Lipid Metabolism Reprogramming in Tumor-Associated Macrophages Modulates Their Function in Primary Liver Cancers
Simple Summary
Abstract
1. Introduction
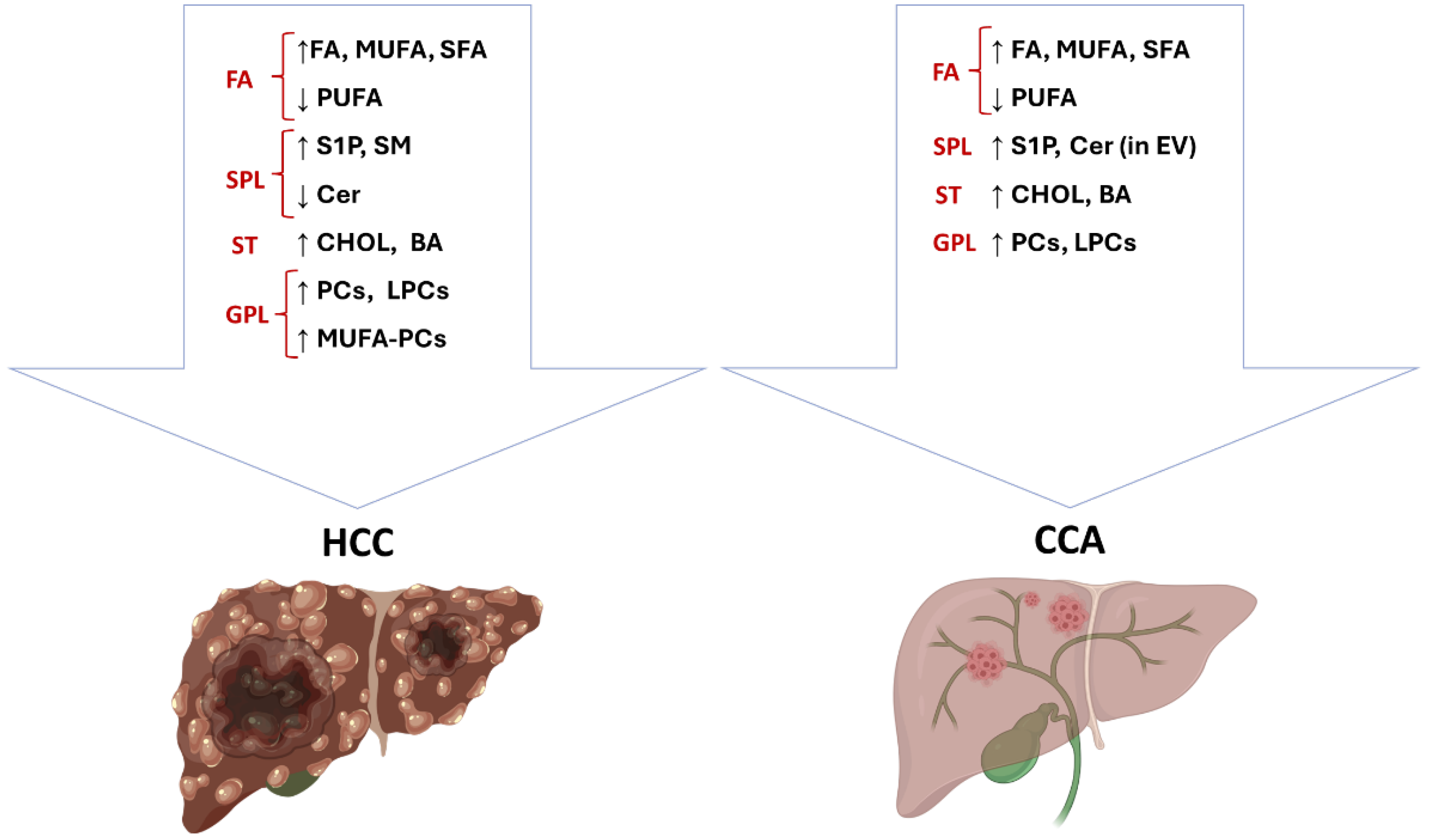
2. Lipid Metabolism in Primary Liver Cancers and Therapeutic Perspectives
3. Liver Macrophages
4. TAM Diversity in Primary Liver Cancer
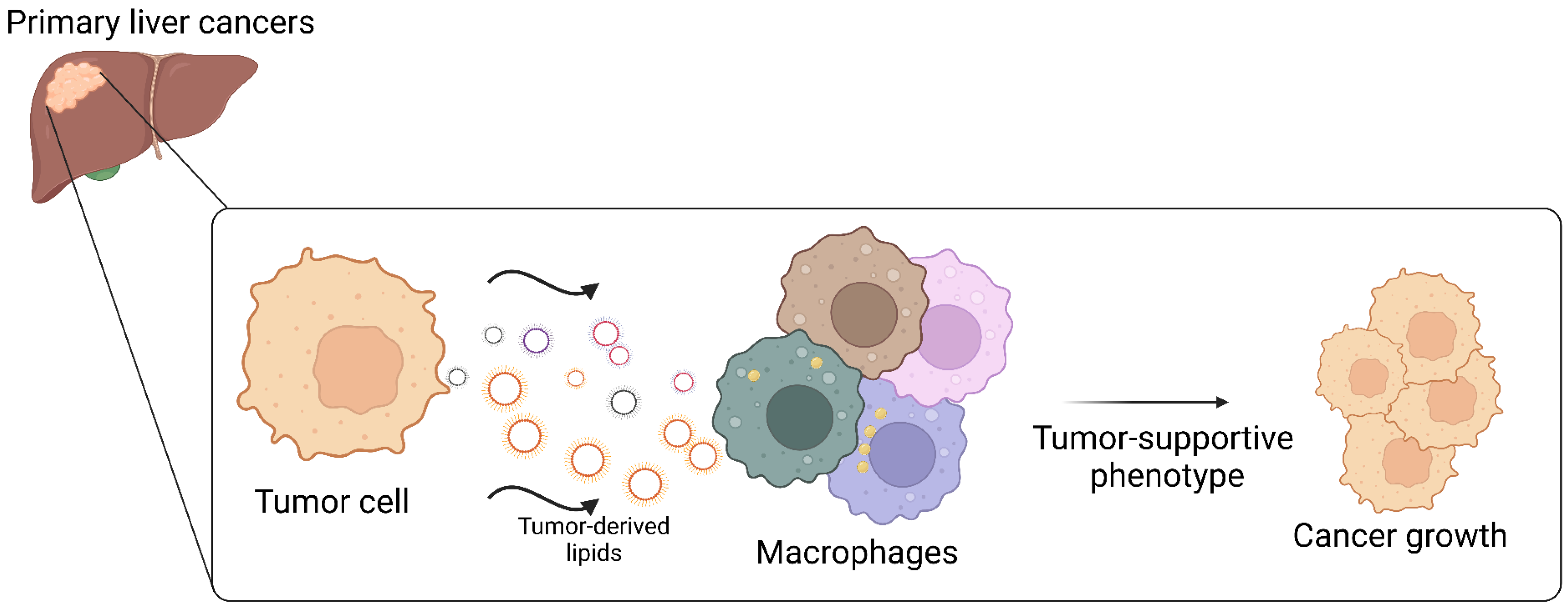
5. Lipid Metabolism Reprogramming in TAM of Primary Liver Cancers
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Villanueva, A. Hepatocellular Carcinoma. N. Engl. J. Med. 2019, 380, 1450–1462. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef]
- Rodriguez, L.A.; Schmittdiel, J.A.; Liu, L.; Macdonald, B.A.; Balasubramanian, S.; Chai, K.P.; Seo, S.I.; Mukhtar, N.; Levin, T.R.; Saxena, V. Hepatocellular Carcinoma in Metabolic Dysfunction-Associated Steatotic Liver Disease. JAMA Netw. Open 2024, 7, e2421019. [Google Scholar] [CrossRef] [PubMed]
- Qin, Y.; Han, S.; Yu, Y.; Qi, D.; Ran, M.; Yang, M.; Liu, Y.; Li, Y.; Lu, L.; Liu, Y.; et al. Lenvatinib in Hepatocellular Carcinoma: Resistance Mechanisms and Strategies for Improved Efficacy. Liver Int. 2024, 44, 1808–1831. [Google Scholar] [CrossRef]
- Forner, A.; Vidili, G.; Rengo, M.; Bujanda, L.; Ponz-Sarvisé, M.; Lamarca, A. Clinical Presentation, Diagnosis and Staging of Cholangiocarcinoma. Liver Int. 2019, 39, 98–107. [Google Scholar] [CrossRef]
- Wei, M.; Lü, L.; Lin, P.; Chen, Z.; Quan, Z.; Tang, Z. Multiple Cellular Origins and Molecular Evolution of Intrahepatic Cholangiocarcinoma. Cancer Lett. 2016, 379, 253–261. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Huang, A.; Kuang, B.; Xiao, Y.; Xiao, Y.; Ma, H. Progress in Radiotherapy for Cholangiocarcinoma. Front. Oncol. 2022, 12, 868034. [Google Scholar] [CrossRef]
- Wu, M.-J.; Shi, L.; Dubrot, J.; Merritt, J.; Vijay, V.; Wei, T.-Y.; Kessler, E.; Olander, K.E.; Adil, R.; Pankaj, A.; et al. Mutant-IDH Inhibits Interferon-TET2 Signaling to Promote Immunoevasion and Tumor Maintenance in Cholangiocarcinoma. Cancer Discov. 2022, 12, 812–835. [Google Scholar] [CrossRef]
- Paul, B.; Lewinska, M.; Andersen, J.B. Lipid Alterations in Chronic Liver Disease and Liver Cancer. JHEP Rep. 2022, 4, 100479. [Google Scholar] [CrossRef]
- Dei Cas, M.; Mantovani, S.; Oliviero, B.; Zulueta, A.; Montavoci, L.; Falleni, M.; Tosi, D.; Morano, C.; Penati, S.; Chiocchetti, A. Cholangiocarcinoma Cells Direct Fatty Acids to Support Membrane Synthesis and Modulate Macrophage Phenotype. Hepatol. Commun. 2025, in press. [Google Scholar] [CrossRef]
- Raggi, C.; Taddei, M.L.; Rae, C.; Braconi, C.; Marra, F. Metabolic Reprogramming in Cholangiocarcinoma. J. Hepatol. 2022, 77, 849–864. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Ruan, K.; Chu, Z.; Wang, X.; Gu, Y.; Jin, H.; Zhang, X.; Liu, Q.; Yang, J. Reprogramming of Fatty Acid Metabolism: A Hidden Force Regulating the Occurrence and Progression of Cholangiocarcinoma. Cell Death Discov. 2025, 11, 72. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.; Zhang, W.; Chen, X.; Guo, H.; Wu, H.; Xu, Y.; He, Q.; Ding, L.; Yang, B. The Impact of Lipids on the Cancer–Immunity Cycle and Strategies for Modulating Lipid Metabolism to Improve Cancer Immunotherapy. Acta Pharm. Sin. B 2023, 13, 1488–1497. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The Tumor Microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Lian, X.; Yang, K.; Li, R.; Li, M.; Zuo, J.; Zheng, B.; Wang, W.; Wang, P.; Zhou, S. Immunometabolic Rewiring in Tumorigenesis and Anti-Tumor Immunotherapy. Mol. Cancer 2022, 21, 27. [Google Scholar] [CrossRef]
- Huang, J.; Pan, H.; Sun, J.; Wu, J.; Xuan, Q.; Wang, J.; Ke, S.; Lu, S.; Li, Z.; Feng, Z.; et al. TMEM147 Aggravates the Progression of HCC by Modulating Cholesterol Homeostasis, Suppressing Ferroptosis, and Promoting the M2 Polarization of Tumor-Associated Macrophages. J. Exp. Clin. Cancer Res. 2023, 42, 286. [Google Scholar] [CrossRef]
