Applications for Circulating Cell-Free DNA in Oral Squamous Cell Carcinoma: A Non-Invasive Approach for Detecting Structural Variants, Fusions, and Oncoviruses
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Methodology for Sample Size Collection
2.2. Sample Collection
2.3. cfDNA Extraction
- Patients diagnosed with histopathologically confirmed OSCC;
- Age ≥ 18 years;
- Availability of complete clinical and demographic data;
- Ability to provide informed consent;
- No prior systemic cancer therapy (chemotherapy or radiotherapy) at the time of sample collection.
- Patients with other concurrent malignancies;
- A history of autoimmune or chronic inflammatory diseases;
- Recent major surgical procedures or trauma within the past month;
- An inadequate sample quality or volume for cfDNA analysis.
2.4. Copy Number Analyses
2.5. Chimera Identification and Validation
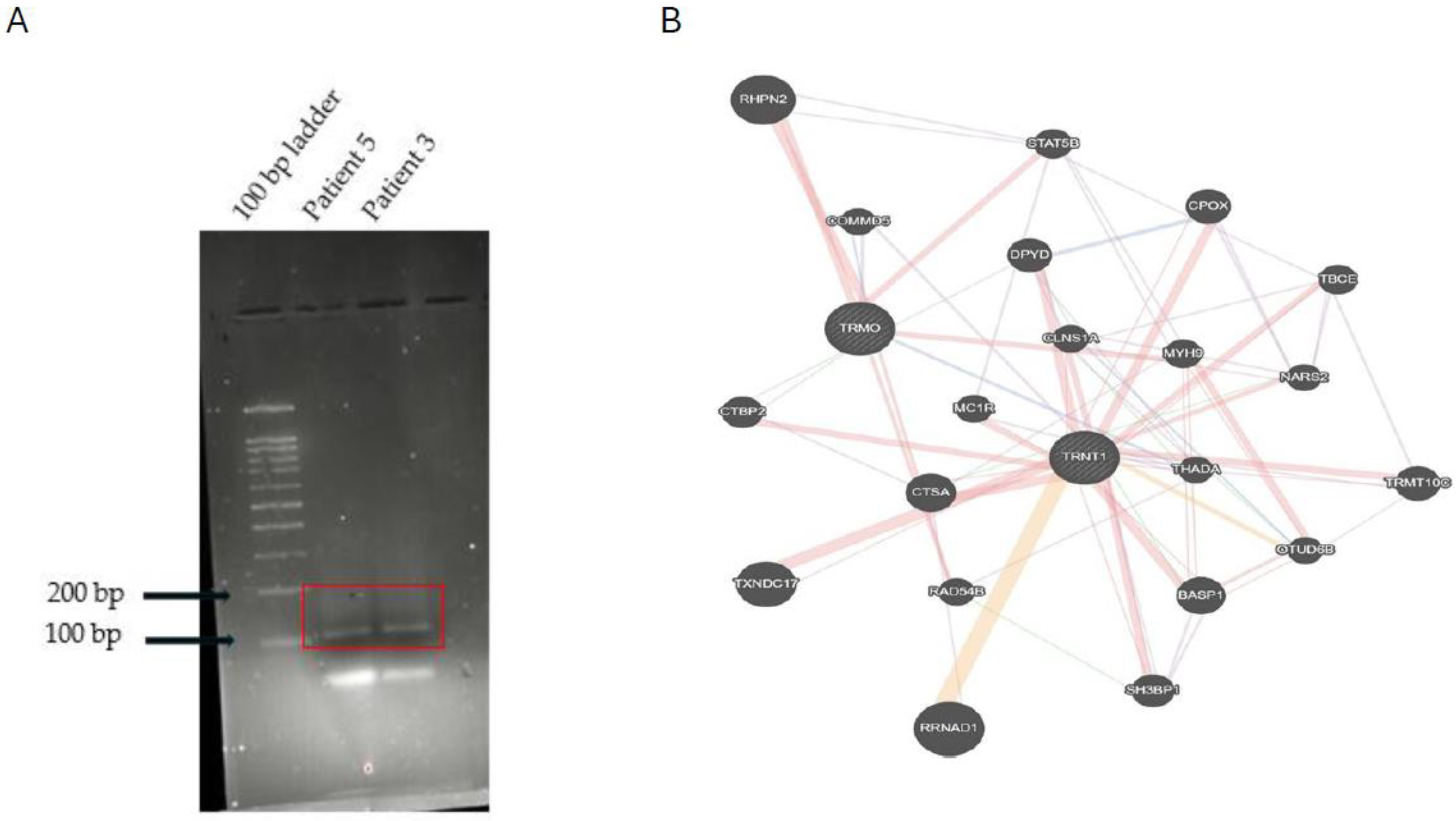
2.6. Gene Interaction Network Analysis
2.7. Identification of Coding Potential
2.8. Virus Detection in cfDNA
2.9. SNV Analysis
2.10. Gene Ontology Analysis
2.11. Statistical Analysis
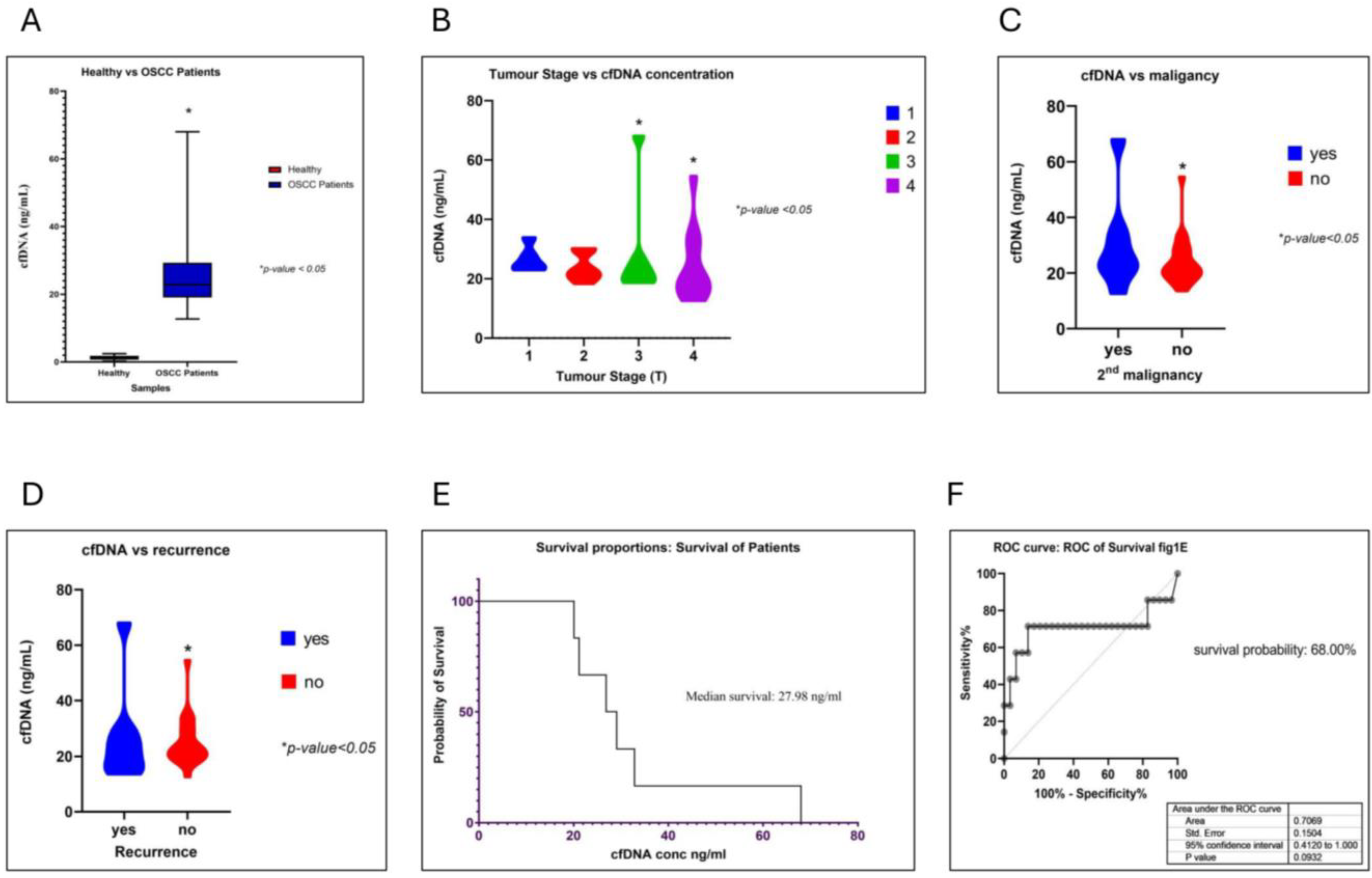
3. Results
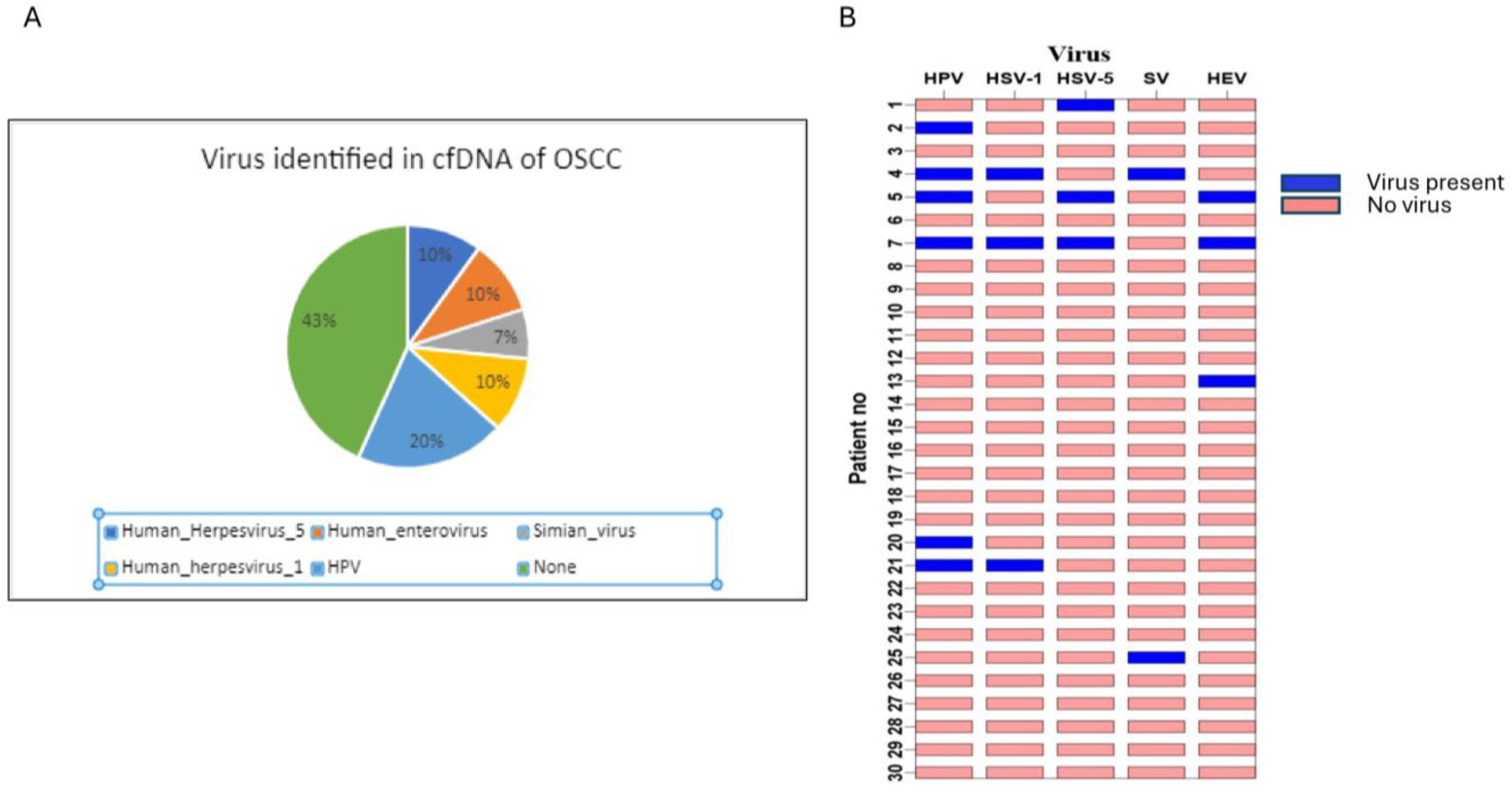
3.1. Oncogenic Viruses Were Identified When Analyzing cfDNA
3.2. A Novel Chimera Identified in cfDNA
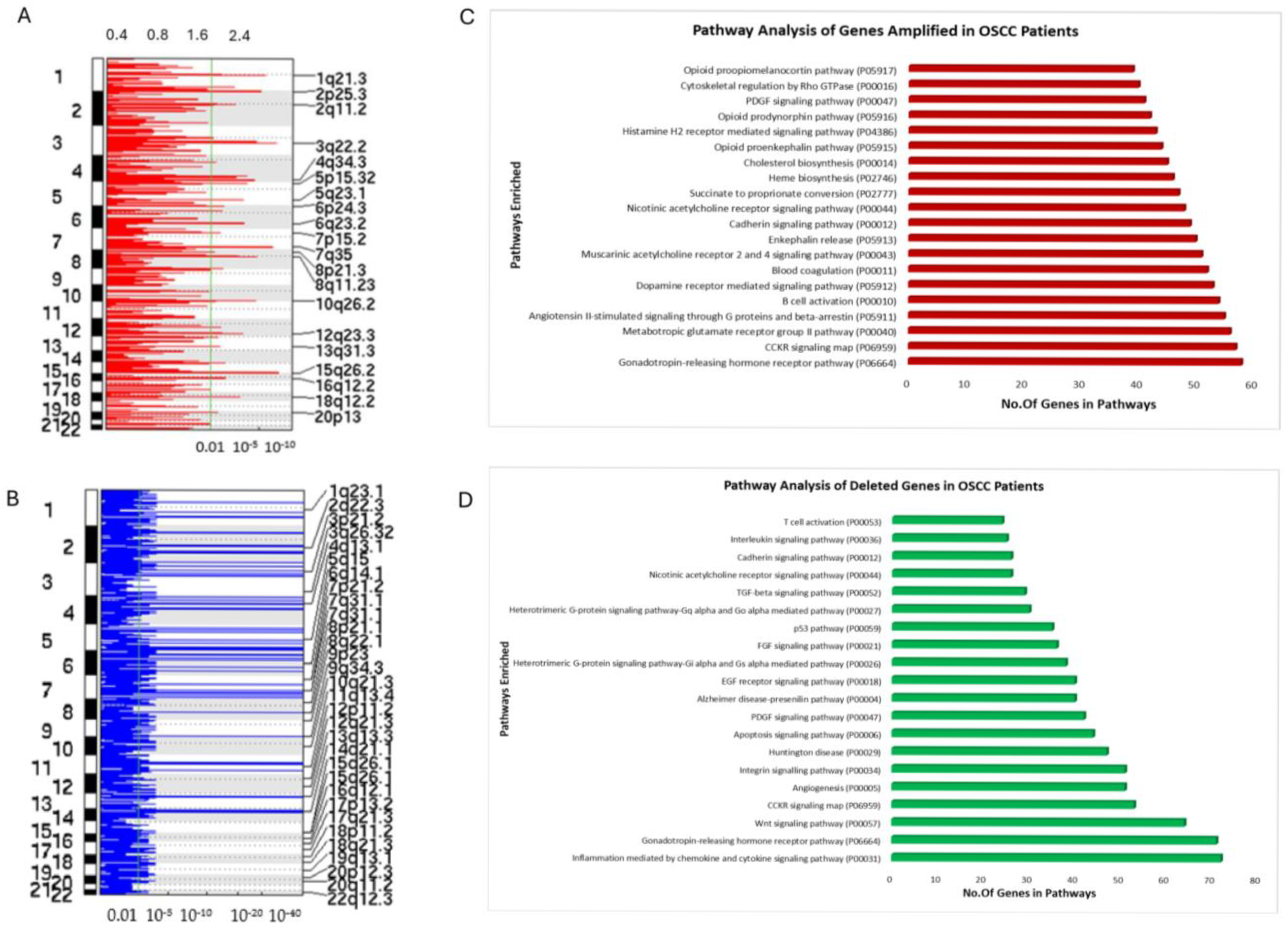
3.3. Copy Number Variation (CNV) Analysis of OSCC Patient cfDNA
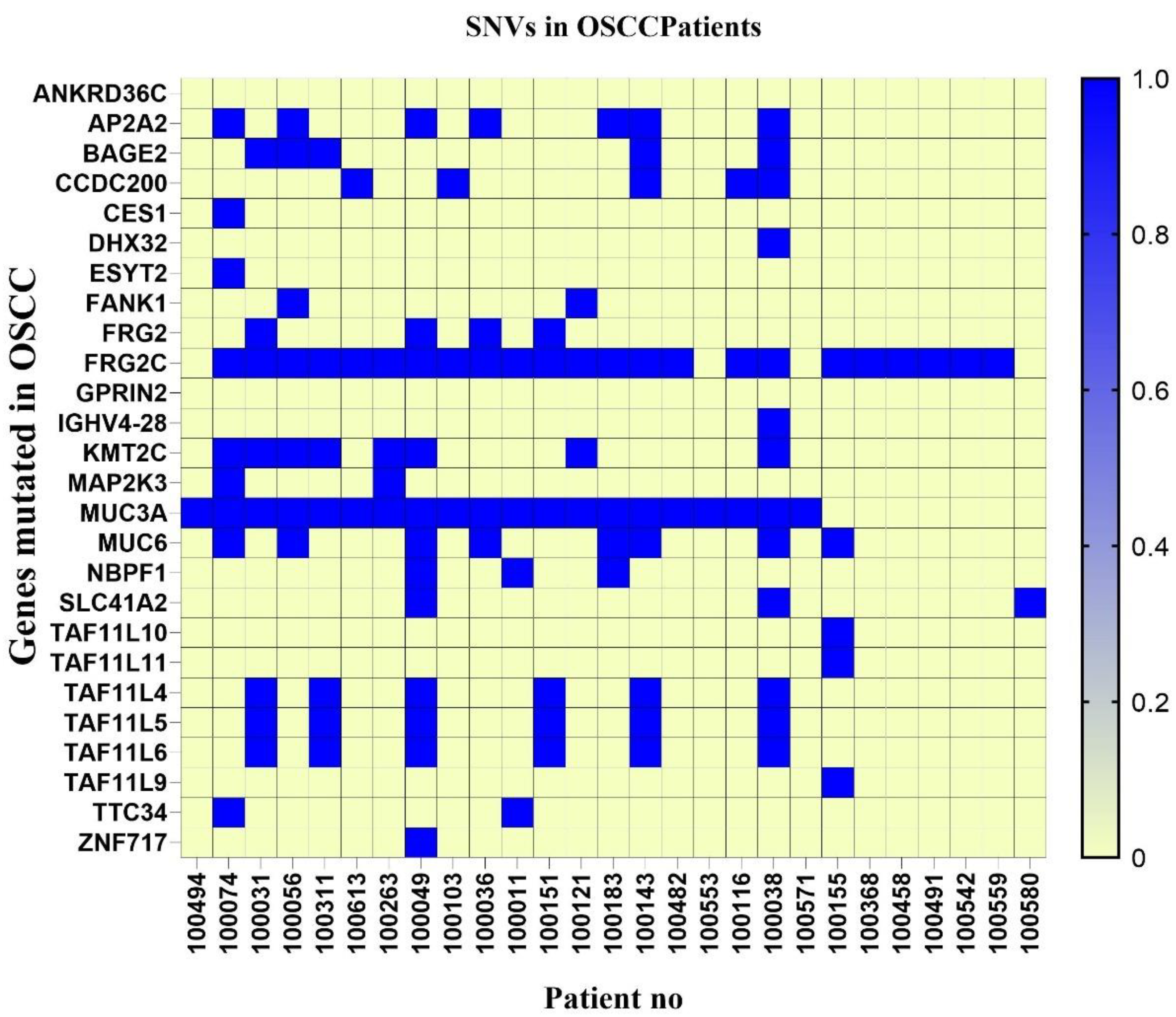
3.4. SNV Analysis of OSCC Patients’ cfDNA
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sinevici, N.; O’sullivan, J. Oral Cancer: Deregulated Molecular Events and Their Use as Biomarkers. Oral. Oncol. 2016, 61, 12–18. [Google Scholar] [CrossRef] [PubMed]
- Montero, P.H.; Patel, S.G. Cancer of the Oral Cavity. Surg. Oncol. Clin. N. Am. 2015, 24, 491–508. [Google Scholar] [CrossRef] [PubMed]
- Inchingolo, F.; Santacroce, L.; Ballini, A.; Topi, S.; Dipalma, G.; Haxhirexha, K.; Bottalico, L.; Charitos, I.A. Oral Cancer: A Historical Review. Int. J. Environ. Res. Public Health 2020, 17, 3168. [Google Scholar] [CrossRef] [PubMed]
- Katirachi, S.K.; Grønlund, M.P.; Jakobsen, K.K.; Grønhøj, C.; von Buchwald, C. The Prevalence of HPV in Oral Cavity Squamous Cell Carcinoma. Viruses 2023, 15, 451. [Google Scholar] [CrossRef]
- Hernandez, B.Y.; Lynch, C.F.; Chan, O.T.M.; Goodman, M.T.; Unger, E.R.; Steinau, M.; Thompson, T.D.; Gillison, M.; Lyu, C.; Saraiya, M. Human Papillomavirus DNA Detection, P16 INK4a, and Oral Cavity Cancer in a U.S. Population. Oral. Oncol. 2019, 91, 92–96. [Google Scholar] [CrossRef]
- Orrù, G.; Mameli, A.; Demontis, C.; Rossi, P.; Ratto, D.; Occhinegro, A.; Piras, V.; Kuqi, L.; Berretta, M.; Taibi, R.; et al. Oral Human Papilloma Virus Infection: An Overview of Clinical-Laboratory Diagnosis and Treatment. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 8148–8157. [Google Scholar]
- Nakagaki, T.; Tamura, M.; Kobashi, K.; Omori, A.; Koyama, R.; Idogawa, M.; Ogi, K.; Hiratsuka, H.; Tokino, T.; Sasaki, Y. Targeted Next-Generation Sequencing of 50 Cancer-Related Genes in Japanese Patients with Oral Squamous Cell Carcinoma. Tumor Biol. 2018, 40, 1010428318800180. [Google Scholar] [CrossRef]
