Improving the Precision of Deep-Learning-Based Head and Neck Target Auto-Segmentation by Leveraging Radiology Reports Using a Large Language Model
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Deep-Learning-Based Auto-Segmentation Model
2.1.1. Dataset
2.1.2. Network Structure and Training
2.2. LLM Prompting for Analyzing Clinical Diagnosis Reports
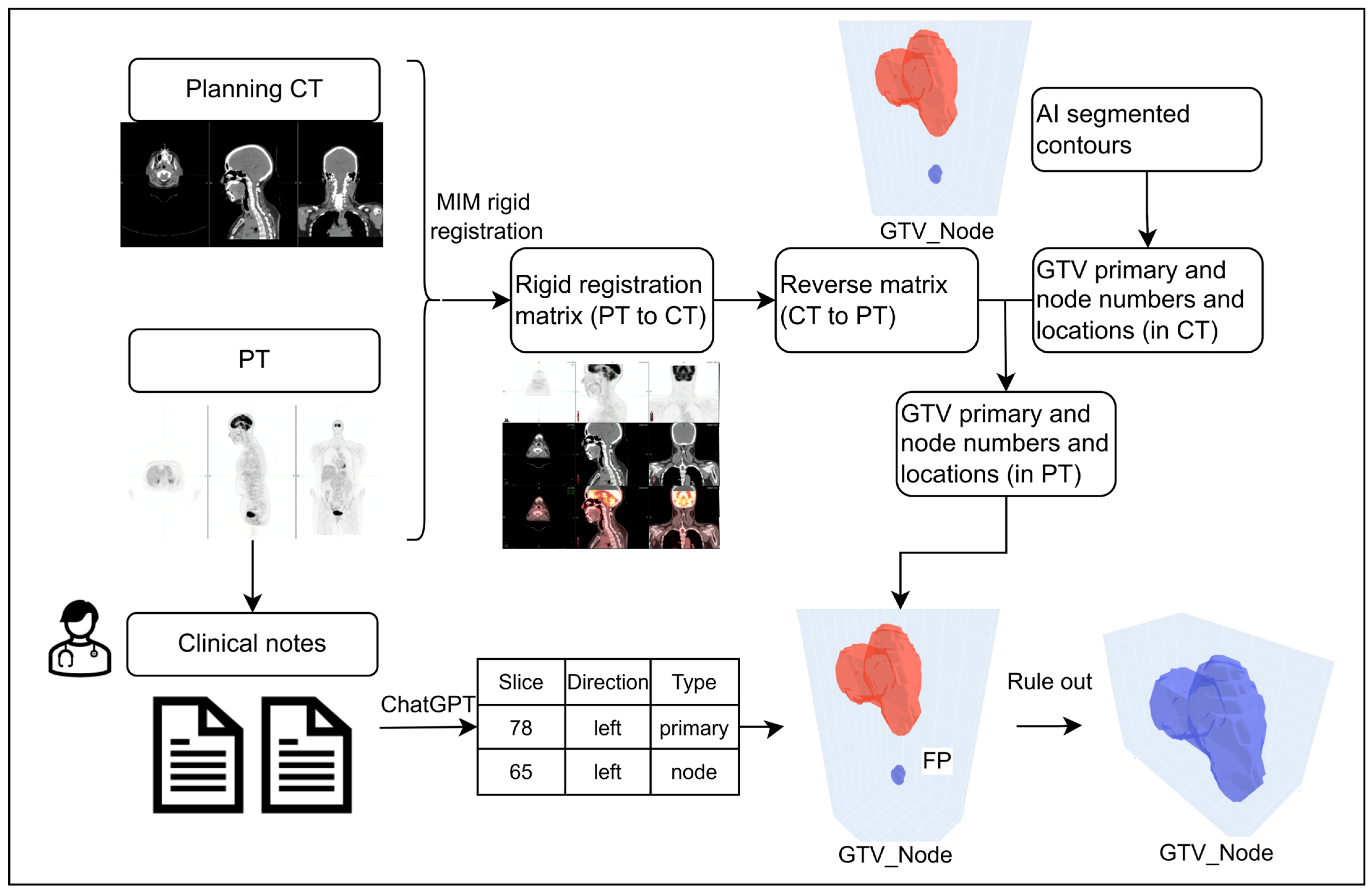
2.3. Workflow to Rule Out False-Positive Errors in DLAS Contours
2.3.1. Scenario 1: Detailed Slice Location-Based Workflow
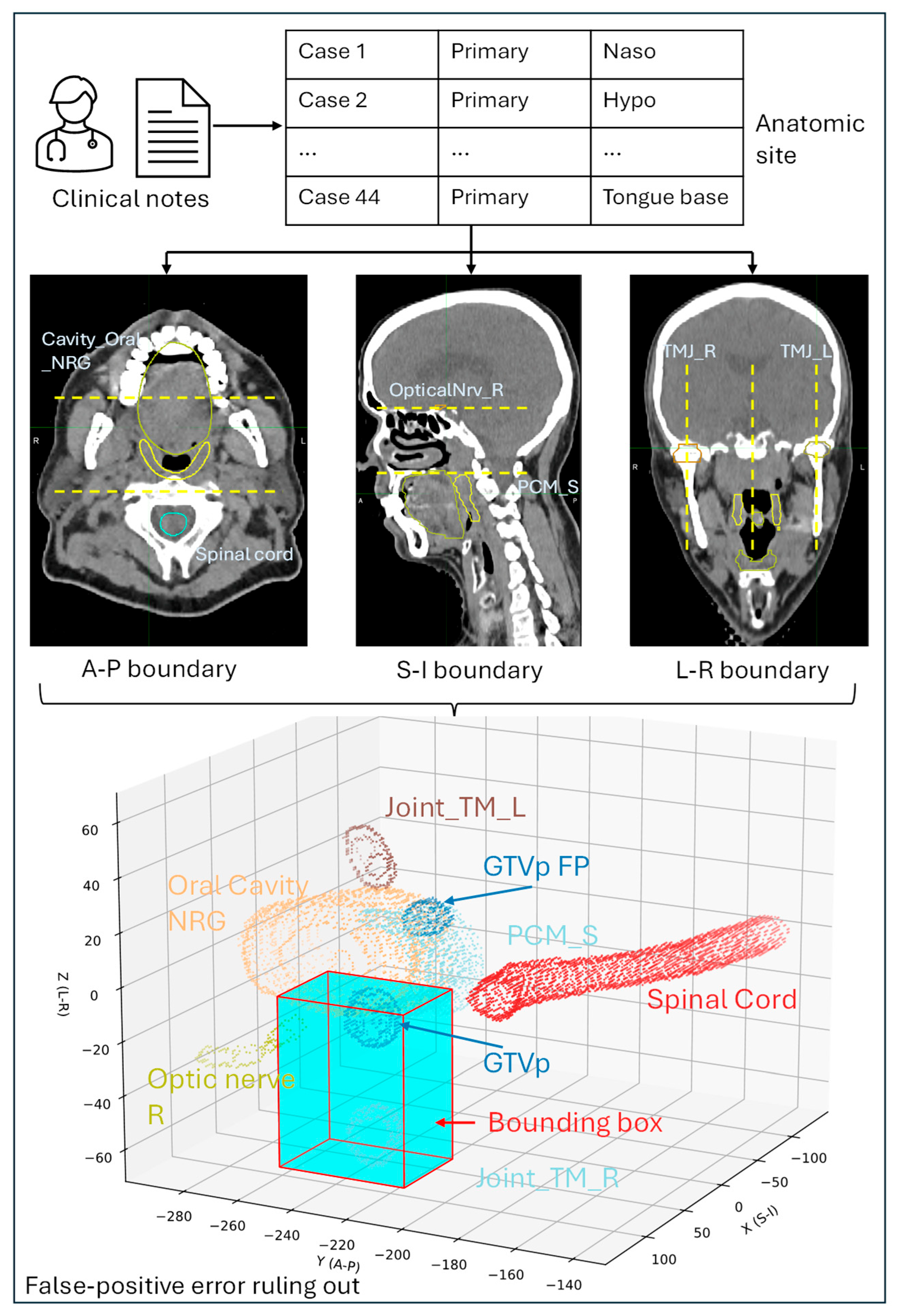
2.3.2. Scenario 2: Anatomic Region-Based Workflow
2.4. Evaluation
3. Results
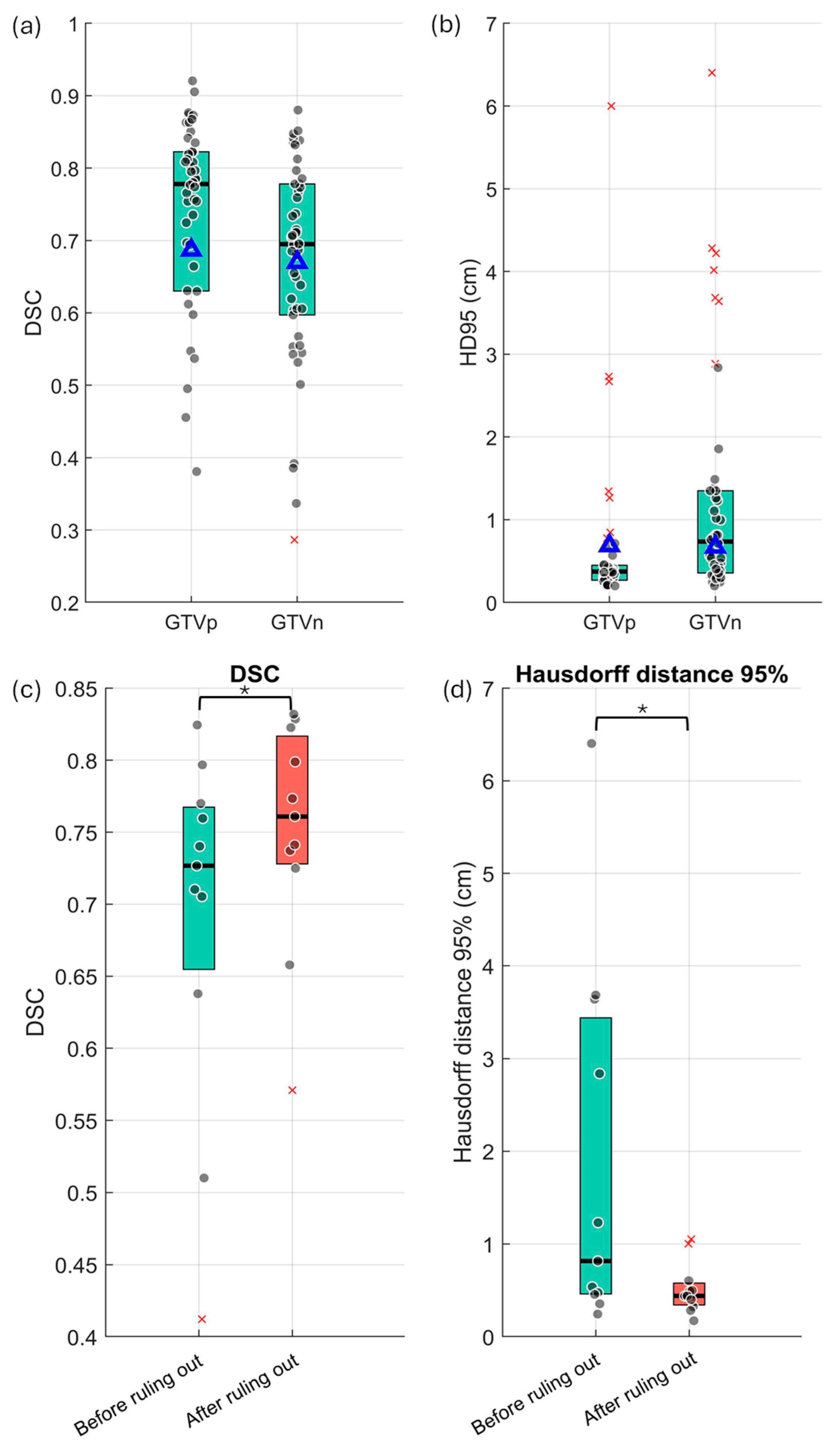
3.1. Baseline Performance of the GTV DLAS Model
3.2. LLM Extraction Results
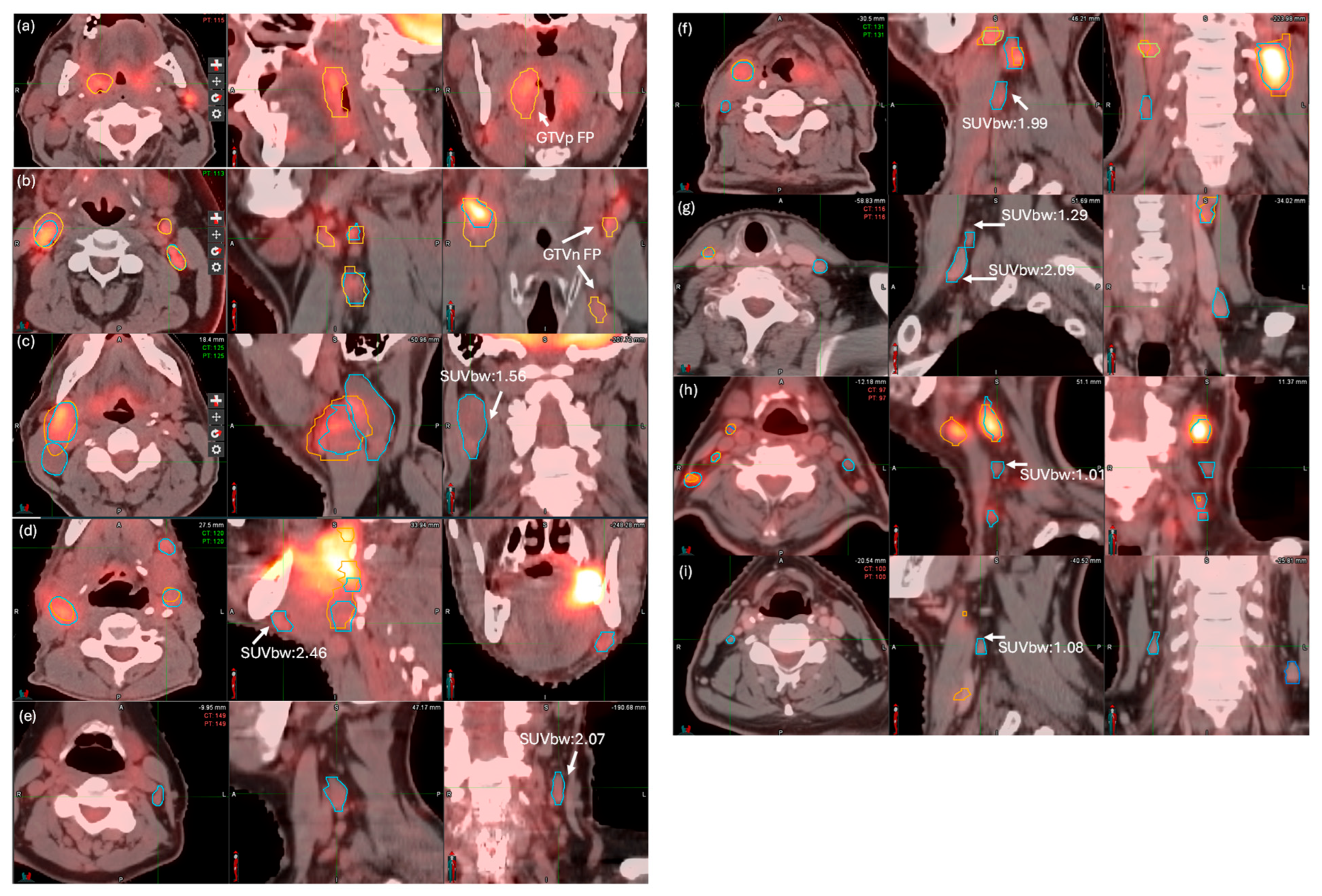
3.3. FP Ruling Out and FN Detection Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Javanbakht, M. Global, Regional, and National Cancer Incidence, Mortality, Years of Life Lost, Years Lived with Disability, and Disability-Adjusted Life-years for 32 Cancer Groups, 1990 to 2015, A Systematic Analysis for the Global Burden of Disease Study. JAMA Oncol. 2017, 3, 524–548. [Google Scholar]
- Andrearczyk, V.; Oreiller, V.; Boughdad, S.; Rest, C.C.L.; Elhalawani, H.; Jreige, M.; Prior, J.O.; Vallières, M.; Visvikis, D.; Hatt, M.; et al. Overview of the HECKTOR Challenge at MICCAI 2021, Automatic Head and Neck Tumor Segmentation and Outcome Prediction in PET/CT Images; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Kihara, S.; Koike, Y.; Takegawa, H.; Anetai, Y.; Nakamura, S.; Tanigawa, N.; Koizumi, M. Clinical target volume segmentation based on gross tumor volume using deep learning for head and neck cancer treatment. Med. Dosim. 2023, 48, 20–24. [Google Scholar] [CrossRef] [PubMed]
