Explainable AI Model Reveals Informative Mutational Signatures for Cancer-Type Classification
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Generating Datasets of Mutational Signatures, Driver Genes and Topological Mutation Information
2.2. Unsupervised Clustering Methods on Mutational Signatures
2.3. Neural Network Architecture for Cancer-Type Prediction
2.4. Cross-Validation and Explainability of Neural Network Models
2.5. Statistical Analysis
2.6. Informative Mutational Signatures
3. Results
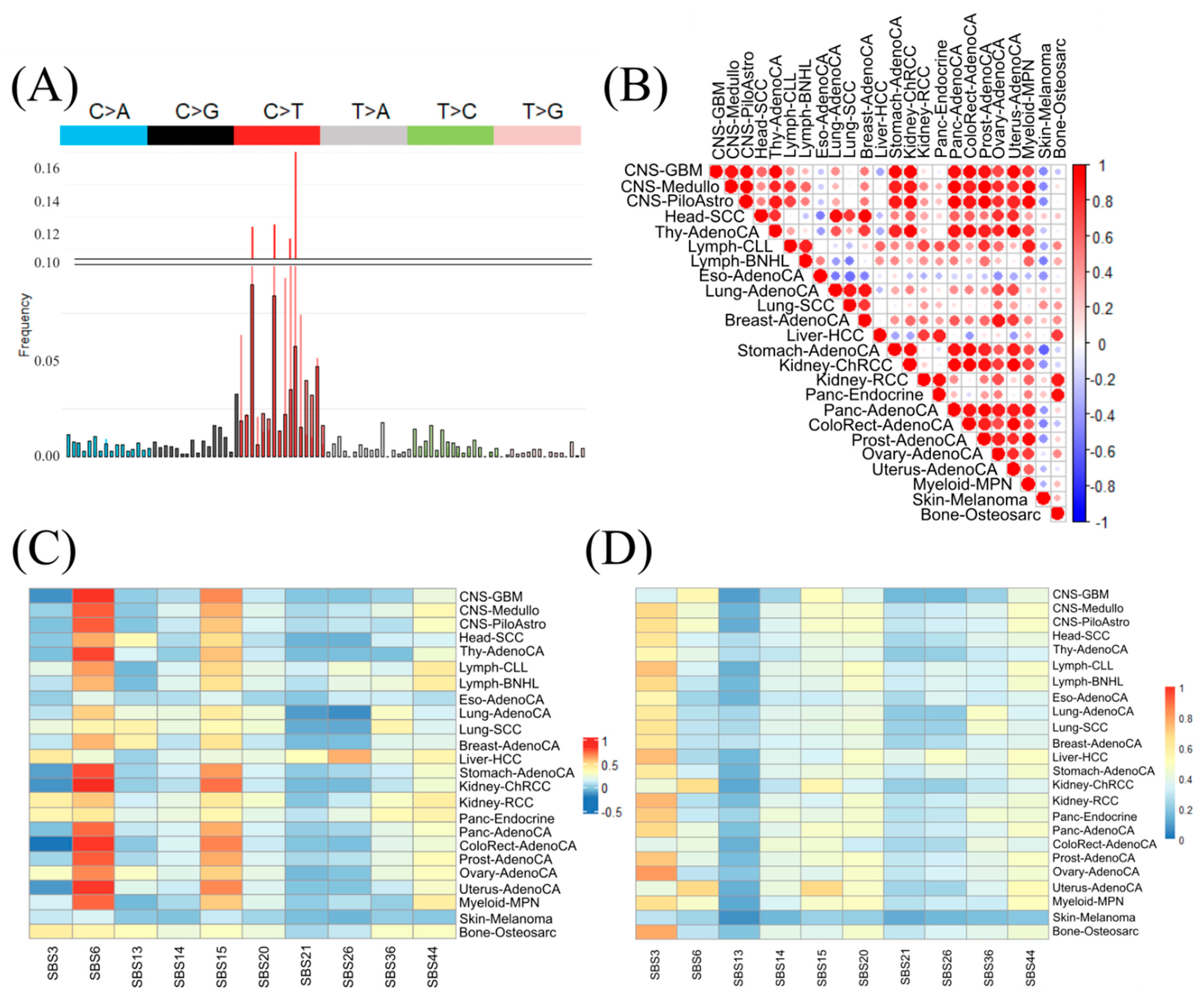
3.1. Challenges to Discriminate Cancer Types Based on Standard Mutational Signatures
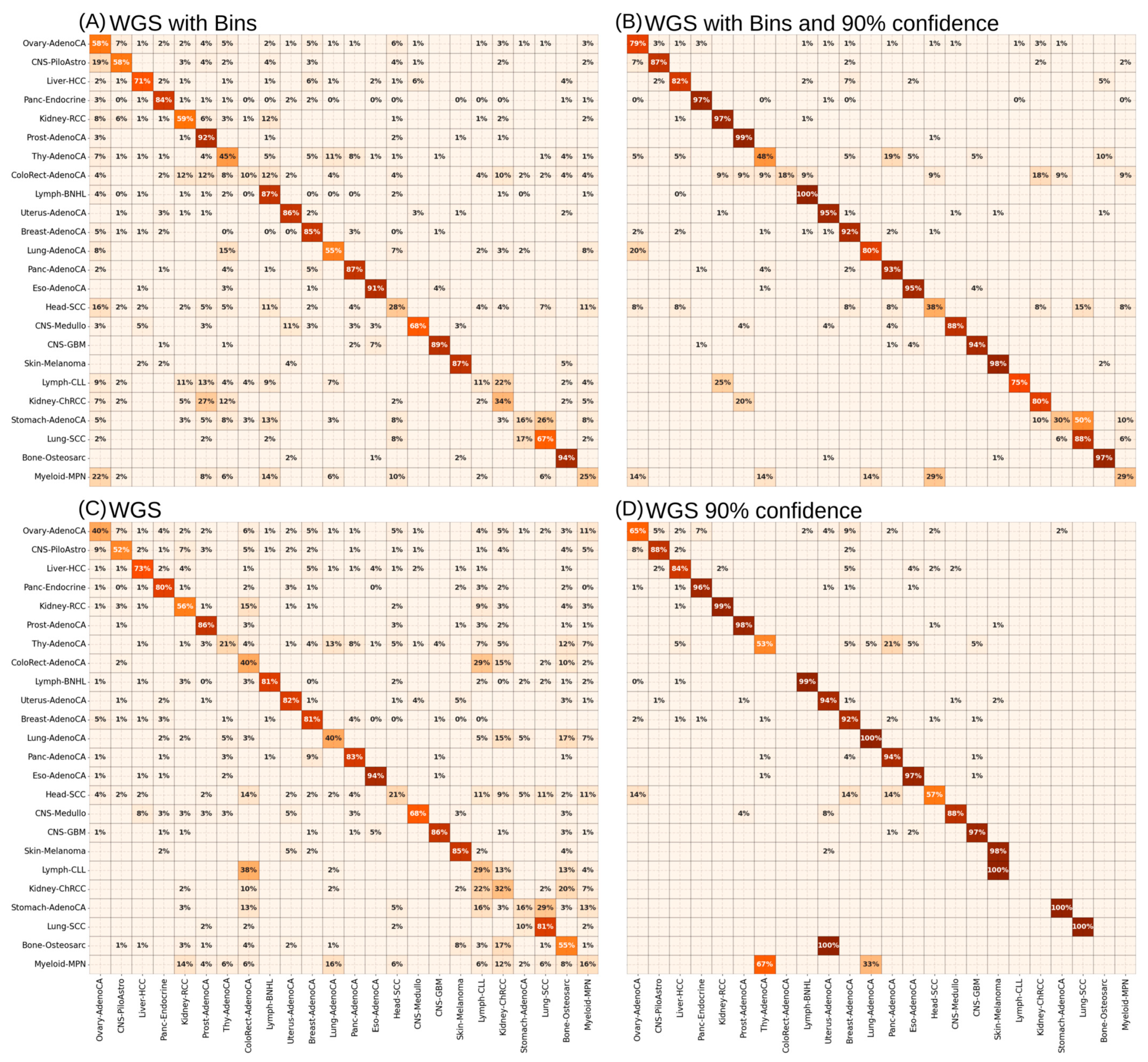
3.2. Supervised ANNs Showed Best Learning Performance on WGS Mutational Signatures
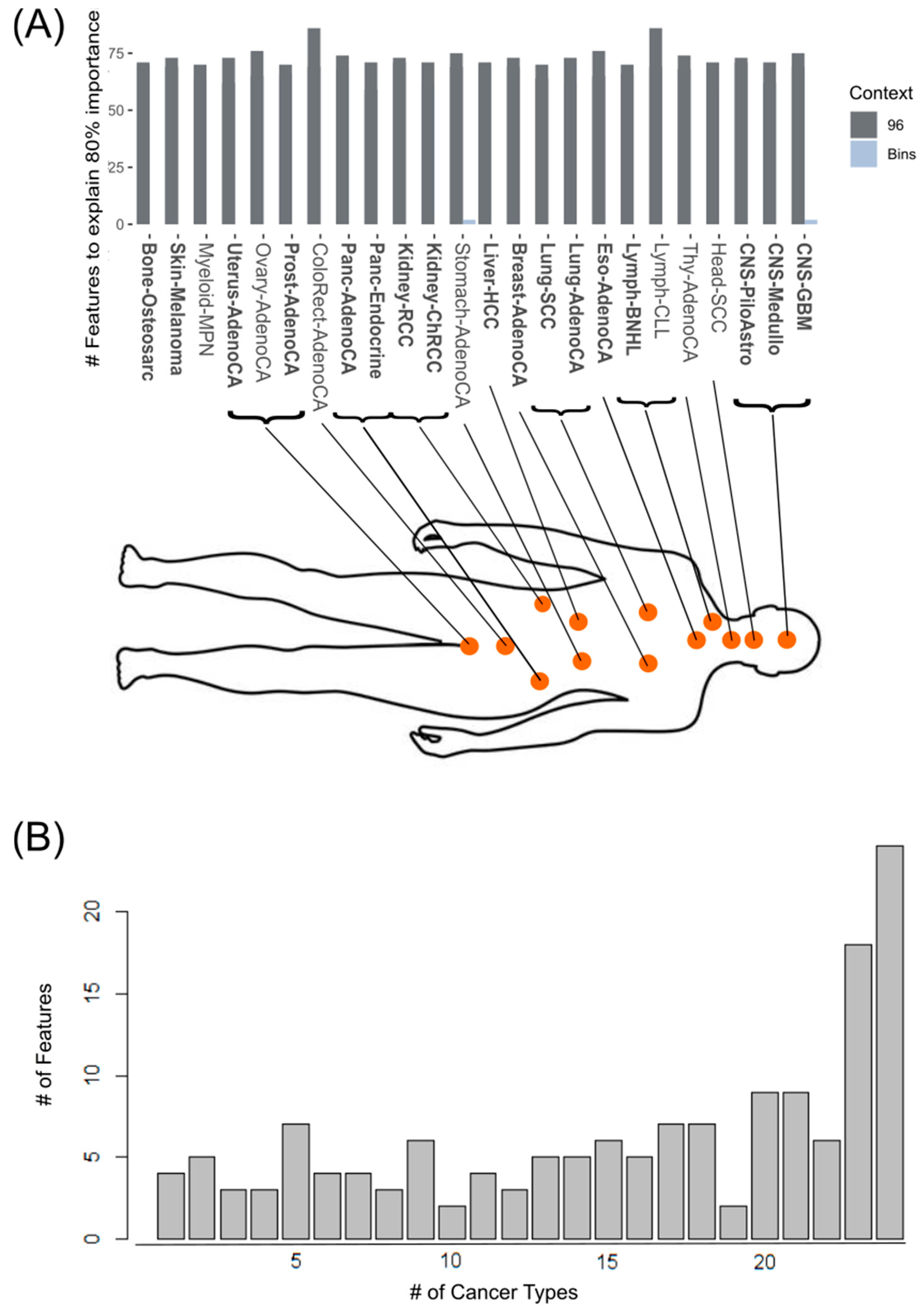
3.3. XAI Models Allow Generation of Informative Mutational Signatures as a Source of Biologically Informed Diagnostics
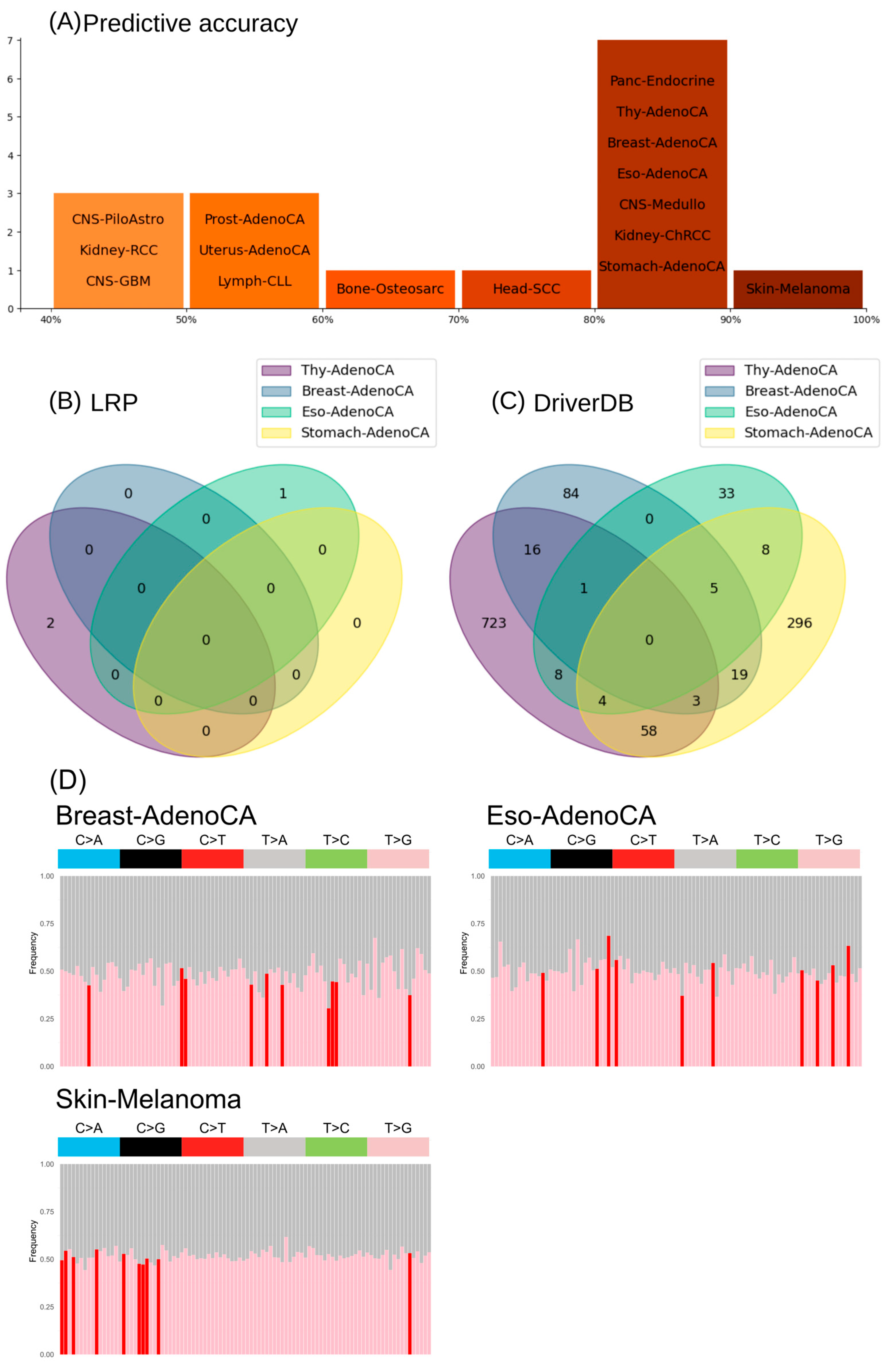
3.4. Informative Mutational Signatures Contain Non-Redundant Information in Comparison to Cancer Driver Gene Mutations
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mebratie, D.Y.; Dagnaw, G.G. Review of Immunohistochemistry Techniques: Applications, Current Status, and Future Perspectives. Semin. Diagn. Pathol. 2024, 41, 154–160. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, A.; Truong, L.D.; Ro, J.Y. Undifferentiated Tumor: True Identity by Immunohistochemistry. Arch. Pathol. Lab. Med. 2008, 132, 326–348. [Google Scholar] [CrossRef] [PubMed]
- Morganti, S.; Tarantino, P.; Ferraro, E.; D’Amico, P.; Duso, B.A.; Curigliano, G. Next Generation Sequencing (NGS): A Revolutionary Technology in Pharmacogenomics and Personalized Medicine in Cancer. In Translational Research and Onco-Omics Applications in the Era of Cancer Personal Genomics; Springer: Cham, Switzerland, 2019; pp. 9–30. [Google Scholar] [CrossRef]
- Ferracin, M.; Pedriali, M.; Veronese, A.; Zagatti, B.; Gafà, R.; Magri, E.; Lunardi, M.; Munerato, G.; Querzoli, G.; Maestri, I.; et al. MicroRNA Profiling for the Identification of Cancers with Unknown Primary Tissue-of-origin. J. Pathol. 2011, 225, 43–53. [Google Scholar] [CrossRef]
