The Telomere Length Signature in Leukemias—From Molecular Mechanisms Underlying Telomere Shortening to Immunotherapeutic Options Against Telomerase
Simple Summary
Abstract
1. Introduction
2. The Molecular Mechanisms Underlying Telomere Shortening in Leukemias
2.1. The Oxidative Stress and Mitochondrial Dysfunction Loop Mediate Telomere Shortening
2.2. The Activation of the DNA Damage Response Through Telomere Shortening and Mitochondrial Dysfunction
2.3. The Association of Telomere Shortening with Metabolism
3. The Interplay Between Oxidative Stress, Telomere Length, and Leukemia Pathogenesis
3.1. The Key Role O of Oxidative Stress in Leukemias
3.2. The Oxidative Stress Drives Telomere Shortening, Increasing the Susceptibility to Leukemias
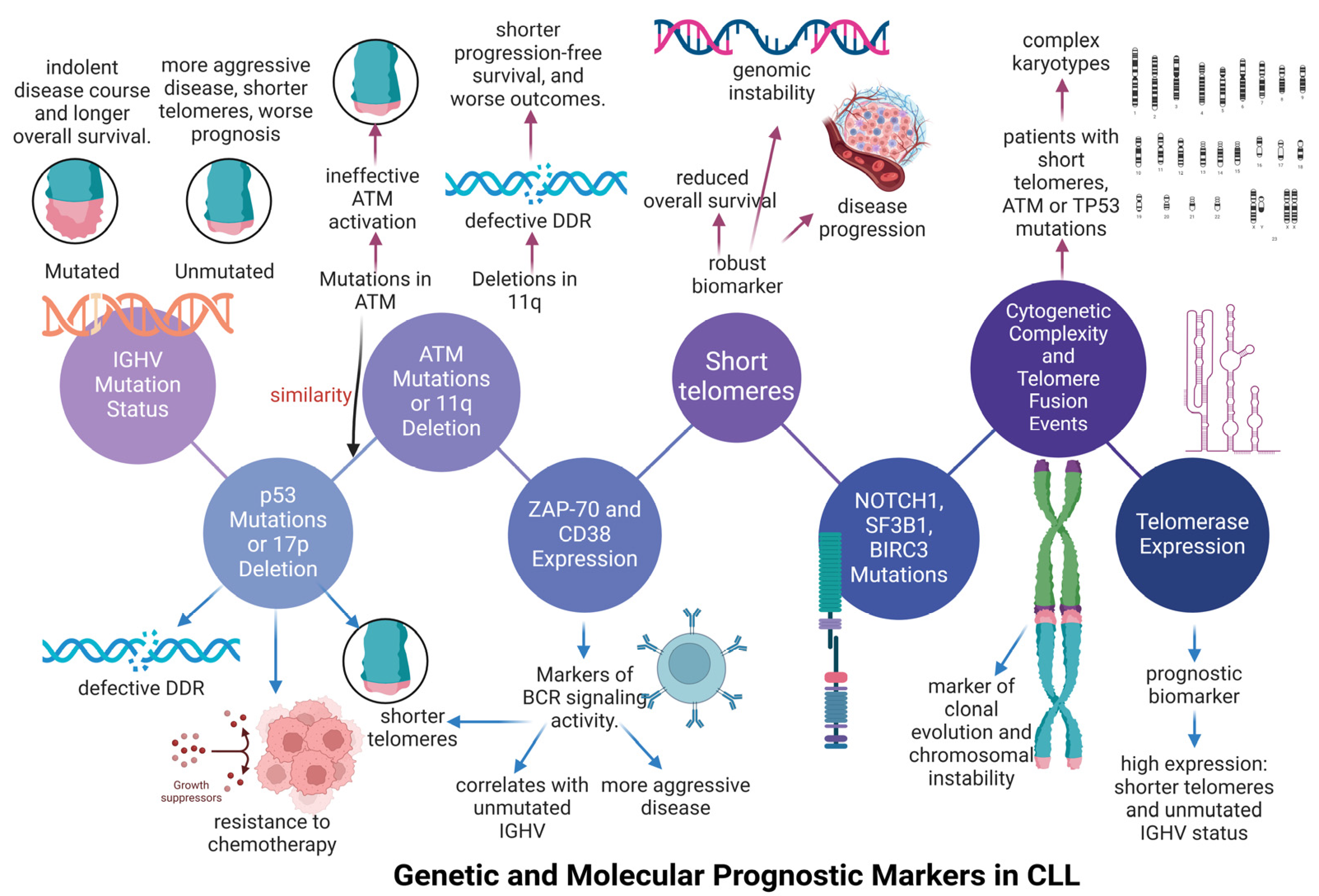
4. The Telomere Length Signature in Chronic Lymphocytic Leukemia (CLL)
4.1. The Association Between Telomere Length and Immunoglobulin Variable Heavy-Chain (IGHV) Status in Chronic Lymphocytic Leukemia (CLL)
4.2. The Association of Telomere Shortening with Genomic Abnormalities Occurred in Chronic Lymphocytic Leukemia (CLL)
4.3. The Inclusion of Telomere Length as a Parameter in Clinical Trial of Chronic Lymphocyic Leukemia (CLL)
5. The Telomere Length Signature in Chronic Myeloid Leukemia (CML)
6. The Telomere Length Signature in Different Types of Acute Leukemias
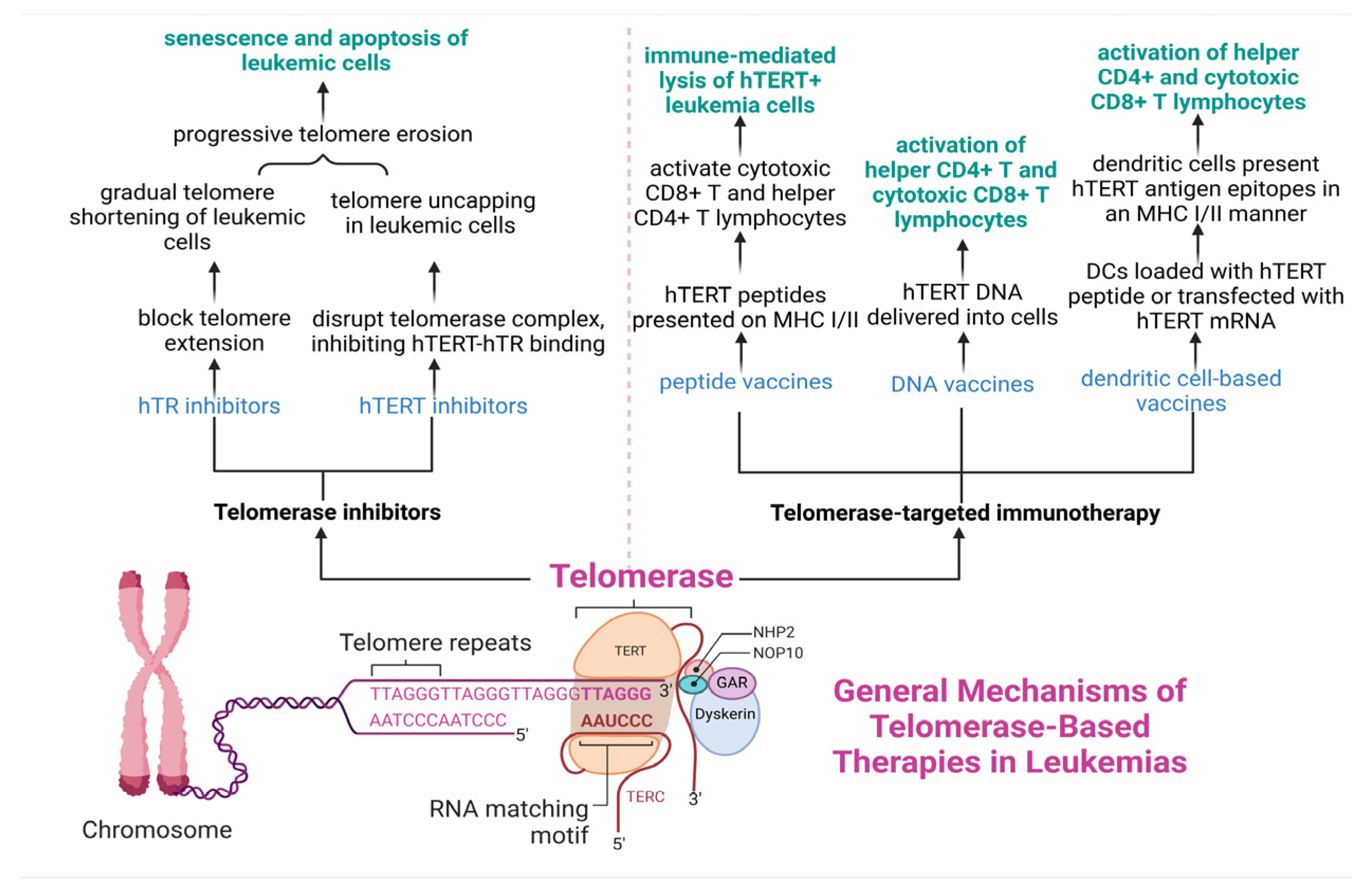
7. Telomerase—A Potential Therapeutic Target in Leukemias
7.1. Telomerase Inhibitors in Leukemias
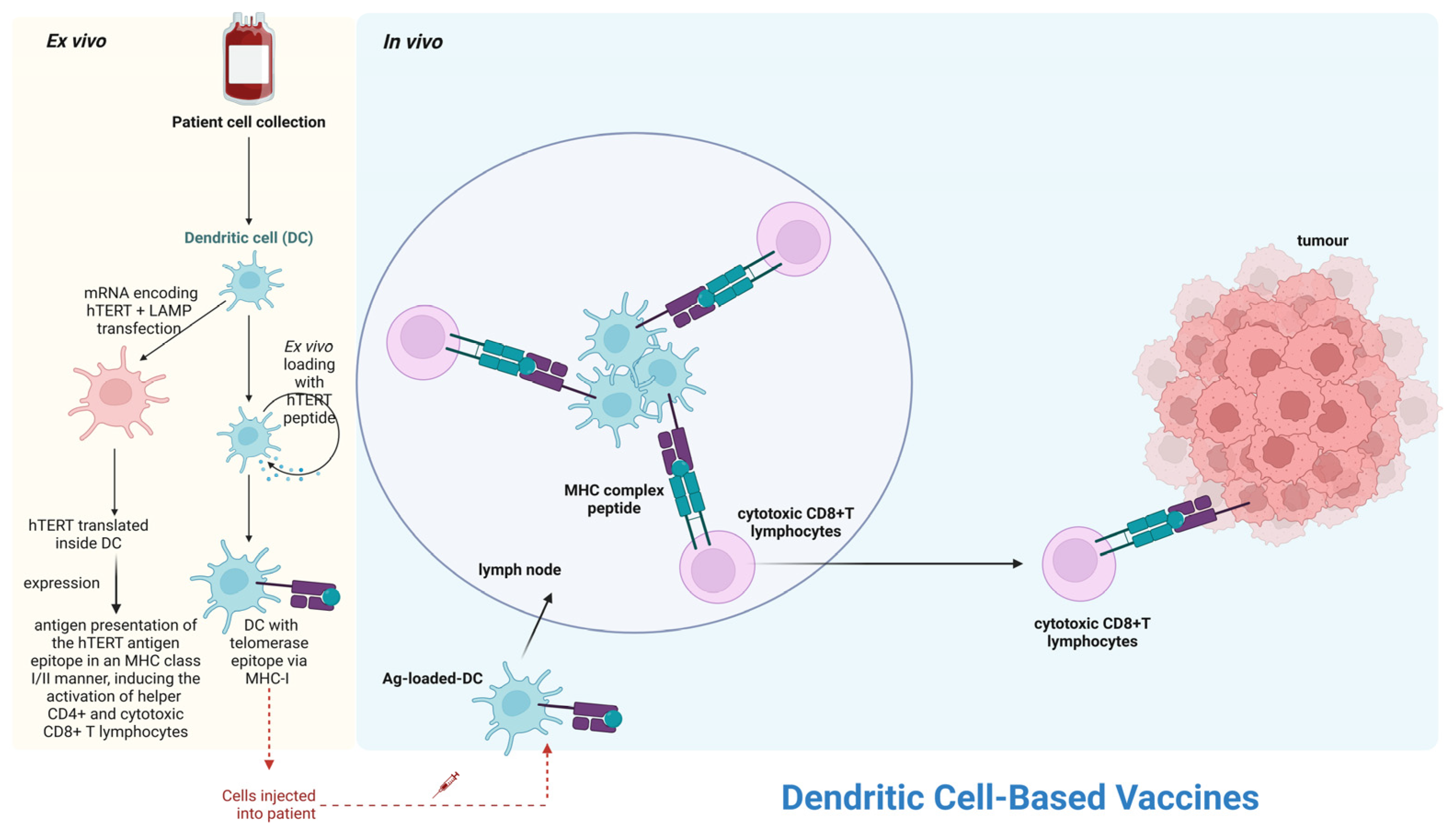
7.2. Telomerase-Based Immunotherapy in Leukemias
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Hockemeyer, D.; Daniels, J.-P.; Takai, H.; de Lange, T. Recent Expansion of the Telomeric Complex in Rodents: Two Distinct POT1 Proteins Protect Mouse Telomeres. Cell 2006, 126, 63–77. [Google Scholar] [CrossRef] [PubMed]
- Blackburn, E.H.; Epel, E.S.; Lin, J. Human Telomere Biology: A Contributory and Interactive Factor in Aging, Disease Risks, and Protection. Science 2015, 350, 1193–1198. [Google Scholar] [CrossRef] [PubMed]
- Palm, W.; De Lange, T. How Shelterin Protects Mammalian Telomeres. Annu. Rev. Genet. 2008, 42, 301–334. [Google Scholar] [CrossRef] [PubMed]
- de Lange, T. Shelterin: The Protein Complex That Shapes and Safeguards Human Telomeres. Genes Dev. 2005, 19, 2100–2110. [Google Scholar] [CrossRef]
- Apetroaei, M.-M.; Fragkiadaki, P.; Velescu, B.Ș.; Baliou, S.; Renieri, E.; Dinu-Pirvu, C.E.; Drăgănescu, D.; Vlăsceanu, A.M.; Nedea, M.I.I.; Udeanu, D.I.; et al. Pharmacotherapeutic Considerations on Telomere Biology: The Positive Effect of Pharmacologically Active Substances on Telomere Length. Int. J. Mol. Sci. 2024, 25, 7694. [Google Scholar] [CrossRef]
- Rossiello, F.; Jurk, D.; Passos, J.F.; d’Adda di Fagagna, F. Telomere Dysfunction in Ageing and Age-Related Diseases. Nat. Cell Biol. 2022, 24, 135–147. [Google Scholar] [CrossRef]
- Gilson, E.; Londoño-Vallejo, A. Telomere Length Profiles in Humans: All Ends Are Not Equal. Cell Cycle 2007, 6, 2486–2494. [Google Scholar] [CrossRef]
- Cesare, A.J.; Karlseder, J. A Three-State Model of Telomere Control over Human Proliferative Boundaries. Curr. Opin. Cell Biol. 2012, 24, 731–738. [Google Scholar] [CrossRef]
- Baliou, S.; Ioannou, P.; Apetroaei, M.-M.; Vakonaki, E.; Fragkiadaki, P.; Kirithras, E.; Tzatzarakis, M.N.; Arsene, A.L.; Docea, A.O.; Tsatsakis, A. The Impact of the Mediterranean Diet on Telomere Biology: Implications for Disease Management-A Narrative Review. Nutrients 2024, 16, 2525. [Google Scholar] [CrossRef]
- d’Adda di Fagagna, F. Living on a Break: Cellular Senescence as a DNA-Damage Response. Nat. Rev. Cancer 2008, 8, 512–522. [Google Scholar] [CrossRef]
- Nassour, J.; Radford, R.; Correia, A.; Fusté, J.M.; Schoell, B.; Jauch, A.; Shaw, R.J.; Karlseder, J. Autophagic Cell Death Restricts Chromosomal Instability during Replicative Crisis. Nature 2019, 565, 659–663. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Sharpless, N.E. Senescence in Health and Disease. Cell 2017, 169, 1000–1011. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The Central Role of DNA Damage in the Ageing Process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Gaidano, G. ATM and Chronic Lymphocytic Leukemia: Mutations, and Not Only Deletions, Matter. Haematologica 2012, 97, 5–8. [Google Scholar] [CrossRef][Green Version]
- McGranahan, N.; Burrell, R.A.; Endesfelder, D.; Novelli, M.R.; Swanton, C. Cancer Chromosomal Instability: Therapeutic and Diagnostic Challenges. EMBO Rep. 2012, 13, 528–538. [Google Scholar] [CrossRef]
- Cuceu, C.; Colicchio, B.; Jeandidier, E.; Junker, S.; Plassa, F.; Shim, G.; Mika, J.; Frenzel, M.; Al Jawhari, M.; Hempel, W.M.; et al. Independent Mechanisms Lead to Genomic Instability in Hodgkin Lymphoma: Microsatellite or Chromosomal Instability. Cancers 2018, 10, 233. [Google Scholar] [CrossRef]
- Tsatsakis, A. (Ed.) Telomeres Biomarkers of a Healthy Life and Successful Aging; Jenny Stanford Publishing: Singapore, 2025; ISBN 978-981-5129-49-6. [Google Scholar]
- Zhang, N.; Wu, J.; Wang, Q.; Liang, Y.; Li, X.; Chen, G.; Ma, L.; Liu, X.; Zhou, F. Global Burden of Hematologic Malignancies and Evolution Patterns over the Past 30 Years. Blood Cancer J. 2023, 13, 82. [Google Scholar] [CrossRef]
- Du, M.; Chen, W.; Liu, K.; Wang, L.; Hu, Y.; Mao, Y.; Sun, X.; Luo, Y.; Shi, J.; Shao, K.; et al. The Global Burden of Leukemia and Its Attributable Factors in 204 Countries and Territories: Findings from the Global Burden of Disease 2019 Study and Projections to 2030. J. Oncol. 2022, 2022, 1612702. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, N.; Zhang, L. The Multi-Omic Prognostic Model of Oxidative Stress-Related Genes in Acute Myeloid Leukemia. Front. Genet. 2021, 12, 722064. [Google Scholar] [CrossRef]
- Dong, C.; Zhang, N.-J.; Zhang, L.-J. Oxidative Stress in Leukemia and Antioxidant Treatment. Chin. Med. J. 2021, 134, 1897–1907. [Google Scholar] [CrossRef]
- Whiteley, A.E.; Price, T.T.; Cantelli, G.; Sipkins, D.A. Leukaemia: A Model Metastatic Disease. Nat. Rev. Cancer 2021, 21, 461–475. [Google Scholar] [CrossRef] [PubMed]
- Juliusson, G.; Hough, R. Leukemia. Prog. Tumor Res. 2016, 43, 87–100. [Google Scholar] [CrossRef] [PubMed]
- Gancarcíková, M.; Zemanová, Z.; Brezinová, J.; Berková, A.; Vcelíková, S.; Smigová, J.; Michalová, K. The Role of Telomeres and Telomerase Complex in Haematological Neoplasia: The Length of Telomeres as a Marker of Carcinogenesis and Prognosis of Disease. Prague Med. Rep. 2010, 111, 91–105. [Google Scholar] [PubMed]
- Jones, C.H.; Pepper, C.; Baird, D.M. Telomere Dysfunction and Its Role in Haematological Cancer. Br. J. Haematol. 2012, 156, 573–587. [Google Scholar] [CrossRef]
- Bryan, T.M.; Englezou, A.; Dalla-Pozza, L.; Dunham, M.A.; Reddel, R.R. Evidence for an Alternative Mechanism for Maintaining Telomere Length in Human Tumors and Tumor-Derived Cell Lines. Nat. Med. 1997, 3, 1271–1274. [Google Scholar] [CrossRef]
- Shay, J.W.; Bacchetti, S. A Survey of Telomerase Activity in Human Cancer. Eur. J. Cancer 1997, 33, 787–791. [Google Scholar] [CrossRef]
- Stanley, S.E.; Armanios, M. The Short and Long Telomere Syndromes: Paired Paradigms for Molecular Medicine. Curr. Opin. Genet. Dev. 2015, 33, 1–9. [Google Scholar] [CrossRef]
- Armando, R.G.; Mengual Gomez, D.L.; Maggio, J.; Sanmartin, M.C.; Gomez, D.E. Telomeropathies: Etiology, Diagnosis, Treatment and Follow-up. Ethical and Legal Considerations. Clin. Genet. 2019, 96, 3–16. [Google Scholar] [CrossRef]
- Kam, M.L.W.; Nguyen, T.T.T.; Ngeow, J.Y.Y. Telomere Biology Disorders. NPJ Genom. Med. 2021, 6, 36. [Google Scholar] [CrossRef]
- McNally, E.J.; Luncsford, P.J.; Armanios, M. Long Telomeres and Cancer Risk: The Price of Cellular Immortality. J. Clin. Investig. 2019, 129, 3474–3481. [Google Scholar] [CrossRef]
- Huang, F.W.; Hodis, E.; Xu, M.J.; Kryukov, G.V.; Chin, L.; Garraway, L.A. Highly Recurrent TERT Promoter Mutations in Human Melanoma. Science 2013, 339, 957–959. [Google Scholar] [CrossRef] [PubMed]
- Heidenreich, B.; Rachakonda, P.S.; Hemminki, K.; Kumar, R. TERT Promoter Mutations in Cancer Development. Curr. Opin. Genet. Dev. 2014, 24, 30–37. [Google Scholar] [CrossRef] [PubMed]
- Chiorazzi, N. Cell Proliferation and Death: Forgotten Features of Chronic Lymphocytic Leukemia B Cells. Best Pract. Res. Clin. Haematol. 2007, 20, 399–413. [Google Scholar] [CrossRef]
- Vasko, T.; Kaifie, A.; Stope, M.; Kraus, T.; Ziegler, P. Telomeres and Telomerase in Hematopoietic Dysfunction: Prognostic Implications and Pharmacological Interventions. IJMS 2017, 18, 2267. [Google Scholar] [CrossRef]
- Olbertova, H.; Plevova, K.; Stranska, K.; Pospisilova, S. Telomere Dynamics in Adult Hematological Malignancies. Biomed. Pap. Med. Fac. Univ. Palacky Olomouc Czech Repub. 2019, 163, 1–7. [Google Scholar] [CrossRef]