- van der Veen, J.; Gulyban, A.; Nuyts, S. Interobserver variability in delineation of target volumes in head and neck cancer. Radiother. Oncol. 2019, 137, 9–15. [Google Scholar] [CrossRef]
- Gudi, S.; Ghosh-Laskar, S.; Agarwal, J.P.; Chaudhari, S.; Rangarajan, V.; Paul, S.N.; Upreti, R.; Murthy, V.; Budrukkar, A.; Gupta, T. Interobserver variability in the delineation of gross tumour volume and specified organs-at-risk during IMRT for head and neck cancers and the impact of FDG-PET/CT on such variability at the primary site. J. Med. Imaging Radiat. Sci. 2017, 48, 184–192. [Google Scholar] [CrossRef]
- Peters, L.J.; O’Sullivan, B.; Giralt, J.; Fitzgerald, T.J.; Trotti, A.; Bernier, J.; Bourhis, J.; Yuen, K.; Fisher, R.; Rischin, D. Critical impact of radiotherapy protocol compliance and quality in the treatment of advanced head and neck cancer: Results from TROG 02.02. J. Clin. Oncol. 2010, 28, 2996–3001. [Google Scholar] [CrossRef]
- Nelms, B.E.; Tomé, W.A.; Robinson, G.; Wheeler, J. Variations in the contouring of organs at risk: Test case from a patient with oropharyngeal cancer. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 368–378. [Google Scholar] [CrossRef]
- Sun, K.Y.; William, H.H.; Mathew, M.; Arthur, B.D.; Vishal, G.; James, A.P.; Allen, M.C. Validating the RTOG-Endorsed Brachial Plexus Contouring Atlas: An Evaluation of Reproducibility Among Patients Treated by Intensity-Modulated Radiotherapy for Head-and-Neck Cancer. Int. J. Radiat. Oncol. Biol. Phys. 2012, 82, 1060–1064. [Google Scholar]
- Andrearczyk, V.; Oreiller, V.; Hatt, M.; Depeursinge, A. Head and Neck Tumor Segmentation and Outcome Prediction; Springer: London, UK, 2022. [Google Scholar]