- Yang, X.; Gao, L.; Zhang, S. Comparative Pan-Cancer DNA Methylation Analysis Reveals Cancer Common and Specific Patterns. Brief Bioinform. 2017, 18, 761–773. [Google Scholar] [CrossRef]
- Rabbani, B.; Tekin, M.; Mahdieh, N. The Promise of Whole-Exome Sequencing in Medical Genetics. J. Hum. Genet. 2014, 59, 5–15. [Google Scholar] [CrossRef]
- Olafsson, S.; Anderson, C.A. Somatic Mutations Provide Important and Unique Insights into the Biology of Complex Diseases. Trends Genet. 2021, 37, 872–881. [Google Scholar] [CrossRef]
- Stratton, M.R.; Campbell, P.J.; Futreal, P.A. The Cancer Genome. Nature 2009, 458, 719–724. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Gao, Y.; Ning, Z.; Lu, Y.; Zhang, X.; Liu, J.; Xie, B.; Xue, Z.; Wang, X.; Yuan, K.; et al. PGG.SNV: Understanding the Evolutionary and Medical Implications of Human Single Nucleotide Variations in Diverse Populations. Genome Biol. 2019, 20, 215. [Google Scholar] [CrossRef]
- Kim, S.; Misra, A. SNP Genotyping: Technologies and Biomedical Applications. Annu. Rev. Biomed. Eng. 2007, 9, 289–320. [Google Scholar] [CrossRef]
- Huang, H.; Cai, M.; Wang, Y.; Liang, B.; Lin, N.; Xu, L. SNP Array as a Tool for Prenatal Diagnosis of Congenital Heart Disease Screened by Echocardiography: Implications for Precision Assessment of Fetal Prognosis. Risk Manag Heal. Policy 2021, 14, 345–355. [Google Scholar] [CrossRef]
- de Haan, H.G.; Bezemer, I.D.; Doggen, C.J.M.; Le Cessie, S.; Reitsma, P.H.; Arellano, A.R.; Tong, C.H.; Devlin, J.J.; Bare, L.A.; Rosendaal, F.R.; et al. Multiple SNP Testing Improves Risk Prediction of First Venous Thrombosis. Blood 2012, 120, 656–663. [Google Scholar] [CrossRef] [PubMed]
- Burke, W.; Daly, M.; Garber, J.; Botkin, J.; Kahn, M.J.; Lynch, P.; McTiernan, A.; Offit, K.; Perlman, J.; Petersen, G.; et al. Recommendations for Follow-up Care of Individuals with an Inherited Predisposition to Cancer. II. BRCA1 and BRCA2. Cancer Genetics Studies Consortium. JAMA 1997, 277, 997–1003. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Guo, J.; Cheng, G.; Wei, Y.; Liu, S.; Qi, Y.; Wang, G.; Xiao, R.; Qi, W.; Qiu, W. Identification and Validation of SNP-Containing Genes With Prognostic Value in Gastric Cancer via Integrated Bioinformatics Analysis. Front. Oncol. 2021, 11, 564296. [Google Scholar] [CrossRef]