- Vulliamy, T.J.; Knight, S.W.; Mason, P.J.; Dokal, I. Very Short Telomeres in the Peripheral Blood of Patients with X-Linked and Autosomal Dyskeratosis Congenita. Blood Cells Mol. Dis. 2001, 27, 353–357. [Google Scholar] [CrossRef]
- Knight, S.W.; Heiss, N.S.; Vulliamy, T.J.; Greschner, S.; Stavrides, G.; Pai, G.S.; Lestringant, G.; Varma, N.; Mason, P.J.; Dokal, I.; et al. X-Linked Dyskeratosis Congenita Is Predominantly Caused by Missense Mutations in the DKC1 Gene. Am. J. Hum. Genet. 1999, 65, 50–58. [Google Scholar] [CrossRef]
- Schmitt, K.; Beier, F.; Panse, J.; Brümmendorf, T.H. [(Pan-)cytopenia as first manifestation of kryptic telomeropathies in adults]. Dtsch. Med. Wochenschr. 2016, 141, 1578–1580. [Google Scholar] [CrossRef]
- Ziegler, P.; Schrezenmeier, H.; Akkad, J.; Brassat, U.; Vankann, L.; Panse, J.; Wilop, S.; Balabanov, S.; Schwarz, K.; Martens, U.M.; et al. Telomere Elongation and Clinical Response to Androgen Treatment in a Patient with Aplastic Anemia and a Heterozygous hTERT Gene Mutation. Ann. Hematol. 2012, 91, 1115–1120. [Google Scholar] [CrossRef]
- Damle, R.N.; Batliwalla, F.M.; Ghiotto, F.; Valetto, A.; Albesiano, E.; Sison, C.; Allen, S.L.; Kolitz, J.; Vinciguerra, V.P.; Kudalkar, P.; et al. Telomere Length and Telomerase Activity Delineate Distinctive Replicative Features of the B-CLL Subgroups Defined by Immunoglobulin V Gene Mutations. Blood 2004, 103, 375–382. [Google Scholar] [CrossRef]
- Jebaraj, B.M.C.; Tausch, E.; Landau, D.A.; Bahlo, J.; Robrecht, S.; Taylor-Weiner, A.N.; Bloehdorn, J.; Scheffold, A.; Mertens, D.; Böttcher, S.; et al. Short Telomeres Are Associated with Inferior Outcome, Genomic Complexity, and Clonal Evolution in Chronic Lymphocytic Leukemia. Leukemia 2019, 33, 2183–2194. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.-M.; Zou, L. Alternative Lengthening of Telomeres: From Molecular Mechanisms to Therapeutic Outlooks. Cell Biosci. 2020, 10, 30. [Google Scholar] [CrossRef] [PubMed]
- Kent, T.; Gracias, D.; Shepherd, S.; Clynes, D. Alternative Lengthening of Telomeres in Pediatric Cancer: Mechanisms to Therapies. Front. Oncol. 2020, 9, 1518. [Google Scholar] [CrossRef]
- Dratwa, M.; Wysoczańska, B.; Łacina, P.; Kubik, T.; Bogunia-Kubik, K. TERT-Regulation and Roles in Cancer Formation. Front. Immunol. 2020, 11, 589929. [Google Scholar] [CrossRef]
- Kusuma, F.K.; Prabhu, A.; Tieo, G.; Ahmed, S.M.; Dakle, P.; Yong, W.K.; Pathak, E.; Madan, V.; Jiang, Y.Y.; Tam, W.L.; et al. Signalling Inhibition by Ponatinib Disrupts Productive Alternative Lengthening of Telomeres (ALT). Nat. Commun. 2023, 14, 1919. [Google Scholar] [CrossRef]
- Liu, L.; Saldanha, S.N.; Pate, M.S.; Andrews, L.G.; Tollefsbol, T.O. Epigenetic Regulation of Human Telomerase Reverse Transcriptase Promoter Activity during Cellular Differentiation. Genes Chromosomes Cancer 2004, 41, 26–37. [Google Scholar] [CrossRef]
- Azouz, A.; Wu, Y.-L.; Hillion, J.; Tarkanyi, I.; Karniguian, A.; Aradi, J.; Lanotte, M.; Chen, G.-Q.; Chehna, M.; Ségal-Bendirdjian, E. Epigenetic Plasticity of hTERT Gene Promoter Determines Retinoid Capacity to Repress Telomerase in Maturation-Resistant Acute Promyelocytic Leukemia Cells. Leukemia 2010, 24, 613–622. [Google Scholar] [CrossRef]
- Smith, J.; Sen, S.; Weeks, R.J.; Eccles, M.R.; Chatterjee, A. Promoter DNA Hypermethylation and Paradoxical Gene Activation. Trends Cancer 2020, 6, 392–406. [Google Scholar] [CrossRef]
- Cantilena, C.R.; Zhao, X.; Kajigaya, S.; Dunavin, N.; Tian, X.; Strickland, S.A.; Savani, B.N.; Mohan, S.R.; Rezvani, K.; Feng, X.; et al. Activity of the Telomerase Inhibitor GRN163L (Imetelstat) on Acute Myeloblastic Leukemia Blasts Is Enhanced by DNA Methyltransferase Inhibitors Irrespective of TERT Promoter Methylation Status. Blood 2015, 126, 1267. [Google Scholar] [CrossRef]
- Zhao, R.-Z.; Jiang, S.; Zhang, L.; Yu, Z.-B. Mitochondrial Electron Transport Chain, ROS Generation and Uncoupling (Review). Int. J. Mol. Med. 2019, 44, 3–15. [Google Scholar] [CrossRef]
- Theurey, P.; Pizzo, P. The Aging Mitochondria. Genes 2018, 9, 22. [Google Scholar] [CrossRef] [PubMed]
- Lesnefsky, E.J.; Moghaddas, S.; Tandler, B.; Kerner, J.; Hoppel, C.L. Mitochondrial Dysfunction in Cardiac Disease: Ischemia--Reperfusion, Aging, and Heart Failure. J. Mol. Cell Cardiol. 2001, 33, 1065–1089. [Google Scholar] [CrossRef] [PubMed]
- Paradies, G.; Ruggiero, F.M.; Quagliariello, E. Age-Dependent Changes in the Activity of Anion Carriers and in the Lipid Composition in Rat Heart Mitochondria. Ann. N. Y. Acad. Sci. 1992, 673, 160–164. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.H.; Park, J.Y.; Lee, H.; Song, E.S.; Kuk, M.U.; Joo, J.; Oh, S.; Kwon, H.W.; Park, J.T.; Park, S.C. Targeting Mitochondrial Metabolism as a Strategy to Treat Senescence. Cells 2021, 10, 3003. [Google Scholar] [CrossRef]
- Wiel, C.; Lallet-Daher, H.; Gitenay, D.; Gras, B.; Le Calvé, B.; Augert, A.; Ferrand, M.; Prevarskaya, N.; Simonnet, H.; Vindrieux, D.; et al. Endoplasmic Reticulum Calcium Release through ITPR2 Channels Leads to Mitochondrial Calcium Accumulation and Senescence. Nat. Commun. 2014, 5, 3792. [Google Scholar] [CrossRef]
- Passos, J.F.; Nelson, G.; Wang, C.; Richter, T.; Simillion, C.; Proctor, C.J.; Miwa, S.; Olijslagers, S.; Hallinan, J.; Wipat, A.; et al. Feedback between P21 and Reactive Oxygen Production Is Necessary for Cell Senescence. Mol. Syst. Biol. 2010, 6, 347. [Google Scholar] [CrossRef]
- Chakravarti, D.; LaBella, K.A.; DePinho, R.A. Telomeres: History, Health, and Hallmarks of Aging. Cell 2021, 184, 306–322. [Google Scholar] [CrossRef]
- Qian, W.; Kumar, N.; Roginskaya, V.; Fouquerel, E.; Opresko, P.L.; Shiva, S.; Watkins, S.C.; Kolodieznyi, D.; Bruchez, M.P.; Van Houten, B. Chemoptogenetic Damage to Mitochondria Causes Rapid Telomere Dysfunction. Proc. Natl. Acad. Sci. USA 2019, 116, 18435–18444. [Google Scholar] [CrossRef]
- Liu, L.; Trimarchi, J.R.; Smith, P.J.S.; Keefe, D.L. Mitochondrial Dysfunction Leads to Telomere Attrition and Genomic Instability. Aging Cell 2002, 1, 40–46. [Google Scholar] [CrossRef]
- Saretzki, G.; Murphy, M.P.; von Zglinicki, T. MitoQ Counteracts Telomere Shortening and Elongates Lifespan of Fibroblasts under Mild Oxidative Stress. Aging Cell 2003, 2, 141–143. [Google Scholar] [CrossRef]
- Gouspillou, G.; Sgarioto, N.; Norris, B.; Barbat-Artigas, S.; Aubertin-Leheudre, M.; Morais, J.A.; Burelle, Y.; Taivassalo, T.; Hepple, R.T. The Relationship between Muscle Fiber Type-Specific PGC-1α Content and Mitochondrial Content Varies between Rodent Models and Humans. PLoS ONE 2014, 9, e103044. [Google Scholar] [CrossRef] [PubMed]
- Sahin, E.; Colla, S.; Liesa, M.; Moslehi, J.; Müller, F.L.; Guo, M.; Cooper, M.; Kotton, D.; Fabian, A.J.; Walkey, C.; et al. Telomere Dysfunction Induces Metabolic and Mitochondrial Compromise. Nature 2011, 470, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Assalve, G.; Lunetti, P.; Rocca, M.S.; Cosci, I.; Di Nisio, A.; Ferlin, A.; Zara, V.; Ferramosca, A. Exploring the Link Between Telomeres and Mitochondria: Mechanisms and Implications in Different Cell Types. Int. J. Mol. Sci. 2025, 26, 993. [Google Scholar] [CrossRef] [PubMed]
- Bian, L.; Meng, Y.; Zhang, M.; Li, D. MRE11-RAD50-NBS1 Complex Alterations and DNA Damage Response: Implications for Cancer Treatment. Mol. Cancer 2019, 18, 169. [Google Scholar] [CrossRef]
- Buchwaldt, J.; Fritsch, T.; Hartmann, M.; Witzel, H.R.; Kloth, M.; Roth, W.; Tagscherer, K.E.; Hartmann, N. Decreased Mitochondrial Transcription Factor A and Mitochondrial DNA Copy Number Promote Cyclin-Dependent Kinase Inhibitor 1A Expression and Reduce Tumorigenic Properties of Colorectal Cancer Cells. Discov. Oncol. 2024, 15, 701. [Google Scholar] [CrossRef]
- Missios, P.; Zhou, Y.; Guachalla, L.M.; von Figura, G.; Wegner, A.; Chakkarappan, S.R.; Binz, T.; Gompf, A.; Hartleben, G.; Burkhalter, M.D.; et al. Glucose Substitution Prolongs Maintenance of Energy Homeostasis and Lifespan of Telomere Dysfunctional Mice. Nat. Commun. 2014, 5, 4924. [Google Scholar] [CrossRef]
- Kruk, J.; Aboul-Enein, H.Y. Reactive Oxygen and Nitrogen Species in Carcinogenesis: Implications of Oxidative Stress on the Progression and Development of Several Cancer Types. Mini Rev. Med. Chem. 2017, 17, 904–919. [Google Scholar] [CrossRef]
- Chen, Y.-C.; Lu, M.-C.; El-Shazly, M.; Lai, K.-H.; Wu, T.-Y.; Hsu, Y.-M.; Lee, Y.-L.; Liu, Y.-C. Breaking down Leukemia Walls: Heteronemin, a Sesterterpene Derivative, Induces Apoptosis in Leukemia Molt4 Cells through Oxidative Stress, Mitochondrial Dysfunction and Induction of Talin Expression. Mar. Drugs 2018, 16, 212. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, M.; Fleming, A.M.; Burrows, C.J.; Wallace, S.S. Neil3 and NEIL1 DNA Glycosylases Remove Oxidative Damages from Quadruplex DNA and Exhibit Preferences for Lesions in the Telomeric Sequence Context. J. Biol. Chem. 2013, 288, 27263–27272. [Google Scholar] [CrossRef]