- Masetti, M.; Carriero, R.; Portale, F.; Marelli, G.; Morina, N.; Pandini, M.; Iovino, M.; Partini, B.; Erreni, M.; Ponzetta, A.; et al. Lipid-Loaded Tumor-Associated Macrophages Sustain Tumor Growth and Invasiveness in Prostate Cancer. J. Exp. Med. 2021, 219, e20210564. [Google Scholar] [CrossRef]
- Wu, Q.; Yang, Y.; Lin, S.; Geller, D.A.; Yan, Y. The Microenvironment in the Development of MASLD-MASH-HCC and Associated Therapeutic in MASH-HCC. Front. Immunol. 2025, 16, 1569915. [Google Scholar] [CrossRef] [PubMed]
- Carli, F.; Della Pepa, G.; Sabatini, S.; Vidal Puig, A.; Gastaldelli, A. Lipid Metabolism in MASLD and MASH: From Mechanism to the Clinic. JHEP Rep. 2024, 6, 101185. [Google Scholar] [CrossRef]
- Li, H.; Chen, Z.; Zhang, Y.; Yuan, P.; Liu, J.; Ding, L.; Ye, Q. MiR-4310 Regulates Hepatocellular Carcinoma Growth and Metastasis through Lipid Synthesis. Cancer Lett. 2021, 519, 161–171. [Google Scholar] [CrossRef]
- Zhang, H.; Zhu, K.; Zhang, R.; Guo, Y.; Wang, J.; Liu, C.; Lu, X.; Zhou, Z.; Wu, W.; Zhang, F.; et al. Oleic Acid-PPARγ-FABP4 Loop Fuels Cholangiocarcinoma Colonization in Lymph Node Metastases Microenvironment. Hepatology 2024, 80, 69–86. [Google Scholar] [CrossRef] [PubMed]
- Hall, Z.; Chiarugi, D.; Charidemou, E.; Leslie, J.; Scott, E.; Pellegrinet, L.; Allison, M.; Mocciaro, G.; Anstee, Q.M.; Evan, G.I.; et al. Lipid Remodeling in Hepatocyte Proliferation and Hepatocellular Carcinoma. Hepatology 2021, 73, 1028. [Google Scholar] [CrossRef]
- Arroyave-Ospina, J.C.; Wu, Z.; Geng, Y.; Moshage, H. Role of Oxidative Stress in the Pathogenesis of Non-Alcoholic Fatty Liver Disease: Implications for Prevention and Therapy. Antioxidants 2021, 10, 174. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Lee, G.; Heo, S.-Y.; Roh, Y.-S. Oxidative Stress Is a Key Modulator in the Development of Nonalcoholic Fatty Liver Disease. Antioxidants 2021, 11, 91. [Google Scholar] [CrossRef] [PubMed]
- Geng, Y.; Faber, K.N.; de Meijer, V.E.; Blokzijl, H.; Moshage, H. How Does Hepatic Lipid Accumulation Lead to Lipotoxicity in Non-Alcoholic Fatty Liver Disease? Hepatol. Int. 2021, 15, 21–35. [Google Scholar] [CrossRef]
- Vinciguerra, M.; Carrozzino, F.; Peyrou, M.; Carlone, S.; Montesano, R.; Benelli, R.; Foti, M. Unsaturated Fatty Acids Promote Hepatoma Proliferation and Progression through Downregulation of the Tumor Suppressor PTEN. J. Hepatol. 2009, 50, 1132–1141. [Google Scholar] [CrossRef]
- Lin, C.-R.; Chu, T.-M.; Luo, A.; Huang, S.-J.; Chou, H.-Y.; Lu, M.-W.; Wu, J.-L. Omega-3 Polyunsaturated Fatty Acids Suppress Metastatic Features of Human Cholangiocarcinoma Cells by Suppressing Twist. J. Nutr. Biochem. 2019, 74, 108245. [Google Scholar] [CrossRef]
- Berkemeyer, S. Primary Liver Cancers: Connecting the Dots of Cellular Studies and Epidemiology with Metabolomics. Int. J. Mol. Sci. 2023, 24, 2409. [Google Scholar] [CrossRef]
- Pastore, M.; Lori, G.; Gentilini, A.; Taddei, M.L.; Di Maira, G.; Campani, C.; Recalcati, S.; Invernizzi, P.; Marra, F.; Raggi, C. Multifaceted Aspects of Metabolic Plasticity in Human Cholangiocarcinoma: An Overview of Current Perspectives. Cells 2020, 9, 596. [Google Scholar] [CrossRef]
- Nakagawa, R.; Hiep, N.C.; Ouchi, H.; Sato, Y.; Harada, K. Expression of Fatty-Acid-Binding Protein 5 in Intrahepatic and Extrahepatic Cholangiocarcinoma: The Possibility of Different Energy Metabolisms in Anatomical Location. Med. Mol. Morphol. 2020, 53, 42–49. [Google Scholar] [CrossRef]
- Li, L.; Che, L.; Tharp, K.M.; Park, H.-M.; Pilo, M.G.; Cao, D.; Cigliano, A.; Latte, G.; Xu, Z.; Ribback, S.; et al. Differential Requirement for de Novo Lipogenesis in Cholangiocarcinoma and Hepatocellular Carcinoma of Mice and Humans. Hepatology 2016, 63, 1900–1913. [Google Scholar] [CrossRef] [PubMed]