- Helleday, T.; Eshtad, S.; Nik-Zainal, S. Mechanisms Underlying Mutational Signatures in Human Cancers. Nat. Rev. Genet. 2014, 15, 585–598. [Google Scholar] [CrossRef]
- Greenman, C.; Stephens, P.; Smith, R.; Dalgliesh, G.L.; Hunter, C.; Bignell, G.; Davies, H.; Teague, J.; Butler, A.; Stevens, C.; et al. Patterns of Somatic Mutation in Human Cancer Genomes. Nature 2007, 446, 153–158. [Google Scholar] [CrossRef]
- Ma, J.; Setton, J.; Lee, N.Y.; Riaz, N.; Powell, S.N. The Therapeutic Significance of Mutational Signatures from DNA Repair Deficiency in Cancer. Nat. Commun. 2018, 9, 3292. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Stratton, M.R. Mutational Signatures: The Patterns of Somatic Mutations Hidden in Cancer Genomes. Curr. Opin. Genet. Dev. 2014, 24, 52–60. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Kim, J.; Haradhvala, N.J.; Huang, M.N.; Tian Ng, A.W.; Wu, Y.; Boot, A.; Covington, K.R.; Gordenin, D.A.; Bergstrom, E.N.; et al. The Repertoire of Mutational Signatures in Human Cancer. Nature 2020, 578, 94–101. [Google Scholar] [CrossRef]
- Islam, S.M.A.; Díaz-Gay, M.; Wu, Y.; Barnes, M.; Vangara, R.; Bergstrom, E.N.; He, Y.; Vella, M.; Wang, J.; Teague, J.W.; et al. Uncovering Novel Mutational Signatures by de Novo Extraction with SigProfilerExtractor. Cell Genom. 2022, 2, 100179. [Google Scholar] [CrossRef]
- Jin, H.; Gulhan, D.C.; Geiger, B.; Ben-Isvy, D.; Geng, D.; Ljungström, V.; Park, P.J. Accurate and Sensitive Mutational Signature Analysis with MuSiCal. Nat. Genet. 2024, 56, 541–552. [Google Scholar] [CrossRef]
- Aaltonen, L.A.; Abascal, F.; Abeshouse, A.; Aburatani, H.; Adams, D.J.; Agrawal, N.; Ahn, K.S.; Ahn, S.-M.; Aikata, H.; Akbani, R.; et al. Pan-Cancer Analysis of Whole Genomes. Nature 2020, 578, 82–93. [Google Scholar] [CrossRef]