- Lousada-Fernandez, F.; Rapado-Gonzalez, O.; Lopez-Cedrun, J.-L.; Lopez-Lopez, R.; Muinelo-Romay, L.; Suarez-Cunqueiro, M. Liquid Biopsy in Oral Cancer. Int. J. Mol. Sci. 2018, 19, 1704. [Google Scholar] [CrossRef]
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating Liquid Biopsies into the Management of Cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548. [Google Scholar] [CrossRef]
- Naito, Y.; Honda, K. Liquid Biopsy for Oral Cancer Diagnosis: Recent Advances and Challenges. J. Pers. Med. 2023, 13, 303. [Google Scholar] [CrossRef]
- Salvi, S.; Gurioli, G.; De Giorgi, U.; Conteduca, V.; Tedaldi, G.; Calistri, D.; Casadio, V. Cell-Free DNA as a Diagnostic Marker for Cancer: Current Insights. Onco Targets Ther. 2016, 9, 6549–6559. [Google Scholar] [CrossRef] [PubMed]
- Di Meo, A.; Bartlett, J.; Cheng, Y.; Pasic, M.D.; Yousef, G.M. Liquid Biopsy: A Step Forward towards Precision Medicine in Urologic Malignancies. Mol. Cancer 2017, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Sumanasuriya, S.; Lambros, M.B.; de Bono, J.S. Application of Liquid Biopsies in Cancer Targeted Therapy. Clin. Pharmacol. Ther. 2017, 102, 745–747. [Google Scholar] [CrossRef] [PubMed]
- Kang, Z.; Stevanovic, S.; Hinrichs, C.S.; Cao, L. Circulating Cell-Free DNA for Metastatic Cervical Cancer Detection, Genotyping, and Monitoring. Clin. Cancer Res. 2017, 23, 6856–6862. [Google Scholar] [CrossRef]
- Arantes, L.M.R.B.; De Carvalho, A.C.; Melendez, M.E.; Carvalho, A.L. Serum, Plasma and Saliva Biomarkers for Head and Neck Cancer. Expert Review of Molecular Diagnostics. Expert Rev. Mol. Diagn. 2017, 18, 85–112. [Google Scholar] [CrossRef]
- Cordeiro Mitchell, C.N.; Murdock, T.; Fader, A.N.; Stone, R.L. Advanced Ovarian Cancer Treated in Pregnancy and Detected by Cell-Free DNA Aneuploidy Screening. Gynecol. Oncol. Rep. 2018, 24, 48–50. [Google Scholar] [CrossRef]
- Wang, X.; Wang, L.; Su, Y.; Yue, Z.; Xing, T.; Zhao, W.; Zhao, Q.; Duan, C.; Huang, C.; Zhang, D.; et al. Plasma Cell-Free DNA Quantification Is Highly Correlated to Tumor Burden in Children with Neuroblastoma. Cancer Med. 2018, 7, 3022–3030. [Google Scholar] [CrossRef]
- Fawzy, A.; Sweify, K.M.; El-Fayoumy, H.M.; Nofal, N. Quantitative Analysis of Plasma Cell-Free DNA and Its DNA Integrity in Patients with Metastatic Prostate Cancer Using ALU Sequence. J. Egypt. Natl. Cancer Inst. 2016, 28, 235–242. [Google Scholar] [CrossRef]
- Grisanti, S.; Almici, C.; Consoli, F.; Buglione, M.; Verardi, R.; Bolzoni-Villaret, A.; Bianchetti, A.; Ciccarese, C.; Mangoni, M.; Ferrari, L.; et al. Circulating Tumor Cells in Patients with Recurrent or Metastatic Head and Neck Carcinoma: Prognostic and Predictive Significance. PLoS ONE 2014, 9, e103918. [Google Scholar] [CrossRef]
- Krebs, M.G.; Metcalf, R.L.; Carter, L.; Brady, G.; Blackhall, F.H.; Dive, C. Molecular Analysis of Circulating Tumour Cells—Biology and Biomarkers. Nat. Rev. Clin. Oncol. 2014, 11, 129–144. [Google Scholar] [CrossRef]
- Yu, M.; Stott, S.; Toner, M.; Maheswaran, S.; Haber, D.A. Circulating Tumor Cells: Approaches to Isolation and Characterization. J. Cell Biol. 2011, 192, 373–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Chang, S.; Li, G.; Sun, Y. Application of Liquid Biopsy in Precision Medicine: Opportunities and Challenges. Front. Med. 2017, 11, 522–527. [Google Scholar] [CrossRef] [PubMed]