- Calvisi, D.F.; Wang, C.; Ho, C.; Ladu, S.; Lee, S.A.; Mattu, S.; Destefanis, G.; Delogu, S.; Zimmermann, A.; Ericsson, J.; et al. Increased Lipogenesis, Induced by AKT-mTORC1-RPS6 Signaling, Promotes Development of Human Hepatocellular Carcinoma. Gastroenterology 2011, 140, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Ho, C.; Wang, C.; Mattu, S.; Destefanis, G.; Ladu, S.; Delogu, S.; Armbruster, J.; Fan, L.; Lee, S.A.; Jiang, L.; et al. AKT (v-Akt Murine Thymoma Viral Oncogene Homolog 1) and N-Ras (Neuroblastoma Ras Viral Oncogene Homolog) Coactivation in the Mouse Liver Promotes Rapid Carcinogenesis by Way of mTOR (Mammalian Target of Rapamycin Complex 1), FOXM1 (Forkhead Box M1)/SKP2, and c-Myc Pathways. Hepatology 2012, 55, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Cheng, C.; Tan, Z.; Li, N.; Tang, M.; Yang, L.; Cao, Y. Emerging Roles of Lipid Metabolism in Cancer Metastasis. Mol. Cancer 2017, 16, 76. [Google Scholar] [CrossRef]
- Du, D.; Liu, C.; Qin, M.; Zhang, X.; Xi, T.; Yuan, S.; Hao, H.; Xiong, J. Metabolic Dysregulation and Emerging Therapeutical Targets for Hepatocellular Carcinoma. Acta Pharm. Sin. B 2022, 12, 558–580. [Google Scholar] [CrossRef]
- Ning, Z.; Guo, X.; Liu, X.; Lu, C.; Wang, A.; Wang, X.; Wang, W.; Chen, H.; Qin, W.; Liu, X.; et al. USP22 Regulates Lipidome Accumulation by Stabilizing PPARγ in Hepatocellular Carcinoma. Nat. Commun. 2022, 13, 2187. [Google Scholar] [CrossRef]
- Bian, Z.; Xu, C.; Wang, X.; Zhang, B.; Xiao, Y.; Liu, L.; Zhao, S.; Huang, N.; Yang, F.; Zhang, Y.; et al. TRIM65/NF2/YAP1 Signaling Coordinately Orchestrates Metabolic and Immune Advantages in Hepatocellular Carcinoma. Adv. Sci. 2024, 11, 2402578. [Google Scholar] [CrossRef]
- Yang, F.; Hilakivi-Clarke, L.; Shaha, A.; Wang, Y.; Wang, X.; Deng, Y.; Lai, J.; Kang, N. Metabolic Reprogramming and Its Clinical Implication for Liver Cancer. Hepatology 2023, 78, 1602–1624. [Google Scholar] [CrossRef]
- Fujiwara, N.; Nakagawa, H.; Enooku, K.; Kudo, Y.; Hayata, Y.; Nakatsuka, T.; Tanaka, Y.; Tateishi, R.; Hikiba, Y.; Misumi, K.; et al. CPT2 Downregulation Adapts HCC to Lipid-Rich Environment and Promotes Carcinogenesis via Acylcarnitine Accumulation in Obesity. Gut 2018, 67, 1493–1504. [Google Scholar] [CrossRef]
- Huang, D.; Li, T.; Li, X.; Zhang, L.; Sun, L.; He, X.; Zhong, X.; Jia, D.; Song, L.; Semenza, G.L.; et al. HIF-1-Mediated Suppression of Acyl-CoA Dehydrogenases and Fatty Acid Oxidation Is Critical for Cancer Progression. Cell Rep. 2014, 8, 1930–1942. [Google Scholar] [CrossRef]
- Iwamoto, H.; Abe, M.; Yang, Y.; Cui, D.; Seki, T.; Nakamura, M.; Hosaka, K.; Lim, S.; Wu, J.; He, X.; et al. Cancer Lipid Metabolism Confers Antiangiogenic Drug Resistance. Cell Metab. 2018, 28, 104–117.e5. [Google Scholar] [CrossRef] [PubMed]
- Lu, G.-D.; Ang, Y.H.; Zhou, J.; Tamilarasi, J.; Yan, B.; Lim, Y.C.; Srivastava, S.; Salto-Tellez, M.; Hui, K.M.; Shen, H.-M.; et al. CCAAT/Enhancer Binding Protein α Predicts Poorer Prognosis and Prevents Energy Starvation–Induced Cell Death in Hepatocellular Carcinoma. Hepatology 2015, 61, 965–978. [Google Scholar] [CrossRef]
- Bi, Y.; Ying, X.; Chen, W.; Wu, J.; Kong, C.; Hu, W.; Fang, S.; Yu, J.; Zhai, M.; Jiang, C.; et al. Glycerophospholipid-Driven Lipid Metabolic Reprogramming as a Common Key Mechanism in the Progression of Human Primary Hepatocellular Carcinoma and Cholangiocarcinoma. Lipids Health Dis. 2024, 23, 326. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.L.W.; Israeli, E.; Ericksen, R.E.; Chow, P.K.H.; Han, W. The Altered Lipidome of Hepatocellular Carcinoma. Semin. Cancer Biol. 2022, 86, 445–456. [Google Scholar] [CrossRef]