- Bhinder, B.; Gilvary, C.; Madhukar, N.S.; Elemento, O. Artificial Intelligence in Cancer Research and Precision Medicine. Cancer Discov. 2021, 11, 900–915. [Google Scholar] [CrossRef] [PubMed]
- Al-Yasriy, H.F.; AL-Husieny, M.S.; Mohsen, F.Y.; Khalil, E.A.; Hassan, Z.S. Diagnosis of Lung Cancer Based on CT Scans Using CNN. IOP Conf. Ser. Mater. Sci. Eng. 2020, 928, 022035. [Google Scholar] [CrossRef]
- Dabeer, S.; Khan, M.M.; Islam, S. Cancer Diagnosis in Histopathological Image: CNN Based Approach. Inform. Med. Unlocked 2019, 16, 100231. [Google Scholar] [CrossRef]
- Esteva, A.; Kuprel, B.; Novoa, R.A.; Ko, J.; Swetter, S.M.; Blau, H.M.; Thrun, S. Dermatologist-Level Classification of Skin Cancer with Deep Neural Networks. Nature 2017, 542, 115–118. [Google Scholar] [CrossRef]
- Khan, J.; Wei, J.S.; Ringnér, M.; Saal, L.H.; Ladanyi, M.; Westermann, F.; Berthold, F.; Schwab, M.; Antonescu, C.R.; Peterson, C.; et al. Classification and Diagnostic Prediction of Cancers Using Gene Expression Profiling and Artificial Neural Networks. Nat. Med. 2001, 7, 673–679. [Google Scholar] [CrossRef]
- Mostavi, M.; Chiu, Y.-C.; Huang, Y.; Chen, Y. Convolutional Neural Network Models for Cancer Type Prediction Based on Gene Expression. BMC Med. Genom. 2020, 13, 44. [Google Scholar] [CrossRef]
- Alanazi, S.A.; Alshammari, N.; Alruwaili, M.; Junaid, K.; Abid, M.R.; Ahmad, F. Integrative Analysis of RNA Expression Data Unveils Distinct Cancer Types through Machine Learning Techniques. Saudi J. Biol. Sci. 2024, 31, 103918. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of Cancer: The Next Generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Shah, A.A.; Malik, H.A.M.; Mohammad, A.; Khan, Y.D.; Alourani, A. Machine Learning Techniques for Identification of Carcinogenic Mutations, Which Cause Breast Adenocarcinoma. Sci. Rep. 2022, 12, 11738. [Google Scholar] [CrossRef]
- Chen, Y.; Sun, J.; Huang, L.-C.; Xu, H.; Zhao, Z. Classification of Cancer Primary Sites Using Machine Learning and Somatic Mutations. Biomed. Res. Int. 2015, 2015, 491502. [Google Scholar] [CrossRef]
