Clinical and Morphological Features of ER-Positive HER2-Negative Breast Tumors with PIK3CA Mutations in Russian Patients
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Samuels, Y.; Wang, Z.; Bardelli, A.; Silliman, N.; Ptak, J.; Szabo, S.; Yan, H.; Gazdar, A.; Powell, S.M.; Riggins, G.J.; et al. High frequency of mutations of the PIK3CA gene in human cancers. Science 2004, 304, 554. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.; Rubovszky, G.; Campone, M.; Loibl, S.; Rugo, H.S.; Iwata, H.; Conte, P.; Mayer, I.A.; Kaufman, B.; et al. Alpelisib for PIK3CA-Mutated, Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2019, 380, 1929–1940. [Google Scholar] [CrossRef] [PubMed]
- André, F.; Ciruelos, E.M.; Juric, D.; Loibl, S.; Campone, M.; Mayer, I.A.; Rubovszky, G.; Yamashita, T.; Kaufman, B.; Lu, Y.S.; et al. Alpelisib plus fulvestrant for PIK3CA-mutated, hormone receptor-positive, human epidermal growth factor receptor-2-negative advanced breast cancer: Final overall survival results from SOLAR-1. Ann. Oncol. 2021, 32, 208–217. [Google Scholar] [CrossRef]
- Turner, N.C.; Oliveira, M.; Howell, S.J.; Dalenc, F.; Cortes, J.; Gomez Moreno, H.L.; Hu, X.; Jhaveri, K.; Krivorotko, P.; Loibl, S.; et al. Capivasertib in Hormone Receptor-Positive Advanced Breast Cancer. N. Engl. J. Med. 2023, 388, 2058–2070. [Google Scholar] [CrossRef] [PubMed]
- Hao, C.; Wei, Y.; Meng, W.; Zhang, J.; Yang, X. PI3K/AKT/mTOR inhibitors for hormone receptor-positive advanced breast cancer. Cancer Treat. Rev. 2025, 132, 102861. [Google Scholar] [CrossRef]
- Jhaveri, K.L.; Accordino, M.K.; Bedard, P.L.; Cervantes, A.; Gambardella, V.; Hamilton, E.; Italiano, A.; Kalinsky, K.; Krop, I.E.; Oliveira, M.; et al. Phase I/Ib Trial of Inavolisib Plus Palbociclib and Endocrine Therapy for PIK3CA-Mutated, Hormone Receptor-Positive, Human Epidermal Growth Factor Receptor 2-Negative Advanced or Metastatic Breast Cancer. J. Clin. Oncol. 2024, 42, 3947–3956. [Google Scholar] [CrossRef]
- Ye, Y.; Huang, Z.; Zhang, M.; Li, J.; Zhang, Y.; Lou, C. Synergistic therapeutic potential of alpelisib in cancers (excluding breast cancer): Preclinical and clinical evidences. Biomed. Pharmacother. 2023, 159, 114183. [Google Scholar] [CrossRef]
- Konstantinopoulos, P.A.; Gonzalez-Martin, A.; Cruz, F.M.; Friedlander, M.; Glasspool, R.; Lorusso, D.; Marth, C.; Monk, B.J.; Kim, J.W.; Hinson, P.; et al. EPIK-O/ENGOT-OV61: Alpelisib plus olaparib vs cytotoxic chemotherapy in high-grade serous ovarian cancer (phase III study). Future Oncol. 2022, 18, 3481–3492. [Google Scholar] [CrossRef]
- Fujimoto, Y.; Morita, T.Y.; Ohashi, A.; Haeno, H.; Hakozaki, Y.; Fujii, M.; Kashima, Y.; Kobayashi, S.S.; Mukohara, T. Combination treatment with a PI3K/Akt/mTOR pathway inhibitor overcomes resistance to anti-HER2 therapy in PIK3CA-mutant HER2-positive breast cancer cells. Sci. Rep. 2020, 10, 21762. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Abramson, V.G.; Sanders, M.E.; Mayer, E.L.; Haddad, T.C.; Nanda, R.; Van Poznak, C.; Storniolo, A.M.; Nangia, J.R.; Gonzalez-Ericsson, P.I.; et al. TBCRC 032 IB/II Multicenter Study: Molecular Insights to AR Antagonist and PI3K Inhibitor Efficacy in Patients with AR(+) Metastatic Triple-Negative Breast Cancer. Clin. Cancer Res. 2020, 26, 2111–2123. [Google Scholar] [CrossRef]
- Berenjeno, I.M.; Piñeiro, R.; Castillo, S.D.; Pearce, W.; McGranahan, N.; Dewhurst, S.M.; Meniel, V.; Birkbak, N.J.; Lau, E.; Sansregret, L.; et al. Oncogenic PIK3CA induces centrosome amplification and tolerance to genome doubling. Nat. Commun. 2017, 8, 1773. [Google Scholar] [CrossRef] [PubMed]
- Sobral-Leite, M.; Salomon, I.; Opdam, M.; Kruger, D.T.; Beelen, K.J.; van der Noort, V.; van Vlierberghe, R.L.P.; Blok, E.J.; Giardiello, D.; Sanders, J.; et al. Cancer-immune interactions in ER-positive breast cancers: PI3K pathway alterations and tumor-infiltrating lymphocytes. Breast Cancer Res. 2019, 21, 90. [Google Scholar] [CrossRef] [PubMed]
- Koundouros, N.; Karali, E.; Tripp, A.; Valle, A.; Inglese, P.; Perry, N.J.S.; Magee, D.J.; Anjomani Virmouni, S.; Elder, G.A.; Tyson, A.L.; et al. Metabolic Fingerprinting Links Oncogenic PIK3CA with Enhanced Arachidonic Acid-Derived Eicosanoids. Cell 2020, 181, 1596–1611.e27. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.R.; Jiang, Y.Z.; Zuo, W.J.; Yu, K.D.; Shao, Z.M. PIK3CA mutations define favorable prognostic biomarkers in operable breast cancer: A systematic review and meta-analysis. Onco Targets Ther. 2014, 7, 543–552. [Google Scholar] [CrossRef]
- Mollon, L.E.; Anderson, E.J.; Dean, J.L.; Warholak, T.L.; Aizer, A.; Platt, E.A.; Tang, D.H.; Davis, L.E. A Systematic Literature Review of the Prognostic and Predictive Value of PIK3CA Mutations in HR+/HER2− Metastatic Breast Cancer. Clin. Breast Cancer 2020, 20, e232–e243. [Google Scholar] [CrossRef]
- Mosele, F.; Stefanovska, B.; Lusque, A.; Tran Dien, A.; Garberis, I.; Droin, N.; Le Tourneau, C.; Sablin, M.P.; Lacroix, L.; Enrico, D.; et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann. Oncol. 2020, 31, 377–386. [Google Scholar] [CrossRef]