- Duan, J.; Tegtmeier, R.C.; Vargas, C.E.; Yu, N.Y.; Laughlin, B.S.; Rwigema, J.M.; Anderson, J.D.; Zhu, L.; Chen, Q.; Rong, Y. Achieving accurate prostate auto-segmentation on CT in the absence of MR imaging. Radiother. Oncol. 2025, 202, 110588. [Google Scholar] [CrossRef]
- Duan, J.; Bernard, M.E.; Rong, Y.; Castle, J.R.; Feng, X.; Johnson, J.D.; Chen, Q. Contour subregion error detection methodology using deep learning auto-segmentation. Med. Phys. 2023, 50, 6673–6683. [Google Scholar] [CrossRef]
- Costea, M.; Zlate, A.; Serre, A.-A.; Racadot, S.; Baudier, T.; Chabaud, S.; Grégoire, V.; Sarrut, D.; Biston, M.-C. Evaluation of different algorithms for automatic segmentation of head-and-neck lymph nodes on CT images. Radiother. Oncol. 2023, 188, 109870. [Google Scholar] [CrossRef]
- Yang, J.; Veeraraghavan, H.; Armato, S.G., 3rd; Farahani, K.; Kirby, J.S.; Kalpathy-Kramer, J.; van Elmpt, W.; Dekker, A.; Han, X.; Feng, X.; et al. Autosegmentation for thoracic radiation treatment planning: A grand challenge at AAPM 2017. Med. Phys. 2018, 45, 4568–4581. [Google Scholar] [CrossRef] [PubMed]
- Myronenko, A.; Siddiquee, M.M.R.; Yang, D.; He, Y.; Xu, D. Automated Head and Neck Tumor Segmentation from 3D PET/CT HECKTOR 2022 Challenge Report. In Head and Neck Tumor Segmentation and Outcome Prediction; Springer Nature: Cham, Switzerland, 2023. [Google Scholar]
- Kaczmarska, M.; Majek, K. 3D Segmentation of Kidneys, Kidney Tumors and Cysts on CT Images—KiTS23 Challenge. In Proceedings of the Kidney and Kidney Tumor Segmentation: MICCAI 2023 Challenge, KiTS 2023, Vancouver, BC, Canada, 8 October 2023; Springer: Berlin/Heidelberg, Germany, 2024; pp. 149–155. [Google Scholar]