- Alpaugh, R.K.; Zhou, L.L.; Dicker, D.T.; Matthew, E.; El-Deiry, W.S. Circulating Tumor Cells: Silent Predictors of Metastasis. F1000Research 2017, 6, 1445. [Google Scholar] [CrossRef]
- Lin, L.H.; Chang, K.W.; Kao, S.Y.; Cheng, H.W.; Liu, C.J. Increased Plasma Circulating Cell-Free DNA Could Be a Potential Marker for Oral Cancer. Int. J. Mol. Sci. 2018, 19, 3303. [Google Scholar] [CrossRef]
- Mazurek, A.M.; Rutkowski, T.; Fiszer-Kierzkowska, A.; Małusecka, E.; Składowski, K. Assessment of the Total CfDNA and HPV16/18 Detection in Plasma Samples of Head and Neck Squamous Cell Carcinoma Patients. Oral. Oncol. 2016, 54, 36–41. [Google Scholar] [CrossRef]
- Morgan, I.M.; Dinardo, L.J.; Windle, B. Integration of Human Papillomavirus Genomes in Head and Neck Cancer: Is It Time to Consider a Paradigm Shift? Viruses 2017, 9, 208. [Google Scholar] [CrossRef]
- Lawrence, M.S.; Sougnez, C.; Lichtenstein, L.; Cibulskis, K.; Lander, E.; Gabriel, S.B.; Getz, G.; Ally, A.; Balasundaram, M.; Birol, I.; et al. Comprehensive Genomic Characterization of Head and Neck Squamous Cell Carcinomas. Nature 2015, 517, 576–582. [Google Scholar] [CrossRef]
- Wang, Y.; Springer, S.; Mulvey, C.L.; Silliman, N.; Schaefer, J.; Sausen, M.; James, N.; Rettig, E.M.; Guo, T.; Pickering, C.R.; et al. Detection of Somatic Mutations and HPV in the Saliva and Plasma of Patients with Head and Neck Squamous Cell Carcinomas. Sci. Transl. Med. 2015, 7, 293ra104. [Google Scholar] [CrossRef]
- Chattopadhyay, I.; Verma, M.; Panda, M. Role of Oral Microbiome Signatures in Diagnosis and Prognosis of Oral Cancer. Technol. Cancer Res. Treat. 2019, 18, 1533033819867354. [Google Scholar] [CrossRef]
- Wołaçewicz, M.; Becht, R.; Grywalska, E.; Niedźwiedzka-Rystwej, P. Herpesviruses in Head and Neck Cancers. Viruses 2020, 12, 172. [Google Scholar] [CrossRef]
- Brown, S.H.; States, V.A.R.; Afghan, A.K.; Satyanarayana, G. Herpes Simplex Virus-Infected Squamous Cell Carcinoma: A Case Report. BMC Infect. Dis. 2022, 22, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Mosmann, J.P.; Talavera, A.D.; Criscuolo, M.I.; Venezuela, R.F.; Kiguen, A.X.; Panico, R.; Ferreyra De Prato, R.; López De Blanc, S.A.; Ré, V.; Cuffini, C.G. Sexually Transmitted Infections in Oral Cavity Lesions: Human Papillomavirus, Chlamydia Trachomatis, and Herpes Simplex Virus. J. Oral. Microbiol. 2019, 11, 1632129. [Google Scholar] [CrossRef] [PubMed]
- Shlien, A.; Malkin, D. Copy Number Variations and Cancer. Genome Med. 2009, 1, 1–9. [Google Scholar] [CrossRef]
- Palande, V.; Siegal, T.; Detroja, R.; Gorohovski, A.; Glass, R.; Flueh, C.; Kanner, A.A.; Laviv, Y.; Har-Nof, S.; Levy-Barda, A.; et al. Detection of Gene Mutations and Gene-Gene Fusions in Circulating Cell-Free DNA of Glioblastoma Patients: An Avenue for Clinically Relevant Diagnostic Analysis. Mol. Oncol. 2022, 16, 2098. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The Genotype-Tissue Expression (GTEx) Project. Nat. Genet. 2013, 45, 580. [Google Scholar] [CrossRef]
- Kulkarni, O.; Sugier, P.E.; Guibon, J.; Boland-Augé, A.; Lonjou, C.; Bacq-Daian, D.; Olaso, R.; Rubino, C.; Souchard, V.; Rachedi, F.; et al. Gene Network and Biological Pathways Associated with Susceptibility to Differentiated Thyroid Carcinoma. Sci. Rep. 2021, 11, 8932. [Google Scholar] [CrossRef]