- Shi, Q.; Xuhong, J.; Tian, H.; Qu, M.; Zhang, Y.; Jiang, J.; Qi, X. Predictive and prognostic value of PIK3CA mutations in HER2-positive breast cancer treated with tyrosine kinase inhibitors: A systematic review and meta-analysis. Biochim. Biophys. Acta Rev. Cancer 2023, 1878, 188847. [Google Scholar] [CrossRef]
- Loibl, S.; Majewski, I.; Guarneri, V.; Nekljudova, V.; Holmes, E.; Bria, E.; Denkert, C.; Schem, C.; Sotiriou, C.; Loi, S.; et al. PIK3CA mutations are associated with reduced pathological complete response rates in primary HER2-positive breast cancer: Pooled analysis of 967 patients from five prospective trials investigating lapatinib and trastuzumab. Ann. Oncol. 2016, 27, 1519–1525. [Google Scholar] [CrossRef]
- Fan, H.; Li, C.; Xiang, Q.; Xu, L.; Zhang, Z.; Liu, Q.; Zhang, T.; Zhou, Y.; Zhao, X.; Cui, Y. PIK3CA mutations and their response to neoadjuvant treatment in early breast cancer: A systematic review and meta-analysis. Thorac. Cancer 2018, 9, 571–579. [Google Scholar] [CrossRef]
- Kim, J.W.; Lim, A.R.; You, J.Y.; Lee, J.H.; Song, S.E.; Lee, N.K.; Jung, S.P.; Cho, K.R.; Kim, C.Y.; Park, K.H. PIK3CA Mutation is Associated with Poor Response to HER2-Targeted Therapy in Breast Cancer Patients. Cancer Res. Treat. 2023, 55, 531–541. [Google Scholar] [CrossRef]
- Zhao, L.; Vogt, P.K. Hot-spot mutations in p110alpha of phosphatidylinositol 3-kinase (pI3K): Differential interactions with the regulatory subunit p85 and with RAS. Cell Cycle 2010, 9, 596–600. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Razavi, P.; Johnson, J.L.; Shao, H.; Shah, H.; Antoine, A.; Ladewig, E.; Gorelick, A.; Lin, T.Y.; Toska, E.; et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kα inhibitors. Science 2019, 366, 714–723. [Google Scholar] [CrossRef] [PubMed]
- Sivakumar, S.; Jin, D.X.; Rathod, R.; Ross, J.; Cantley, L.C.; Scaltriti, M.; Chen, J.W.; Hutchinson, K.E.; Wilson, T.R.; Sokol, E.S.; et al. Genetic Heterogeneity and Tissue-specific Patterns of Tumors with Multiple PIK3CA Mutations. Clin. Cancer Res. 2023, 29, 1125–1136. [Google Scholar] [CrossRef] [PubMed]
- Garay, J.P.; Smith, R.; Devlin, K.; Hollern, D.P.; Liby, T.; Liu, M.; Boddapati, S.; Watson, S.S.; Esch, A.; Zheng, T.; et al. Sensitivity to targeted therapy differs between HER2-amplified breast cancer cells harboring kinase and helical domain mutations in PIK3CA. Breast Cancer Res. 2021, 23, 81. [Google Scholar] [CrossRef]
- Guo, S.; Loibl, S.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Denkert, C. PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane-Based Neoadjuvant Chemotherapy. Cancer Res. Treat. 2020, 52, 689–696. [Google Scholar] [CrossRef]
- Barbareschi, M.; Buttitta, F.; Felicioni, L.; Cotrupi, S.; Barassi, F.; Del Grammastro, M.; Ferro, A.; Dalla Palma, P.; Galligioni, E.; Marchetti, A. Different prognostic roles of mutations in the helical and kinase domains of the PIK3CA gene in breast carcinomas. Clin. Cancer Res. 2007, 13, 6064–6069. [Google Scholar] [CrossRef]
- Papaxoinis, G.; Kotoula, V.; Alexopoulou, Z.; Kalogeras, K.T.; Zagouri, F.; Timotheadou, E.; Gogas, H.; Pentheroudakis, G.; Christodoulou, C.; Koutras, A.; et al. Significance of PIK3CA Mutations in Patients with Early Breast Cancer Treated with Adjuvant Chemotherapy: A Hellenic Cooperative Oncology Group (HeCOG) Study. PLoS ONE 2015, 10, e0140293. [Google Scholar] [CrossRef][Green Version]
- Dogruluk, T.; Tsang, Y.H.; Espitia, M.; Chen, F.; Chen, T.; Chong, Z.; Appadurai, V.; Dogruluk, A.; Eterovic, A.K.; Bonnen, P.E.; et al. Identification of Variant-Specific Functions of PIK3CA by Rapid Phenotyping of Rare Mutations. Cancer Res. 2015, 75, 5341–5354. [Google Scholar] [CrossRef]
- Spangle, J.M.; Von, T.; Pavlick, D.C.; Khotimsky, A.; Zhao, J.J.; Roberts, T.M. PIK3CA C-terminal frameshift mutations are novel oncogenic events that sensitize tumors to PI3K-α inhibition. Proc. Natl. Acad. Sci. USA 2020, 117, 24427–24433. [Google Scholar] [CrossRef]
- Rugo, H.S.; Raskina, K.; Schrock, A.B.; Madison, R.W.; Graf, R.P.; Sokol, E.S.; Sivakumar, S.; Lee, J.K.; Fisher, V.; Oxnard, G.R.; et al. Biology and Targetability of the Extended Spectrum of PIK3CA Mutations Detected in Breast Carcinoma. Clin. Cancer Res. 2023, 29, 1056–1067. [Google Scholar] [CrossRef]
- Ademuyiwa, F.O.; Tao, Y.; Luo, J.; Weilbaecher, K.; Ma, C.X. Differences in the mutational landscape of triple-negative breast cancer in African Americans and Caucasians. Breast Cancer Res. Treat. 2017, 161, 491–499. [Google Scholar] [CrossRef] [PubMed]
- Omilian, A.R.; Wei, L.; Hong, C.C.; Bandera, E.V.; Liu, S.; Khoury, T.; Ambrosone, C.B.; Yao, S. Somatic mutations of triple-negative breast cancer: A comparison between Black and White women. Breast Cancer Res. Treat. 2020, 182, 503–509. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.W.; Murugesan, K.; Newberg, J.Y.; Sokol, E.S.; Savage, H.M.; Stout, T.J.; Maund, S.L.; Hutchinson, K.E. Comparison of PIK3CA Mutation Prevalence in Breast Cancer Across Predicted Ancestry Populations. JCO Precis. Oncol. 2022, 6, e2200341. [Google Scholar] [CrossRef] [PubMed]
- Russian Unified Interdepartmental Information and Statistical System. Available online: https://www.fedstat.ru/indicator/31517 (accessed on 5 February 2025).
