A High Performing Biomarker Signature for Detecting Early-Stage Pancreatic Ductal Adenocarcinoma in High-Risk Individuals
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design and Participants
2.2. Candidate Protein Selection and Measurement
2.3. Statistical Analyses
3. Results
3.1. Patient Characteristics
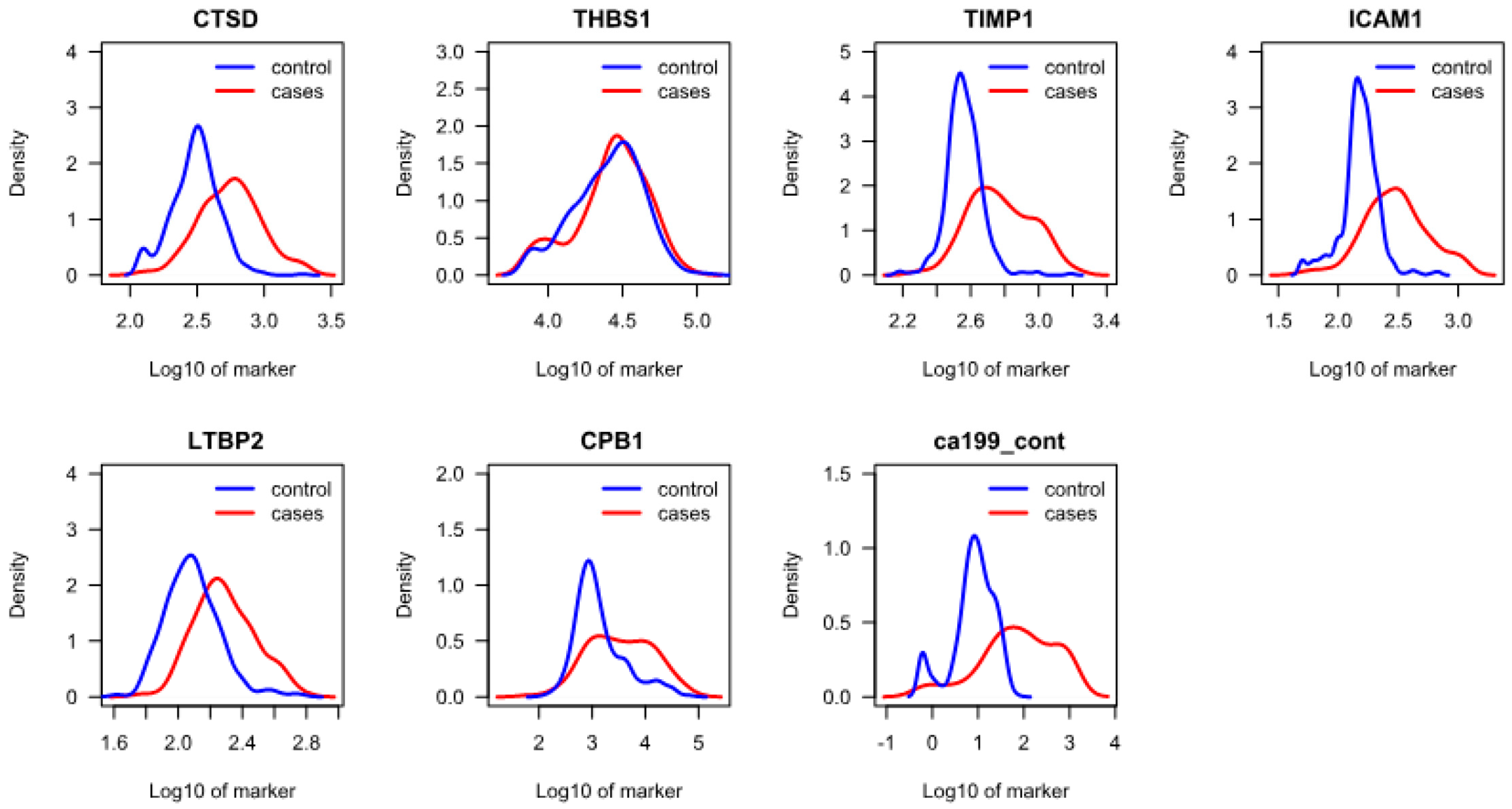
3.2. Protein Analysis and Selection of Final Candidates
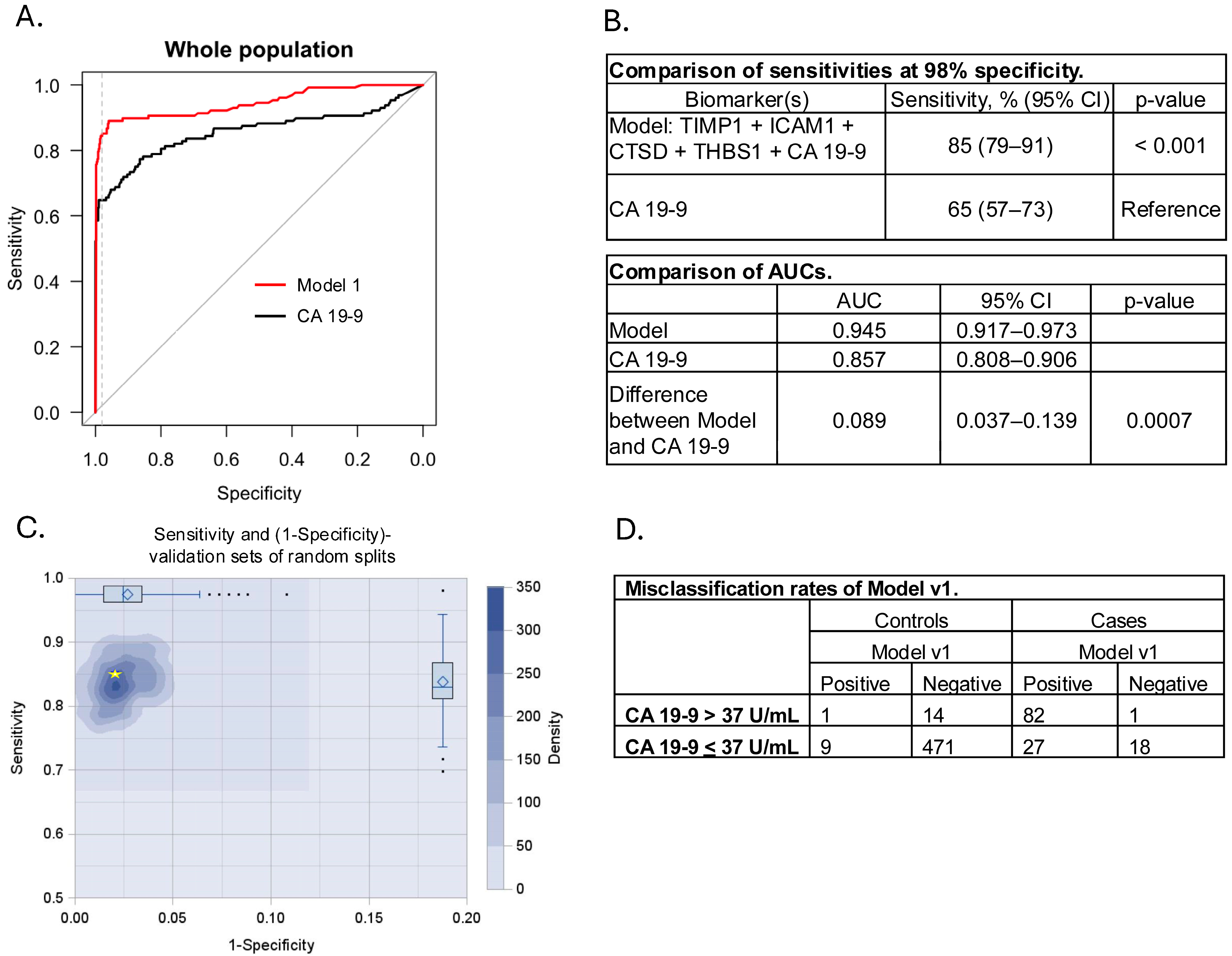
3.3. Model Development and Cross Validation
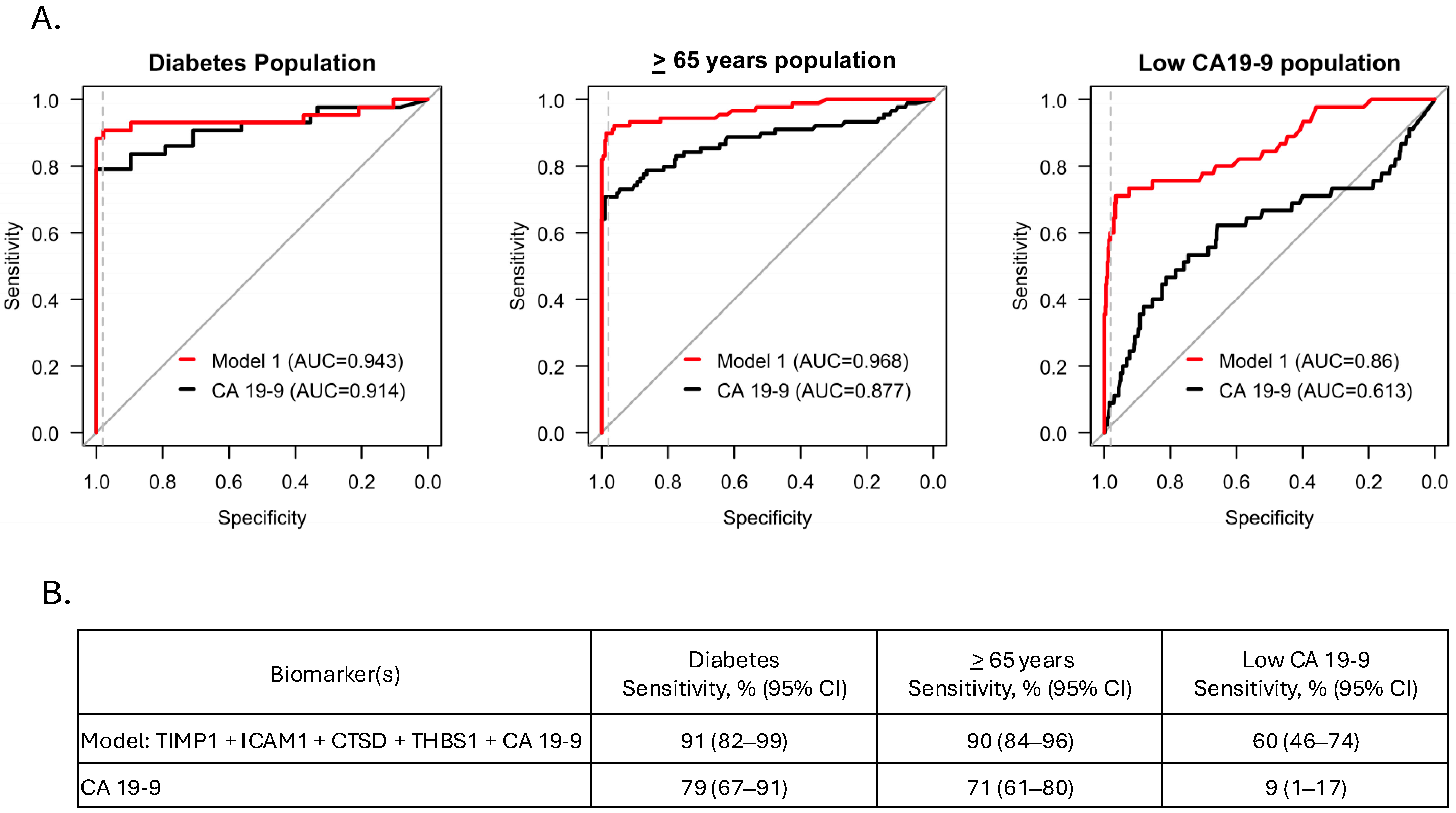
3.4. Model Performance in Sub-Populations
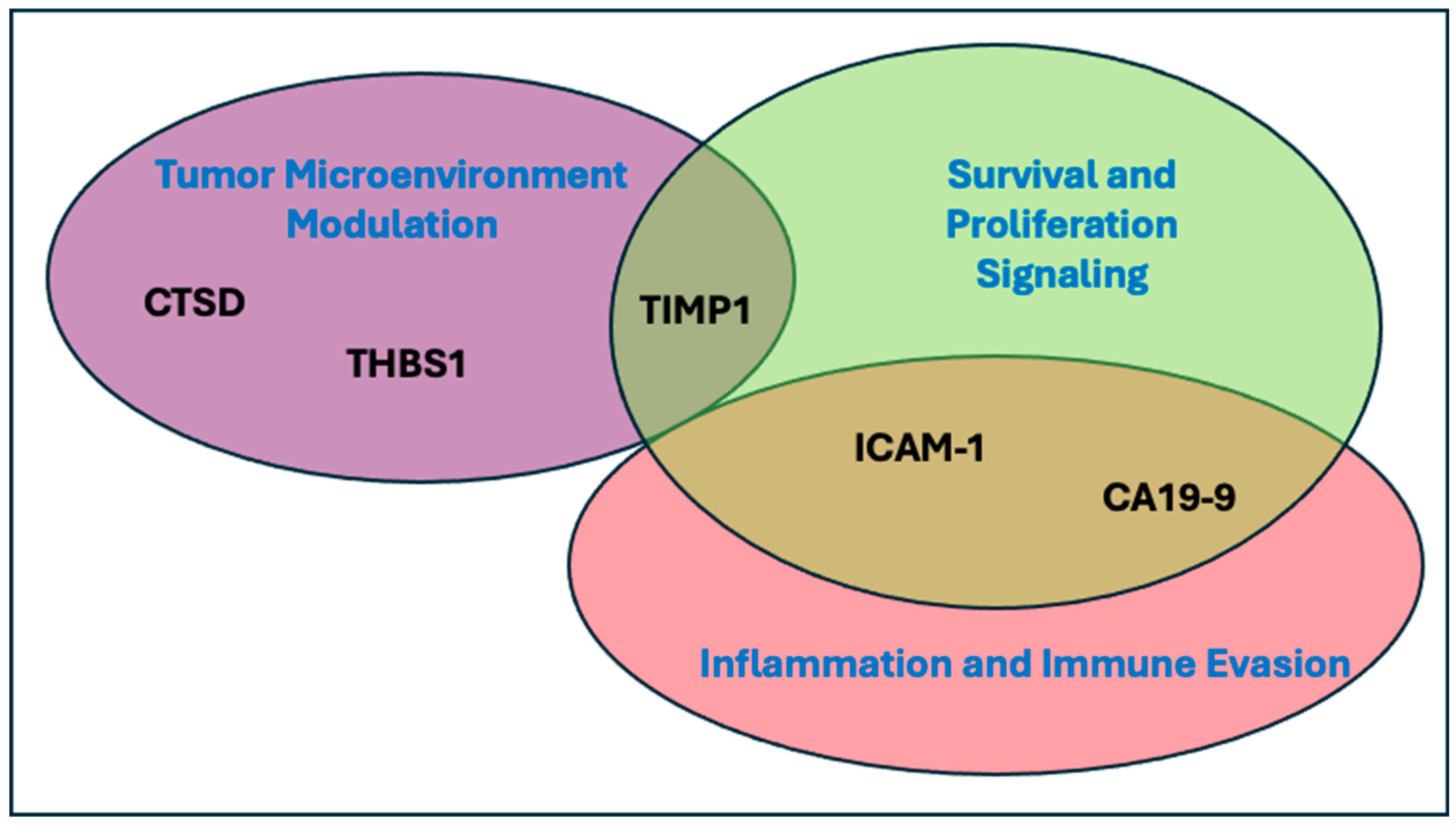
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CA 19-9 | Carbohydrate Antigen 19-9 |