- Wedatilake, Y.; Niazi, R.; Fassone, E.; Powell, C.A.; Pearce, S.; Plagnol, V.; Saldanha, J.W.; Kleta, R.; Chong, W.K.; Footitt, E.; et al. TRNT1 Deficiency: Clinical, Biochemical and Molecular Genetic Features. Orphanet J. Rare Dis. 2016, 11, 1–14. [Google Scholar] [CrossRef]
- Adalsteinsson, V.A.; Ha, G.; Freeman, S.S.; Choudhury, A.D.; Stover, D.G.; Parsons, H.A.; Gydush, G.; Reed, S.C.; Rotem, D.; Rhoades, J.; et al. Scalable Whole-Exome Sequencing of Cell-Free DNA Reveals High Concordance with Metastatic Tumors. Nat. Commun. 2017, 8, 1324. [Google Scholar] [CrossRef]
- Mermel, C.H.; Schumacher, S.E.; Hill, B.; Meyerson, M.L.; Beroukhim, R.; Getz, G. GISTIC2.0 Facilitates Sensitive and Confident Localization of the Targets of Focal Somatic Copy-Number Alteration in Human Cancers. Genome Biol. 2011, 12, R41. [Google Scholar] [CrossRef]
- Detroja, R.; Gorohovski, A.; Giwa, O.; Baum, G.; Frenkel-Morgenstern, M. ChiTaH: A Fast and Accurate Tool for Identifying Known Human Chimeric Sequences from High-Throughput Sequencing Data. NAR Genom. Bioinform. 2021, 3, lqab112. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The GeneMANIA Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 2010, 38, W214–W220. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Park, H.J.; Dasari, S.; Wang, S.; Kocher, J.P.; Li, W. CPAT: Coding-Potential Assessment Tool Using an Alignment-Free Logistic Regression Model. Nucleic Acids Res. 2013, 41, e74. [Google Scholar] [CrossRef]
- Guo, J.C.; Fang, S.S.; Wu, Y.; Zhang, J.H.; Chen, Y.; Liu, J.; Wu, B.; Wu, J.R.; Li, E.M.; Xu, L.Y.; et al. CNIT: A Fast and Accurate Web Tool for Identifying Protein-Coding and Long Non-Coding Transcripts Based on Intrinsic Sequence Composition. Nucleic Acids Res. 2019, 47, W516–W522. [Google Scholar] [CrossRef]
- Wang, Q.; Jia, P.; Zhao, Z. VirusFinder: Software for Efficient and Accurate Detection of Viruses and Their Integration Sites in Host Genomes through next Generation Sequencing Data. PLoS ONE 2013, 8, e64465. [Google Scholar] [CrossRef]
- Koboldt, D.C.; Chen, K.; Wylie, T.; Larson, D.E.; Mclellan, M.D.; Mardis, E.R.; Weinstock, G.M.; Wilson, R.K.; Ding, L. VarScan: Variant Detection in Massively Parallel Sequencing of Individual and Pooled Samples. Bioinform. Appl. Note 2009, 25, 2283–2285. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional Annotation of Genetic Variants from High-Throughput Sequencing Data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Poplin, R.; Chang, P.C.; Alexander, D.; Schwartz, S.; Colthurst, T.; Ku, A.; Newburger, D.; Dijamco, J.; Nguyen, N.; Afshar, P.T.; et al. A Universal SNP and Small-Indel Variant Caller Using Deep Neural Networks. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef] [PubMed]
- Benjamin, D.; Sato, T.; Cibulskis, K.; Getz, G.; Stewart, C.; Lichtenstein, L. Calling Somatic SNVs and Indels with Mutect2. Nat. Biotechnol. 2018, 36, 983–987. [Google Scholar] [CrossRef]
- Mi, H.; Ebert, D.; Muruganujan, A.; Mills, C.; Albou, L.P.; Mushayamaha, T.; Thomas, P.D. PANTHER Version 16: A Revised Family Classification, Tree-Based Classification Tool, Enhancer Regions and Extensive API. Nucleic Acids Res. 2021, 49, D394–D403. [Google Scholar] [CrossRef]