- Battisti, V.; Maders, L.D.K.; Bagatini, M.D.; Santos, K.F.; Spanevello, R.M.; Maldonado, P.A.; Brulé, A.O.; Araújo, M.d.C.; Schetinger, M.R.C.; Morsch, V.M. Measurement of Oxidative Stress and Antioxidant Status in Acute Lymphoblastic Leukemia Patients. Clin. Biochem. 2008, 41, 511–518. [Google Scholar] [CrossRef]
- Austin, C. Does Oxidative Damage Contribute to the Generation of Leukemia? Leuk. Res. 2009, 33, 1297. [Google Scholar] [CrossRef] [PubMed]
- Hole, P.S.; Darley, R.L.; Tonks, A. Do Reactive Oxygen Species Play a Role in Myeloid Leukemias? Blood 2011, 117, 5816–5826. [Google Scholar] [CrossRef] [PubMed]
- Zhou, F.-L.; Zhang, W.-G.; Wei, Y.-C.; Meng, S.; Bai, G.-G.; Wang, B.-Y.; Yang, H.-Y.; Tian, W.; Meng, X.; Zhang, H.; et al. Involvement of Oxidative Stress in the Relapse of Acute Myeloid Leukemia. J. Biol. Chem. 2010, 285, 15010–15015. [Google Scholar] [CrossRef]
- Mimura, K.; Kua, L.-F.; Shimasaki, N.; Shiraishi, K.; Nakajima, S.; Siang, L.K.; Shabbir, A.; So, J.; Yong, W.-P.; Kono, K. Upregulation of Thioredoxin-1 in Activated Human NK Cells Confers Increased Tolerance to Oxidative Stress. Cancer Immunol. Immunother. 2017, 66, 605–613. [Google Scholar] [CrossRef]
- Zhou, F.; Pan, Y.; Wei, Y.; Zhang, R.; Bai, G.; Shen, Q.; Meng, S.; Le, X.-F.; Andreeff, M.; Claret, F.X. Jab1/Csn5-Thioredoxin Signaling in Relapsed Acute Monocytic Leukemia under Oxidative Stress. Clin. Cancer Res. 2017, 23, 4450–4461. [Google Scholar] [CrossRef]
- Fan, C.; Yang, X.; Yan, L.; Shi, Z. Oxidative Stress Is Two-Sided in the Treatment of Acute Myeloid Leukemia. Cancer Med. 2024, 13, e6806. [Google Scholar] [CrossRef]
- Rodrigues, M.S.; Reddy, M.M.; Sattler, M. Cell Cycle Regulation by Oncogenic Tyrosine Kinases in Myeloid Neoplasias: From Molecular Redox Mechanisms to Health Implications. Antioxid. Redox Signal. 2008, 10, 1813–1848. [Google Scholar] [CrossRef]
- Nieborowska-Skorska, M.; Kopinski, P.K.; Ray, R.; Hoser, G.; Ngaba, D.; Flis, S.; Cramer, K.; Reddy, M.M.; Koptyra, M.; Penserga, T.; et al. Rac2-MRC-cIII-Generated ROS Cause Genomic Instability in Chronic Myeloid Leukemia Stem Cells and Primitive Progenitors. Blood 2012, 119, 4253–4263. [Google Scholar] [CrossRef]
- Navrkalova, V.; Kafkova, L.R.; Divoky, V.; Pospisilova, S. Oxidative Stress as a Therapeutic Perspective for ATM-Deficient Chronic Lymphocytic Leukemia Patients. Haematologica 2015, 100, 994–996. [Google Scholar] [CrossRef]
- Salimi, A.; Roudkenar, M.H.; Sadeghi, L.; Mohseni, A.; Seydi, E.; Pirahmadi, N.; Pourahmad, J. Selective Anticancer Activity of Acacetin Against Chronic Lymphocytic Leukemia Using Both In Vivo and In Vitro Methods: Key Role of Oxidative Stress and Cancerous Mitochondria. Nutr. Cancer 2016, 68, 1404–1416. [Google Scholar] [CrossRef]
- Koczula, K.M.; Ludwig, C.; Hayden, R.; Cronin, L.; Pratt, G.; Parry, H.; Tennant, D.; Drayson, M.; Bunce, C.M.; Khanim, F.L.; et al. Metabolic Plasticity in CLL: Adaptation to the Hypoxic Niche. Leukemia 2016, 30, 65–73. [Google Scholar] [CrossRef] [PubMed]
- Mikhelson, V.M.; Gamaley, I.A. Telomere Shortening Is a Sole Mechanism of Aging in Mammals. Curr. Aging Sci. 2012, 5, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Reichert, S.; Stier, A. Does Oxidative Stress Shorten Telomeres in vivo? A Review. Biol. Lett. 2017, 13, 20170463. [Google Scholar] [CrossRef]
- Schutte, N.S.; Malouff, J.M. The Relationship Between Perceived Stress and Telomere Length: A Meta-analysis. Stress Health 2016, 32, 313–319. [Google Scholar] [CrossRef]
- Pepper, G.V.; Bateson, M.; Nettle, D. Telomeres as Integrative Markers of Exposure to Stress and Adversity: A Systematic Review and Meta-Analysis. R. Soc. Open Sci. 2018, 5, 180744. [Google Scholar] [CrossRef]
- Niveta, J.P.S.; Kumar, M.A.; Parvathi, V.D. Telomere Attrition and Inflammation: The Chicken and the Egg Story. Egypt. J. Med. Hum. Genet. 2022, 23, 131. [Google Scholar] [CrossRef]
- Barnes, R.P.; Fouquerel, E.; Opresko, P.L. The Impact of Oxidative DNA Damage and Stress on Telomere Homeostasis. Mech. Ageing Dev. 2019, 177, 37–45. [Google Scholar] [CrossRef]
- Epel, E.S.; Blackburn, E.H.; Lin, J.; Dhabhar, F.S.; Adler, N.E.; Morrow, J.D.; Cawthon, R.M. Accelerated Telomere Shortening in Response to Life Stress. Proc. Natl. Acad. Sci. USA 2004, 101, 17312–17315. [Google Scholar] [CrossRef]
- Kawanishi, S.; Oikawa, S. Mechanism of Telomere Shortening by Oxidative Stress. Ann. N. Y. Acad. Sci. 2004, 1019, 278–284. [Google Scholar] [CrossRef]
- Cadet, J.; Wagner, J.R. DNA Base Damage by Reactive Oxygen Species, Oxidizing Agents, and UV Radiation. Cold Spring Harb. Perspect. Biol. 2013, 5, a012559. [Google Scholar] [CrossRef]
- von Zglinicki, T. Oxidative Stress Shortens Telomeres. Trends Biochem. Sci. 2002, 27, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Fischer, A.; Reagan, J.D.; Yan, L.J.; Ames, B.N. Oxidative DNA Damage and Senescence of Human Diploid Fibroblast Cells. Proc. Natl. Acad. Sci. USA 1995, 92, 4337–4341. [Google Scholar] [CrossRef] [PubMed]
- Salehi, F.; Behboudi, H.; Kavoosi, G.; Ardestani, S.K. Oxidative DNA Damage Induced by ROS-Modulating Agents with the Ability to Target DNA: A Comparison of the Biological Characteristics of Citrus Pectin and Apple Pectin. Sci. Rep. 2018, 8, 13902. [Google Scholar] [CrossRef] [PubMed]
- Fouquerel, E.; Lormand, J.; Bose, A.; Lee, H.-T.; Kim, G.S.; Li, J.; Sobol, R.W.; Freudenthal, B.D.; Myong, S.; Opresko, P.L. Oxidative Guanine Base Damage Regulates Human Telomerase Activity. Nat. Struct. Mol. Biol. 2016, 23, 1092–1100. [Google Scholar] [CrossRef]
- Fouquerel, E.; Barnes, R.P.; Uttam, S.; Watkins, S.C.; Bruchez, M.P.; Opresko, P.L. Targeted and Persistent 8-Oxoguanine Base Damage at Telomeres Promotes Telomere Loss and Crisis. Mol. Cell 2019, 75, 117–130.e6. [Google Scholar] [CrossRef]
- Burton, G.J.; Jauniaux, E. Oxidative Stress. Best Pract. Res. Clin. Obstet. Gynaecol. 2011, 25, 287–299. [Google Scholar] [CrossRef]
- Opresko, P.L.; Fan, J.; Danzy, S.; Wilson, D.M.; Bohr, V.A. Oxidative Damage in Telomeric DNA Disrupts Recognition by TRF1 and TRF2. Nucleic Acids Res. 2005, 33, 1230–1239. [Google Scholar] [CrossRef]
- Fischer, K.; Bahlo, J.; Fink, A.M.; Goede, V.; Herling, C.D.; Cramer, P.; Langerbeins, P.; von Tresckow, J.; Engelke, A.; Maurer, C.; et al. Long-Term Remissions after FCR Chemoimmunotherapy in Previously Untreated Patients with CLL: Updated Results of the CLL8 Trial. Blood 2016, 127, 208–215. [Google Scholar] [CrossRef]
- Rio-Machin, A.; Vulliamy, T.; Hug, N.; Walne, A.; Tawana, K.; Cardoso, S.; Ellison, A.; Pontikos, N.; Wang, J.; Tummala, H.; et al. The Complex Genetic Landscape of Familial MDS and AML Reveals Pathogenic Germline Variants. Nat. Commun. 2020, 11, 1044. [Google Scholar] [CrossRef]
- Herling, C.D.; Klaumünzer, M.; Rocha, C.K.; Altmüller, J.; Thiele, H.; Bahlo, J.; Kluth, S.; Crispatzu, G.; Herling, M.; Schiller, J.; et al. Complex Karyotypes and KRAS and POT1 Mutations Impact Outcome in CLL after Chlorambucil-Based Chemotherapy or Chemoimmunotherapy. Blood 2016, 128, 395–404. [Google Scholar] [CrossRef]
- Celli, G.B.; de Lange, T. DNA Processing Is Not Required for ATM-Mediated Telomere Damage Response after TRF2 Deletion. Nat. Cell Biol. 2005, 7, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Takai, H.; Smogorzewska, A.; de Lange, T. DNA Damage Foci at Dysfunctional Telomeres. Curr. Biol. 2003, 13, 1549–1556. [Google Scholar] [CrossRef] [PubMed]
- Rhee, D.B.; Ghosh, A.; Lu, J.; Bohr, V.A.; Liu, Y. Factors That Influence Telomeric Oxidative Base Damage and Repair by DNA Glycosylase OGG1. DNA Repair 2011, 10, 34–44. [Google Scholar] [CrossRef] [PubMed]
- Markkanen, E. Not Breathing Is Not an Option: How to Deal with Oxidative DNA Damage. DNA Repair 2017, 59, 82–105. [Google Scholar] [CrossRef]
- Ahmed, S.; Passos, J.F.; Birket, M.J.; Beckmann, T.; Brings, S.; Peters, H.; Birch-Machin, M.A.; Von Zglinicki, T.; Saretzki, G. Telomerase Does Not Counteract Telomere Shortening but Protects Mitochondrial Function under Oxidative Stress. J. Cell Sci. 2008, 121, 1046–1053. [Google Scholar] [CrossRef]
- Ceccaldi, R.; Rondinelli, B.; D’Andrea, A.D. Repair Pathway Choices and Consequences at the Double-Strand Break. Trends Cell Biol. 2016, 26, 52–64. [Google Scholar] [CrossRef]
- Kuzminov, A. Single-Strand Interruptions in Replicating Chromosomes Cause Double-Strand Breaks. Proc. Natl. Acad. Sci. USA 2001, 98, 8241–8246. [Google Scholar] [CrossRef]
- Fouquerel, E.; Parikh, D.; Opresko, P. DNA Damage Processing at Telomeres: The Ends Justify the Means. DNA Repair 2016, 44, 159–168. [Google Scholar] [CrossRef]