- Kaprin, A.D.; Starinsky, V.V.; Shakhzadova, A.O. Malignant Tumors in Russia in 2023 (Morbidity and Mortality); P.A. Gertsen Moscow Research Institute of Oncology—Branch of the Federal State Budgetary Institution «Scientific Medical Research Centre of Radiology» of the Ministry of Health of Russia: Moscow, Russia, 2024; 276p. (In Russian) [Google Scholar]
- Lambertini, M.; Blondeaux, E.; Bisagni, G.; Mura, S.; De Placido, S.; De Laurentiis, M.; Fabi, A.; Rimanti, A.; Michelotti, A.; Mansutti, M.; et al. Prognostic and clinical impact of the endocrine resistance/sensitivity classification according to international consensus guidelines for advanced breast cancer: An individual patient-level analysis from the Mammella InterGruppo (MIG) and Gruppo Italiano Mammella (GIM) studies. EClinicalMedicine 2023, 59, 101931. [Google Scholar] [CrossRef]
- Anderson, E.J.; Mollon, L.E.; Dean, J.L.; Warholak, T.L.; Aizer, A.; Platt, E.A.; Tang, D.H.; Davis, L.E. A Systematic Review of the Prevalence and Diagnostic Workup of PIK3CA Mutations in HR+/HER2− Metastatic Breast Cancer. Int. J. Breast Cancer 2020, 2020, 3759179. [Google Scholar] [CrossRef]
- Martínez-Sáez, O.; Chic, N.; Pascual, T.; Adamo, B.; Vidal, M.; González-Farré, B.; Sanfeliu, E.; Schettini, F.; Conte, B.; Brasó-Maristany, F.; et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020, 22, 45. [Google Scholar] [CrossRef]
- Cancer Genome Atlas Network. Comprehensive molecular portraits of human breast tumours. Nature 2012, 490, 61–70. [Google Scholar] [CrossRef]
- Rajadurai, P.; Semiglazova, T.; Hegmane, A.; Karak, F.; Chiu, J.; Gupta, S.; Azim, H.; Kitzen, J.; Arnaud, A.; Haftchenary, S.; et al. PIK3CA Registry: A Non-interventional, Descriptive, Retrospective Cohort Study of PIK3CA Mutations in Patients With Hormone Receptor-Positive (HR+), Human Epidermal Growth Factor Receptor 2-Negative (HER2−) Advanced Breast Cancer (ABC). Cancer Res. 2022, 82, 5–13. [Google Scholar] [CrossRef]
- Gymnopoulos, M.; Elsliger, M.A.; Vogt, P.K. Rare cancer-specific mutations in PIK3CA show gain of function. Proc. Natl. Acad. Sci. USA 2007, 104, 5569–5574. [Google Scholar] [CrossRef]
- Jin, N.; Keam, B.; Cho, J.; Lee, M.J.; Kim, H.R.; Torosyan, H.; Jura, N.; Ng, P.K.; Mills, G.B.; Li, H.; et al. Therapeutic implications of activating noncanonical PIK3CA mutations in head and neck squamous cell carcinoma. J. Clin. Investig. 2021, 131, e150335. [Google Scholar] [CrossRef]
- Ikenoue, T.; Kanai, F.; Hikiba, Y.; Obata, T.; Tanaka, Y.; Imamura, J.; Ohta, M.; Jazag, A.; Guleng, B.; Tateishi, K.; et al. Functional analysis of PIK3CA gene mutations in human colorectal cancer. Cancer Res. 2005, 65, 4562–4567. [Google Scholar] [CrossRef] [PubMed]
- Ng, P.K.; Li, J.; Jeong, K.J.; Shao, S.; Chen, H.; Tsang, Y.H.; Sengupta, S.; Wang, Z.; Bhavana, V.H.; Tran, R.; et al. Systematic Functional Annotation of Somatic Mutations in Cancer. Cancer Cell 2018, 33, 450–462.e10. [Google Scholar] [CrossRef] [PubMed]