- Zhang, N.; Wang, M.; Zhang, P.; Huang, T. Classification of Cancers Based on Copy Number Variation Landscapes. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 2750–2755. [Google Scholar] [CrossRef]
- Tan, E.S.; Knepper, T.C.; Wang, X.; Permuth, J.B.; Wang, L.; Fleming, J.B.; Xie, H. Copy Number Alterations as Novel Biomarkers and Therapeutic Targets in Colorectal Cancer. Cancers 2022, 14, 2223. [Google Scholar] [CrossRef] [PubMed]
- Kaveh, F.; Baumbusch, L.O.; Nebdal, D.; Børresen-Dale, A.L.; Lingjærde, O.C.; Edvardsen, H.; Kristensen, V.N.; Solvang, H.K. A Systematic Comparison of Copy Number Alterations in Four Types of Female Cancer. BMC Cancer 2016, 16, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Bailey, M.H.; Tokheim, C.; Porta-Pardo, E.; Sengupta, S.; Bertrand, D.; Weerasinghe, A.; Colaprico, A.; Wendl, M.C.; Kim, J.; Reardon, B.; et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell 2018, 173, 371–385.e18. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-I.; Kao, H.-K.; Chen, T.-W.; Huang, Y.; Cheng, H.-W.; Yi, J.-S.; Hung, S.-Y.; Wu, C.-S.; Lee, Y.-S.; Chang, K.-P. Characterization of Copy Number Variations in Oral Cavity Squamous Cell Carcinoma Reveals a Novel Role for MLLT3 in Cell Invasiveness. Oncologist 2019, 24, e1388–e1400. [Google Scholar] [CrossRef]
- Rhie, A.; Park, W.S.; Choi, M.K.; Kim, J.H.; Ryu, J.; Ryu, C.H.; Kim, J.I.; Jung, Y.S. Genomic Copy Number Variations Characterize the Prognosis of Both P16-Positive and P16-Negative Oropharyngeal Squamous Cell Carcinoma after Curative Resection. Medicine 2015, 94, e2187. [Google Scholar] [CrossRef]
- Rapado-González, Ó.; López-Cedrún, J.L.; Lago-Lestón, R.M.; Abalo, A.; Rubin-Roger, G.; Salgado-Barreira, Á.; López-López, R.; Muinelo-Romay, L.; Suárez-Cunqueiro, M.M. Integrity and Quantity of Salivary Cell-Free DNA as a Potential Molecular Biomarker in Oral Cancer: A Preliminary Study. J. Oral. Pathol. Med. 2022, 51, 429–435. [Google Scholar] [CrossRef]
- van Ginkel, J.H.; Slieker, F.J.B.; de Bree, R.; van Es, R.J.J.; Van Cann, E.M. Cell-Free Nucleic Acids in Body Fluids as Biomarkers for the Prediction and Early Detection of Recurrent Head and Neck Cancer: A Systematic Review of the Literature. Oral. Oncol. 2017, 75, 8–15. [Google Scholar] [CrossRef]
- Johann, D.J.; Steliga, M.; Shin, I.J.; Yoon, D.; Arnaoutakis, K.; Hutchins, L.; Liu, M.; Liem, J.; Walker, K.; Pereira, A.; et al. Liquid Biopsy and Its Role in an Advanced Clinical Trial for Lung Cancer. Exp. Biol. Med. 2018, 243, 262–271. [Google Scholar] [CrossRef] [PubMed]
- Hamana, K.; Uzawa, K.; Ogawara, K.; Shiiba, M.; Bukawa, H.; Yokoe, H.; Tanzawa, H. Monitoring of Circulating Tumour-Associated DNA as a Prognostic Tool for Oral Squamous Cell Carcinoma. Br. J. Cancer 2005, 92, 2181–2184. [Google Scholar] [CrossRef]
- Serrano, B.; Brotons, M.; Bosch, F.X.; Bruni, L. Epidemiology and Burden of HPV-Related Disease. Best. Pr. Pract. Res. Clin. Obs. Obstet. Gynaecol. 2018, 47, 14–26. [Google Scholar] [CrossRef]
- Bodelon, C.; Untereiner, M.E.; Machiela, M.J.; Vinokurova, S.; Wentzensen, N. Genomic Characterization of Viral Integration Sites in HPV-Related Cancers. Int. J. Cancer 2016, 139, 2001–2011. [Google Scholar] [CrossRef] [PubMed]