- Guri, Y.; Colombi, M.; Dazert, E.; Hindupur, S.K.; Roszik, J.; Moes, S.; Jenoe, P.; Heim, M.H.; Riezman, I.; Riezman, H.; et al. mTORC2 Promotes Tumorigenesis via Lipid Synthesis. Cancer Cell 2017, 32, 807–823.e12. [Google Scholar] [CrossRef]
- Lewinska, M.; Santos-Laso, A.; Arretxe, E.; Alonso, C.; Zhuravleva, E.; Jimenez-Agüero, R.; Eizaguirre, E.; Pareja, M.J.; Romero-Gómez, M.; Jimenez, M.A.; et al. The Altered Serum Lipidome and Its Diagnostic Potential for Non-Alcoholic Fatty Liver (NAFL)-Associated Hepatocellular Carcinoma. eBioMedicine 2021, 73, 103661. [Google Scholar] [CrossRef] [PubMed]
- Hirose, Y.; Nagahashi, M.; Katsuta, E.; Yuza, K.; Miura, K.; Sakata, J.; Kobayashi, T.; Ichikawa, H.; Shimada, Y.; Kameyama, H.; et al. Generation of Sphingosine-1-Phosphate Is Enhanced in Biliary Tract Cancer Patients and Is Associated with Lymphatic Metastasis. Sci. Rep. 2018, 8, 10814. [Google Scholar] [CrossRef]
- Cheng, J.-C.; Wang, E.Y.; Yi, Y.; Thakur, A.; Tsai, S.-H.; Hoodless, P.A. S1P Stimulates Proliferation by Upregulating CTGF Expression through S1PR2-Mediated YAP Activation. Mol. Cancer Res. 2018, 16, 1543–1555. [Google Scholar] [CrossRef]
- Zeng, Y.; Yao, X.; Chen, L.; Yan, Z.; Liu, J.; Zhang, Y.; Feng, T.; Wu, J.; Liu, X. Sphingosine-1-Phosphate Induced Epithelial-Mesenchymal Transition of Hepatocellular Carcinoma via an MMP-7/Syndecan-1/TGF-β Autocrine Loop. Oncotarget 2016, 7, 63324–63337. [Google Scholar] [CrossRef]
- Lu, Y.; Chen, J.; Huang, C.; Li, N.; Zou, L.; Chia, S.E.; Chen, S.; Yu, K.; Ling, Q.; Cheng, Q.; et al. Comparison of Hepatic and Serum Lipid Signatures in Hepatocellular Carcinoma Patients Leads to the Discovery of Diagnostic and Prognostic Biomarkers. Oncotarget 2017, 9, 5032–5043. [Google Scholar] [CrossRef]
- Oliviero, B.; Dei Cas, M.; Zulueta, A.; Maiello, R.; Villa, A.; Martinelli, C.; Del Favero, E.; Falleni, M.; Montavoci, L.; Varchetta, S.; et al. Ceramide Present in Cholangiocarcinoma-Derived Extracellular Vesicle Induces a pro-Inflammatory State in Monocytes. Sci. Rep. 2023, 13, 7766. [Google Scholar] [CrossRef]
- Eggens, I.; Ekström, T.J.; Aberg, F. Studies on the Biosynthesis of Polyisoprenols, Cholesterol and Ubiquinone in Highly Differentiated Human Hepatomas. J. Exp. Pathol. 1990, 71, 219–232. [Google Scholar]
- Sharif, A.W.; Williams, H.R.T.; Lampejo, T.; Khan, S.A.; Bansi, D.S.; Westaby, D.; Thillainayagam, A.V.; Thomas, H.C.; Cox, I.J.; Taylor-Robinson, S.D. Metabolic Profiling of Bile in Cholangiocarcinoma Using in Vitro Magnetic Resonance Spectroscopy. HPB 2010, 12, 396–402. [Google Scholar] [CrossRef] [PubMed]
- Jia, W.; Xie, G.; Jia, W. Bile Acid–Microbiota Cross-Talk in Gastrointestinal Inflammation and Carcinogenesis. Nat. Rev. Gastroenterol. Hepatol. 2018, 15, 111–128. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Lv, G.; Li, R.; Liu, W.; Zong, C.; Ye, F.; Li, X.; Yang, X.; Jiang, J.; Hou, X.; et al. Glycochenodeoxycholate Promotes Hepatocellular Carcinoma Invasion and Migration by AMPK/mTOR Dependent Autophagy Activation. Cancer Lett. 2019, 454, 215–223. [Google Scholar] [CrossRef]
- Labib, P.L.; Goodchild, G.; Pereira, S.P. Molecular Pathogenesis of Cholangiocarcinoma. BMC Cancer 2019, 19, 185. [Google Scholar] [CrossRef]
- Liu, R.; Zhao, R.; Zhou, X.; Liang, X.; Campbell, D.J.; Zhang, X.; Zhang, L.; Shi, R.; Wang, G.; Pandak, W.M.; et al. Conjugated Bile Acids Promote Cholangiocarcinoma Cell Invasive Growth through Activation of Sphingosine 1-Phosphate Receptor 2. Hepatology 2014, 60, 908–918. [Google Scholar] [CrossRef]
- Lo Re, O.; Douet, J.; Buschbeck, M.; Fusilli, C.; Pazienza, V.; Panebianco, C.; Castracani, C.C.; Mazza, T.; Li Volti, G.; Vinciguerra, M. Histone Variant macroH2A1 Rewires Carbohydrate and Lipid Metabolism of hepatocellular Carcinoma Cells Towards Cancer Stem Cells. Available online: https://colab.ws/articles/10.1080%2F15592294.2018.1514239 (accessed on 13 May 2025).
