Multiple Instance Learning for the Detection of Lymph Node and Omental Metastases in Carcinoma of the Ovaries, Fallopian Tubes and Peritoneum
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ovarian Carcinoma Clinical and Pathology Data
2.2. Whole-Slide Image Classification
2.3. Model Evaluation
3. Results
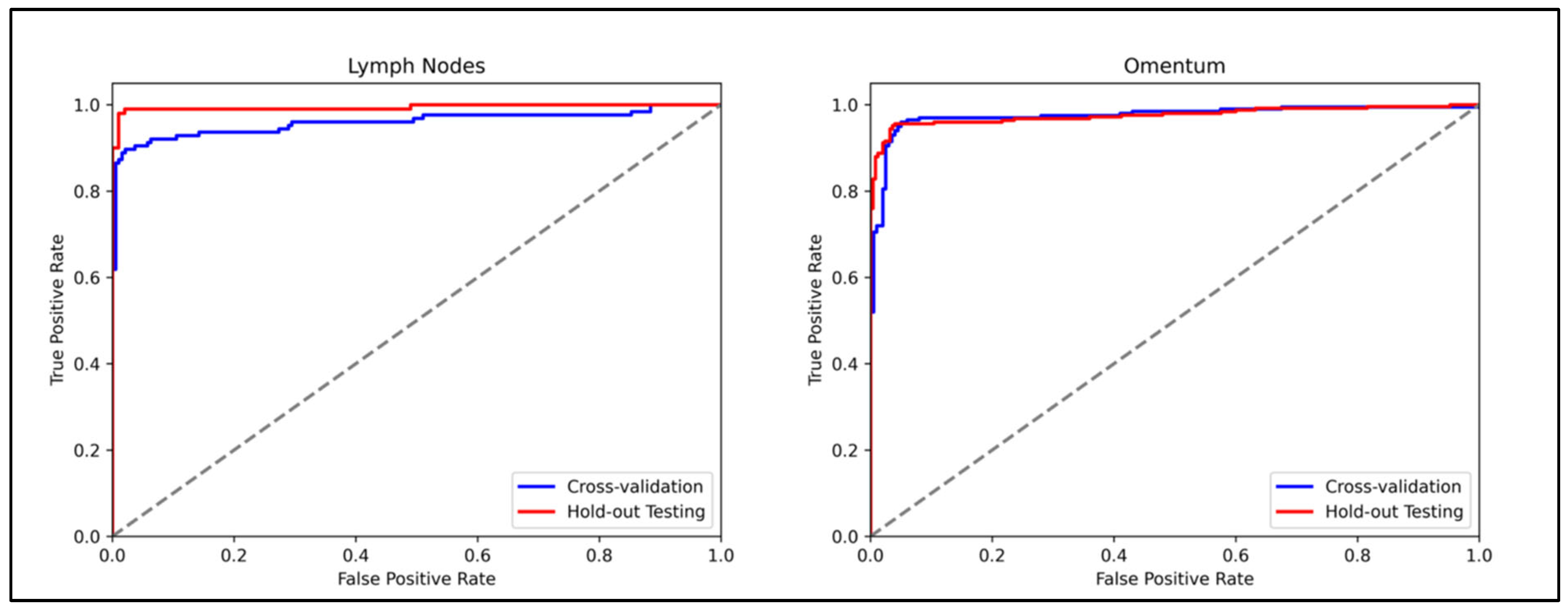
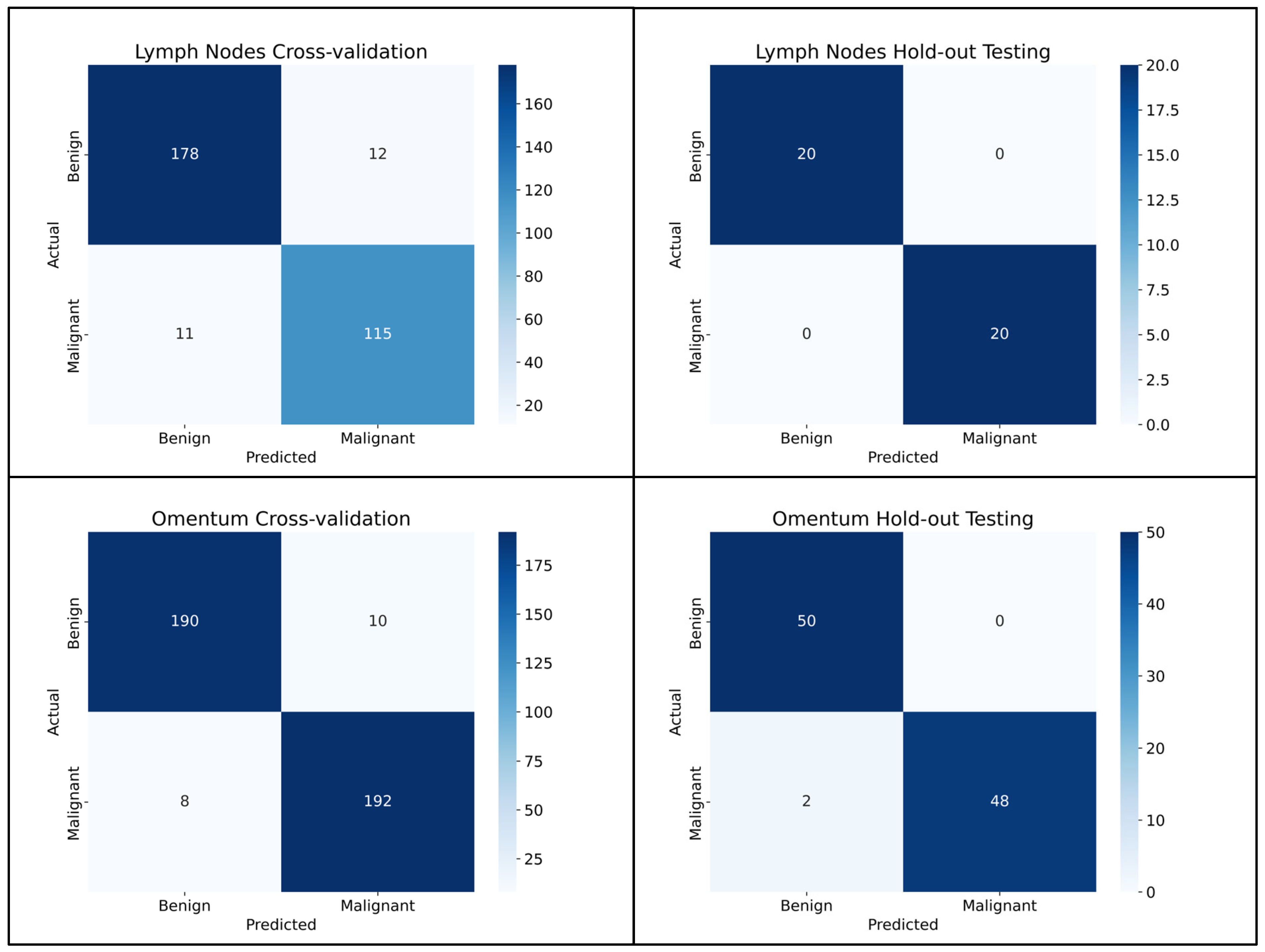
3.1. Lymph Node Metastasis Evaluation
3.2. Omentum Metastasis Evaluation
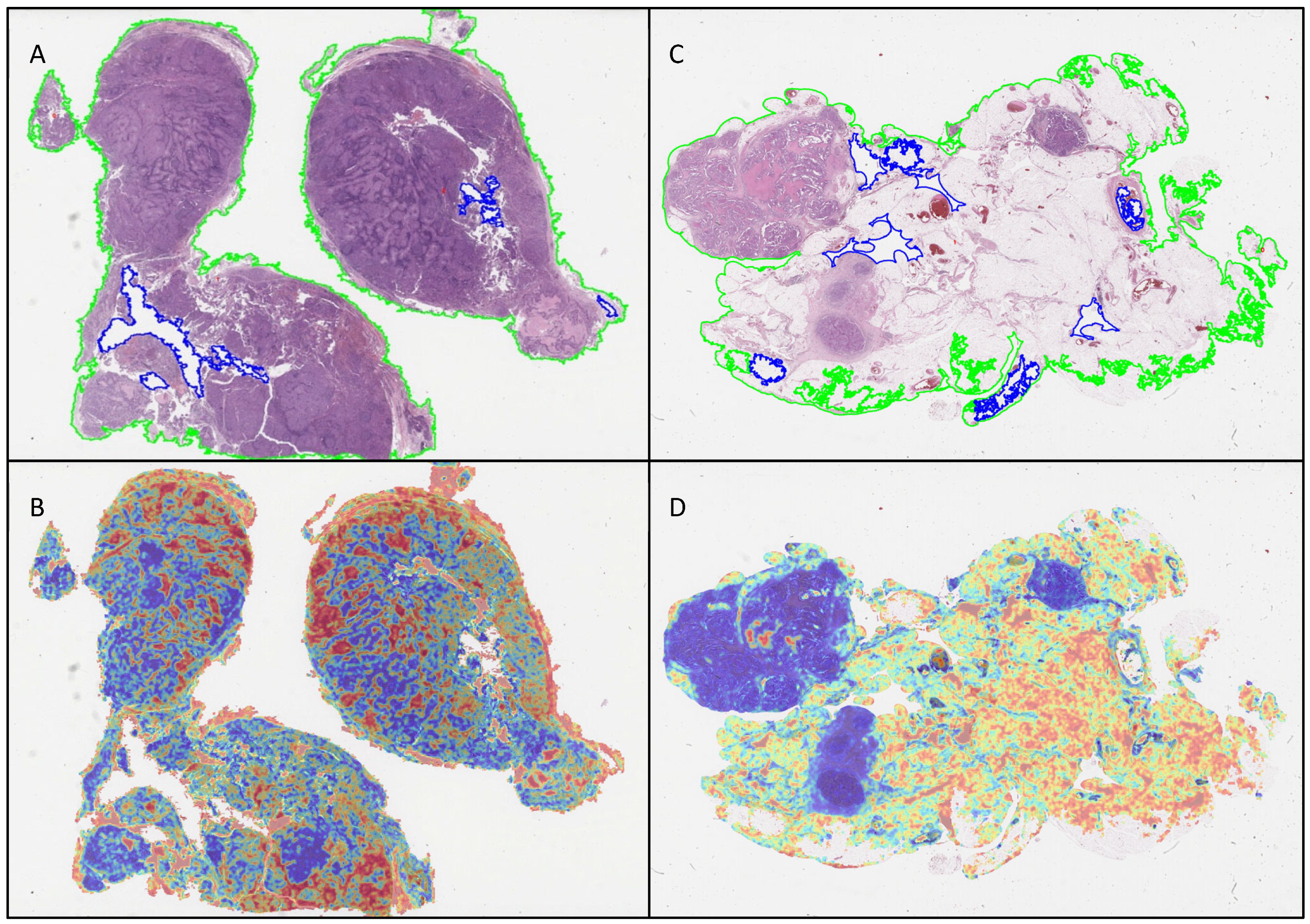
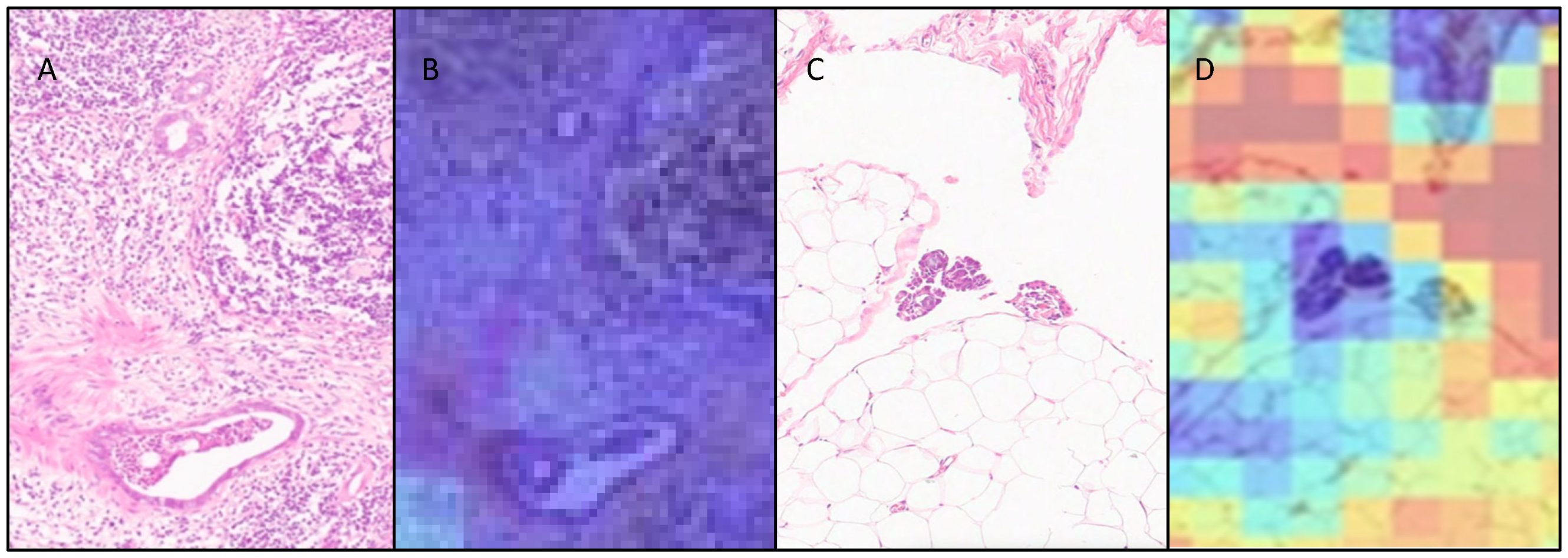
3.3. Attention Heatmaps