| CI | Confidence Interval |
| CPB1 | Carboxypeptidase B1 |
| CTSD | Cathepsin D |
| GPNMB | Glycoprotein Non-Metastatic Melanoma Protein B |
| ICAM1 | Intracellular Adhesion Molecule 1 |
| KLK10 | Kallikrein Related Peptidase 10 |
| LTBP2 | Latent Transforming Growth Factor-Beta Binding Protein 2 |
| MERTK | Mer Tyrosine Kinase |
| MMP-9 | Matrix Metalloproteinase 9 |
| NET | Neutrophil Extracellular Trap |
| PDAC | Pancreatic Ductal Adenocarcinoma |
| PLA2G1B | Phospholipase A2 group 1B |
| TGF-β1 | Transforming Growth Factor-Beta 1 |
| THBS1 | Thrombospondin 1 |
| TIMP1 | Tissue Inhibitor of Metalloproteinase 1 |
References
- Institute, National Cancer Cancer Stat Facts: Common Cancer Sites. 2024. Available online: https://seer.cancer.gov/statfacts/html/common.html (accessed on 1 March 2025).
- Society, A.C. Survival Rates for Pancreatic Cancer. 2024. Available online: https://www.cancer.org/cancer/types/pancreatic-cancer/detection-diagnosis-staging/survival-rates.html (accessed on 1 March 2025).
- da Costa, W.L., Jr.; Oluyomi, A.O.; Thrift, A.P. Trends in the Incidence of Pancreatic Adenocarcinoma in All 50 United States Examined Through an Age-Period-Cohort Analysis. JNCI Cancer Spectr. 2020, 4, pkaa033. [Google Scholar] [CrossRef] [PubMed]
- Klein, A.P.; Lindstrom, S.; Mendelsohn, J.B.; Steplowski, E.; Arslan, A.A.; Bueno-de-Mesquita, H.B.; Fuchs, C.S.; Gallinger, S.; Gross, M.; Helzlsouer, K.; et al. An absolute risk model to identify individuals at elevated risk for pancreatic cancer in the general population. PLoS ONE 2013, 8, e72311. [Google Scholar] [CrossRef] [PubMed]
- Ryan, D.P.; Hong, T.S.; Bardeesy, N. Pancreatic adenocarcinoma. N. Engl. J. Med. 2014, 371, 1039–1049. [Google Scholar] [CrossRef] [PubMed]
- Society, A.C. Cancer Facts & Figures 2025. 2025. Available online: https://www.cancer.org/research/cancer-facts-statistics/all-cancer-facts-figures/2025-cancer-facts-figures.html (accessed on 10 March 2025).