- Meng, H.; Shen, M.; Li, J.; Zhang, R.; Li, X.; Zhao, L.; Huang, G.; Liu, J. Novel SREBP1 Inhibitor Cinobufotalin Suppresses Proliferation of Hepatocellular Carcinoma by Targeting Lipogenesis. Eur. J. Pharmacol. 2021, 906, 174280. [Google Scholar] [CrossRef]
- Huang, C.-J.; Zhang, C.-Y.; Zhao, Y.-K.; Wang, D.; Zhuang, L.; Qian, L.; Xie, L.; Zhu, Y.; Meng, Z.-Q. Bufalin Inhibits Tumorigenesis and SREBP-1-Mediated Lipogenesis in Hepatocellular Carcinoma via Modulating the ATP1A1/CA2 Axis. Am. J. Chin. Med. 2023, 51, 461–485. [Google Scholar] [CrossRef]
- Cheng, L.; Deepak, R.N.V.K.; Wang, G.; Meng, Z.; Tao, L.; Xie, M.; Chi, W.; Zhang, Y.; Yang, M.; Liao, Y.; et al. Hepatic Mitochondrial NAD+ Transporter SLC25A47 Activates AMPKα Mediating Lipid Metabolism and Tumorigenesis. Hepatology 2023, 78, 1828–1842. [Google Scholar] [CrossRef]
- Zhou, X.; Li, Y.; Yang, C.; Chen, D.; Wang, T.; Liu, T.; Yan, W.; Su, Z.; Peng, B.; Ren, X. Cordycepin Reprogramming Lipid Metabolism to Block Metastasis and EMT via ERO1A/mTOR/SREBP1 Axis in Cholangiocarcinoma. Life Sci. 2023, 327, 121698. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-D.; Wu, H.; Fu, G.-B.; Zhang, H.-L.; Zhou, X.; Tang, L.; Dong, L.-W.; Qin, C.-J.; Huang, S.; Zhao, L.-H.; et al. Acetyl-Coenzyme A Carboxylase Alpha Promotion of Glucose-Mediated Fatty Acid Synthesis Enhances Survival of Hepatocellular Carcinoma in Mice and Patients. Hepatology 2016, 63, 1272–1286. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Pilo, G.M.; Li, X.; Cigliano, A.; Latte, G.; Che, L.; Joseph, C.; Mela, M.; Wang, C.; Jiang, L.; et al. Inactivation of Fatty Acid Synthase Impairs Hepatocarcinogenesis Driven by AKT in Mice and Humans. J. Hepatol. 2016, 64, 333–341. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Zhou, Y.; Xu, H.; Wang, X.; Zhang, Y.; Shang, R.; O’Farrell, M.; Roessler, S.; Sticht, C.; Stahl, A.; et al. Therapeutic Efficacy of FASN Inhibition in Preclinical Models of HCC. Hepatology 2022, 76, 951. [Google Scholar] [CrossRef]
- Terry, A.R.; Hay, N. Emerging Targets in Lipid Metabolism for Cancer Therapy. Trends Pharmacol. Sci. 2024, 45, 537–551. [Google Scholar] [CrossRef]
- Loomba, R.; Mohseni, R.; Lucas, K.J.; Gutierrez, J.A.; Perry, R.G.; Trotter, J.F.; Rahimi, R.S.; Harrison, S.A.; Ajmera, V.; Wayne, J.D.; et al. TVB-2640 (FASN Inhibitor) for the Treatment of Nonalcoholic Steatohepatitis: FASCINATE-1, a Randomized, Placebo-Controlled Phase 2a Trial. Gastroenterology 2021, 161, 1475–1486. [Google Scholar] [CrossRef]
- Li, X.; He, W.; Chen, X.; Zhang, Y.; Zhang, J.; Liu, F.; Li, J.; Zhao, D.; Xia, P.; Ma, W.; et al. TRIM45 Facilitates NASH-Progressed HCC by Promoting Fatty Acid Synthesis via Catalyzing FABP5 Ubiquitylation. Oncogene 2024, 43, 2063–2077. [Google Scholar] [CrossRef]
- Zhou, Y.; Guo, Y.; Zhu, Y.; Sun, Y.; Li, W.; Li, Z.; Wei, L. Dual PPARγ/ɑ Agonist Oroxyloside Suppresses Cell Cycle Progression by Glycolipid Metabolism Switch-Mediated Increase of Reactive Oxygen Species Levels. Free Radic. Biol. Med. 2021, 167, 205–217. [Google Scholar] [CrossRef]
- Pan, Y.; Li, Y.; Fan, H.; Cui, H.; Chen, Z.; Wang, Y.; Jiang, M.; Wang, G. Roles of the Peroxisome Proliferator-Activated Receptors (PPARs) in the Pathogenesis of Hepatocellular Carcinoma (HCC). Biomed. Pharmacother. 2024, 177, 117089. [Google Scholar] [CrossRef]
- Yarchoan, M.; Powderly, J.D.; Bastos, B.R.; Karasic, T.B.; Crysler, O.V.; Munster, P.N.; McKean, M.A.; Emens, L.A.; Saenger, Y.M.; Ged, Y.; et al. First-in-Human Phase I Trial of TPST-1120, an Inhibitor of PPARα, as Monotherapy or in Combination with Nivolumab, in Patients with Advanced Solid Tumors. Cancer Res. Commun. 2024, 4, 1100–1110. [Google Scholar] [CrossRef]
- Lu, X.; Paliogiannis, P.; Calvisi, D.F.; Chen, X. Role of the mTOR Pathway in Liver Cancer: From Molecular Genetics to Targeted Therapies. Hepatology 2021, 73, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Wu, Z. Sphingosine Kinase 1 Overexpression Is Associated with Poor Prognosis and Oxaliplatin Resistance in Hepatocellular Carcinoma. Exp. Ther. Med. 2018, 15, 5371–5376. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Zhang, S.; Ma, D.; Yan, D.; Zhang, G.; Cao, Y.; Wang, Z.; Wu, J.; Jiang, C. Targeting SphK2 Reverses Acquired Resistance of Regorafenib in Hepatocellular Carcinoma. Front. Oncol. 2020, 10, 694. [Google Scholar] [CrossRef] [PubMed]
- Britten, C.D.; Garrett-Mayer, E.; Chin, S.H.; Shirai, K.; Ogretmen, B.; Bentz, T.A.; Brisendine, A.; Anderton, K.; Cusack, S.L.; Maines, L.W.; et al. A Phase I Study of ABC294640, a First-in-Class Sphingosine Kinase-2 Inhibitor, in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2017, 23, 4642–4650. [Google Scholar] [CrossRef]
- Yokohama, K.; Fukunishi, S.; Ii, M.; Nakamura, K.; Ohama, H.; Tsuchimoto, Y.; Asai, A.; Tsuda, Y.; Higuchi, K. Rosuvastatin as a Potential Preventive Drug for the Development of Hepatocellular Carcinoma Associated with Non-Alcoholic Fatty Liver Disease in Mice. Int. J. Mol. Med. 2016, 38, 1499–1506. [Google Scholar] [CrossRef]
- Cheng, Y.; Luo, R.C.; Zheng, H.; Wang, B.; Liu, Y.; Liu, D.L.; Chen, J.; Xu, W.; Li, A.; Zhu, Y. Synergistic Anti-Tumor Efficacy of Sorafenib and Fluvastatin in Hepatocellular Carcinoma. Oncotarget 2017, 8, 23265–23276. [Google Scholar] [CrossRef]