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| AJCC | American Joint Committee on Cancer |
| AUROC | Area under the receiver operating characteristic curve |
| AI | Artificial intelligence |
| ABMIL | Attention-based multiple-instance learning |
| BRCA | Breast cancer gene |
| CLAM | Clustering constrained-attention multiple-instance learning |
| CAD | Computer-aided diagnosis |
| CI | Confidence intervals |
| EHR | Electronic health record |
| FFPE | Formalin-fixed paraffin-embedded |
| HRD | Homologous recombination deficiency |
| FIGO | International Federation of Gynecology and Obstetrics |
| IDS | Interval debulking surgery |
| LIMS | Laboratory information management systems |
| NACT | Neoadjuvant chemotherapy |
| PDS | Primary debulking surgery |
| STIC | Serous tubal intraepithelial carcinoma |
| TCGA | The Cancer Genome Atlas |
| WSI | Whole-slide image |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Cancer Research UK Early Diagnosis Hub. Available online: https://crukcancerintelligence.shinyapps.io/EarlyDiagnosis/ (accessed on 13 February 2025).
- Gaitskell, K.; Hermon, C.; Barnes, I.; Pirie, K.; Floud, S.; Green, J.; Beral, V.; Reeves, G.K. Ovarian cancer survival by stage, histotype, and pre-diagnostic lifestyle factors, in the prospective UK Million Women Study. Cancer Epidemiol. 2021, 76, 102074. [Google Scholar] [CrossRef] [PubMed]
- de Oliveira, L.R.L.B.; Horvat, N.; Andrieu, P.I.C.; Panizza, P.S.B.; Cerri, G.G.; Viana, P.C.C. Ovarian cancer staging: What the surgeon needs to know. Br. J. Radiol. 2021, 94, 20210091. [Google Scholar] [CrossRef]
- The Royal College of Pathologists Meeting Pathology Demand Histopathology Workforce Census. Available online: https://www.rcpath.org/profession/workforce-planning/our-workforce-research/histopathology-workforce-survey-2018.html (accessed on 13 February 2025).
