R.; Oszutowska, D.; Castaneda, J.; Kurzawski, G.; Suchy, J.; Nej-Wołosiak, K.; Cybulski, C.; Gronwald, J.; Dębniak, T.; Górski, B.; et al. Prevalence of the NOD2 3020insC mutation in aggregations of breast and lung cancer. Breast Cancer Res. Treat. 2006, 95, 141–145. [Google Scholar] [CrossRef]
- Forrest, M.S.; Skibola, C.F.; Lightfoot, T.J.; Bracci, P.M.; Willett, E.M.; Smith, M.T.; Evenson, T.M.; Holly, E.A. Polymorphisms in innate immunity genes and risk of non-Hodgkin lymphoma. Br. J. Haematol. 2006, 134, 180–183. [Google Scholar] [CrossRef] [PubMed]
- Kupka, T.; Simova, J.; Dvorackova, J.; Martinek, L.; Motyka, O.; Uvirova, M.; Vasku, A.; Horak, M.; Zapletalova, J.; Douda, L.; et al. Crohn’s disease—Genetic factors and progress of the disease. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czechoslov. 2018, 162, 139–143. [Google Scholar] [CrossRef]
- Ozbayer, C.; Kurt, H.; Bayramoglu, A.; Gunes, H.V.; Metintas, M.; Degirmenci, İ.; Bilgin, S. The role of NOD1/CARD4 and NOD2/CARD15 genetic variations in lung cancer risk. Inflamm. Res. 2015, 64, 775–779. [Google Scholar] [CrossRef]
- Feki, S.; Abida, O.; Bouzid, D.; Masmoudi, A.; ben Ayed, M.; Turki, H.; Ammar, O.; Baccouche, N. Is there any Association of NOD2/CARD15 Gene Polymorphism with Tunisian Pemphigus Foliaceus? J. Dermatol. Plast. Surg. 2018, 3, 1019. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).