- Oda, K.; Okada, J.; Timmerman, L.; Rodriguez-Viciana, P.; Stokoe, D.; Shoji, K.; Taketani, Y.; Kuramoto, H.; Knight, Z.A.; Shokat, K.M.; et al. PIK3CA cooperates with other phosphatidylinositol 3′-kinase pathway mutations to effect oncogenic transformation. Cancer Res. 2008, 68, 8127–8136. [Google Scholar] [CrossRef] [PubMed]
- Miled, N.; Yan, Y.; Hon, W.C.; Perisic, O.; Zvelebil, M.; Inbar, Y.; Schneidman-Duhovny, D.; Wolfson, H.J.; Backer, J.M.; Williams, R.L. Mechanism of two classes of cancer mutations in the phosphoinositide 3-kinase catalytic subunit. Science 2007, 317, 239–242. [Google Scholar] [CrossRef]
- Rosin, J.; Svegrup, E.; Valachis, A.; Zerdes, I. Discordance of PIK3CA mutational status between primary and metastatic breast cancer: A systematic review and meta-analysis. Breast Cancer Res. Treat. 2023, 201, 161–169. [Google Scholar] [CrossRef]
- Dumont, A.G.; Dumont, S.N.; Trent, J.C. The favorable impact of PIK3CA mutations on survival: An analysis of 2587 patients with breast cancer. Chin. J. Cancer 2012, 31, 327–334. [Google Scholar] [CrossRef]
- Zardavas, D.; Te Marvelde, L.; Milne, R.L.; Fumagalli, D.; Fountzilas, G.; Kotoula, V.; Razis, E.; Papaxoinis, G.; Joensuu, H.; Moynahan, M.E.; et al. Tumor PIK3CA Genotype and Prognosis in Early-Stage Breast Cancer: A Pooled Analysis of Individual Patient Data. J. Clin. Oncol. 2018, 36, 981–990. [Google Scholar] [CrossRef]
- Fillbrunn, M.; Signorovitch, J.; André, F.; Wang, I.; Lorenzo, I.; Ridolfi, A.; Park, J.; Dua, A.; Rugo, H.S. PIK3CA mutation status, progression and survival in advanced HR+/HER2− breast cancer: A meta-analysis of published clinical trials. BMC Cancer 2022, 22, 1002. [Google Scholar] [CrossRef]
- Cizkova, M.; Susini, A.; Vacher, S.; Cizeron-Clairac, G.; Andrieu, C.; Driouch, K.; Fourme, E.; Lidereau, R.; Bièche, I. PIK3CA mutation impact on survival in breast cancer patients and in ERα, PR and ERBB2-based subgroups. Breast Cancer Res. 2012, 14, R28. [Google Scholar] [CrossRef]
- Kalinsky, K.; Jacks, L.M.; Heguy, A.; Patil, S.; Drobnjak, M.; Bhanot, U.K.; Hedvat, C.V.; Traina, T.A.; Solit, D.; Gerald, W.; et al. PIK3CA mutation associates with improved outcome in breast cancer. Clin. Cancer Res. 2009, 15, 5049–5059. [Google Scholar] [CrossRef]
- Wu, H.; Wang, W.; Du, J.; Li, H.; Wang, H.; Huang, L.; Xiang, H.; Xie, J.; Liu, X.; Li, H.; et al. The distinct clinicopathological and prognostic implications of PIK3CA mutations in breast cancer patients from Central China. Cancer Manag. Res. 2019, 11, 1473–1492. [Google Scholar] [CrossRef] [PubMed]
- Lv, W.; Du, C.; Zhang, Y.; Wu, F.; Jin, Y.; Chen, X.; Liu, X.; Feng, C.; Ma, X.; Zhang, S. Clinicopathological characteristics and prognostic analysis of PIK3CA mutation in breast cancer patients in Northwest China. Pathol. Res. Pract. 2022, 238, 154063. [Google Scholar] [CrossRef] [PubMed]