- Marquard, A.M.; Birkbak, N.J.; Thomas, C.E.; Favero, F.; Krzystanek, M.; Lefebvre, C.; Ferté, C.; Jamal-Hanjani, M.; Wilson, G.A.; Shafi, S.; et al. TumorTracer: A Method to Identify the Tissue of Origin from the Somatic Mutations of a Tumor Specimen. BMC Med. Genom. 2015, 8, 58. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Shi, Y.; Li, C.; Kim, J.; Cai, W.; Han, Z.; Feng, D.D. DeepGene: An Advanced Cancer Type Classifier Based on Deep Learning and Somatic Point Mutations. BMC Bioinform. 2016, 17, 476. [Google Scholar] [CrossRef]
- Jiao, W.; Atwal, G.; Polak, P.; Karlic, R.; Cuppen, E.; Al-Shahrour, F.; Atwal, G.; Bailey, P.J.; Biankin, A.V.; Boutros, P.C.; et al. A Deep Learning System Accurately Classifies Primary and Metastatic Cancers Using Passenger Mutation Patterns. Nat. Commun. 2020, 11, 728. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y. A Novel Ensemble Machine Learning for Robust Microarray Data Classification. Comput. Biol. Med. 2006, 36, 553–573. [Google Scholar] [CrossRef]
- Liu, B.; Liu, Y.; Pan, X.; Li, M.; Yang, S.; Li, S.C. DNA Methylation Markers for Pan-Cancer Prediction by Deep Learning. Genes 2019, 10, 778. [Google Scholar] [CrossRef]
- Kim, B.-H.; Yu, K.; Lee, P.C.W. Cancer Classification of Single-Cell Gene Expression Data by Neural Network. Bioinformatics 2020, 36, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Zelli, V.; Manno, A.; Compagnoni, C.; Ibraheem, R.O.; Zazzeroni, F.; Alesse, E.; Rossi, F.; Arbib, C.; Tessitore, A. Classification of Tumor Types Using XGBoost Machine Learning Model: A Vector Space Transformation of Genomic Alterations. J. Transl. Med. 2023, 21, 836. [Google Scholar] [CrossRef]
- Darmofal, M.; Suman, S.; Atwal, G.; Toomey, M.; Chen, J.-F.; Chang, J.C.; Vakiani, E.; Varghese, A.M.; Balakrishnan Rema, A.; Syed, A.; et al. Deep-Learning Model for Tumor-Type Prediction Using Targeted Clinical Genomic Sequencing Data. Cancer Discov. 2024, 14, 1064–1081. [Google Scholar] [CrossRef]
- Berisha, V.; Krantsevich, C.; Hahn, P.R.; Hahn, S.; Dasarathy, G.; Turaga, P.; Liss, J. Digital Medicine and the Curse of Dimensionality. NPJ Digit. Med. 2021, 4, 153. [Google Scholar] [CrossRef]