- Bychkov, A.; Schubert, M. Constant Demand, Patchy Supply. Pathologist 2023, 88, 18–27. [Google Scholar]
- Walsh, E.; Orsi, N.M. The current troubled state of the global pathology workforce: A concise review. Diagn. Pathol. 2024, 19, 163. [Google Scholar] [CrossRef]
- Baxi, V.; Edwards, R.; Montalto, M.; Saha, S. Digital pathology and artificial intelligence in translational medicine and clinical practice. Mod. Pathol. 2021, 35, 23–32. [Google Scholar] [CrossRef]
- Breen, J.; Allen, K.; Zucker, K.; Adusumilli, P.; Scarsbrook, A.; Hall, G.; Orsi, N.M.; Ravikumar, N. Artificial intelligence in ovarian cancer histopathology: A systematic review. NPJ Precis. Oncol. 2023, 7, 83. [Google Scholar] [CrossRef]
- Holback, C.; Jarosz, R.; Prior, F.; Mutch, D.G.; Bhosale, P.; Garcia, K.; Lee, Y.; Kirk, S.; Sadow, C.A.; Levine, S.; et al. The Cancer Genome Atlas Ovarian Cancer Collection (TCGA-OV) (Version 4) 2016, the Cancer Imaging Archive. Available online: https://www.cancerimagingarchive.net/collection/tcga-ov/ (accessed on 13 February 2025).
- Zeng, H.; Chen, L.; Zhang, M.; Luo, Y.; Ma, X. Integration of histopathological images and multi-dimensional omics analyses predicts molecular features and prognosis in high-grade serous ovarian cancer. Gynecol. Oncol. 2021, 163, 171–180. [Google Scholar] [CrossRef]
- Ho, D.J.; Chui, M.H.; Vanderbilt, C.M.; Jung, J.; Robson, M.E.; Park, C.-S.; Roh, J.; Fuchs, T.J. Deep Interactive Learning-based ovarian cancer segmentation of H&E-stained whole slide images to study morphological patterns of BRCA mutation. J. Pathol. Inform. 2022, 14, 100160. [Google Scholar] [CrossRef]
- Nero, C.; Boldrini, L.; Lenkowicz, J.; Giudice, M.T.; Piermattei, A.; Inzani, F.; Pasciuto, T.; Minucci, A.; Fagotti, A.; Zannoni, G.; et al. Deep-Learning to Predict BRCA Mutation and Survival from Digital H&E Slides of Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2022, 23, 11326. [Google Scholar] [CrossRef]
- Lazard, T.; Bataillon, G.; Naylor, P.; Popova, T.; Bidard, F.-C.; Stoppa-Lyonnet, D.; Stern, M.-H.; Decencière, E.; Walter, T.; Vincent-Salomon, A. Deep learning identifies morphological patterns of homologous recombination deficiency in luminal breast cancers from whole slide images. Cell Rep. Med. 2022, 3, 100872. [Google Scholar] [CrossRef] [PubMed]
- Bogaerts, J.M.A.; Bokhorst, J.-M.; Simons, M.; van Bommel, M.H.D.; Steenbeek, M.P.; de Hullu, J.A.; Linmans, J.; Bart, J.; Bentz, J.L.; Bosse, T.; et al. Deep learning detects premalignant lesions in the Fallopian tube. NPJ Women’s Health 2024, 2, 11. [Google Scholar] [CrossRef]
- Yaar, A.; Asif, A.; Ahmed Raza, S.E.; Rajpoot, N.; Minhas, F. Cross-Domain Knowledge Transfer for Prediction of Chemosensitivity in Ovarian Cancer Patients. In Proceedings of the 2020 IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops (CVPRW), Seattle, WA, USA, 14–19 June 2020; pp. 4020–4025. [Google Scholar]
- Yu, K.-H.; Hu, V.; Wang, F.; Matulonis, U.A.; Mutter, G.L.; Golden, J.A.; Kohane, I.S. Deciphering serous ovarian carcinoma histopathology and platinum response by convolutional neural networks. BMC Med. 2020, 18, 236. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-W.; Chang, C.-C.; Lee, Y.-C.; Lin, Y.-J.; Lo, S.-C.; Hsu, P.-C.; Liou, Y.-A.; Wang, C.-H.; Chao, T.-K. Weakly supervised deep learning for prediction of treatment effectiveness on ovarian cancer from histopathology images. Comput. Med Imaging Graph. 2022, 99, 102093. [Google Scholar] [CrossRef]