- Reinhardt, K.; Stückrath, K.; Hartung, C.; Kaufhold, S.; Uleer, C.; Hanf, V.; Lantzsch, T.; Peschel, S.; John, J.; Pöhler, M.; et al. PIK3CA-mutations in breast cancer. Breast Cancer Res. Treat. 2022, 196, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Toska, E.; Scaltriti, M. Overview of the relevance of PI3K pathway in HR-positive breast cancer. Ann. Oncol. 2019, 30 (Suppl. 10), x3–x11. [Google Scholar] [CrossRef]
- Beelen, K.; Hoefnagel, L.D.; Opdam, M.; Wesseling, J.; Sanders, J.; Vincent, A.D.; van Diest, P.J.; Linn, S.C. PI3K/AKT/mTOR pathway activation in primary and corresponding metastatic breast tumors after adjuvant endocrine therapy. Int. J. Cancer 2014, 135, 1257–1263. [Google Scholar] [CrossRef]
- Presti, D.; Quaquarini, E. The PI3K/AKT/mTOR and CDK4/6 Pathways in Endocrine Resistant HR+/HER2− Metastatic Breast Cancer: Biological Mechanisms and New Treatments. Cancers 2019, 11, 1242. [Google Scholar] [CrossRef]
- Stemke-Hale, K.; Gonzalez-Angulo, A.M.; Lluch, A.; Neve, R.M.; Kuo, W.L.; Davies, M.; Carey, M.; Hu, Z.; Guan, Y.; Sahin, A.; et al. An integrative genomic and proteomic analysis of PIK3CA, PTEN, and AKT mutations in breast cancer. Cancer Res. 2008, 68, 6084–6091. [Google Scholar] [CrossRef]
- Ramirez-Ardila, D.E.; Helmijr, J.C.; Look, M.P.; Lurkin, I.; Ruigrok-Ritstier, K.; van Laere, S.; Dirix, L.; Sweep, F.C.; Span, P.N.; Linn, S.C.; et al. Hotspot mutations in PIK3CA associate with first-line treatment outcome for aromatase inhibitors but not for tamoxifen. Breast Cancer Res. Treat. 2013, 139, 39–49. [Google Scholar] [CrossRef]
- Beelen, K.; Opdam, M.; Severson, T.M.; Koornstra, R.H.; Vincent, A.D.; Wesseling, J.; Muris, J.J.; Berns, E.M.; Vermorken, J.B.; van Diest, P.J.; et al. PIK3CA mutations, phosphatase and tensin homolog, human epidermal growth factor receptor 2, and insulin-like growth factor 1 receptor and adjuvant tamoxifen resistance in postmenopausal breast cancer patients. Breast Cancer Res. 2014, 16, R13. [Google Scholar] [CrossRef]
- Reinhardt, K.; Vetter, M.; Kaufhold, S.; Kantelhardt, E.; Thomssen, C. Adjuvant aromatase inhibitors in patients with PIK3CA mutation early breast cancer. Ann. Oncol. 2023, 34 (Suppl. 2), S278–S324. [Google Scholar] [CrossRef]
- Vasudevan, K.M.; Barbie, D.A.; Davies, M.A.; Rabinovsky, R.; McNear, C.J.; Kim, J.J.; Hennessy, B.T.; Tseng, H.; Pochanard, P.; Kim, S.Y.; et al. AKT-independent signaling downstream of oncogenic PIK3CA mutations in human cancer. Cancer Cell 2009, 16, 21–32. [Google Scholar] [CrossRef]
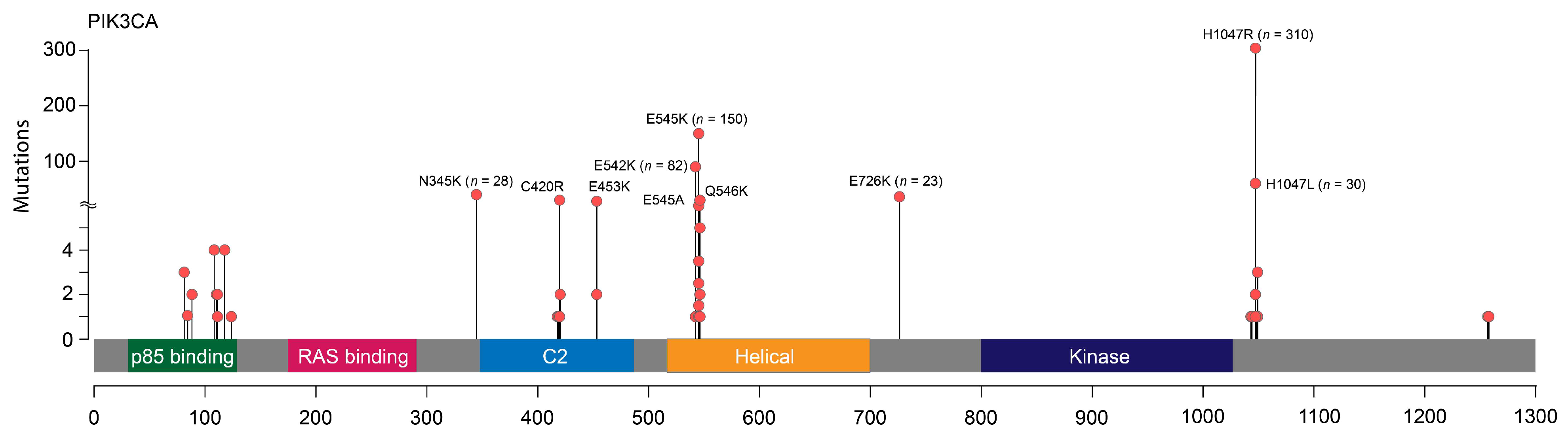
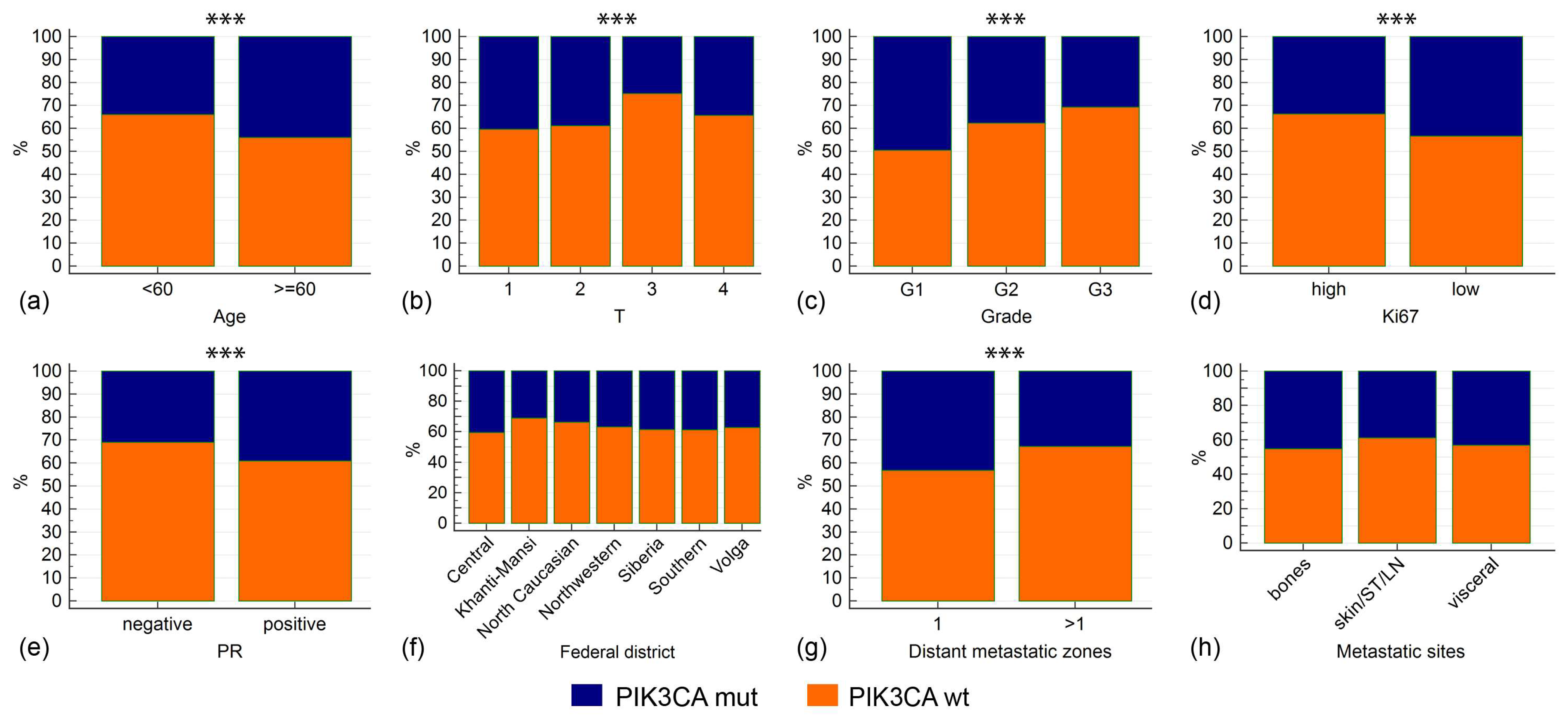
| Characteristics | PIK3CA WT (n = 1179) | PIK3CA MUT (n = 693) | p-Value (PIK3CA WT vs. MUT) | PIK3CA MUT (Exon 10, n = 257) | PIK3CA MUT (Exon 20, n = 323) | p-Value (Exon 10 vs. Exon 20 Mutations) |
|---|---|---|---|---|---|---|