- Andrearczyk, V.; Oreiller, V.; Jreige, M.; Vallieres, M.; Castelli, J.; Elhalawani, H.; Boughdad, S.; Prior, J.O.; Depeursinge, A. Overview of the HECKTOR challenge at MICCAI 2020, automatic head and neck tumor segmentation in PET/CT. In Proceedings of the Head and Neck Tumor Segmentation: First Challenge, HECKTOR 2020, Lima, Peru, 4 October 2020; Proceedings 1. Springer: Berlin/Heidelberg, Germany, 2021. [Google Scholar]
- Voter, A.F.; Meram, E.; Garrett, J.W.; John-Paul, J.Y. Diagnostic accuracy and failure mode analysis of a deep learning algorithm for the detection of intracranial hemorrhage. J. Am. Coll. Radiol. 2021, 18, 1143–1152. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.; Otles, E.; Donnelly, J.P.; Krumm, A.; McCullough, J.; DeTroyer-Cooley, O.; Pestrue, J.; Phillips, M.; Konye, J.; Penoza, C. External validation of a widely implemented proprietary sepsis prediction model in hospitalized patients. JAMA Intern. Med. 2021, 181, 1065–1070. [Google Scholar] [CrossRef] [PubMed]
- Mackay, K.; Bernstein, D.; Glocker, B.; Kamnitsas, K.; Taylor, A. A Review of the Metrics Used to Assess Auto-Contouring Systems in Radiotherapy. Clin. Oncol. 2023, 35, 354–369. [Google Scholar] [CrossRef]
- Jordy, P.P.; Pieter, H.N.; Thomas, C.K.; Andor, W.J.M.G.; Riemer, H.J.A.S.; Lars, C.G. Limitations and Pitfalls of FDG-PET/CT in Infection and Inflammation. Semin. Nucl. Med. 2021, 51, 633–645. [Google Scholar]
- Mamlouk, M.D.; Chang, P.C.; Saket, R.R. Contextual Radiology Reporting: A New Approach to Neuroradiology Structured Templates. AJNR Am. J. Neuroradiol. 2018, 39, 1406–1414. [Google Scholar] [CrossRef] [PubMed]