- Goggins, M.; Overbeek, K.A.; Brand, R.; Syngal, S.; Del Chiaro, M.; Bartsch, D.K.; Bassi, C.; Carrato, A.; Farrell, J.; Fishman, E.K.; et al. Management of patients with increased risk for familial pancreatic cancer: Updated recommendations from the International Cancer of the Pancreas Screening (CAPS) Consortium. Gut 2020, 69, 7–17. [Google Scholar] [CrossRef]
- Daly, M.B.; Pal, T.; Maxwell, K.N.; Churpek, J.; Kohlmann, W.; AlHilli, Z.; Arun, B.; Buys, S.S.; Cheng, H.; Domchek, S.M.; et al. NCCN Guidelines(R) Insights: Genetic/Familial High-Risk Assessment: Breast, Ovarian, and Pancreatic, Version 2.2024. J. Natl. Compr. Cancer Netw. 2023, 21, 1000–1010. [Google Scholar] [CrossRef]
- Sawhney, M.S.; Calderwood, A.H.; Thosani, N.C.; Rebbeck, T.R.; Wani, S.; Canto, M.I.; Fishman, D.S.; Golan, T.; Hidalgo, M.; Kwon, R.S.; et al. ASGE guideline on screening for pancreatic cancer in individuals with genetic susceptibility: Summary and recommendations. Gastrointest. Endosc. 2022, 95, 817–826. [Google Scholar] [CrossRef]
- Zhang, L.; Sanagapalli, S.; Stoita, A. Challenges in diagnosis of pancreatic cancer. World J. Gastroenterol. 2018, 24, 2047–2060. [Google Scholar] [CrossRef]
- Locker, G.Y.; Hamilton, S.; Harris, J.; Jessup, J.M.; Kemeny, N.; Macdonald, J.S.; Somerfield, M.R.; Hayes, D.F.; Bast, R.C., Jr. ASCO 2006 update of recommendations for the use of tumor markers in gastrointestinal cancer. J. Clin. Oncol. 2006, 24, 5313–5327. [Google Scholar] [CrossRef]
- Mann, D.V.; Edwards, R.; Ho, S.; Lau, W.Y.; Glazer, G. Elevated tumour marker CA19-9: Clinical interpretation and influence of obstructive jaundice. Eur. J. Surg. Oncol. 2000, 26, 474–479. [Google Scholar] [CrossRef]
- Katsanos, K.H.; Kitsanou, M.; Christodoulou, D.K.; Tsianos, E.V. High CA 19-9 levels in benign biliary tract diseases. Report of four cases and review of the literature. Eur. J. Intern. Med. 2002, 13, 132–135. [Google Scholar] [CrossRef]
- Koprowski, H.; Herlyn, M.; Steplewski, Z.; Sears, H.F. Specific antigen in serum of patients with colon carcinoma. Science 1981, 212, 53–55. [Google Scholar] [CrossRef] [PubMed]
- Orntoft, T.F.; Vestergaard, E.M.; Holmes, E.; Jakobsen, J.S.; Grunnet, N.; Mortensen, M.; Johnson, P.; Bross, P.; Gregersen, N.; Skorstengaard, K.; et al. Influence of Lewis alpha1-3/4-L-fucosyltransferase (FUT3) gene mutations on enzyme activity, erythrocyte phenotyping, and circulating tumor marker sialyl-Lewis a levels. J. Biol. Chem. 1996, 271, 32260–32268. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.M.; Bai, S.M.; Zhang, G.M.; Ma, X.B. Reference intervals of carbohydrate antigen 19-9 in the apparently healthy adult population. J. Clin. Lab. Anal. 2018, 32, e22380. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.P.; Sung, I.K.; Kim, J.H.; Lee, S.Y.; Park, H.S.; Shim, C.S. Usefulness of Carbohydrate Antigen 19-9 Test in Healthy People and Necessity of Medical Follow-up in Individuals with Elevated Carbohydrate Antigen 19-9 Level. Korean J. Fam. Med. 2019, 40, 314–322. [Google Scholar] [CrossRef]
- Kane, L.E.; Mellotte, G.S.; Mylod, E.; O’Brien, R.M.; O’Connell, F.; Buckley, C.E.; Arlow, J.; Nguyen, K.; Mockler, D.; Meade, A.D.; et al. Diagnostic Accuracy of Blood-based Biomarkers for Pancreatic Cancer: A Systematic Review and Meta-analysis. Cancer Res. Commun. 2022, 2, 1229–1243. [Google Scholar] [CrossRef]
- Nene, N.R.; Ney, A.; Nazarenko, T.; Blyuss, O.; Johnston, H.E.; Whitwell, H.J.; Sedlak, E.; Gentry-Maharaj, A.; Apostolidou, S.; Costello, E.; et al. Serum biomarker-based early detection of pancreatic ductal adenocarcinomas with ensemble learning. Commun. Med. 2023, 3, 10. [Google Scholar] [CrossRef]
- Al-Shaheri, F.N.; Alhamdani, M.S.S.; Bauer, A.S.; Giese, N.; Buchler, M.W.; Hackert, T.; Hoheisel, J.D. Blood biomarkers for differential diagnosis and early detection of pancreatic cancer. Cancer Treat. Rev. 2021, 96, 102193. [Google Scholar] [CrossRef]
- Zhang, W.H.; Wang, W.Q.; Han, X.; Gao, H.L.; Li, T.J.; Xu, S.S.; Li, S.; Xu, H.X.; Li, H.; Ye, L.Y.; et al. Advances on diagnostic biomarkers of pancreatic ductal adenocarcinoma: A systems biology perspective. Comput. Struct. Biotechnol. J. 2020, 18, 3606–3614. [Google Scholar] [CrossRef]
- Athanasiou, A.; Kureshi, N.; Wittig, A.; Sterner, M.; Huber, R.; Palma, N.A.; King, T.; Schiess, R. Biomarker Discovery for Early Detection of Pancreatic Ductal Adenocarcinoma (PDAC) Using Multiplex Proteomics Technology. J. Proteome Res. 2025, 24, 315–322. [Google Scholar] [CrossRef]
- Bujang, M.A.; Adnan, T.H. Requirements for Minimum Sample Size for Sensitivity and Specificity Analysis. J. Clin. Diagn. Res. 2016, 10, YE01–YE06. [Google Scholar] [CrossRef]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef] [PubMed]
- Sundjaja, J.H.; Shrestha, R.; Krishan, K. McNemar And Mann-Whitney U Tests. 2024. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560699/ (accessed on 1 February 2025).