| Age at diagnosis (mean ± SD), range | 50.67 (±11.13); range: 20–83 | 52.77 (±11.55); range: 24–88) | 0.0002 | 53.57 (±11.33); (range: 29–88) | 52.07 (±11.22); (range: 24–85) | 0.110 |
| Histologic type | ||||||
| Invasive cancer of no special type (NST) (n = 1448) | 903 (86.2%) | 545 (86.1%) | 0.909 | 191 (83.0%) | 267 (89.0%) | 0.423 |
| Invasive lobular cancer (ILC) (n = 156) | 96 (9.2%) | 60 (9.5%) | (NST vs. ILC) | 23 (10.0%) | 24 (8.0%) | (NST vs. ILC) |
| Other (n = 77) | 49 (4.7%) | 28 (4.4%) | 16 (7.0%) | 9 (3.0%) | ||
| ND (n = 191) | ||||||
| Primary tumor size (T) | ||||||
| T1 (n = 386) | 230 (21.1%) | 156 (24.3%) | 0.005 | 60 (25.0%) | 77 (26.1%) | 0.114 |
| T2 (n = 809) | 495 (45.4%) | 314 (48.8%) | (T1–T2 vs. T3–T4) | 109 (45.4%) | 149 (50.5%) | (T1–T2 vs. T3–T4) |
| T3 (n = 125) | 94 (8.6%) | 31 (4.8%) | 13 (5.4%) | 11 (3.7%) | ||
| T4 (n = 414) | 272 (24.9%) | 142 (22.1%) | 58 (24.2%) | 58 (19.7%) | ||
| Other/ND (n = 138) | ||||||
| Lymph node involvement (N) | ||||||
| N0 (n = 485) | 302 (27.9%) | 183 (29.1%) | 0.780 | 63 (27.0%) | 91 (31.4%) | 0.713 |
| N1 (n = 588) | 371 (34.3%) | 217 (34.5%) | 86 (36.9%) | 97 (33.4%) | ||
| N2 (n = 323) | 212 (19.6%) | 111 (17.6%) | 42 (18.0%) | 49 (16.9%) | ||
| N3 (n = 314) | 196 (18.1%) | 118 (18.8%) | 42 (18.0%) | 53 (18.3%) | ||
| ND (n = 162) | ||||||
| Distant metastasis (M) | ||||||
| M0 (n = 1240) | 769 (79.5%) | 471 (81.9%) | 0.253 | 174 (81.7%) | 215 (82.4%) | 0.847 |
| M1 (n = 302) | 198 (20.5%) | 104 (18.1%) | 39 (18.3%) | 46 (17.6%) | ||
| ND (n = 330) | ||||||
| Bilateral/unilateral disease | ||||||
| Unilateral BC (n = 1785) | 1118 (94.8%) | 667 (96.2%) | 0.194 | 247 (96.1%) | 307 (95.3%) | 0.606 |
| Bilateral BC (n = 87) | 61 (5.2%) | 26 (3.8%) | 10 (3.9%) | 15 (4.7%) | ||
| Stage | ||||||
| I (n = 184) | 115 (10.6%) | 69 (11.0%) | 0.536 | 22 (9.6%) | 38 (13.1%) | 0.557 |
| II (n = 576) | 353 (32.7%) | 223 (35.6%) | 80 (34.8%) | 104 (36.0%) | ||
| III (n = 619) | 396 (36.6%) | 223 (35.6%) | 86 (37.4%) | 97 (33.6%) | ||
| IV (n = 329) | 217 (20.1%) | 112 (17.9%) | 42 (18.3%) | 50 (17.3%) | ||
| ND (n = 164) | ||||||
| Grade | ||||||
| G1 (n = 99) | 50 (7.6%) | 49 (12.4%) | 0.005 | 14 (9.7%) | 26 (14.0%) | 0.673 |
| G2 (n = 726) | 452 (68.4%) | 274 (69.5%) | 103 (71.5%) | 124 (66.7%) | ||
| G3 (n = 230) | 159 (24.1%) | 71 (18.0%) | 27 (18.8%) | 36 (19.4%) | ||
| ND (n = 817) | ||||||
| PR status | ||||||
| Negative (n = 485) | 335 (28.9%) | 150 (22.2%) | 0.002 | 59 (23.5%) | 69 (21.7%) | 0.608 |
| Positive (n = 1350) | 823 (71.1%) | 527 (77.8%) | 192 (76.5%) | 249 (78.3%) | ||
| ND (n = 37) | ||||||
| Ki67 | ||||||
| Low (<20%) (n = 563) | 319 (32.8%) | 244 (42.4%) | 0.0001 | 79 (36.7%) | 126 (47.2%) | 0.021 |
| High (≥20%) (n = 984) | 653 (67.2%) | 331 (57.6%) | 136 (63.3%) | 141 (52.8%) | ||
| ND (n = 325) | ||||||
| Number of metastatic sites | ||||||