- Ghoniem, R.M.; Algarni, A.D.; Refky, B.; Ewees, A.A. Multi-Modal Evolutionary Deep Learning Model for Ovarian Cancer Diagnosis. Symmetry 2021, 13, 643. [Google Scholar] [CrossRef]
- Pinto, D.G.; Bychkov, A.; Tsuyama, N.; Fukuoka, J.; Eloy, C. Real-World Implementation of Digital Pathology: Results from an Intercontinental Survey. Mod. Pathol. 2023, 103, 100261. [Google Scholar] [CrossRef]
- Jansen, P.; Baguer, D.O.; Duschner, N.; Arrastia, J.L.; Schmidt, M.; Landsberg, J.; Wenzel, J.; Schadendorf, D.; Hadaschik, E.; Maass, P.; et al. Deep learning detection of melanoma metastases in lymph nodes. Eur. J. Cancer 2023, 188, 161–170. [Google Scholar] [CrossRef]
- Marmé, F.; Krieghoff-Henning, E.; Gerber, B.; Schmitt, M.; Zahm, D.-M.; Bauerschlag, D.; Forstbauer, H.; Hildebrandt, G.; Ataseven, B.; Brodkorb, T.; et al. Deep learning to predict breast cancer sentinel lymph node status on INSEMA histological images. Eur. J. Cancer 2023, 195, 113390. [Google Scholar] [CrossRef]
- Bejnordi, B.E.; Veta, M.; Van Diest, P.J.; Van Ginneken, B.; Karssemeijer, N.; Litjens, G.; Van Der Laak, J.A.W.M.; Hermsen, M.; Manson, Q.F.; Balkenhol, M.; et al. Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women with Breast Cancer. JAMA 2017, 318, 2199–2210. [Google Scholar] [CrossRef]
- Challa, B.; Tahir, M.; Hu, Y.; Kellough, D.; Lujan, G.; Sun, S.; Parwani, A.V.; Li, Z. Artificial Intelligence–Aided Diagnosis of Breast Cancer Lymph Node Metastasis on Histologic Slides in a Digital Workflow. Mod. Pathol. 2023, 36, 100216. [Google Scholar] [CrossRef]
- Campanella, G.; Hanna, M.G.; Geneslaw, L.; Miraflor, A.; Silva, V.W.K.; Busam, K.J.; Brogi, E.; Reuter, V.E.; Klimstra, D.S.; Fuchs, T.J. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat. Med. 2019, 25, 1301–1309. [Google Scholar] [CrossRef] [PubMed]
- Retamero, J.A.M.M.; Gulturk, E.M.; Bozkurt, A.M.; Liu, S.; Gorgan, M.; Moral, L.; Horton, M.; Parke, A.; Malfroid, K.M.; Sue, J.; et al. Artificial Intelligence Helps Pathologists Increase Diagnostic Accuracy and Efficiency in the Detection of Breast Cancer Lymph Node Metastases. Am. J. Surg. Pathol. 2024, 48, 846–854. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Hong, G.; Xu, A.; Zeng, H.; Chen, X.; Wang, Y.; Luo, Y.; Wu, P.; Liu, C.; Jiang, N.; et al. Artificial intelligence-based model for lymph node metastases detection on whole slide images in bladder cancer: A retrospective, multicentre, diagnostic study. Lancet Oncol. 2023, 24, 360–370. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Su, F.; Dong, K.; Wang, X.; Zhao, X.; Jiang, Y.; Li, J.; Ji, J.; Sun, Y. Deep learning system for lymph node quantification and metastatic cancer identification from whole-slide pathology images. Gastric Cancer 2021, 24, 868–877. [Google Scholar] [CrossRef]
- Huang, S.-C.; Lan, J.; Hsieh, T.-Y.; Chuang, H.-C.; Chien, M.-Y.; Ou, T.-S.; Chen, K.-H.; Wu, R.-C.; Liu, Y.-J.; Cheng, C.-T.; et al. Deep neural network trained on gigapixel images improves lymph node metastasis detection in clinical settings. Nat. Commun. 2022, 13, 3347. [Google Scholar] [CrossRef]
- DeepPath Lydia. Available online: https://www.deeppath.io/deeppath-lydia/ (accessed on 6 March 2025).