- McBride, A.A.; Warburton, A. The Role of Integration in Oncogenic Progression of HPV-Associated Cancers. PLoS Pathog. 2017, 13, e1006211. [Google Scholar] [CrossRef]
- Nees, M.; Van Wijngaarden, E.; Bakos, E.; Schneider, A.; Dürst, M. Identification of Novel Molecular Markers Which Correlate with HPV-Induced Tumor Progression. Oncogene 1998, 16, 2447–2458. [Google Scholar] [CrossRef]
- Perri, F.; Longo, F.; Della Vittoria Scarpati, G.; Pisconti, S.; Longo, V.; Addeo, R.; Carducci, F.; Buonerba, C.; Fulciniti, F.; Solla, R. Metastatic HPV-Related Oropharyngeal Carcinoma Cured with Chemoradiotherapy: Importance of Pretherapy Biomolecular Assessment. Clin. Case Rep. 2018, 6, 56–62. [Google Scholar] [CrossRef]
- Ngan, H.L.; Wang, L.; Lo, K.W.; Lui, V.W.Y. Genomic Landscapes of EBV-Associated Nasopharyngeal Carcinoma vs. HPV-Associated Head and Neck Cancer. Cancers 2018, 10, 210. [Google Scholar] [CrossRef] [PubMed]
- Sastre-Garau, X.; Diop, M.; Martin, F.; Dolivet, G.; Marchal, F.; Charra-Brunaud, C.; Peiffert, D.; Leufflen, L.; Dembele, B.; Demange, J.; et al. A NGS-Based Blood Test for the Diagnosis of Invasive HPV-Associated Carcinomas with Extensive Viral Genomic Characterization. Clin. Cancer Res. 2021, 27, 5307–5316. [Google Scholar] [CrossRef] [PubMed]
- Panda, B.; Krishnan, N.; Gupta, S.; Palve, V.; Varghese, L.; Pattnaik, S.; Jain, P.; Khyriem, C.; Hariharan, A.; Dhas, K.; et al. Integrated Analysis of Oral Tongue Squamous Cell Carcinoma Identifies Key Variants and Pathways Linked to Risk Habits, HPV, Clinical Parameters and Tumor Recurrence. F1000Research 2015, 4, 1215. [Google Scholar] [CrossRef]
- Emmett, S.; Whiteman, D.C.; Panizza, B.J.; Antonsson, A. An Update on Cellular MicroRNA Expression in Human Papillomavirus-Associated Head and Neck Squamous Cell Carcinoma. Oncology 2018, 95, 193–201. [Google Scholar] [CrossRef]
- Ryoo, N.-K.; Kim, J.-E.; Choung, H.-K.; Kim, N.; Lee, M.-J.; Khwarg, S.-I. Human Papilloma Virus in Retinoblastoma Tissues from Korean Patients. Korean J. Ophthalmol. 2013, 27, 368. [Google Scholar] [CrossRef]
- Zhang, X.; Wang, H.; Sun, Y.; Qi, M.; Li, W.; Zhang, Z.; Zhang, X.E.; Cui, Z. Enterovirus A71 Oncolysis of Malignant Gliomas. Mol. Ther. 2020, 28, 1533–1546. [Google Scholar] [CrossRef]
- Shah, K.V.; Galloway, D.A.; Knowles, W.A.; Viscidi, R.P. Simian Virus 40 (SV40) and Human Cancer: A Review of the Serological Data. Rev. Med. Virol. 2004, 14, 231–239. [Google Scholar] [CrossRef] [PubMed]
- Mazzoni, E.; Benassi, M.S.; Corallini, A.; Barbanti-Brodano, G.; Taronna, A.; Picci, P.; Guerra, G.; D’Agostino, A.; Trevisiol, L.; Nocini, P.F.; et al. Significant Association between Human Osteosarcoma and Simian Virus 40. Cancer 2015, 121, 708–715. [Google Scholar] [CrossRef] [PubMed]
- De Sanjose, S.; Shah, K.V.; Domingo-Domenech, E.; Engels, E.A.; De Sevilla, A.F.; Alvaro, T.; Garcia-Villanueva, M.; Romagosa, V.; Gonzalez-Barca, E.; Viscidi, R.P. Lack of Serological Evidence for an Association between Simian Virus 40 and Lymphoma. Int. J. Cancer 2003, 104, 522–524. [Google Scholar] [CrossRef]