- Kawata, S.; Yamasaki, E.; Nagase, T.; Inui, Y.; Ito, N.; Matsuda, Y.; Inada, M.; Tamura, S.; Noda, S.; Imai, Y.; et al. Effect of Pravastatin on Survival in Patients with Advanced Hepatocellular Carcinoma. A Randomized Controlled Trial. Br. J. Cancer 2001, 84, 886–891. [Google Scholar] [CrossRef]
- Peng, Y.-C.; Lin, C.-L.; Hsu, W.-Y.; Chang, C.-S.; Yeh, H.-Z.; Tung, C.-F.; Wu, Y.-L.; Sung, F.-C.; Kao, C.-H. Statins Are Associated with a Reduced Risk of Cholangiocarcinoma: A Population-Based Case–Control Study. Br. J. Clin. Pharmacol. 2015, 80, 755–761. [Google Scholar] [CrossRef]
- Liu, Z.; Alsaggaf, R.; McGlynn, K.A.; Anderson, L.; Tsai, H.-T.; Zhu, B.; Zhu, Y.; Mbulaiteye, S.M.; Gadalla, S.M.; Koshiol, J. Statin Use and Reduced Risk of Biliary Tract Cancers in the United Kingdom Clinical Practice Research Datalink. Gut 2019, 68, 1458–1464. [Google Scholar] [CrossRef]
- Kamigaki, M.; Sasaki, T.; Serikawa, M.; Inoue, M.; Kobayashi, K.; Itsuki, H.; Minami, T.; Yukutake, M.; Okazaki, A.; Ishigaki, T.; et al. Statins Induce Apoptosis and Inhibit Proliferation in Cholangiocarcinoma Cells. Int. J. Oncol. 2011, 39, 561–568. [Google Scholar] [CrossRef][Green Version]
- Kitagawa, K.; Moriya, K.; Kaji, K.; Saikawa, S.; Sato, S.; Nishimura, N.; Namisaki, T.; Akahane, T.; Mitoro, A.; Yoshiji, H. Atorvastatin Augments Gemcitabine-Mediated Anti-Cancer Effects by Inhibiting Yes-Associated Protein in Human Cholangiocarcinoma Cells. Int. J. Mol. Sci. 2020, 21, 7588. [Google Scholar] [CrossRef] [PubMed]
- Krenkel, O.; Tacke, F. Liver Macrophages in Tissue Homeostasis and Disease. Nat. Rev. Immunol. 2017, 17, 306–321. [Google Scholar] [CrossRef] [PubMed]
- Blériot, C.; Ginhoux, F. Understanding the Heterogeneity of Resident Liver Macrophages. Front. Immunol. 2019, 10, 2694. [Google Scholar] [CrossRef] [PubMed]
- Guillot, A.; Tacke, F. Liver Macrophages: Old Dogmas and New Insights. Hepatol. Commun. 2019, 3, 730–743. [Google Scholar] [CrossRef] [PubMed]
- Wen, Y.; Lambrecht, J.; Ju, C.; Tacke, F. Hepatic Macrophages in Liver Homeostasis and Diseases-Diversity, Plasticity and Therapeutic Opportunities. Cell Mol. Immunol. 2021, 18, 45–56. [Google Scholar] [CrossRef]
- MacParland, S.A.; Liu, J.C.; Ma, X.-Z.; Innes, B.T.; Bartczak, A.M.; Gage, B.K.; Manuel, J.; Khuu, N.; Echeverri, J.; Linares, I.; et al. Single Cell RNA Sequencing of Human Liver Reveals Distinct Intrahepatic Macrophage Populations. Nat. Commun. 2018, 9, 4383. [Google Scholar] [CrossRef]
- Andrews, T.S.; Nakib, D.; Perciani, C.T.; Ma, X.Z.; Liu, L.; Winter, E.; Camat, D.; Chung, S.W.; Lumanto, P.; Manuel, J.; et al. Single-Cell, Single-Nucleus, and Spatial Transcriptomics Characterization of the Immunological Landscape in the Healthy and PSC Human Liver. J. Hepatol. 2024, 80, 730–743. [Google Scholar] [CrossRef]
- de Visser, K.E.; Joyce, J.A. The Evolving Tumor Microenvironment: From Cancer Initiation to Metastatic Outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumor-Associated Macrophages as Treatment Targets in Oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Christofides, A.; Strauss, L.; Yeo, A.; Cao, C.; Charest, A.; Boussiotis, V.A. The Complex Role of Tumor-Infiltrating Macrophages. Nat. Immunol. 2022, 23, 1148–1156. [Google Scholar] [CrossRef]
- Basak, U.; Sarkar, T.; Mukherjee, S.; Chakraborty, S.; Dutta, A.; Dutta, S.; Nayak, D.; Kaushik, S.; Das, T.; Sa, G. Tumor-Associated Macrophages: An Effective Player of the Tumor Microenvironment. Front. Immunol. 2023, 14, 1295257. [Google Scholar] [CrossRef] [PubMed]
- Donadon, M.; Torzilli, G.; Cortese, N.; Soldani, C.; Di Tommaso, L.; Franceschini, B.; Carriero, R.; Barbagallo, M.; Rigamonti, A.; Anselmo, A.; et al. Macrophage Morphology Correlates with Single-Cell Diversity and Prognosis in Colorectal Liver Metastasis. J. Exp. Med. 2020, 217, e20191847. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Sica, A.; Mantovani, A.; Locati, M. Macrophage Activation and Polarization. Front. Biosci. 2008, 13, 453–461. [Google Scholar] [CrossRef]
- Wu, K.; Lin, K.; Li, X.; Yuan, X.; Xu, P.; Ni, P.; Xu, D. Redefining Tumor-Associated Macrophage Subpopulations and Functions in the Tumor Microenvironment. Front. Immunol. 2020, 11, 1731. [Google Scholar] [CrossRef] [PubMed]
- Locati, M.; Curtale, G.; Mantovani, A. Diversity, Mechanisms and Significance of Macrophage Plasticity. Annu. Rev. Pathol. 2020, 15, 123–147. [Google Scholar] [CrossRef]
- Gao, J.; Liang, Y.; Wang, L. Shaping Polarization of Tumor-Associated Macrophages in Cancer Immunotherapy. Front. Immunol. 2022, 13, 888713. [Google Scholar] [CrossRef]
- Wang, J.; Zhu, N.; Su, X.; Gao, Y.; Yang, R. Novel Tumor-Associated Macrophage Populations and Subpopulations by Single Cell RNA Sequencing. Front. Immunol. 2024, 14, 1264774. [Google Scholar] [CrossRef]
- Zhang, Q.; He, Y.; Luo, N.; Patel, S.J.; Han, Y.; Gao, R.; Modak, M.; Carotta, S.; Haslinger, C.; Kind, D.; et al. Landscape and Dynamics of Single Immune Cells in Hepatocellular Carcinoma. Cell 2019, 179, 829–845.e20. [Google Scholar] [CrossRef]
- Giraud, J.; Chalopin, D.; Ramel, E.; Boyer, T.; Zouine, A.; Derieppe, M.-A.; Larmonier, N.; Adotevi, O.; Le Bail, B.; Blanc, J.-F.; et al. THBS1+ Myeloid Cells Expand in SLD Hepatocellular Carcinoma and Contribute to Immunosuppression and Unfavorable Prognosis through TREM1. Cell Rep. 2024, 43, 113773. [Google Scholar] [CrossRef]