- Poetsch, A.R. The Genomics of Oxidative DNA Damage, Repair, and Resulting Mutagenesis. Comput. Struct. Biotechnol. J. 2020, 18, 207–219. [Google Scholar] [CrossRef]
- Chen, Y.; Geng, A.; Zhang, W.; Qian, Z.; Wan, X.; Jiang, Y.; Mao, Z. Fight to the Bitter End: DNA Repair and Aging. Ageing Res. Rev. 2020, 64, 101154. [Google Scholar] [CrossRef]
- Guidarelli, A.; Clementi, E.; Sciorati, C.; Cantoni, O. The Mechanism of the Nitric Oxide-Mediated Enhancement of Tert-Butylhydroperoxide-Induced DNA Single Strand Breakage. Br. J. Pharmacol. 1998, 125, 1074–1080. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, H.; Villani, R.M.; Wang, H.; Simpson, M.J.; Roberts, M.S.; Tang, M.; Liang, X. The Role of Cellular Reactive Oxygen Species in Cancer Chemotherapy. J. Exp. Clin. Cancer Res. 2018, 37, 266. [Google Scholar] [CrossRef] [PubMed]
- Haendeler, J.; Hoffmann, J.; Brandes, R.P.; Zeiher, A.M.; Dimmeler, S. Hydrogen Peroxide Triggers Nuclear Export of Telomerase Reverse Transcriptase via Src Kinase Family-Dependent Phosphorylation of Tyrosine 707. Mol. Cell Biol. 2003, 23, 4598–4610. [Google Scholar] [CrossRef] [PubMed]
- Santos, J.H.; Meyer, J.N.; Skorvaga, M.; Annab, L.A.; Van Houten, B. Mitochondrial hTERT Exacerbates Free-Radical-Mediated mtDNA Damage. Aging Cell 2004, 3, 399–411. [Google Scholar] [CrossRef]
- Eichhorst, B.; Hallek, M. Prognostication of Chronic Lymphocytic Leukemia in the Era of New Agents. Hematol. Am. Soc. Hematol. Educ. Progr. 2016, 2016, 149–155. [Google Scholar] [CrossRef]
- Paiva, R.M.A.; Calado, R.T. Telomere Dysfunction and Hematologic Disorders. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2014; Volume 125, pp. 133–157. ISBN 978-0-12-397898-1. [Google Scholar]
- Ropio, J.; Merlio, J.-P.; Soares, P.; Chevret, E. Telomerase Activation in Hematological Malignancies. Genes 2016, 7, 61. [Google Scholar] [CrossRef]
- Rampazzo, E.; Bonaldi, L.; Trentin, L.; Visco, C.; Keppel, S.; Giunco, S.; Frezzato, F.; Facco, M.; Novella, E.; Giaretta, I.; et al. Telomere Length and Telomerase Levels Delineate Subgroups of B-Cell Chronic Lymphocytic Leukemia with Different Biological Characteristics and Clinical Outcomes. Haematologica 2012, 97, 56–63. [Google Scholar] [CrossRef]
- Bechter, O.E.; Eisterer, W.; Pall, G.; Hilbe, W.; Kühr, T.; Thaler, J. Telomere Length and Telomerase Activity Predict Survival in Patients with B Cell Chronic Lymphocytic Leukemia. Cancer Res. 1998, 58, 4918–4922. [Google Scholar]
- Ricca, I.; Rocci, A.; Drandi, D.; Francese, R.; Compagno, M.; Lobetti Bodoni, C.; De Marco, F.; Astolfi, M.; Monitillo, L.; Vallet, S.; et al. Telomere Length Identifies Two Different Prognostic Subgroups among VH-Unmutated B-Cell Chronic Lymphocytic Leukemia Patients. Leukemia 2007, 21, 697–705. [Google Scholar] [CrossRef]
- Roos, G.; Kröber, A.; Grabowski, P.; Kienle, D.; Bühler, A.; Döhner, H.; Rosenquist, R.; Stilgenbauer, S. Short Telomeres Are Associated with Genetic Complexity, High-Risk Genomic Aberrations, and Short Survival in Chronic Lymphocytic Leukemia. Blood 2008, 111, 2246–2252. [Google Scholar] [CrossRef]
- Rossi, D.; Lobetti Bodoni, C.; Genuardi, E.; Monitillo, L.; Drandi, D.; Cerri, M.; Deambrogi, C.; Ricca, I.; Rocci, A.; Ferrero, S.; et al. Telomere Length Is an Independent Predictor of Survival, Treatment Requirement and Richter’s Syndrome Transformation in Chronic Lymphocytic Leukemia. Leukemia 2009, 23, 1062–1072. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, L.; Grabowski, P.; Degerman, S.; Svenson, U.; Gunnarsson, R.; Cahill, N.; Smedby, K.E.; Geisler, C.; Juliusson, G.; Roos, G.; et al. Short Telomere Length Is Associated with NOTCH1/SF3B1/TP53 Aberrations and Poor Outcome in Newly Diagnosed Chronic Lymphocytic Leukemia Patients. Am. J. Hematol. 2013, 88, 647–651. [Google Scholar] [CrossRef]
- Strefford, J.C.; Kadalayil, L.; Forster, J.; Rose-Zerilli, M.J.J.; Parker, A.; Lin, T.T.; Heppel, N.; Norris, K.; Gardiner, A.; Davies, Z.; et al. Telomere Length Predicts Progression and Overall Survival in Chronic Lymphocytic Leukemia: Data from the UK LRF CLL4 Trial. Leukemia 2015, 29, 2411–2414. [Google Scholar] [CrossRef]
- Hultdin, M.; Rosenquist, R.; Thunberg, U.; Tobin, G.; Norrback, K.-F.; Johnson, A.; Sundström, C.; Roos, G. Association between Telomere Length and V(H) Gene Mutation Status in Chronic Lymphocytic Leukaemia: Clinical and Biological Implications. Br. J. Cancer 2003, 88, 593–598. [Google Scholar] [CrossRef][Green Version]
- Terrin, L.; Trentin, L.; Degan, M.; Corradini, I.; Bertorelle, R.; Carli, P.; Maschio, N.; Bo, M.D.; Noventa, F.; Gattei, V.; et al. Telomerase Expression in B-Cell Chronic Lymphocytic Leukemia Predicts Survival and Delineates Subgroups of Patients with the Same igVH Mutation Status and Different Outcome. Leukemia 2007, 21, 965–972. [Google Scholar] [CrossRef]
- Adam, R.; Díez-González, L.; Ocaña, A.; Šeruga, B.; Amir, E.; Templeton, A.J. Prognostic Role of Telomere Length in Malignancies: A Meta-Analysis and Meta-Regression. Exp. Mol. Pathol. 2017, 102, 455–474. [Google Scholar] [CrossRef]
- Grabowski, P.; Hultdin, M.; Karlsson, K.; Tobin, G.; Aleskog, A.; Thunberg, U.; Laurell, A.; Sundström, C.; Rosenquist, R.; Roos, G. Telomere Length as a Prognostic Parameter in Chronic Lymphocytic Leukemia with Special Reference to VH Gene Mutation Status. Blood 2005, 105, 4807–4812. [Google Scholar] [CrossRef]
- Jebaraj, B.M.C.; Stilgenbauer, S. Telomere Dysfunction in Chronic Lymphocytic Leukemia. Front. Oncol. 2020, 10, 612665. [Google Scholar] [CrossRef]
- Ladetto, M.; Compagno, M.; Ricca, I.; Pagano, M.; Rocci, A.; Astolfi, M.; Drandi, D.; Di Celle, P.F.; Dell’Aquila, M.; Mantoan, B.; et al. Telomere Length Correlates with Histopathogenesis According to the Germinal Center in Mature B-Cell Lymphoproliferative Disorders. Blood 2004, 103, 4644–4649. [Google Scholar] [CrossRef]
- Ladetto, M. Telomere Disrupts, CLL Progresses. Blood 2010, 116, 1821–1822. [Google Scholar] [CrossRef][Green Version]
- Lin, T.T.; Letsolo, B.T.; Jones, R.E.; Rowson, J.; Pratt, G.; Hewamana, S.; Fegan, C.; Pepper, C.; Baird, D.M. Telomere Dysfunction and Fusion during the Progression of Chronic Lymphocytic Leukemia: Evidence for a Telomere Crisis. Blood 2010, 116, 1899–1907. [Google Scholar] [CrossRef] [PubMed]
- Steinbrecher, D.; Jebaraj, B.M.C.; Schneider, C.; Edelmann, J.; Cymbalista, F.; Leblond, V.; Delmer, A.; Ibach, S.; Tausch, E.; Scheffold, A.; et al. Telomere Length in Poor-Risk Chronic Lymphocytic Leukemia: Associations with Disease Characteristics and Outcome. Leuk. Lymphoma 2018, 59, 1614–1623. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.A.; Tam, C.S.; O’Brien, S.M.; Wierda, W.G.; Stingo, F.; Plunkett, W.; Smith, S.C.; Kantarjian, H.M.; Freireich, E.J.; Keating, M.J. Fludarabine, Cyclophosphamide, and Rituximab Treatment Achieves Long-Term Disease-Free Survival in IGHV-Mutated Chronic Lymphocytic Leukemia. Blood 2016, 127, 303–309. [Google Scholar] [CrossRef] [PubMed]
- Rossi, D.; Terzi-di-Bergamo, L.; De Paoli, L.; Cerri, M.; Ghilardi, G.; Chiarenza, A.; Bulian, P.; Visco, C.; Mauro, F.R.; Morabito, F.; et al. Molecular Prediction of Durable Remission after First-Line Fludarabine-Cyclophosphamide-Rituximab in Chronic Lymphocytic Leukemia. Blood 2015, 126, 1921–1924. [Google Scholar] [CrossRef]
- Lazarian, G.; Cymbalista, F.; Baran-Marszak, F. Impact of Low-Burden TP53 Mutations in the Management of CLL. Front. Oncol. 2022, 12, 841630. [Google Scholar] [CrossRef]
- Lin, T.T.; Norris, K.; Heppel, N.H.; Pratt, G.; Allan, J.M.; Allsup, D.J.; Bailey, J.; Cawkwell, L.; Hills, R.; Grimstead, J.W.; et al. Telomere Dysfunction Accurately Predicts Clinical Outcome in Chronic Lymphocytic Leukaemia, Even in Patients with Early Stage Disease. Br. J. Haematol. 2014, 167, 214–223. [Google Scholar] [CrossRef]
- Norris, K.; Hillmen, P.; Rawstron, A.; Hills, R.; Baird, D.M.; Fegan, C.D.; Pepper, C. Telomere Length Predicts for Outcome to FCR Chemotherapy in CLL. Leukemia 2019, 33, 1953–1963. [Google Scholar] [CrossRef]
- Dos Santos, P.; Panero, J.; Palau Nagore, V.; Stanganelli, C.; Bezares, R.F.; Slavutsky, I. Telomere Shortening Associated with Increased Genomic Complexity in Chronic Lymphocytic Leukemia. Tumour Biol. 2015, 36, 8317–8324. [Google Scholar] [CrossRef]
- Hoxha, M.; Fabris, S.; Agnelli, L.; Bollati, V.; Cutrona, G.; Matis, S.; Recchia, A.G.; Gentile, M.; Cortelezzi, A.; Morabito, F.; et al. Relevance of Telomere/Telomerase System Impairment in Early Stage Chronic Lymphocytic Leukemia. Genes Chromosomes Cancer 2014, 53, 612–621. [Google Scholar] [CrossRef]
- Zenz, T.; Mertens, D.; Döhner, H.; Stilgenbauer, S. Importance of Genetics in Chronic Lymphocytic Leukemia. Blood Rev. 2011, 25, 131–137. [Google Scholar] [CrossRef]