- Oldenburg, J.; Wagner, J.; Troschke-Meurer, S.; Plietz, J.; Kaderali, L.; Völzke, H.; Nauck, M.; Homuth, G.; Völker, U.; Simm, S. XModNN: Explainable Modular Neural Network to Identify Clinical Parameters and Disease Biomarkers in Transcriptomic Datasets. Biomolecules 2024, 14, 1501. [Google Scholar] [CrossRef] [PubMed]
- Lapuschkin, S.; Wäldchen, S.; Binder, A.; Montavon, G.; Samek, W.; Müller, K.-R. Unmasking Clever Hans Predictors and Assessing What Machines Really Learn. Nat. Commun. 2019, 10, 1096. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.-H.; Lai, Y.-L.; Shen, P.-C.; Liu, H.-C.; Tsai, M.-H.; Wang, Y.-D.; Lin, W.-J.; Chen, F.-H.; Li, C.-Y.; Wang, S.-C.; et al. DriverDBv4: A Multi-Omics Integration Database for Cancer Driver Gene Research. Nucleic Acids Res. 2024, 52, D1246–D1252. [Google Scholar] [CrossRef]
- Bach, S.; Binder, A.; Montavon, G.; Klauschen, F.; Müller, K.-R.; Samek, W. On Pixel-Wise Explanations for Non-Linear Classifier Decisions by Layer-Wise Relevance Propagation. PLoS ONE 2015, 10, e0130140. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex Heatmaps Reveal Patterns and Correlations in Multidimensional Genomic Data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Lawrence, M.S.; Stojanov, P.; Mermel, C.H.; Robinson, J.T.; Garraway, L.A.; Golub, T.R.; Meyerson, M.; Gabriel, S.B.; Lander, E.S.; Getz, G. Discovery and Saturation Analysis of Cancer Genes across 21 Tumour Types. Nature 2014, 505, 495–501. [Google Scholar] [CrossRef]
- Samek, W.; Binder, A.; Montavon, G.; Lapuschkin, S.; Muller, K.-R. Evaluating the Visualization of What a Deep Neural Network Has Learned. IEEE Trans. Neural Netw. Learn. Syst. 2017, 28, 2660–2673. [Google Scholar] [CrossRef]
- Sondka, Z.; Dhir, N.B.; Carvalho-Silva, D.; Jupe, S.; Madhumita; McLaren, K.; Starkey, M.; Ward, S.; Wilding, J.; Ahmed, M.; et al. COSMIC: A Curated Database of Somatic Variants and Clinical Data for Cancer. Nucleic Acids Res. 2024, 52, D1210–D1217. [Google Scholar] [CrossRef]