| N = 1 (n = 531) | 302 (32.9%) | 229 (43.2%) | 0.0001 | 91 (44.8%) | 100 (42.6%) | 0.633 |
| N > 1 (n = 917) | 616 (67.1%) | 301 (56.8%) | 112 (55.2%) | 135 (57.4%) | ||
| ND (n = 424) | ||||||
| Distant metastases | ||||||
| Bones | ||||||
| Yes (n = 874) | 548 (59.5%) | 326 (61.2%) | 0.389 | 115 (56.7%) | 152 (64.1%) | 0.110 |
| No (n = 580) | 373 (40.5%) | 207 (38.8%) | 88 (43.3%) | 85 (35.9%) | ||
| Brain | ||||||
| Yes (n = 46) | 34 (3.7%) | 12 (2.3%) | 0.133 | 5 (2.5%) | 5 (2.1%) | 0.810 |
| No (n = 1405) | 886 (96.3%) | 519 (97.7%) | 198 (97.5%) | 231 (97.9%) | ||
| Liver | ||||||
| Yes (n = 453) | 291(31.6%) | 162 (30.4%) | 0.240 | 67 (33.0%) | 71 (30.0%) | 0.493 |
| No (n = 1000) | 629 (68.4%) | 371 (69.6%) | 136 (67.0%) | 166 (70.0%) | ||
| Lung | ||||||
| Yes (n = 457) | 295 (31.0%) | 162 (29.3%) | 0.474 | 55 (26.7%) | 81 (32.3%) | 0.218 |
| No (n = 1046) | 656 (69.0%) | 390 (70.7%) | 151 (73.3%) | 170 (67.7%) | ||
| Lymph nodes | ||||||
| Yes (n = 579) | 383 (41.5%) | 196 (36.8%) | 0.084 | 83 (40.7%) | 80 (33.9%) | 0.142 |
| No (n = 877) | 541 (58.5%) | 336 (63.2%) | 121 (59.3%) | 156 (66.1%) | ||
| Peritoneum | ||||||
| Yes (n = 38) | 28 (3.0%) | 10 (1.9%) | 0.181 | 4 (2.0%) | 5 (2.1%) | 0.918 |
| No (n = 1414) | 892 (97.0%) | 522 (98.1%) | 199 (98.0%) | 232 (97.9%) | ||
| Pleura | ||||||
| Yes (n = 160) | 100 (10.9%) | 60 (11.3%) | 0.801 | 18 (8.9%) | 30 (12.7%) | 0.199 |
| No (n = 1291) | 820 (89.1%) | 471 (88.7%) | 185 (91.1%) | 206 (87.3%) | ||
| Skin | ||||||
| Yes (n = 118) | 84 (9.1%) | 34 (6.4%) | 0.069 | 10 (4.9%) | 18 (7.6%) | 0.249 |
| No (n = 1335) | 838 (90.9%) | 497 (93.6%) | 193 (95.1%) | 218 (92.4%) | ||
| Soft tissues | ||||||
| Yes (n = 114) | 82 (8.9%) | 32 (6.0%) | 0.049 | 13 (6.4%) | 16 (6.8%) | 0.875 |
| No (n = 1336) | 837 (91.1%) | 499 (94.0%) | 190 (93.6%) | 220 (93.2%) |
| Federal District | PIK3CA WT (n = 1179) | PIK3CA MUT (n = 693) | p-Value (PIK3CA WT vs. MUT) | PIK3CA MUT (Exon 10, n = 257) | PIK3CA MUT (Exon 20, n = 323) | p-Value (Exon 10 vs. Exon 20 Mutations) |
|---|---|---|---|---|---|---|
| Northwestern (n = 713) | 451 (63.3%) | 262 (36.7%) | 0.800 | 103 (46.0%) | 121 (54.0%) | 0.400 |
| Southern (n = 217) | 133 (61.3%) | 84 (38.7%) | 29 (42.0%) | 40 (58.0%) | ||
| Central (n = 185) | 110 (59.5%) | 75 (40.5%) | 33 (52.4%) | 30 (47.6%) | ||
| North Caucasian (n = 175) | 116 (66.3%) | 59 (33.7%) | 27 (49.1%) | 28 (50.9%) | ||
| Volga (n = 129) | 81 (62.8%) | 48 (37.2%) | 14 (33.3%) | 28 (66.7%) | ||
| Khanty-Mansi (n = 113) | 78 (69.0%) | 35 (31.0%) | 9 (32.1%) | 19 (67.9%) | ||
| Siberian (n = 109) | 67 (61.5%) | 42 (38.5%) | 17 (51.5%) | 16 (48.4%) | ||
| Urals (n = 88) | 54 (61.4%) | 34 (38.6%) | 11 (45.8%) | 13 (54.2%) | ||
| Far Eastern (n = 88) | 52 (59.1%) | 36 (40.9%) | 8 (29.6%) | 19 (70.4%) | ||
| Yamalo-Nenets (n = 13) | 6 (46.2%) | 7 (53.8%) | 2 (33.3%) | 4 (66.7%) | ||
| ND (n = 42) |