- Landau, D.A.; Tausch, E.; Taylor-Weiner, A.N.; Stewart, C.; Reiter, J.G.; Bahlo, J.; Kluth, S.; Bozic, I.; Lawrence, M.; Böttcher, S.; et al. Mutations Driving CLL and Their Evolution in Progression and Relapse. Nature 2015, 526, 525–530. [Google Scholar] [CrossRef] [PubMed]
- Song, D.Y.; Kim, J.-A.; Jeong, D.; Yun, J.; Kim, S.-M.; Lim, K.; Park, S.N.; Im, K.; Choi, S.; Yoon, S.-S.; et al. Telomere Length and Its Correlation with Gene Mutations in Chronic Lymphocytic Leukemia in a Korean Population. PLoS ONE 2019, 14, e0220177. [Google Scholar] [CrossRef] [PubMed]
- Thomay, K.; Fedder, C.; Hofmann, W.; Kreipe, H.; Stadler, M.; Titgemeyer, J.; Zander, I.; Schlegelberger, B.; Göhring, G. Telomere Shortening, TP53 Mutations and Deletions in Chronic Lymphocytic Leukemia Result in Increased Chromosomal Instability and Breakpoint Clustering in Heterochromatic Regions. Ann. Hematol. 2017, 96, 1493–1500. [Google Scholar] [CrossRef]
- M’kacher, R.; Colicchio, B.; Borie, C.; Junker, S.; Marquet, V.; Heidingsfelder, L.; Soehnlen, K.; Najar, W.; Hempel, W.M.; Oudrhiri, N.; et al. Telomere and Centromere Staining Followed by M-FISH Improves Diagnosis of Chromosomal Instability and Its Clinical Utility. Genes 2020, 11, 475. [Google Scholar] [CrossRef]
- Olbertova, H.; Plevova, K.; Pavlova, S.; Malcikova, J.; Kotaskova, J.; Stranska, K.; Spunarova, M.; Trbusek, M.; Navrkalova, V.; Dvorackova, B.; et al. Evolution of TP53 Abnormalities during CLL Disease Course Is Associated with Telomere Length Changes. BMC Cancer 2022, 22, 137. [Google Scholar] [CrossRef]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; de Lange, T. Chromothripsis and Kataegis Induced by Telomere Crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Dewhurst, S.M. Chromothripsis and Telomere Crisis: Engines of Genome Instability. Curr. Opin. Genet. Dev. 2020, 60, 41–47. [Google Scholar] [CrossRef]
- Wan Mohamad Zamri, W.N.; Mohd Yunus, N.; Abdul Aziz, A.A.; Zulkipli, N.N.; Sulong, S. Perspectives on the Application of Cytogenomic Approaches in Chronic Lymphocytic Leukaemia. Diagnostics 2023, 13, 964. [Google Scholar] [CrossRef]
- Pepper, A.G.S.; Zucchetto, A.; Norris, K.; Tissino, E.; Polesel, J.; Soe, Z.; Allsup, D.; Hockaday, A.; Ow, P.L.; Hillmen, P.; et al. Combined Analysis of IGHV Mutations, Telomere Length and CD49d Identifies Long-Term Progression-Free Survivors in TP53 Wild-Type CLL Treated with FCR-Based Therapies. Leukemia 2022, 36, 271–274. [Google Scholar] [CrossRef]
- Champlin, R.E.; Golde, D.W. Chronic Myelogenous Leukemia: Recent Advances. Blood 1985, 65, 1039–1047. [Google Scholar] [CrossRef]
- Lobo, N.A.; Shimono, Y.; Qian, D.; Clarke, M.F. The Biology of Cancer Stem Cells. Annu. Rev. Cell Dev. Biol. 2007, 23, 675–699. [Google Scholar] [CrossRef] [PubMed]
- Deininger, M.; Buchdunger, E.; Druker, B.J. The Development of Imatinib as a Therapeutic Agent for Chronic Myeloid Leukemia. Blood 2005, 105, 2640–2653. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, K.; Ohyashiki, J.H.; Iwama, H.; Hayashi, S.; Shay, J.W.; Toyama, K. Telomerase Activity and Cytogenetic Changes in Chronic Myeloid Leukemia with Disease Progression. Leukemia 1997, 11, 190–194. [Google Scholar] [CrossRef]
- Brümmendorf, T.H.; Holyoake, T.L.; Rufer, N.; Barnett, M.J.; Schulzer, M.; Eaves, C.J.; Eaves, A.C.; Lansdorp, P.M. Prognostic Implications of Differences in Telomere Length between Normal and Malignant Cells from Patients with Chronic Myeloid Leukemia Measured by Flow Cytometry. Blood 2000, 95, 1883–1890. [Google Scholar] [CrossRef]
- Boultwood, J.; Peniket, A.; Watkins, F.; Shepherd, P.; McGale, P.; Richards, S.; Fidler, C.; Littlewood, T.J.; Wainscoat, J.S. Telomere Length Shortening in Chronic Myelogenous Leukemia Is Associated with Reduced Time to Accelerated Phase. Blood 2000, 96, 358–361. [Google Scholar] [CrossRef]
- Drummond, M.; Lennard, A.; Brûmmendorf, T.; Holyoake, T. Telomere Shortening Correlates with Prognostic Score at Diagnosis and Proceeds Rapidly during Progression of Chronic Myeloid Leukemia. Leuk. Lymphoma 2004, 45, 1775–1781. [Google Scholar] [CrossRef]
- Wenn, K.; Tomala, L.; Wilop, S.; Vankann, L.; Hasenbank, C.; Frank, O.; Hochhaus, A.; Giles, F.J.; Lange, T.; Müller, M.C.; et al. Telomere Length at Diagnosis of Chronic Phase Chronic Myeloid Leukemia (CML-CP) Identifies a Subgroup with Favourable Prognostic Parameters and Molecular Response According to the ELN Criteria after 12 Months of Treatment with Nilotinib. Leukemia 2015, 29, 2402–2404. [Google Scholar] [CrossRef]
- Brümmendorf, T.H.; Ersöz, I.; Hartmann, U.; Bartolovic, K.; Balabanov, S.; Wahl, A.; Paschka, P.; Kreil, S.; Lahaye, T.; Berger, U.; et al. Telomere Length in Peripheral Blood Granulocytes Reflects Response to Treatment with Imatinib in Patients with Chronic Myeloid Leukemia. Blood 2003, 101, 375. [Google Scholar] [CrossRef]
- Broccoli, D.; Young, J.W.; de Lange, T. Telomerase Activity in Normal and Malignant Hematopoietic Cells. Proc. Natl. Acad. Sci. USA 1995, 92, 9082–9086. [Google Scholar] [CrossRef]
- Engelhardt, M.; Mackenzie, K.; Drullinsky, P.; Silver, R.T.; Moore, M.A. Telomerase Activity and Telomere Length in Acute and Chronic Leukemia, Pre- and Post-Ex Vivo Culture. Cancer Res. 2000, 60, 610–617. [Google Scholar]
- Schmidlechner, L.; Nagel, I.; Vater, I.; Cascorbi, I.; Kaehler, M. BTK Acts as a Modulator of the Response to Imatinib in Chronic Myeloid Leukemia. Oncol. Lett. 2024, 28, 424. [Google Scholar] [CrossRef] [PubMed]
- Mandal, M.; Kumar, R. Bcl-2 Modulates Telomerase Activity. J. Biol. Chem. 1997, 272, 14183–14187. [Google Scholar] [CrossRef] [PubMed]
- Peng, X.; Tang, F.; Li, Y.; Bai, J.; Li, L.; Zhang, L. Combination of BCL-2 Inhibitors and Immunotherapy: A Promising Therapeutic Strategy for Hematological Malignancies. Discov. Oncol. 2024, 15, 311. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Dong, X.; Huang, D.C.S.; Xu, P.; Zhao, Q.; Chen, B. Current Advances and Future Strategies for BCL-2 Inhibitors: Potent Weapons against Cancers. Cancers 2023, 15, 4957. [Google Scholar] [CrossRef]
- Jabbour, E.; Kantarjian, H.; Cortes, J. Use of Second- and Third-Generation Tyrosine Kinase Inhibitors in the Treatment of Chronic Myeloid Leukemia: An Evolving Treatment Paradigm. Clin. Lymphoma Myeloma Leuk. 2015, 15, 323–334. [Google Scholar] [CrossRef]
- Bouillon, A.-S.; Ventura Ferreira, M.S.; Awad, S.A.; Richter, J.; Hochhaus, A.; Kunzmann, V.; Dengler, J.; Janssen, J.; Ossenkoppele, G.; Westerweel, P.E.; et al. Telomere Shortening Correlates with Leukemic Stem Cell Burden at Diagnosis of Chronic Myeloid Leukemia. Blood Adv. 2018, 2, 1572–1579. [Google Scholar] [CrossRef]
- Brummendorf, T.H.; Ersoz, I.; Hartmann, U.; Balabanov, S.; Wolke, H.; Paschka, P.; Lahaye, T.; Berner, B.; Bartolovic, K.; Kreil, S.; et al. Normalization of Previously Shortened Telomere Length under Treatment with Imatinib Argues against a Preexisting Telomere Length Deficit in Normal Hematopoietic Stem Cells from Patients with Chronic Myeloid Leukemia. Ann. N. Y. Acad. Sci. 2003, 996, 26–38. [Google Scholar] [CrossRef]
- Lobetti-Bodoni, C.; Ferrero, D.; Genuardi, E.; Passera, R.; Bernocco, E.; Sia, D.; Grignani, G.; Crisà, E.; Monitillo, L.; Rocci, A.; et al. Telomere Loss in Philadelphia-Negative Hematopoiesis after Successful Treatment of Chronic Myeloid Leukemia: Evidence for Premature Aging of the Myeloid Compartment. Mech. Ageing Dev. 2012, 133, 479–488. [Google Scholar] [CrossRef]
- Inaba, H.; Mullighan, C.G. Pediatric Acute Lymphoblastic Leukemia. Haematologica 2020, 105, 2524–2539. [Google Scholar] [CrossRef]
- Iacobucci, I.; Mullighan, C.G. Genetic Basis of Acute Lymphoblastic Leukemia. JCO 2017, 35, 975–983. [Google Scholar] [CrossRef]
- da Mota, T.H.A.; Camargo, R.; Biojone, E.R.; Guimarães, A.F.R.; Pittella-Silva, F.; de Oliveira, D.M. The Relevance of Telomerase and Telomere-Associated Proteins in B-Acute Lymphoblastic Leukemia. Genes 2023, 14, 691. [Google Scholar] [CrossRef] [PubMed]
- Iacobucci, I.; Kimura, S.; Mullighan, C.G. Biologic and Therapeutic Implications of Genomic Alterations in Acute Lymphoblastic Leukemia. JCM 2021, 10, 3792. [Google Scholar] [CrossRef] [PubMed]
- Malard, F.; Mohty, M. Acute Lymphoblastic Leukaemia. Lancet 2020, 395, 1146–1162. [Google Scholar] [CrossRef]
- Lagunas-Rangel, F.A.; Chávez-Valencia, V.; Gómez-Guijosa, M.Á.; Cortes-Penagos, C. Acute Myeloid Leukemia-Genetic Alterations and Their Clinical Prognosis. Int. J. Hematol. Oncol. Stem Cell Res. 2017, 11, 328–339. [Google Scholar]
- DiNardo, C.D.; Stone, R.M.; Medeiros, B.C. Novel Therapeutics in Acute Myeloid Leukemia. Am. Soc. Clin. Oncol. Educ. Book 2017, 37, 495–503. [Google Scholar] [CrossRef]
- Rubnitz, J.E.; Kaspers, G.J.L. How I Treat Pediatric Acute Myeloid Leukemia. Blood 2021, 138, 1009–1018. [Google Scholar] [CrossRef]