- Secrier, M.; Li, X.; de Silva, N.; Eldridge, M.D.; Contino, G.; Bornschein, J.; MacRae, S.; Grehan, N.; O’Donovan, M.; Miremadi, A.; et al. Mutational Signatures in Esophageal Adenocarcinoma Define Etiologically Distinct Subgroups with Therapeutic Relevance. Nat. Genet. 2016, 48, 1131–1141. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Jones, P.H.; Wedge, D.C.; Sale, J.E.; Campbell, P.J.; Nik-Zainal, S.; Stratton, M.R. Clock-like Mutational Processes in Human Somatic Cells. Nat. Genet. 2015, 47, 1402–1407. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.J.R.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Børresen-Dale, A.-L.; et al. Signatures of Mutational Processes in Human Cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Bignold, L.P. Typing, Grading, and Staging of Cases of Tumor. In Principles of Tumors; Elsevier: Amsterdam, The Netherlands, 2020; pp. 279–315. [Google Scholar] [CrossRef]
- McPhail, S.; Johnson, S.; Greenberg, D.; Peake, M.; Rous, B. Stage at Diagnosis and Early Mortality from Cancer in England. Br. J. Cancer 2015, 112, S108–S115. [Google Scholar] [CrossRef] [PubMed]
- Badgwell, D.; Bast, R.C. Early Detection of Ovarian Cancer. Dis. Markers 2007, 23, 397–410. [Google Scholar] [CrossRef]
- Blandin Knight, S.; Crosbie, P.A.; Balata, H.; Chudziak, J.; Hussell, T.; Dive, C. Progress and Prospects of Early Detection in Lung Cancer. Open Biol. 2017, 7, 170070. [Google Scholar] [CrossRef]
- Meropol, N.J.; Schulman, K.A. Cost of Cancer Care: Issues and Implications. J. Clin. Oncol. 2007, 25, 180–186. [Google Scholar] [CrossRef]
- Dvortsin, E.; Gout-Zwart, J.; Eijssen, E.-L.M.; van Brussel, J.; Postma, M.J. Comparative Cost-Effectiveness of Drugs in Early versus Late Stages of Cancer; Review of the Literature and a Case Study in Breast Cancer. PLoS ONE 2016, 11, e0146551. [Google Scholar] [CrossRef][Green Version]
- Sheahan, K.; O’Keane, J.C.; Abramowitz, A.; Carlson, J.A.; Burke, B.; Gottlieb, L.S.; O’Brien, M.J. Metastatic Adenocarcinoma of an Unknown Primary Site: A Comparison of the Relative Contributions of Morphology, Minimal Essential Clinical Data and CEA Immunostaining Status. Am. J. Clin. Pathol. 1993, 99, 729–735. [Google Scholar] [CrossRef] [PubMed]