- Solomon, B.; Young, R.J.; Rischin, D. Head and Neck Squamous Cell Carcinoma: Genomics and Emerging Biomarkers for Immunomodulatory Cancer Treatments. Semin. Cancer Biol. 2018, 52, 228–240. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Du, Y.; Hou, X.; Cheng, W. A Prognostic Model for Oral Squamous Cell Carcinoma Using 7 Genes Related to Tumor Mutational Burden. BMC Oral. Health 2022, 22, 1–15. [Google Scholar] [CrossRef]
- Chen, R.; Davydov, E.V.; Sirsota, M.; Butte, A.J. Non-Synonymous and Synonymous Coding SNPS Show Similar Likelihood and Effect Size of Human Disease Association. PLoS ONE 2010, 5, e13574. [Google Scholar] [CrossRef]
- Stransky, N.; Egloff, A.M.; Tward, A.D.; Kostic, A.D.; Cibulskis, K.; Sivachenko, A.; Kryukov, G.V.; Lawrence, M.S.; Sougnez, C.; McKenna, A.; et al. The Mutational Landscape of Head and Neck Squamous Cell Carcinoma. Science 2011, 333, 1157–1160. [Google Scholar] [CrossRef]
- Perdomo, S.; Avogbe, P.H.; Foll, M.; Abedi-Ardekani, B.; Lescher Facciolla, V.; Anantharaman, D.; Chopard, P.; Le Calvez-Kelm, F.; Vilensky, M.; Polesel, J.; et al. Circulating Tumor DNA Detection in Head and Neck Cancer: Evaluation of Two Different Detection Approaches. Oncotarget 2017, 8, 72621–72632. [Google Scholar] [CrossRef]
- Ghasemi, F.; Prokopec, S.D.; MacNeil, D.; Mundi, N.; Gameiro, S.F.; Howlett, C.; Stecho, W.; Plantinga, P.; Pinto, N.; Ruicci, K.M.; et al. Mutational Analysis of Head and Neck Squamous Cell Carcinoma Stratified by Smoking Status. JCI Insight 2019, 4, e123443. [Google Scholar] [CrossRef]
- Maitra, A.; Biswas, N.K.; Amin, K.; Kowtal, P.; Kumar, S.; Das, S.; Sarin, R.; Majumder, P.P.; Bagchi, I.; Bairagya, B.B.; et al. Mutational Landscape of Gingivo-Buccal Oral Squamous Cell Carcinoma Reveals New Recurrently-Mutated Genes and Molecular Subgroups. Nat. Commun. 2013, 4, 2873. [Google Scholar] [CrossRef]
- Su, S.C.; Chang, L.C.; Lin, C.W.; Chen, M.K.; Yu, C.P.; Chung, W.H.; Yang, S.F. Mutational Signatures and Mutagenic Impacts Associated with Betel Quid Chewing in Oral Squamous Cell Carcinoma. Hum. Genet. 2019, 138, 1379–1389. [Google Scholar] [CrossRef] [PubMed]
- John, S.; Sharma, K.; Singh, P.; Chandra, S.; Singh, G.; Gupta, S. Quantitative Proteogenomic Characterization in MUC1 and MUC4 in Oral Squamous Cell Carcinoma, Oral Potentially Malignant Disorders, and Normal Oral Mucosa in Carcinogenesis. Eur. Arch. Oto-Rhino-Laryngol. 2024, 282, 1–10. [Google Scholar] [CrossRef]
- Xia, P.; Choi, A.H.; Deng, Z.; Yang, Y.; Zhao, J.; Wang, Y.; Hardwidge, P.R.; Zhu, G. Cell Membrane-Anchored MUC4 Promotes Tumorigenicity in Epithelial Carcinomas. Oncotarget 2017, 8, 14147–14157. [Google Scholar] [CrossRef]
- Lu, H.; Liang, D.; Zhu, Y.; Xu, W.; Zhou, K.; Liu, L.; Liu, S.; Yang, W. Prognostic and Clinicopathological Significance of MUC Expression in Head and Neck Cancer: A Systematic Review and Meta-Analysis. Oncotarget 2017, 8, 96359. [Google Scholar] [CrossRef] [PubMed]
- Mahomed, F. Recent Advances in Mucin Immunohistochemistry in Salivary Gland Tumors and Head and Neck Squamous Cell Carcinoma. Oral. Oncol. 2011, 47, 797–803. [Google Scholar] [CrossRef] [PubMed]
- Hua, C.H.; Chuang, C.Y.; Chien, Y.C.; Su, C.W.; Chen, S.C.; Liu, L.C.; Yang, S.F.; Yu, Y.L. Analysis of MUC6 Polymorphisms on the Clinicopathologic Characteristics of Asian Patients with Oral Squamous Cell Carcinoma. J. Cell Mol. Med. 2023, 27, 2594–2602. [Google Scholar] [CrossRef]