- Gertz, R.J.; Dratsch, T.; Bunck, A.C.; Lennartz, S.; Iuga, A.-I.; Hellmich, M.G.; Persigehl, T.; Pennig, L.; Gietzen, C.H.; Fervers, P. Potential of GPT-4 for Detecting Errors in Radiology Reports: Implications for Reporting Accuracy. Radiology 2024, 311, e232714. [Google Scholar] [CrossRef]
- Hanzhou, L.; John, T.M.; Deepak, I.; Patricia, B.; Elizabeth, A.K.; Zachary, L.B.; Janice, M.N.; Imon, B.; Judy, W.G.; Hari, M.T. Decoding radiology reports: Potential application of OpenAI ChatGPT to enhance patient understanding of diagnostic reports. Clin. Imaging 2023, 101, 137–141. [Google Scholar]
- Mitsuyama, Y.; Tatekawa, H.; Takita, H.; Sasaki, F.; Tashiro, A.; Oue, S.; Walston, S.L.; Nonomiya, Y.; Shintani, A.; Miki, Y. Comparative analysis of GPT-4-based ChatGPT’s diagnostic performance with radiologists using real-world radiology reports of brain tumors. Eur. Radiol. 2025, 35, 1938–1947. [Google Scholar] [CrossRef]
- Kim, S.H.; Schramm, S.; Adams, L.C.; Braren, R.; Bressem, K.K.; Keicher, M.; Zimmer, C.; Hedderich, D.M.; Wiestler, B. Performance of Open-Source LLMs in Challenging Radiological Cases—A Benchmark Study on 1933 Eurorad Case Reports. medRxiv 2024. [Google Scholar] [CrossRef]
- Zhu, L.; Rong, Y.; McGee, L.A.; Rwigema, J.M.; Patel, S.H. Testing and Validation of a Custom Retrained Large Language Model for the Supportive Care of HN Patients with External Knowledge Base. Cancers 2024, 16, 2311. [Google Scholar] [CrossRef] [PubMed]
- Rong, Y.; Chen, Q.; Fu, Y.; Yang, X.; Al-Hallaq, H.A.; Wu, Q.J.; Yuan, L.; Xiao, Y.; Cai, B.; Latifi, K.; et al. NRG Oncology Assessment of Artificial Intelligence Deep Learning–Based Auto-segmentation for Radiation Therapy: Current Developments, Clinical Considerations, and Future Directions. Int. J. Radiat. Oncol. Biol. Phys. 2024, 119, 261–280. [Google Scholar] [CrossRef] [PubMed]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Zhao, L.