- Ho, D.W.-H.; Tsui, Y.-M.; Chan, L.-K.; Sze, K.M.-F.; Zhang, X.; Cheu, J.W.-S.; Chiu, Y.-T.; Lee, J.M.-F.; Chan, A.C.-Y.; Cheung, E.T.-Y.; et al. Single-Cell RNA Sequencing Shows the Immunosuppressive Landscape and Tumor Heterogeneity of HBV-Associated Hepatocellular Carcinoma. Nat. Commun. 2021, 12, 3684. [Google Scholar] [CrossRef]
- Sun, Y.; Wu, L.; Zhong, Y.; Zhou, K.; Hou, Y.; Wang, Z.; Zhang, Z.; Xie, J.; Wang, C.; Chen, D.; et al. Single-Cell Landscape of the Ecosystem in Early-Relapse Hepatocellular Carcinoma. Cell 2021, 184, 404–421.e16. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zheng, Z.; Liu, H.; Zhang, Y.; Kang, J.; Kong, X.; Rong, D.; Sun, G.; Sun, G.; Liu, L.; et al. Inhibition of APOC1 Promotes the Transformation of M2 into M1 Macrophages via the Ferroptosis Pathway and Enhances Anti-PD1 Immunotherapy in Hepatocellular Carcinoma Based on Single-Cell RNA Sequencing. Redox Biol. 2022, 56, 102463. [Google Scholar] [CrossRef]
- Lu, Y.; Yang, A.; Quan, C.; Pan, Y.; Zhang, H.; Li, Y.; Gao, C.; Lu, H.; Wang, X.; Cao, P.; et al. A Single-Cell Atlas of the Multicellular Ecosystem of Primary and Metastatic Hepatocellular Carcinoma. Nat. Commun. 2022, 13, 4594. [Google Scholar] [CrossRef]
- Liu, Y.; Xun, Z.; Ma, K.; Liang, S.; Li, X.; Zhou, S.; Sun, L.; Liu, Y.; Du, Y.; Guo, X.; et al. Identification of a Tumour Immune Barrier in the HCC Microenvironment That Determines the Efficacy of Immunotherapy. J. Hepatol. 2023, 78, 770–782. [Google Scholar] [CrossRef]
- Li, K.; Zhang, R.; Wen, F.; Zhao, Y.; Meng, F.; Li, Q.; Hao, A.; Yang, B.; Lu, Z.; Cui, Y.; et al. Single-Cell Dissection of the Multicellular Ecosystem and Molecular Features Underlying Microvascular Invasion in HCC. Hepatology 2024, 79, 1293–1309. [Google Scholar] [CrossRef] [PubMed]
- Zhou, K.-Q.; Zhong, Y.-C.; Song, M.-F.; Sun, Y.-F.; Zhu, W.; Cheng, J.-W.; Xu, Y.; Zhang, Z.-F.; Wang, P.-X.; Tang, Z.; et al. Distinct Immune Microenvironment of Venous Tumor Thrombus in Hepatocellular Carcinoma at Single-Cell Resolution. Hepatology 2024, 10.1097/HEP.0000000000001182. [Google Scholar] [CrossRef]
- Jiang, S.; Lu, H.; Pan, Y.; Yang, A.; Aikemu, A.; Li, H.; Hao, R.; Huang, Q.; Qi, X.; Tao, Z.; et al. Characterization of the Distinct Immune Microenvironments between Hepatocellular Carcinoma and Intrahepatic Cholangiocarcinoma. Cancer Lett. 2024, 588, 216799. [Google Scholar] [CrossRef] [PubMed]
- Tomlinson, J.L.; Valle, J.W.; Ilyas, S.I. Immunobiology of Cholangiocarcinoma. J. Hepatol. 2023, 79, 867–875. [Google Scholar] [CrossRef]
- Dong, Z.-R.; Zhang, M.-Y.; Qu, L.-X.; Zou, J.; Yang, Y.-H.; Ma, Y.-L.; Yang, C.-C.; Cao, X.-L.; Wang, L.-Y.; Zhang, X.-L.; et al. Spatial Resolved Transcriptomics Reveals Distinct Cross-Talk between Cancer Cells and Tumor-Associated Macrophages in Intrahepatic Cholangiocarcinoma. Biomark. Res. 2024, 12, 100. [Google Scholar] [CrossRef]
- Kim, J.H.; Oh, S.-H.; Kim, E.-J.; Park, S.J.; Hong, S.P.; Cheon, J.H.; Kim, T.I.; Kim, W.H. The Role of Myofibroblasts in Upregulation of S100A8 and S100A9 and the Differentiation of Myeloid Cells in the Colorectal Cancer Microenvironment. Biochem. Biophys. Res. Commun. 2012, 423, 60–66. [Google Scholar] [CrossRef]
- Yuan, H.; Lin, Z.; Liu, Y.; Jiang, Y.; Liu, K.; Tu, M.; Yao, N.; Qu, C.; Hong, J. Intrahepatic Cholangiocarcinoma Induced M2-Polarized Tumor-Associated Macrophages Facilitate Tumor Growth and Invasiveness. Cancer Cell Int. 2020, 20, 586. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.; Tao, C.; Li, L.; Xie, T.; Tang, L.; Han, X.; Shi, Y. The Co-Location of MARCO+ Tumor-Associated Macrophages and CTSE+ Tumor Cells Determined the Poor Prognosis in Intrahepatic Cholangiocarcinoma. Hepatology 2024, 10.1097/HEP.0000000000001138. [Google Scholar] [CrossRef]
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and Risk Factors. Liver Int. 2019, 39, 19–31. [Google Scholar] [CrossRef]
- Clements, O.; Eliahoo, J.; Kim, J.U.; Taylor-Robinson, S.D.; Khan, S.A. Risk Factors for Intrahepatic and Extrahepatic Cholangiocarcinoma: A Systematic Review and Meta-Analysis. J. Hepatol. 2020, 72, 95–103. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Y.; Lin, Z.; Deng, X.; Ren, X.; Huang, M.; Li, S.; Zhou, Q.; Fang, F.; Yang, Q.; et al. FASN-Mediated Fatty Acid Biosynthesis Remodels Immune Environment in Clonorchis sinensis Infection-Related Intrahepatic Cholangiocarcinoma. J. Hepatol. 2024, 81, 265–277. [Google Scholar] [CrossRef]
- Alvisi, G.; Termanini, A.; Soldani, C.; Portale, F.; Carriero, R.; Pilipow, K.; Costa, G.; Polidoro, M.; Franceschini, B.; Malenica, I.; et al. Multimodal Single-Cell Profiling of Intrahepatic Cholangiocarcinoma Defines Hyperactivated Tregs as a Potential Therapeutic Target. J. Hepatol. 2022, 77, 1359–1372. [Google Scholar] [CrossRef]
- Vitale, I.; Manic, G.; Coussens, L.M.; Kroemer, G.; Galluzzi, L. Macrophages and Metabolism in the Tumor Microenvironment. Cell Metab. 2019, 30, 36–50. [Google Scholar] [CrossRef] [PubMed]
- Batista-Gonzalez, A.; Vidal, R.; Criollo, A.; Carreño, L.J. New Insights on the Role of Lipid Metabolism in the Metabolic Reprogramming of Macrophages. Front. Immunol. 2020, 10, 2993. [Google Scholar] [CrossRef]