- Tran, T.H.; Hunger, S.P. The Genomic Landscape of Pediatric Acute Lymphoblastic Leukemia and Precision Medicine Opportunities. Semin. Cancer Biol. 2022, 84, 144–152. [Google Scholar] [CrossRef]
- Samra, B.; Jabbour, E.; Ravandi, F.; Kantarjian, H.; Short, N.J. Evolving Therapy of Adult Acute Lymphoblastic Leukemia: State-of-the-Art Treatment and Future Directions. J. Hematol. Oncol. 2020, 13, 70. [Google Scholar] [CrossRef]
- Aberuyi, N.; Rahgozar, S.; Ghodousi, E.S.; Ghaedi, K. Drug Resistance Biomarkers and Their Clinical Applications in Childhood Acute Lymphoblastic Leukemia. Front. Oncol. 2019, 9, 1496. [Google Scholar] [CrossRef]
- Liu, Y.-F.; Wang, B.-Y.; Zhang, W.-N.; Huang, J.-Y.; Li, B.-S.; Zhang, M.; Jiang, L.; Li, J.-F.; Wang, M.-J.; Dai, Y.-J.; et al. Genomic Profiling of Adult and Pediatric B-Cell Acute Lymphoblastic Leukemia. EBioMedicine 2016, 8, 173–183. [Google Scholar] [CrossRef]
- Waanders, E.; Gu, Z.; Dobson, S.M.; Antić, Ž.; Crawford, J.C.; Ma, X.; Edmonson, M.N.; Payne-Turner, D.; van de Vorst, M.; Jongmans, M.C.J.; et al. Mutational Landscape and Patterns of Clonal Evolution in Relapsed Pediatric Acute Lymphoblastic Leukemia. Blood Cancer Discov. 2020, 1, 96–111. [Google Scholar] [CrossRef] [PubMed]
- Moorman, A.V. New and Emerging Prognostic and Predictive Genetic Biomarkers in B-Cell Precursor Acute Lymphoblastic Leukemia. Haematologica 2016, 101, 407–416. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Fang, M.; Sun, X.; Sun, J. Telomerase Activity and Telomere Length in Acute Leukemia: Correlations with Disease Progression, Subtypes and Overall Survival. Int. J. Lab. Hematol. 2010, 32, 230–238. [Google Scholar] [CrossRef]
- Capraro, V.; Zane, L.; Poncet, D.; Perol, D.; Galia, P.; Preudhomme, C.; Bonnefoy-Berard, N.; Gilson, E.; Thomas, X.; El-Hamri, M.; et al. Telomere Deregulations Possess Cytogenetic, Phenotype, and Prognostic Specificities in Acute Leukemias. Exp. Hematol. 2011, 39, 195–202.e2. [Google Scholar] [CrossRef]
- Asfour, I.A.; Fayek, M.H.; El-Kourashy, S.A.-E.A.; Youssef, S.R.; El-Gohary, G.M.T.; Mohamed, O.F. Correlation of Telomerase Activity to Apoptosis and Survival in Adult Acute Lymphoblastic Leukemia: An Egyptian Single-Center Study. Ann. Hematol. 2008, 87, 213–221. [Google Scholar] [CrossRef]
- Guilleret, I.; Yan, P.; Grange, F.; Braunschweig, R.; Bosman, F.T.; Benhattar, J. Hypermethylation of the Human Telomerase Catalytic Subunit (hTERT) Gene Correlates with Telomerase Activity. Int. J. Cancer 2002, 101, 335–341. [Google Scholar] [CrossRef]
- Borssén, M.; Cullman, I.; Norén-Nyström, U.; Sundström, C.; Porwit, A.; Forestier, E.; Roos, G. hTERT Promoter Methylation and Telomere Length in Childhood Acute Lymphoblastic Leukemia: Associations with Immunophenotype and Cytogenetic Subgroup. Exp. Hematol. 2011, 39, 1144–1151. [Google Scholar] [CrossRef]
- Babor, F.; Peters, C.; Manser, A.R.; Glogova, E.; Sauer, M.; Pötschger, U.; Ahlmann, M.; Cario, G.; Feuchtinger, T.; Gruhn, B.; et al. Presence of Centromeric but Absence of Telomeric Group B KIR Haplotypes in Stem Cell Donors Improve Leukaemia Control after HSCT for Childhood ALL. Bone Marrow Transplant. 2019, 54, 1847–1858. [Google Scholar] [CrossRef]
- Mrózek, K.; Heerema, N.A.; Bloomfield, C.D. Cytogenetics in Acute Leukemia. Blood Rev. 2004, 18, 115–136. [Google Scholar] [CrossRef]
- Xu, D.; Gruber, A.; Peterson, C.; Pisa, P. Telomerase Activity and the Expression of Telomerase Components in Acute Myelogenous Leukaemia. Br. J. Haematol. 1998, 102, 1367–1375. [Google Scholar] [CrossRef]
- Hartmann, U.; Brümmendorf, T.H.; Balabanov, S.; Thiede, C.; Illme, T.; Schaich, M. Telomere Length and hTERT Expression in Patients with Acute Myeloid Leukemia Correlates with Chromosomal Abnormalities. Haematologica 2005, 90, 307–316. [Google Scholar] [PubMed]
- Cogulu, O.; Kosova, B.; Karaca, E.; Gunduz, C.; Ozkinay, F.; Aksoylar, S.; Gulen, H.; Kantar, M.; Oniz, H.; Karapinar, D.; et al. Evaluation of Telomerase mRNA (hTERT) in Childhood Acute Leukemia. Leuk. Lymphoma 2004, 45, 2477–2480. [Google Scholar] [CrossRef] [PubMed]
- Watts, J.M.; Dumitriu, B.; Hilden, P.; Kishtagari, A.; Rapaport, F.; Chen, C.; Ahn, J.; Devlin, S.M.; Stein, E.M.; Rampal, R.; et al. Telomere Length and Associations with Somatic Mutations and Clinical Outcomes in Acute Myeloid Leukemia. Leuk. Res. 2016, 49, 62–65. [Google Scholar] [CrossRef]
- Patel, J.P.; Gönen, M.; Figueroa, M.E.; Fernandez, H.; Sun, Z.; Racevskis, J.; Van Vlierberghe, P.; Dolgalev, I.; Thomas, S.; Aminova, O.; et al. Prognostic Relevance of Integrated Genetic Profiling in Acute Myeloid Leukemia. N. Engl. J. Med. 2012, 366, 1079–1089. [Google Scholar] [CrossRef]
- Warny, M.; Helby, J.; Sengeløv, H.; Nordestgaard, B.G.; Birgens, H.; Bojesen, S.E. Bone Marrow Mononuclear Cell Telomere Length in Acute Myeloid Leukaemia and High-Risk Myelodysplastic Syndrome. Eur. J. Haematol. 2019, 102, 218–226. [Google Scholar] [CrossRef]
- Song, B.; Lou, J.; Mu, L.; Lu, X.; Sun, J.; Tang, B. An Innovative Telomere-Associated Prognosis Model in AML: Predicting Immune Infiltration and Treatment Responsiveness. Curr. Med. Chem. 2024. Online ahead of print. [Google Scholar] [CrossRef]
- Alter, B.P.; Giri, N.; Savage, S.A.; Rosenberg, P.S. Cancer in the National Cancer Institute Inherited Bone Marrow Failure Syndrome Cohort after Fifteen Years of Follow-Up. Haematologica 2018, 103, 30–39. [Google Scholar] [CrossRef]
- Schratz, K.E.; Haley, L.; Danoff, S.K.; Blackford, A.L.; DeZern, A.E.; Gocke, C.D.; Duffield, A.S.; Armanios, M. Cancer Spectrum and Outcomes in the Mendelian Short Telomere Syndromes. Blood 2020, 135, 1946–1956. [Google Scholar] [CrossRef]
- Mangaonkar, A.A.; Patnaik, M.M. Short Telomere Syndromes in Clinical Practice: Bridging Bench and Bedside. Mayo Clin. Proc. 2018, 93, 904–916. [Google Scholar] [CrossRef]
- Chakraborty, S.; Sun, C.-L.; Francisco, L.; Sabado, M.; Li, L.; Chang, K.L.; Forman, S.; Bhatia, S.; Bhatia, R. Accelerated Telomere Shortening Precedes Development of Therapy-Related Myelodysplasia or Acute Myelogenous Leukemia after Autologous Transplantation for Lymphoma. J. Clin. Oncol. 2009, 27, 791–798. [Google Scholar] [CrossRef]
- Ohyashiki, K.; Iwama, H.; Yahata, N.; Tauchi, T.; Kawakubo, K.; Shimamoto, T.; Ohyashiki, J.H. Telomere Dynamics in Myelodysplastic Syndromes and Acute Leukemic Transformation. Leuk. Lymphoma 2001, 42, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Terasaki, Y.; Okumura, H.; Ohtake, S.; Nakao, S. Accelerated Telomere Length Shortening in Granulocytes: A Diagnostic Marker for Myeloproliferative Diseases. Exp. Hematol. 2002, 30, 1399–1404. [Google Scholar] [CrossRef] [PubMed]
- Junko, T.; Ly, H.; Yamaguchi, H.; Carroll, K.A.; Fumiko, K.; Kazuhiro, S.; Yoshio, M.; Ayako, W.; Seiji, G.; Koiti, I.; et al. Identification and Functional Characterization of Novel Telomerase Variant Alleles in Japanese Patients with Bone-Marrow Failure Syndromes. Blood Cells Mol. Dis. 2008, 40, 185–191. [Google Scholar] [CrossRef][Green Version]
- Mason, P.J. Stem Cells, Telomerase and Dyskeratosis Congenita. Bioessays 2003, 25, 126–133. [Google Scholar] [CrossRef]
- Catto, L.F.B.; Zanelatto, L.C.; Donaires, F.S.; de Carvalho, V.S.; Santana, B.A.; Pinto, A.L.; Fantacini, D.; de Souza, L.E.B.; Fonseca, N.P.; Telho, B.S.; et al. Telomeric Repeat-Containing RNA Is Dysregulated in Acute Myeloid Leukemia. Blood Adv. 2023, 7, 7067–7078. [Google Scholar] [CrossRef]
- Caslini, C.; Serna, A. Telomere Transcription in MLL-Rearranged Leukemia Cell Lines: Increased Levels of TERRA Associate with Lymphoid Lineage and Are Independent of Telomere Length and Ploidy. Biomedicines 2023, 11, 925. [Google Scholar] [CrossRef]
- Morin, G.B. The Human Telomere Terminal Transferase Enzyme Is a Ribonucleoprotein That Synthesizes TTAGGG Repeats. Cell 1989, 59, 521–529. [Google Scholar] [CrossRef]
- Mohamad Kamal, N.S.; Safuan, S.; Shamsuddin, S.; Foroozandeh, P. Aging of the Cells: Insight into Cellular Senescence and Detection Methods. Eur. J. Cell Biol. 2020, 99, 151108. [Google Scholar] [CrossRef]
- Stewart, S.A.; Weinberg, R.A. Telomerase and Human Tumorigenesis. Semin. Cancer Biol. 2000, 10, 399–406. [Google Scholar] [CrossRef]
- Bruedigam, C.; Lane, S.W. Telomerase in Hematologic Malignancies. Curr. Opin. Hematol. 2016, 23, 346–353. [Google Scholar] [CrossRef]
- Fernández-Marcelo, T.; Gómez, A.; Pascua, I.; de Juan, C.; Head, J.; Hernando, F.; Jarabo, J.-R.; Calatayud, J.; Torres-García, A.-J.; Iniesta, P. Telomere Length and Telomerase Activity in Non-Small Cell Lung Cancer Prognosis: Clinical Usefulness of a Specific Telomere Status. J. Exp. Clin. Cancer Res. 2015, 34, 78. [Google Scholar] [CrossRef] [PubMed]