-M.; Zhang, H.; Kim, D.D.; Ghimire, K.; Hu, R.; Kargilis, D.C.; Tang, L.; Meng, S.; Chen, Q.; Liao, W.-H. Head and neck tumor segmentation convolutional neural network robust to missing PET/CT modalities using channel dropout. Phys. Med. Biol. 2023, 68, 095011. [Google Scholar] [CrossRef]
- Ghimire, K.; Chen, Q.; Feng, X. Head and Neck Tumor Segmentation with Deeply-Supervised 3D UNet and Progression-Free Survival Prediction with Linear Model. In Head and Neck Tumor Segmentation and Outcome Prediction; Springer International Publishing: Cham, Switzerland, 2022. [Google Scholar]
- Gai, Z.; Tong, M.; Li, J. Chuanhu Chat (Software). 2023. Available online: https://github.com/GaiZhenbiao/ChuanhuChatGPT (accessed on 30 October 2024).
- Dubey, A.; Jauhri, A.; Pandey, A.; Kadian, A.; Al-Dahle, A.; Letman, A.; Mathur, A.; Schelten, A.; Yang, A.; Fan, A. The llama 3 herd of models. arXiv 2024, arXiv:2407.21783. [Google Scholar]
- van den Bosch, S.; Dijkema, T.; Verhoef, L.C.; Zwijnenburg, E.M.; Janssens, G.O.; Kaanders, J.H. Patterns of Recurrence in Electively Irradiated Lymph Node Regions After Definitive Accelerated Intensity Modulated Radiation Therapy for Head and Neck Squamous Cell Carcinoma. Int. J. Radiat. Oncol. Biol. Phys. 2016, 94, 766–774. [Google Scholar] [CrossRef]
- Kaur, H.; Pannu, H.S.; Malhi, A.K. A systematic review on imbalanced data challenges in machine learning: Applications and solutions. ACM Comput. Surv. (CSUR) 2019, 52, 1–36. [Google Scholar] [CrossRef]