- Allen, K.E.; Breen, J.; Hall, G.; Zucker, K.; Ravikumar, N.; Orsi, N.M. #900 Comparative evaluation of ovarian carcinoma subtyping in primary versus interval debulking surgery specimen whole slide images using artificial intelligence. Int. J. Gynecol. Cancer 2023, 33, A429–A430. [Google Scholar]
- Ilse, M.; Tomczak, J.; Welling, M. Attention-Based Deep Multiple Instance Learning. In Proceedings of the 35th International Conference on Machine Learning, Stockholm, Sweden, 10–15 July 2018; Dy, J., Krause, A., Eds.; PMLR: Stockholm, Sweden, 2018; pp. 2127–2136. [Google Scholar]
- Lu, M.Y.; Williamson, D.F.K.; Chen, T.Y.; Chen, R.J.; Barbieri, M.; Mahmood, F. Data-efficient and weakly supervised computational pathology on whole-slide images. Nat. Biomed. Eng. 2021, 5, 555–570. [Google Scholar] [CrossRef]
- Breen, J.; Allen, K.; Zucker, K.; Godson, L.; Orsi, N.M.; Ravikumar, N. A comprehensive evaluation of histopathology foundation models for ovarian cancer subtype classification. NPJ Precis. Oncol. 2025, 9, 33. [Google Scholar] [CrossRef]
- Breen, J.; Ravikumar, N.; Allen, K.; Zucker, K.; Orsi, N.M. Reducing Histopathology Slide Magnification Improves the Accuracy and Speed of Ovarian Cancer Subtyping. In Proceedings of the 2024 IEEE International Symposium on Biomedical Imaging (ISBI), Athens, Greece, 27–30 May 2024; pp. 1–5. [Google Scholar]
- Chen, R.J.; Ding, T.; Lu, M.Y.; Williamson, D.F.K.; Jaume, G.; Song, A.H.; Chen, B.; Zhang, A.; Shao, D.; Shaban, M.; et al. Towards a general-purpose foundation model for computational pathology. Nat. Med. 2024, 30, 850–862. [Google Scholar] [CrossRef]
- Liu, Y.; Kohlberger, T.; Norouzi, M.; Dahl, G.E.; Smith, J.L.; Mohtashamian, A.; Olson, N.; Peng, L.H.; Hipp, J.D.; Stumpe, M.C. Artificial Intelligence–Based Breast Cancer Nodal Metastasis Detection: Insights into the Black Box for Pathologists. Arch. Pathol. Lab. Med. 2018, 143, 859–868. [Google Scholar] [CrossRef]
- Köbel, M.; Kalloger, S.E.; Baker, P.M.; Ewanowich, C.A.; Arseneau, J.; Zherebitskiy, V.; Abdulkarim, S.; Leung, S.; Duggan, M.A.; Fontaine, D.; et al. Diagnosis of Ovarian Carcinoma Cell Type is Highly Reproducible. Am. J. Surg. Pathol. 2010, 34, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Asadi-Aghbolaghi, M.; Farahani, H.; Zhang, A.; Akbari, A.; Kim, S.; Chow, A.; Dane, S.; Huntsman, D.G.; Gilks, C.B.; Ramus, S.; et al. Machine Learning-Driven Histotype Diagnosis of Ovarian Carcinoma: Insights from the OCEAN AI Challenge 2024. medRxiv 2024. medRxiv:2024.04.19.24306099. [Google Scholar]
- Prat, J.; FIGO Committee on Gynecologic Oncology. Staging classification for cancer of the ovary, fallopian tube, and peritoneum. Int. J. Gynecol. Obstet. 2013, 124, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Edge, S.B.; Byrd, D.R.; Compton, C.C.; Fritz, A.G.; Greene, F.L. Trotti A AJCC Cancer Staging Manual, 7th ed.; Springer: New York, NY, USA, 2010. [Google Scholar]