- Karow, A.; Haubitz, M.; Oppliger Leibundgut, E.; Helsen, I.; Preising, N.; Steiner, D.; Dantonello, T.M.; Ammann, R.A.; Roessler, J.; Kartal-Kaess, M.; et al. Targeting Telomere Biology in Acute Lymphoblastic Leukemia. Int. J. Mol. Sci. 2021, 22, 6653. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Piatyszek, M.A.; Kobayashi, T.; Estey, E.; Andreeff, M.; Deisseroth, A.B.; Wright, W.E.; Shay, J.W. Telomerase Activity in Human Acute Myelogenous Leukemia: Inhibition of Telomerase Activity by Differentiation-Inducing Agents. Clin. Cancer Res. 1996, 2, 799–803. [Google Scholar] [PubMed]
- Marian, C.O.; Shay, J.W. Prostate Tumor-Initiating Cells: A New Target for Telomerase Inhibition Therapy? Biochim. Biophys. Acta 2009, 1792, 289–296. [Google Scholar] [CrossRef]
- Baylie, T.; Jemal, M.; Baye, G.; Getinet, M.; Amare, G.A.; Adugna, A.; Abebaw, D.; Hibstu, Z.; Tegegne, B.A.; Gugsa, E.; et al. The Role of Telomere and Telomerase in Cancer and Novel Therapeutic Target: Narrative Review. Front. Oncol. 2025, 15, 1542930. [Google Scholar] [CrossRef]
- Trybek, T.; Kowalik, A.; Góźdź, S.; Kowalska, A. Telomeres and Telomerase in Oncogenesis. Oncol. Lett. 2020, 20, 1015–1027. [Google Scholar] [CrossRef]
- Folini, M.; Gandellini, P.; Zaffaroni, N. Targeting the Telosome: Therapeutic Implications. Biochim. Biophys. Acta 2009, 1792, 309–316. [Google Scholar] [CrossRef]
- Ouellette, M.M.; Wright, W.E.; Shay, J.W. Targeting Telomerase-Expressing Cancer Cells. J. Cell Mol. Med. 2011, 15, 1433–1442. [Google Scholar] [CrossRef]
- Guterres, A.N.; Villanueva, J. Targeting Telomerase for Cancer Therapy. Oncogene 2020, 39, 5811–5824. [Google Scholar] [CrossRef]
- Herbert, B.-S.; Pongracz, K.; Shay, J.W.; Gryaznov, S.M. Oligonucleotide N3′-->P5′ Phosphoramidates as Efficient Telomerase Inhibitors. Oncogene 2002, 21, 638–642. [Google Scholar] [CrossRef]
- Rafat, A.; Dizaji Asl, K.; Mazloumi, Z.; Movassaghpour, A.A.; Talebi, M.; Shanehbandi, D.; Farahzadi, R.; Nejati, B.; Nozad Charoudeh, H. Telomerase Inhibition on Acute Myeloid Leukemia Stem Cell Induced Apoptosis with Both Intrinsic and Extrinsic Pathways. Life Sci. 2022, 295, 120402. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.-H.; Bhatia, R. Pushing the Limits: Defeating Leukemia Stem Cells by Depleting Telomerase. Cell Stem Cell 2014, 15, 673–675. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Lennox, A.L.; Huang, F.; Behrs, M.K.; González-Sales, M.; Bhise, N.; Wan, Y.; Sun, L.; Berry, T.; Feller, F.; Morcos, P.N. Imetelstat, a Novel, First-in-Class Telomerase Inhibitor: Mechanism of Action, Clinical, and Translational Science. Clin. Transl. Sci. 2024, 17, e70076. [Google Scholar] [CrossRef] [PubMed]
- Thomas, X. Examining the Safety and Efficacy of Imetelstat in Low-Risk Myelodysplastic Syndrome. Expert Opin. Pharmacother. 2025, 26, 525–533. [Google Scholar] [CrossRef]
- Steensma, D.P.; Fenaux, P.; Van Eygen, K.; Raza, A.; Santini, V.; Germing, U.; Font, P.; Diez-Campelo, M.; Thepot, S.; Vellenga, E.; et al. Imetelstat Achieves Meaningful and Durable Transfusion Independence in High Transfusion-Burden Patients with Lower-Risk Myelodysplastic Syndromes in a Phase II Study. J. Clin. Oncol. 2021, 39, 48–56. [Google Scholar] [CrossRef]
- Imetelstat FDA Approval. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-Imetelstat-low-intermediate-1-risk-myelodysplastic-syndromes-transfusion-dependent (accessed on 1 June 2025).
- Waksal, J.A.; Bruedigam, C.; Komrokji, R.S.; Jamieson, C.H.M.; Mascarenhas, J.O. Telomerase-Targeted Therapies in Myeloid Malignancies. Blood Adv. 2023, 7, 4302–4314. [Google Scholar] [CrossRef]
- Tefferi, A.; Lasho, T.L.; Begna, K.H.; Patnaik, M.M.; Zblewski, D.L.; Finke, C.M.; Laborde, R.R.; Wassie, E.; Schimek, L.; Hanson, C.A.; et al. A Pilot Study of the Telomerase Inhibitor Imetelstat for Myelofibrosis. N. Engl. J. Med. 2015, 373, 908–919. [Google Scholar] [CrossRef]
- Armanios, M.; Greider, C.W. Treating Myeloproliferation--On Target or Off? N. Engl. J. Med. 2015, 373, 965–966. [Google Scholar] [CrossRef]
- Baerlocher, G.M.; Oppliger Leibundgut, E.; Ottmann, O.G.; Spitzer, G.; Odenike, O.; McDevitt, M.A.; Röth, A.; Daskalakis, M.; Burington, B.; Stuart, M.; et al. Telomerase Inhibitor Imetelstat in Patients with Essential Thrombocythemia. N. Engl. J. Med. 2015, 373, 920–928. [Google Scholar] [CrossRef]
- Bashash, D.; Zareii, M.; Safaroghli-Azar, A.; Omrani, M.D.; Ghaffari, S.H. Inhibition of Telomerase Using BIBR1532 Enhances Doxorubicin-Induced Apoptosis in Pre-B Acute Lymphoblastic Leukemia Cells. Hematology 2017, 22, 330–340. [Google Scholar] [CrossRef]
- Li, Y.; Gu, J.; Ding, Y.; Gao, H.; Li, Y.; Sun, Y.; He, M.; Zhang, W.; Yin, J.; Bai, C.; et al. A Small Molecule Compound IX Inhibits Telomere and Attenuates Oncogenesis of Drug-Resistant Leukemia Cells. FASEB J. 2020, 34, 8843–8857. [Google Scholar] [CrossRef] [PubMed]
- Tauchi, T.; Ohyashiki, J.H.; Ohyashiki, K. Telomerase Inhibition Combined with Other Chemotherapeutic Reagents to Enhance Anti-Cancer Effect. Methods Mol. Biol. 2007, 405, 181–189. [Google Scholar] [CrossRef] [PubMed]
- Ghasemimehr, N.; Farsinejad, A.; Mirzaee Khalilabadi, R.; Yazdani, Z.; Fatemi, A. The Telomerase Inhibitor MST-312 Synergistically Enhances the Apoptotic Effect of Doxorubicin in Pre-B Acute Lymphoblastic Leukemia Cells. Biomed. Pharmacother. 2018, 106, 1742–1750. [Google Scholar] [CrossRef] [PubMed]
- Jafri, M.A.; Ansari, S.A.; Alqahtani, M.H.; Shay, J.W. Roles of Telomeres and Telomerase in Cancer, and Advances in Telomerase-Targeted Therapies. Genome Med. 2016, 8, 69. [Google Scholar] [CrossRef]
- Nair, S.K.; Heiser, A.; Boczkowski, D.; Majumdar, A.; Naoe, M.; Lebkowski, J.S.; Vieweg, J.; Gilboa, E. Induction of Cytotoxic T Cell Responses and Tumor Immunity against Unrelated Tumors Using Telomerase Reverse Transcriptase RNA Transfected Dendritic Cells. Nat. Med. 2000, 6, 1011–1017. [Google Scholar] [CrossRef]
- Frolkis, M.; Fischer, M.B.; Wang, Z.; Lebkowski, J.S.; Chiu, C.-P.; Majumdar, A.S. Dendritic Cells Reconstituted with Human Telomerase Gene Induce Potent Cytotoxic T-Cell Response against Different Types of Tumors. Cancer Gene Ther. 2003, 10, 239–249. [Google Scholar] [CrossRef]
- Vahidi, S.; Zabeti Touchaei, A. Telomerase-Based Vaccines: A Promising Frontier in Cancer Immunotherapy. Cancer Cell Int. 2024, 24, 421. [Google Scholar] [CrossRef]
- Kailashiya, C.; Sharma, H.B.; Kailashiya, J. Telomerase Based Anticancer Immunotherapy and Vaccines Approaches. Vaccine 2017, 35, 5768–5775. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, X.; Chen, G.; Shao, M. Clinical Research Progress of Telomerase Targeted Cancer Immunotherapy: A Literature Review. Transl. Cancer Res. 2024, 13, 3904–3921. [Google Scholar] [CrossRef]
- Pateras, I.S.; Kotsakis, A.; Avgeris, M.; Baliou, E.; Kouroupakis, P.; Patsea, E.; Georgoulias, V.; Menez-Jamet, J.; Kinet, J.-P.; Kosmatopoulos, K. Clinical Activity of an hTERT-Specific Cancer Vaccine (Vx-001) in “Immune Desert” NSCLC. Cancers 2021, 13, 1658. [Google Scholar] [CrossRef]
- Haakensen, V.D.; Nowak, A.K.; Ellingsen, E.B.; Farooqi, S.J.; Bjaanæs, M.M.; Horndalsveen, H.; Mcculloch, T.; Grundberg, O.; Cedres, S.M.; Helland, Å. NIPU: A Randomised, Open-Label, Phase II Study Evaluating Nivolumab and Ipilimumab Combined with UV1 Vaccination as Second Line Treatment in Patients with Malignant Mesothelioma. J. Transl. Med. 2021, 19, 232. [Google Scholar] [CrossRef] [PubMed]
- Aamdal, E.; Inderberg, E.M.; Ellingsen, E.B.; Rasch, W.; Brunsvig, P.F.; Aamdal, S.; Heintz, K.-M.; Vodák, D.; Nakken, S.; Hovig, E.; et al. Combining a Universal Telomerase Based Cancer Vaccine with Ipilimumab in Patients with Metastatic Melanoma—Five-Year Follow Up of a Phase I/IIa Trial. Front. Immunol. 2021, 12, 663865. [Google Scholar] [CrossRef] [PubMed]
- Rebucci-Peixoto, M.; Vienot, A.; Adotevi, O.; Jacquin, M.; Ghiringhelli, F.; de la Fouchardière, C.; You, B.; Maurina, T.; Kalbacher, E.; Bazan, F.; et al. A Phase II Study Evaluating the Interest to Combine UCPVax, a Telomerase CD4 TH1-Inducer Cancer Vaccine, and Atezolizumab for the Treatment of HPV Positive Cancers: VolATIL Study. Front. Oncol. 2022, 12, 957580. [Google Scholar] [CrossRef]
- Greten, T.F.; Forner, A.; Korangy, F.; N’Kontchou, G.; Barget, N.; Ayuso, C.; Ormandy, L.A.; Manns, M.P.; Beaugrand, M.; Bruix, J. A Phase II Open Label Trial Evaluating Safety and Efficacy of a Telomerase Peptide Vaccination in Patients with Advanced Hepatocellular Carcinoma. BMC Cancer 2010, 10, 209. [Google Scholar] [CrossRef]
- Bernhardt, S.L.; Gjertsen, M.K.; Trachsel, S.; Møller, M.; Eriksen, J.A.; Meo, M.; Buanes, T.; Gaudernack, G. Telomerase Peptide Vaccination of Patients with Non-Resectable Pancreatic Cancer: A Dose Escalating Phase I/II Study. Br. J. Cancer 2006, 95, 1474–1482. [Google Scholar] [CrossRef]