- Chawla, N.V.; Japkowicz, N.; Kotcz, A. Special issue on learning from imbalanced data sets. ACM SIGKDD Explor. Newsl. 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Kotsiantis, S.; Kanellopoulos, D.; Pintelas, P. Handling imbalanced datasets: A review. GESTS Int. Trans. Comput. Sci. Eng. 2006, 30, 25–36. [Google Scholar]
- Ford, E.C.; Kinahan, P.E.; Hanlon, L.; Alessio, A.; Rajendran, J.; Schwartz, D.L.; Phillips, M. Tumor delineation using PET in head and neck cancers: Threshold contouring and lesion volumes. Med. Phys. 2006, 33, 4280–4288. [Google Scholar] [CrossRef]
- Zaidi, H.; El Naqa, I. PET-guided delineation of radiation therapy treatment volumes: A survey of image segmentation techniques. Eur. J. Nucl. Med. Mol. Imaging 2010, 37, 2165–2187. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Thorstad, W.L.; Biehl, K.J.; Laforest, R.; Su, Y.; Shoghi, K.I.; Donnelly, E.D.; Low, D.A.; Lu, W. A novel PET tumor delineation method based on adaptive region-growing and dual-front active contours. Med. Phys. 2008, 35, 3711–3721. [Google Scholar] [CrossRef] [PubMed]
- Caillol, H.; Pieczynski, W.; Hillion, A. Estimation of fuzzy Gaussian mixture and unsupervised statistical image segmentation. IEEE Trans. Image Process. 1997, 6, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Ng, S.P.; Dyer, B.A.; Kalpathy-Cramer, J.; Mohamed, A.S.R.; Awan, M.J.; Gunn, G.B.; Phan, J.; Zafereo, M.; Debnam, J.M.; Lewis, C.M. A prospective in silico analysis of interdisciplinary and interobserver spatial variability in post-operative target delineation of high-risk oral cavity cancers: Does physician specialty matter? Clin. Transl. Radiat. Oncol. 2018, 12, 40–46. [Google Scholar] [CrossRef]