| LYMPH NODES | OMENTUM | |||
|---|---|---|---|---|
| Training WSIs (Patients) | Hold-Out Testing WSIs (Patients) | Training WSIs (Patients) | Hold-Out Testing WSIs (Patients) | |
| Benign | 189 (48) | 20 (20) | 200 (57) | 50 (50) |
| Malignant | 126 (65) | 20 (20) | 200 (106) | 50 (50) |
| Overall | 315 (113) | 40 (40) | 400 (161) | 100 (100) |
| Characteristic | Lymph Node Cohort (n = 113) | Omentum Cohort (n = 161) | ||
|---|---|---|---|---|
| Age | Mean (SD) | 59.8 (12.5) | 61.2 (13.1) | |
| Median (IQR) | 60.0 (51.5–68.5) | 62.0 (52.0–71.0) | ||
| FIGO * stage | 1 | 30 | 37 | |
| 2 | 6 | 9 | ||
| 3 | 61 | 94 | ||
| 4 | 16 | 21 | ||
| Surgery type | PDS | 85 | 122 | |
| IDS | 27 | 39 | ||
| Morphological subtype and grade | High-grade serous carcinoma | 66 | 93 | |
| Low-grade serous carcinoma | 3 | 3 | ||
| Clear-cell carcinoma | 11 | 16 | ||
| Endometrioid carcinoma | G1 | 5 | 7 | |
| G2 | 6 | 7 | ||
| G3 | 7 | 10 | ||
| Mucinous carcinoma | G1 | 2 | 4 | |
| G2 | 2 | 5 | ||
| G3 | 0 | 0 | ||
| UG | 3 | 4 | ||
| Mixed | HG | 5 | 6 | |
| LG | 0 | 0 | ||
| Carcinosarcoma | 4 | 6 | ||
| LYMPH NODES | OMENTUM | |||
|---|---|---|---|---|
| METRIC | Cross-Validation (95% CI) | Hold-Out Testing (95% CI) | Cross-Validation (95% CI) | Hold-Out Testing (95% CI) |
| AUROC | 0.959 (0.929–0.983) | 0.998 (0.985–1.0) | 0.975 (0.958–0.989) | 0.963 (0.911–0.999) |
| Accuracy | 92.7% (89.6–95.3%) | 100.0% (100.0–100.0%) | 95.4% (93.5–97.5%) | 98.0 (95.0–100.0%) |
| Balanced Accuracy | 92.4% (89.2–95.3%) | 100.0% (100.0–100.0%) | 95.5% (93.4–97.5%) | 98.0% (94.8–100.0%) |
| F1 | 0.908 (0.868–0.943) | 1.0 (1.0–1.0) | 0.955 (0.933–0.975) | 0.979 (0.945–1.0) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Allen, K.E.; Breen, J.; Hall, G.; Mappa, G.; Zucker, K.; Ravikumar, N.; Orsi, N.M. Multiple Instance Learning for the Detection of Lymph Node and Omental Metastases in Carcinoma of the Ovaries, Fallopian Tubes and Peritoneum. Cancers 2025, 17, 1789. https://doi.org/10.3390/cancers17111789
Allen KE, Breen J, Hall G, Mappa G, Zucker K, Ravikumar N, Orsi NM. Multiple Instance Learning for the Detection of Lymph Node and Omental Metastases in Carcinoma of the Ovaries, Fallopian Tubes and Peritoneum. Cancers. 2025; 17(11):1789. https://doi.org/10.3390/cancers17111789
Chicago/Turabian StyleAllen, Katie E., Jack Breen, Geoff Hall, Georgia Mappa, Kieran Zucker, Nishant Ravikumar, and Nicolas M. Orsi. 2025. "Multiple Instance Learning for the Detection of Lymph Node and Omental Metastases in Carcinoma of the Ovaries, Fallopian Tubes and Peritoneum" Cancers 17, no. 11: 1789. https://doi.org/10.3390/cancers17111789
APA StyleAllen, K. E., Breen, J., Hall, G., Mappa, G., Zucker, K., Ravikumar, N., & Orsi, N. M. (2025). Multiple Instance Learning for the Detection of Lymph Node and Omental Metastases in Carcinoma of the Ovaries, Fallopian Tubes and Peritoneum. Cancers, 17(11), 1789. https://doi.org/10.3390/cancers17111789