- Zheng, X.; Sun, Y.; Li, Y.; Ma, J.; Lv, Y.; Hu, Y.; Zhou, Y.; Zhang, J. A Novel Tongue Squamous Cell Carcinoma Cell Line Escapes from Immune Recognition Due to Genetic Alterations in HLA Class I Complex. Cells 2023, 12, 35. [Google Scholar] [CrossRef]
- Zhang, R.; Li, C.; Wan, Z.; Qin, J.; Li, Y.; Wang, Z.; Zheng, Q.; Kang, X.; Chen, X.; Li, Y.; et al. Comparative Genomic Analysis of Esophageal Squamous Cell Carcinoma among Different Geographic Regions. Front. Oncol. 2023, 12, 999424. [Google Scholar] [CrossRef]
- Farah, C.S.; Shearston, K.; Melton, P.E.; Fox, S.A. Genome-Wide Characterization of the Mutational Landscape of Proliferative Verrucous Leukoplakia. Oral. Surg. Oral. Med. Oral. Pathol. Oral. Radiol. 2024, 138, 99–111. [Google Scholar] [CrossRef]
- Ghias, K.; Rehmani, S.S.; Razzak, S.A.; Madhani, S.; Azim, M.K.; Ahmed, R.; Khan, M.J. Mutational Landscape of Head and Neck Squamous Cell Carcinomas in a South Asian Population. Genet. Mol. Biol. 2019, 42, 526–542. [Google Scholar] [CrossRef]
- Expression of cancer-testis antigens as possible targets for antigen-specific immunotherapy in head and neck squamous cell carcinoma. Cancer Biol. Ther. 2006, 5, 1218–1225. [CrossRef] [PubMed]
- Shin, J.; Nile, A.; Oh, J.W. Role of Adaptin Protein Complexes in Intracellular Trafficking and Their Impact on Diseases. Bioengineered 2021, 12, 8259–8278. [Google Scholar] [CrossRef] [PubMed]
| Mutations | Observed Frequencies |
|---|---|
| Deletions | 4% |
| Non-synonymous | 20% |
| Frameshift | 1% |
| Insertion | 0.1% |
| Transversion | 1.2% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bhattacharya, M.; Yaniv, D.; D’Souza, D.P.; Yosefof, E.; Tzelnick, S.; Detroja, R.; Wax, T.; Levy-Barda, A.; Baum, G.; Mizrachi, A.; et al. Applications for Circulating Cell-Free DNA in Oral Squamous Cell Carcinoma: A Non-Invasive Approach for Detecting Structural Variants, Fusions, and Oncoviruses. Cancers 2025, 17, 1901. https://doi.org/10.3390/cancers17121901
Bhattacharya M, Yaniv D, D’Souza DP, Yosefof E, Tzelnick S, Detroja R, Wax T, Levy-Barda A, Baum G, Mizrachi A, et al. Applications for Circulating Cell-Free DNA in Oral Squamous Cell Carcinoma: A Non-Invasive Approach for Detecting Structural Variants, Fusions, and Oncoviruses. Cancers. 2025; 17(12):1901. https://doi.org/10.3390/cancers17121901
Chicago/Turabian StyleBhattacharya, Mahua, Dan Yaniv, Dylan P. D’Souza, Eyal Yosefof, Sharon Tzelnick, Rajesh Detroja, Tal Wax, Adva Levy-Barda, Gideon Baum, Aviram Mizrachi, and et al. 2025. "Applications for Circulating Cell-Free DNA in Oral Squamous Cell Carcinoma: A Non-Invasive Approach for Detecting Structural Variants, Fusions, and Oncoviruses" Cancers 17, no. 12: 1901. https://doi.org/10.3390/cancers17121901
APA StyleBhattacharya, M., Yaniv, D., D’Souza, D. P., Yosefof, E., Tzelnick, S., Detroja, R., Wax, T., Levy-Barda, A., Baum, G., Mizrachi, A., Bachar, G., & Frenkel Morgenstern, M. (2025). Applications for Circulating Cell-Free DNA in Oral Squamous Cell Carcinoma: A Non-Invasive Approach for Detecting Structural Variants, Fusions, and Oncoviruses. Cancers, 17(12), 1901. https://doi.org/10.3390/cancers17121901