- Riegel, A.C.; Berson, A.M.; Destian, S.; Ng, T.; Tena, L.B.; Mitnick, R.J.; Wong, P.S. Variability of gross tumor volume delineation in head-and-neck cancer using CT and PET/CT fusion. Int. J. Radiat. Oncol. Biol. Phys. 2006, 65, 726–732. [Google Scholar] [CrossRef]

| Metric | GTVn | GTVp | Overall | |||
|---|---|---|---|---|---|---|
| Before | After | Before | After | Before | After | |
| FP error (count) | 17/111 | 0/111 | 5/40 | 0/40 | 22/151 | 0/151 |
| FN error (count) | 8/111 | 8/111 | 0/40 | 0/40 | 8/151 | 8/151 |
| Precision | 0.83 | 1.00 | 0.88 | 1.00 | 0.85 | 1.00 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, L.; Rwigema, J.-C.M.; Feng, X.; Ansari, B.; Duan, J.; Rong, Y.; Chen, Q. Improving the Precision of Deep-Learning-Based Head and Neck Target Auto-Segmentation by Leveraging Radiology Reports Using a Large Language Model. Cancers 2025, 17, 1935. https://doi.org/10.3390/cancers17121935
Zhu L, Rwigema J-CM, Feng X, Ansari B, Duan J, Rong Y, Chen Q. Improving the Precision of Deep-Learning-Based Head and Neck Target Auto-Segmentation by Leveraging Radiology Reports Using a Large Language Model. Cancers. 2025; 17(12):1935. https://doi.org/10.3390/cancers17121935
Chicago/Turabian StyleZhu, Libing, Jean-Claude M. Rwigema, Xue Feng, Bilaal Ansari, Jingwei Duan, Yi Rong, and Quan Chen. 2025. "Improving the Precision of Deep-Learning-Based Head and Neck Target Auto-Segmentation by Leveraging Radiology Reports Using a Large Language Model" Cancers 17, no. 12: 1935. https://doi.org/10.3390/cancers17121935
APA StyleZhu, L., Rwigema, J.-C. M., Feng, X., Ansari, B., Duan, J., Rong, Y., & Chen, Q. (2025). Improving the Precision of Deep-Learning-Based Head and Neck Target Auto-Segmentation by Leveraging Radiology Reports Using a Large Language Model. Cancers, 17(12), 1935. https://doi.org/10.3390/cancers17121935