- Brunsvig, P.F.; Aamdal, S.; Gjertsen, M.K.; Kvalheim, G.; Markowski-Grimsrud, C.J.; Sve, I.; Dyrhaug, M.; Trachsel, S.; Møller, M.; Eriksen, J.A.; et al. Telomerase Peptide Vaccination: A Phase I/II Study in Patients with Non-Small Cell Lung Cancer. Cancer Immunol. Immunother. 2006, 55, 1553–1564. [Google Scholar] [CrossRef]
- Kim, Y.; Lee, D.; Jo, H.; Go, C.; Yang, J.; Kang, D.; Kang, J.S. GV1001 Interacts with Androgen Receptor to Inhibit Prostate Cell Proliferation in Benign Prostatic Hyperplasia by Regulating Expression of Molecules Related to Epithelial-Mesenchymal Transition. Aging (Albany N. Y.) 2021, 13, 3202–3217. [Google Scholar] [CrossRef]
- Brower, V. Telomerase-Based Therapies Emerging Slowly. J. Natl. Cancer Inst. 2010, 102, 520–521. [Google Scholar] [CrossRef]
- Inderberg-Suso, E.-M.; Trachsel, S.; Lislerud, K.; Rasmussen, A.-M.; Gaudernack, G. Widespread CD4+ T-Cell Reactivity to Novel hTERT Epitopes Following Vaccination of Cancer Patients with a Single hTERT Peptide GV1001. Oncoimmunology 2012, 1, 670–686. [Google Scholar] [CrossRef]
- Buseman, C.M.; Wright, W.E.; Shay, J.W. Is Telomerase a Viable Target in Cancer? Mutat. Res. 2012, 730, 90–97. [Google Scholar] [CrossRef]
- Galati, D.; Zanotta, S. Empowering Dendritic Cell Cancer Vaccination: The Role of Combinatorial Strategies. Cytotherapy 2018, 20, 1309–1323. [Google Scholar] [CrossRef] [PubMed]
- González, F.E.; Gleisner, A.; Falcón-Beas, F.; Osorio, F.; López, M.N.; Salazar-Onfray, F. Tumor Cell Lysates as Immunogenic Sources for Cancer Vaccine Design. Hum. Vaccin. Immunother. 2014, 10, 3261–3269. [Google Scholar] [CrossRef] [PubMed]
- Pang, J.-X. [Human telomerase reverse transcriptase as a novel target for tumor immunotherapy]. Ai Zheng 2003, 22, 893–895. [Google Scholar]
- Aloysius, M.M.; Mc Kechnie, A.J.; Robins, R.A.; Verma, C.; Eremin, J.M.; Farzaneh, F.; Habib, N.A.; Bhalla, J.; Hardwick, N.R.; Satthaporn, S.; et al. Generation in Vivo of Peptide-Specific Cytotoxic T Cells and Presence of Regulatory T Cells during Vaccination with hTERT (Class I and II) Peptide-Pulsed DCs. J. Transl. Med. 2009, 7, 18. [Google Scholar] [CrossRef]
- Khoury, H.J.; Collins, R.H.; Blum, W.; Stiff, P.S.; Elias, L.; Lebkowski, J.S.; Reddy, A.; Nishimoto, K.P.; Sen, D.; Wirth, E.D.; et al. Immune Responses and Long-Term Disease Recurrence Status after Telomerase-Based Dendritic Cell Immunotherapy in Patients with Acute Myeloid Leukemia. Cancer 2017, 123, 3061–3072. [Google Scholar] [CrossRef]
- Sekhri, K. Telomeres and Telomerase: Understanding Basic Structure and Potential New Therapeutic Strategies Targeting It in the Treatment of Cancer. J. Postgrad. Med. 2014, 60, 303–308. [Google Scholar] [CrossRef]
- Vonderheide, R.H.; Hahn, W.C.; Schultze, J.L.; Nadler, L.M. The Telomerase Catalytic Subunit Is a Widely Expressed Tumor-Associated Antigen Recognized by Cytotoxic T Lymphocytes. Immunity 1999, 10, 673–679. [Google Scholar] [CrossRef]
- Minev, B.; Hipp, J.; Firat, H.; Schmidt, J.D.; Langlade-Demoyen, P.; Zanetti, M. Cytotoxic T Cell Immunity against Telomerase Reverse Transcriptase in Humans. Proc. Natl. Acad. Sci. USA 2000, 97, 4796–4801. [Google Scholar] [CrossRef]
- Kokhaei, P.; Palma, M.; Hansson, L.; Osterborg, A.; Mellstedt, H.; Choudhury, A. Telomerase (hTERT 611-626) Serves as a Tumor Antigen in B-Cell Chronic Lymphocytic Leukemia and Generates Spontaneously Antileukemic, Cytotoxic T Cells. Exp. Hematol. 2007, 35, 297–304. [Google Scholar] [CrossRef]
- Barbullushi, K.; Rampi, N.; Serpenti, F.; Sciumè, M.; Fabris, S.; De Roberto, P.; Fracchiolla, N.S. Vaccination Therapy for Acute Myeloid Leukemia: Where Do We Stand? Cancers 2022, 14, 2994. [Google Scholar] [CrossRef]
- Su, Z.; Dannull, J.; Yang, B.K.; Dahm, P.; Coleman, D.; Yancey, D.; Sichi, S.; Niedzwiecki, D.; Boczkowski, D.; Gilboa, E.; et al. Telomerase mRNA-Transfected Dendritic Cells Stimulate Antigen-Specific CD8+ and CD4+ T Cell Responses in Patients with Metastatic Prostate Cancer. J. Immunol. 2005, 174, 3798–3807. [Google Scholar] [CrossRef] [PubMed]
- Kyte, J.A.; Gaudernack, G.; Faane, A.; Lislerud, K.; Inderberg, E.M.; Brunsvig, P.; Aamdal, S.; Kvalheim, G.; Wälchli, S.; Pule, M. T-Helper Cell Receptors from Long-Term Survivors after Telomerase Cancer Vaccination for Use in Adoptive Cell Therapy. Oncoimmunology 2016, 5, e1249090. [Google Scholar] [CrossRef] [PubMed]
- Vonderheide, R.H.; Domchek, S.M.; Schultze, J.L.; George, D.J.; Hoar, K.M.; Chen, D.-Y.; Stephans, K.F.; Masutomi, K.; Loda, M.; Xia, Z.; et al. Vaccination of Cancer Patients against Telomerase Induces Functional Antitumor CD8+ T Lymphocytes. Clin. Cancer Res. 2004, 10, 828–839. [Google Scholar] [CrossRef] [PubMed]
- Dupont, J.; Latouche, J.-B.; Ma, C.; Sadelain, M. Artificial Antigen-Presenting Cells Transduced with Telomerase Efficiently Expand Epitope-Specific, Human Leukocyte Antigen-Restricted Cytotoxic T Cells. Cancer Res. 2005, 65, 5417–5427. [Google Scholar] [CrossRef]
- Schmidtke, A.; Weinacker, B. [Suicide rates, suicide methods and uncertain cause of death in the elderly]. Z. Gerontol. 1991, 24, 3–11. [Google Scholar]
- Khoury, H.J.; Collins, R.H.; Blum, W.; Maness, L.; Stiff, P.; Kelsey, S.M.; Reddy, A.; Smith, J.A.; DiPersio, J.F. Prolonged Administration of the Telomerase Vaccine GRNVAC1 Is Well Tolerated and Appears to Be Associated with Favorable Outcomes In High-Risk Acute Myeloid Leukemia (AML). Blood 2010, 116, 2190. [Google Scholar] [CrossRef]
- Ruden, M.; Puri, N. Novel Anticancer Therapeutics Targeting Telomerase. Cancer Treat. Rev. 2013, 39, 444–456. [Google Scholar] [CrossRef]
- Mehrotra, S.; Britten, C.D.; Chin, S.; Garrett-Mayer, E.; Cloud, C.A.; Li, M.; Scurti, G.; Salem, M.L.; Nelson, M.H.; Thomas, M.B.; et al. Vaccination with Poly(IC:LC) and Peptide-Pulsed Autologous Dendritic Cells in Patients with Pancreatic Cancer. J. Hematol. Oncol. 2017, 10, 82. [Google Scholar] [CrossRef]
- Yan, J.; Pankhong, P.; Shin, T.H.; Obeng-Adjei, N.; Morrow, M.P.; Walters, J.N.; Khan, A.S.; Sardesai, N.Y.; Weiner, D.B. Highly Optimized DNA Vaccine Targeting Human Telomerase Reverse Transcriptase Stimulates Potent Antitumor Immunity. Cancer Immunol. Res. 2013, 1, 179–189. [Google Scholar] [CrossRef]
- Thalmensi, J.; Pliquet, E.; Liard, C.; Escande, M.; Bestetti, T.; Julithe, M.; Kostrzak, A.; Pailhes-Jimenez, A.-S.; Bourges, E.; Loustau, M.; et al. Anticancer DNA Vaccine Based on Human Telomerase Reverse Transcriptase Generates a Strong and Specific T Cell Immune Response. Oncoimmunology 2016, 5, e1083670. [Google Scholar] [CrossRef]
- Yang, B.; Jeang, J.; Yang, A.; Wu, T.C.; Hung, C.-F. DNA Vaccine for Cancer Immunotherapy. Hum. Vaccin. Immunother. 2014, 10, 3153–3164. [Google Scholar] [CrossRef] [PubMed]
- Lu, B.; Lim, J.M.; Yu, B.; Song, S.; Neeli, P.; Sobhani, N.; K, P.; Bonam, S.R.; Kurapati, R.; Zheng, J.; et al. The Next-Generation DNA Vaccine Platforms and Delivery Systems: Advances, Challenges and Prospects. Front. Immunol. 2024, 15, 1332939. [Google Scholar] [CrossRef] [PubMed]
- Pandya, A.; Shah, Y.; Kothari, N.; Postwala, H.; Shah, A.; Parekh, P.; Chorawala, M.R. The Future of Cancer Immunotherapy: DNA Vaccines Leading the Way. Med. Oncol. 2023, 40, 200. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Baliou, S.; Pelagiadis, I.; Apetroaei, M.-M.; Vakonaki, E.; Arsene, A.L.; Hatzidaki, E.; Tzatzarakis, M.N.; Ioannou, P.; Tsatsakis, A.; Stiakaki, E. The Telomere Length Signature in Leukemias—From Molecular Mechanisms Underlying Telomere Shortening to Immunotherapeutic Options Against Telomerase. Cancers 2025, 17, 1936. https://doi.org/10.3390/cancers17121936
Baliou S, Pelagiadis I, Apetroaei M-M, Vakonaki E, Arsene AL, Hatzidaki E, Tzatzarakis MN, Ioannou P, Tsatsakis A, Stiakaki E. The Telomere Length Signature in Leukemias—From Molecular Mechanisms Underlying Telomere Shortening to Immunotherapeutic Options Against Telomerase. Cancers. 2025; 17(12):1936. https://doi.org/10.3390/cancers17121936
Chicago/Turabian StyleBaliou, Stella, Iordanis Pelagiadis, Miruna-Maria Apetroaei, Elena Vakonaki, Andreea Letiția Arsene, Eleftheria Hatzidaki, Manolis N. Tzatzarakis, Petros Ioannou, Aristides Tsatsakis, and Eftichia Stiakaki. 2025. "The Telomere Length Signature in Leukemias—From Molecular Mechanisms Underlying Telomere Shortening to Immunotherapeutic Options Against Telomerase" Cancers 17, no. 12: 1936. https://doi.org/10.3390/cancers17121936
APA StyleBaliou, S., Pelagiadis, I., Apetroaei, M.-M., Vakonaki, E., Arsene, A. L., Hatzidaki, E., Tzatzarakis, M. N., Ioannou, P., Tsatsakis, A., & Stiakaki, E. (2025). The Telomere Length Signature in Leukemias—From Molecular Mechanisms Underlying Telomere Shortening to Immunotherapeutic Options Against Telomerase. Cancers, 17(12), 1936. https://doi.org/10.3390/cancers17121936