- The R Project for Statistical Computing. 2024. Available online: https://www.r-project.org/ (accessed on 1 February 2025).
- Silva-Santisteban, A.; Hernandez Woodbine, M.J.; Noriega, M.A.; Rabinowitz, L.G.; Grimshaw, A.; Farrell, J.J.; Chhoda, A.; Sawhney, M.S. Disparities in race, ethnicity, sex, and age inclusion in pancreatic cancer screening studies: A systematic review and meta-analysis. Gastrointest. Endosc. 2024, 100, 1–16.e20. [Google Scholar] [CrossRef] [PubMed]
- Ferlay, J.; Colombet, M.; Soerjomataram, I.; Parkin, D.M.; Pineros, M.; Znaor, A.; Bray, F. Cancer statistics for the year 2020: An overview. Int. J. Cancer 2021, 149, 778–789. [Google Scholar] [CrossRef] [PubMed]
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Network, National Comprehensive Cancer NCCN Guidelines Pancreatic Adenocarcenoma. 2024. Available online: https://www.nccn.org/guidelines/guidelines-detail?category=1&id=1455 (accessed on 10 March 2025).
- Poruk, K.E.; Firpo, M.A.; Scaife, C.L.; Adler, D.G.; Emerson, L.L.; Boucher, K.M.; Mulvihill, S.J. Serum osteopontin and tissue inhibitor of metalloproteinase 1 as diagnostic and prognostic biomarkers for pancreatic adenocarcinoma. Pancreas 2013, 42, 193–197. [Google Scholar] [CrossRef]
- Tian, Z.; Ou, G.; Su, M.; Li, R.; Pan, L.; Lin, X.; Zou, J.; Chen, S.; Li, Y.; Huang, K.; et al. TIMP1 derived from pancreatic cancer cells stimulates Schwann cells and promotes the occurrence of perineural invasion. Cancer Lett. 2022, 546, 215863. [Google Scholar] [CrossRef]
- Huang, C.; Li, N.; Li, Z.; Chang, A.; Chen, Y.; Zhao, T.; Li, Y.; Wang, X.; Zhang, W.; Wang, Z.; et al. Tumour-derived Interleukin 35 promotes pancreatic ductal adenocarcinoma cell extravasation and metastasis by inducing ICAM1 expression. Nat. Commun. 2017, 8, 14035. [Google Scholar] [CrossRef]
- Roland, C.L.; Dineen, S.P.; Toombs, J.E.; Carbon, J.G.; Smith, C.W.; Brekken, R.A.; Barnett, C.C., Jr. Tumor-derived intercellular adhesion molecule-1 mediates tumor-associated leukocyte infiltration in orthotopic pancreatic xenografts. Exp. Biol. Med. 2010, 235, 263–270. [Google Scholar] [CrossRef]
- Grunwald, B.; Harant, V.; Schaten, S.; Fruhschutz, M.; Spallek, R.; Hochst, B.; Stutzer, K.; Berchtold, S.; Erkan, M.; Prokopchuk, O.; et al. Pancreatic Premalignant Lesions Secrete Tissue Inhibitor of Metalloproteinases-1, Which Activates Hepatic Stellate Cells Via CD63 Signaling to Create a Premetastatic Niche in the Liver. Gastroenterology 2016, 151, 1011–1024.e7. [Google Scholar] [CrossRef]
- Schoeps, B.; Eckfeld, C.; Prokopchuk, O.; Bottcher, J.; Haussler, D.; Steiger, K.; Demir, I.E.; Knolle, P.; Soehnlein, O.; Jenne, D.E.; et al. TIMP1 Triggers Neutrophil Extracellular Trap Formation in Pancreatic Cancer. Cancer Res. 2021, 81, 3568–3579. [Google Scholar] [CrossRef]
- Liou, G.Y.; Doppler, H.; Necela, B.; Edenfield, B.; Zhang, L.; Dawson, D.W.; Storz, P. Mutant KRAS-induced expression of ICAM-1 in pancreatic acinar cells causes attraction of macrophages to expedite the formation of precancerous lesions. Cancer Discov. 2015, 5, 52–63. [Google Scholar] [CrossRef] [PubMed]