| Parameter | Univariable | Multivariable | ||
|---|---|---|---|---|
| Hazard Ratio [95% CI] | p-Value | Hazard Ratio [95% CI] | p-Value | |
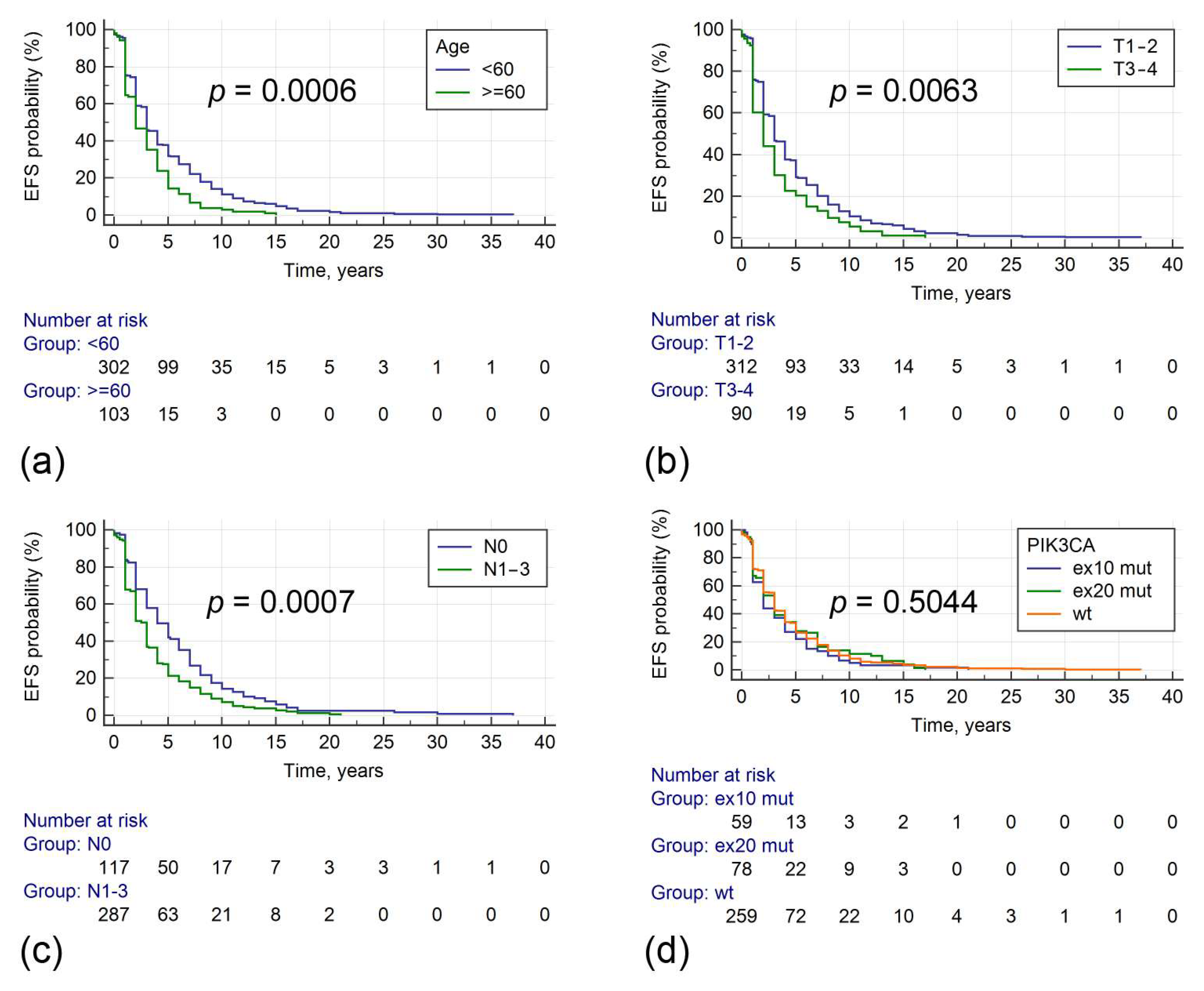
| Age (<60 vs. ≥60 years) (n = 415) | 1.473 [1.151–1.886] (≥60) | 0.0006 | 1.482 [1.178–1.866] | 0.0008 |
| Grade (G1–2 vs. G3) (n = 216) | 1.126 [0.818–1.550] (G3) | 0.384 | ||
| Ki67 (<20% vs. ≥20%) (n = 342) | 1.075 [0.860–1.344] (≥20%) | 0.529 | ||
| Stage (I–III) (n = 431) | 1.401 [1.213–1.619] * | <0.0001 | ||
| Tumor size (T1–2 vs. T3–4) (n = 412) | 1.378 [1.068–1.777] (T3–4) | 0.0063 | 1.301 [1.024–1.653] | 0.031 |
| Nodal involvement (N0 vs. N1–3) (n = 414) | 0.687 [0.562–0.838] (N0) | 0.0005 | 0.722 [0.577–0.903] | 0.004 |
| PR status (positive vs. negative) (n = 418) | 1.020 [0.822–1.266] (PR-) | 0.856 | ||
| PIK3CA (wt vs. mut) (n = 431) | 1.055 [0.867–1.283] (mut) | 0.592 | ||
| Type of Adjuvant Therapy | Primary Resistance | Secondary Resistance | Sensitivity | p-Value |
|---|---|---|---|---|
| Tamoxifen (n = 331) | 66/163 (40.5%) | 28/103 (27.2%) | 28/65 (43.1%) | 0.046 |
| Aromatase inhibitors (n = 82) | 19/42 (45.2%) | 11/28 (39.3%) | 4/12 (33.3%) | 0.730 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sokolova, T.N.; Yanus, G.A.; Aleksakhina, S.N.; Belysheva, Y.V.; Chernyakova, A.P.; Zharnakova, Y.S.; Nikitina, A.S.; Stebneva, T.M.; Martianov, A.S.; Goryainova, A.Y.; et al. Clinical and Morphological Features of ER-Positive HER2-Negative Breast Tumors with PIK3CA Mutations in Russian Patients. Cancers 2025, 17, 1833. https://doi.org/10.3390/cancers17111833
Sokolova TN, Yanus GA, Aleksakhina SN, Belysheva YV, Chernyakova AP, Zharnakova YS, Nikitina AS, Stebneva TM, Martianov AS, Goryainova AY, et al. Clinical and Morphological Features of ER-Positive HER2-Negative Breast Tumors with PIK3CA Mutations in Russian Patients. Cancers. 2025; 17(11):1833. https://doi.org/10.3390/cancers17111833
Chicago/Turabian StyleSokolova, Tatyana N., Grigory A. Yanus, Svetlana N. Aleksakhina, Yana V. Belysheva, Aleksandra P. Chernyakova, Yulia S. Zharnakova, Alisa S. Nikitina, Tatyana M. Stebneva, Aleksandr S. Martianov, Alla Yu. Goryainova, and et al. 2025. "Clinical and Morphological Features of ER-Positive HER2-Negative Breast Tumors with PIK3CA Mutations in Russian Patients" Cancers 17, no. 11: 1833. https://doi.org/10.3390/cancers17111833
APA StyleSokolova, T. N., Yanus, G. A., Aleksakhina, S. N., Belysheva, Y. V., Chernyakova, A. P., Zharnakova, Y. S., Nikitina, A. S., Stebneva, T. M., Martianov, A. S., Goryainova, A. Y., Gluzman, M. I., Orlova, R. V., Stukan’, A. I., Zyuzyukina, A. V., Zukov, R. A., Korzun, P. R., Binnatova, J. O., Abuzova, A. S., Murunova, Y. N., ... Imyanitov, E. N. (2025). Clinical and Morphological Features of ER-Positive HER2-Negative Breast Tumors with PIK3CA Mutations in Russian Patients. Cancers, 17(11), 1833. https://doi.org/10.3390/cancers17111833