- Yang, H.; Kim, C.; Zou, W. Metabolism and Macrophages in the Tumor Microenvironment. Curr. Opin. Immunol. 2024, 91, 102491. [Google Scholar] [CrossRef]
- Furuhashi, M.; Hotamisligil, G.S. Fatty Acid-Binding Proteins: Role in Metabolic Diseases and Potential as Drug Targets. Nat. Rev. Drug Discov. 2008, 7, 489. [Google Scholar] [CrossRef]
- Storch, J.; Corsico, B. The Multifunctional Family of Mammalian Fatty Acid–Binding Proteins. Annu. Rev. Nutr. 2023, 43, 25–54. [Google Scholar] [CrossRef] [PubMed]
- Tang, W.; Sun, G.; Ji, G.-W.; Feng, T.; Zhang, Q.; Cao, H.; Wu, W.; Zhang, X.; Liu, C.; Liu, H.; et al. Single-Cell RNA-Sequencing Atlas Reveals an FABP1-Dependent Immunosuppressive Environment in Hepatocellular Carcinoma. J. Immunother. Cancer 2023, 11, e007030. [Google Scholar] [CrossRef]
- Liu, J.; Sun, B.; Guo, K.; Yang, Z.; Zhao, Y.; Gao, M.; Yin, Z.; Jiang, K.; Dong, C.; Gao, Z.; et al. Lipid-Related FABP5 Activation of Tumor-Associated Monocytes Fosters Immune Privilege via PD-L1 Expression on Treg Cells in Hepatocellular Carcinoma. Cancer Gene Ther. 2022, 29, 1951–1960. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Deng, B.; Zhao, W.; Guo, Y.; Wan, Y.; Wu, Z.; Su, S.; Gu, J.; Hu, X.; Feng, W.; et al. FABP5+ Lipid-Loaded Macrophages Process Tumour-Derived Unsaturated Fatty Acid Signal to Suppress T-Cell Antitumour Immunity. J. Hepatol. 2025, 82, 676–689. [Google Scholar] [CrossRef]
- Sun, J.; Esplugues, E.; Bort, A.; Cardelo, M.P.; Ruz-Maldonado, I.; Fernández-Tussy, P.; Wong, C.; Wang, H.; Ojima, I.; Kaczocha, M.; et al. Fatty Acid Binding Protein 5 Suppression Attenuates Obesity-Induced Hepatocellular Carcinoma by Promoting Ferroptosis and Intratumoral Immune Rewiring. Nat. Metab. 2024, 6, 741–763. [Google Scholar] [CrossRef]
- Yang, P.; Qin, H.; Li, Y.; Xiao, A.; Zheng, E.; Zeng, H.; Su, C.; Luo, X.; Lu, Q.; Liao, M.; et al. CD36-Mediated Metabolic Crosstalk between Tumor Cells and Macrophages Affects Liver Metastasis. Nat. Commun. 2022, 13, 5782. [Google Scholar] [CrossRef]
- Murai, H.; Kodama, T.; Maesaka, K.; Tange, S.; Motooka, D.; Suzuki, Y.; Shigematsu, Y.; Inamura, K.; Mise, Y.; Saiura, A.; et al. Multiomics Identifies the Link between Intratumor Steatosis and the Exhausted Tumor Immune Microenvironment in Hepatocellular Carcinoma. Hepatology 2023, 77, 77–91. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Chen, W.; Qiao, S.; Zou, H.; Yu, X.; Yang, Y.; Li, Z.; Wang, J.; Chen, M.; Xu, J.; et al. Lipid Droplet Accumulation Mediates Macrophage Survival and Treg Recruitment via the CCL20/CCR6 Axis in Human Hepatocellular Carcinoma. Cell Mol. Immunol. 2024, 21, 1120–1130. [Google Scholar] [CrossRef]
- Li, Z.; Wang, Y.; Xing, R.; Zeng, H.; Yu, X.-J.; Zhang, Y.-J.; Xu, J.; Zheng, L. Cholesterol Efflux Drives the Generation of Immunosuppressive Macrophages to Promote the Progression of Human Hepatocellular Carcinoma. Cancer Immunol. Res. 2023, 11, 1400–1413. [Google Scholar] [CrossRef]
- Keawvilai, P.; Kueanjinda, P.; Klomsing, J.; Palaga, T. Coculturing Liver Cancer Cells and Monocytes in Spheroids Conditions Monocytes to Adopt Tumor-Associated Macrophage Phenotypes That Favor Tumor Growth via Cholesterol Metabolism. J. Leukoc. Biol. 2024, 115, 344–357. [Google Scholar] [CrossRef]
- Shmarakov, I.O.; Jiang, H.; Liu, J.; Fernandez, E.J.; Blaner, W.S. Hepatic Stellate Cell Activation: A Source for Bioactive Lipids. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 629–642. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, T.; Friedman, S.L. Mechanisms of Hepatic Stellate Cell Activation. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 397–411. [Google Scholar] [CrossRef] [PubMed]
- Jeong, J.-M.; Choi, S.E.; Shim, Y.-R.; Kim, H.-H.; Lee, Y.-S.; Yang, K.; Kim, K.; Kim, M.J.; Chung, K.P.S.; Kim, S.-H.; et al. CX3CR1+ Macrophages Interact with HSCs to Promote HCC through CD8+ T-Cell Suppression. Hepatology 2024, 10.1097/HEP.0000000000001021. [Google Scholar] [CrossRef]
- Chen, J.; Tang, Y.; Qin, D.; Yu, X.; Tong, H.; Tang, C.; Tang, Z. ALOX5 Acts as a Key Role in Regulating the Immune Microenvironment in Intrahepatic Cholangiocarcinoma, Recruiting Tumor-Associated Macrophages through PI3K Pathway. J. Transl. Med. 2023, 21, 923. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Oliviero, B.; Caretti, A.; Mondelli, M.U.; Mantovani, S. Lipid Metabolism Reprogramming in Tumor-Associated Macrophages Modulates Their Function in Primary Liver Cancers. Cancers 2025, 17, 1858. https://doi.org/10.3390/cancers17111858
Oliviero B, Caretti A, Mondelli MU, Mantovani S. Lipid Metabolism Reprogramming in Tumor-Associated Macrophages Modulates Their Function in Primary Liver Cancers. Cancers. 2025; 17(11):1858. https://doi.org/10.3390/cancers17111858
Chicago/Turabian StyleOliviero, Barbara, Anna Caretti, Mario U. Mondelli, and Stefania Mantovani. 2025. "Lipid Metabolism Reprogramming in Tumor-Associated Macrophages Modulates Their Function in Primary Liver Cancers" Cancers 17, no. 11: 1858. https://doi.org/10.3390/cancers17111858
APA StyleOliviero, B., Caretti, A., Mondelli, M. U., & Mantovani, S. (2025). Lipid Metabolism Reprogramming in Tumor-Associated Macrophages Modulates Their Function in Primary Liver Cancers. Cancers, 17(11), 1858. https://doi.org/10.3390/cancers17111858