- Nomura, T.; Katunuma, N. Involvement of cathepsins in the invasion, metastasis and proliferation of cancer cells. J. Med. Investig. 2005, 52, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Dumartin, L.; Whiteman, H.J.; Weeks, M.E.; Hariharan, D.; Dmitrovic, B.; Iacobuzio-Donahue, C.A.; Brentnall, T.A.; Bronner, M.P.; Feakins, R.M.; Timms, J.F.; et al. AGR2 is a novel surface antigen that promotes the dissemination of pancreatic cancer cells through regulation of cathepsins B and D. Cancer Res. 2011, 71, 7091–7102. [Google Scholar] [CrossRef] [PubMed]
- Albo, D.; Berger, D.H.; Tuszynski, G.P. The effect of thrombospondin-1 and TGF-beta 1 on pancreatic cancer cell invasion. J. Surg. Res. 1998, 76, 86–90. [Google Scholar] [CrossRef]
- Bocci, G.; Fioravanti, A.; Orlandi, P.; Di Desidero, T.; Natale, G.; Fanelli, G.; Viacava, P.; Naccarato, A.G.; Francia, G.; Danesi, R. Metronomic ceramide analogs inhibit angiogenesis in pancreatic cancer through up-regulation of caveolin-1 and thrombospondin-1 and down-regulation of cyclin D1. Neoplasia 2012, 14, 833–845. [Google Scholar] [CrossRef]
- McElroy, M.K.; Kaushal, S.; Tran Cao, H.S.; Moossa, A.R.; Talamini, M.A.; Hoffman, R.M.; Bouvet, M. Upregulation of thrombospondin-1 and angiogenesis in an aggressive human pancreatic cancer cell line selected for high metastasis. Mol. Cancer Ther. 2009, 8, 1779–1786. [Google Scholar] [CrossRef]
- Qian, X.; Rothman, V.L.; Nicosia, R.F.; Tuszynski, G.P. Expression of thrombospondin-1 in human pancreatic adenocarcinomas: Role in matrix metalloproteinase-9 production. Pathol. Oncol. Res. 2001, 7, 251–259. [Google Scholar] [CrossRef]
- Matsumura, K.; Hayashi, H.; Uemura, N.; Ogata, Y.; Zhao, L.; Sato, H.; Shiraishi, Y.; Kuroki, H.; Kitamura, F.; Kaida, T.; et al. Thrombospondin-1 overexpression stimulates loss of Smad4 and accelerates malignant behavior via TGF-beta signal activation in pancreatic ductal adenocarcinoma. Transl. Oncol. 2022, 26, 101533. [Google Scholar] [CrossRef]
- mohamed, A.; Saad, Y.; Saleh, D.; Elawady, R.; Eletreby, R.; Kharalla, A.S.; Badr, E. Can Serum ICAM 1 distinguish pancreatic cancer from chronic pancreatitis? Asian Pac. J. Cancer Prev. 2016, 17, 4671–4675. [Google Scholar]
- Park, H.D.; Kang, E.S.; Kim, J.W.; Lee, K.T.; Lee, K.H.; Park, Y.S.; Park, J.O.; Lee, J.; Heo, J.S.; Choi, S.H.; et al. Serum CA19-9, cathepsin D, and matrix metalloproteinase-7 as a diagnostic panel for pancreatic ductal adenocarcinoma. Proteomics 2012, 12, 3590–3597. [Google Scholar] [CrossRef]
- Jenkinson, C.; Elliott, V.L.; Evans, A.; Oldfield, L.; Jenkins, R.E.; O’Brien, D.P.; Apostolidou, S.; Gentry-Maharaj, A.; Fourkala, E.O.; Jacobs, I.J.; et al. Decreased Serum Thrombospondin-1 Levels in Pancreatic Cancer Patients Up to 24 Months Prior to Clinical Diagnosis: Association with Diabetes Mellitus. Clin. Cancer Res. 2016, 22, 1734–1743. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Liu, C.; Guo, M.; Cheng, H.; Lu, Y.; Jin, K.; Liu, L.; Long, J.; Xu, J.; Lu, R.; et al. Potential Biomarkers in Lewis Negative Patients With Pancreatic Cancer. Ann. Surg. 2017, 265, 800–805. [Google Scholar] [CrossRef] [PubMed]
- Ando, Y.; Dbouk, M.; Yoshida, T.; Saba, H.; Abou Diwan, E.; Yoshida, K.; Dbouk, A.; Blackford, A.L.; Lin, M.T.; Lennon, A.M.; et al. Using Tumor Marker Gene Variants to Improve the Diagnostic Accuracy of DUPAN-2 and Carbohydrate Antigen 19-9 for Pancreatic Cancer. J. Clin. Oncol. 2024, 42, 2196–2206. [Google Scholar] [CrossRef] [PubMed]
- Luo, G.; Guo, M.; Jin, K.; Liu, Z.; Liu, C.; Cheng, H.; Lu, Y.; Long, J.; Liu, L.; Xu, J.; et al. Optimize CA19-9 in detecting pancreatic cancer by Lewis and Secretor genotyping. Pancreatology 2016, 16, 1057–1062. [Google Scholar] [CrossRef]
- Katona, B.W.; Klute, K.; Brand, R.E.; Everett, J.N.; Farrell, J.J.; Hawthorne, K.; Kaul, V.; Kupfer, S.S.; Paiella, S.; Simeone, D.M.; et al. Racial, Ethnic, and Sex-based Disparities among High-risk Individuals Undergoing Pancreatic Cancer Surveillance. Cancer Prev. Res. 2023, 16, 343–352. [Google Scholar] [CrossRef]
| Clinical Variable | All (n = 623) | Cases (n = 128) | Controls (n = 495) |
|---|---|---|---|
| Age, median (min-max) | 64 (40–95) | 68 (46–95) | 63 (40–83) |
| CA 19-9 U/mL, median (min-max) | 10 (0.6–1000) * | 78 (0.6–1000) | 9 (0.6–70) |
| Sex male, n (% of column heading) | 244 (39%) | 69 (54%) | 175 (35%) |
| PDAC Stage I, n (% of column heading) | 88 (14%) | 88 (69%) | N/A |
| PDAC Stage II, n (% of column heading) | 40 (6%) | 40 (31%) | N/A |
| White (% of column heading) | 481 (77%) | 52 (41%) | 429 (86%) |
| Not White (% of column heading) | 28 (4%) | 4 (3%) | 28 (22%) |
| Not reported (% of column heading) | 114 (18%) | 72 (56%) | 42 (33%) |
| Marker | p-Value (Controls vs. Cases) | Concentration Means, Cases (CA19-9, U/mL; All Others, ng/mL) | Concentration Means, Controls (CA19-9, U/mL; All Others, ng/mL) | Sensitivity at 98% Specificity, % (95% CI) |
|---|---|---|---|---|
| CA 19-9 | <0.001 | 248 | 12 | 65 (57–73) |
| ICAM1 | <0.001 | 363 | 161 | 52 (44–61) |
| TIMP1 | <0.001 | 657 | 375 | 48 (39–56) |
| CTSD | <0.001 | 646 | 333 | 31 (23–39) |
| LTBP2 | <0.001 | 215 | 133 | 12 (6–17) |
| CPB1 | <0.001 | 8139 | 3199 | 6 (2–10) |
| THBS1 | 0.220 | 30613 | 28844 | 2 (0–5) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Palma, N.A.; Lucas, A.L.; Katona, B.W.; Athanasiou, A.; Kureshi, N.M.; Ford, L.; Keller, T.; Weber, S.; Schiess, R.; King, T.; et al. A High Performing Biomarker Signature for Detecting Early-Stage Pancreatic Ductal Adenocarcinoma in High-Risk Individuals. Cancers 2025, 17, 1866. https://doi.org/10.3390/cancers17111866
Palma NA, Lucas AL, Katona BW, Athanasiou A, Kureshi NM, Ford L, Keller T, Weber S, Schiess R, King T, et al. A High Performing Biomarker Signature for Detecting Early-Stage Pancreatic Ductal Adenocarcinoma in High-Risk Individuals. Cancers. 2025; 17(11):1866. https://doi.org/10.3390/cancers17111866
Chicago/Turabian StylePalma, Norma A., Aimee L. Lucas, Bryson W. Katona, Alcibiade Athanasiou, Natasha M. Kureshi, Lisa Ford, Thomas Keller, Stephen Weber, Ralph Schiess, Thomas King, and et al. 2025. "A High Performing Biomarker Signature for Detecting Early-Stage Pancreatic Ductal Adenocarcinoma in High-Risk Individuals" Cancers 17, no. 11: 1866. https://doi.org/10.3390/cancers17111866
APA StylePalma, N. A., Lucas, A. L., Katona, B. W., Athanasiou, A., Kureshi, N. M., Ford, L., Keller, T., Weber, S., Schiess, R., King, T., Simeone, D. M., & Brand, R. (2025). A High Performing Biomarker Signature for Detecting Early-Stage Pancreatic Ductal Adenocarcinoma in High-Risk Individuals. Cancers, 17(11), 1866. https://doi.org/10.3390/cancers17111866