- Magaki, S.; Hojat, S.A.; Wei, B.; So, A.; Yong, W.H. An Introduction to the Performance of Immunohistochemistry; Springer: New York, NY, USA, 2019; pp. 289–298. [Google Scholar]
- Duraiyan, J.; Govindarajan, R.; Kaliyappan, K.; Palanisamy, M. Applications of Immunohistochemistry. J. Pharm. Bioallied Sci. 2012, 4, 307. [Google Scholar] [CrossRef]
- Handorf, C.R.; Kulkarni, A.; Grenert, J.P.; Weiss, L.M.; Rogers, W.M.; Kim, O.S.; Monzon, F.A.; Halks-Miller, M.; Anderson, G.G.; Walker, M.G.; et al. A Multicenter Study Directly Comparing the Diagnostic Accuracy of Gene Expression Profiling and Immunohistochemistry for Primary Site Identification in Metastatic Tumors. Am. J. Surg. Pathol. 2013, 37, 1067–1075. [Google Scholar] [CrossRef]
- Monzon, F.A.; Koen, T.J. Diagnosis of Metastatic Neoplasms: Molecular Approaches for Identification of Tissue of Origin. Arch. Pathol. Lab. Med. 2010, 134, 216–224. [Google Scholar] [CrossRef]
- Lu, J.; Getz, G.; Miska, E.A.; Alvarez-Saavedra, E.; Lamb, J.; Peck, D.; Sweet-Cordero, A.; Ebert, B.L.; Mak, R.H.; Ferrando, A.A.; et al. MicroRNA Expression Profiles Classify Human Cancers. Nature 2005, 435, 834–838. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Elmas, A.; Huang, K. Mutation Impact on MRNA Versus Protein Expression across Human Cancers. GigaScience 2025, 14, giae113. [Google Scholar] [CrossRef] [PubMed]
- Sample, I. Routine DNA tests will put NHS at the ‘forefront of medicine’. The Guardian, 3 July 2018. [Google Scholar]
- Tothill, R.W.; Li, J.; Mileshkin, L.; Doig, K.; Siganakis, T.; Cowin, P.; Fellowes, A.; Semple, T.; Fox, S.; Byron, K.; et al. Massively-parallel Sequencing Assists the Diagnosis and Guided Treatment of Cancers of Unknown Primary. J. Pathol. 2013, 231, 413–423. [Google Scholar] [CrossRef] [PubMed]
- Soh, K.P.; Szczurek, E.; Sakoparnig, T.; Beerenwinkel, N. Predicting Cancer Type from Tumour DNA Signatures. Genome Med. 2017, 9, 104. [Google Scholar] [CrossRef]
- Liu, H.; Qiu, C.; Wang, B.; Bing, P.; Tian, G.; Zhang, X.; Ma, J.; He, B.; Yang, J. Evaluating DNA Methylation, Gene Expression, Somatic Mutation, and Their Combinations in Inferring Tumor Tissue-of-Origin. Front. Cell Dev. Biol. 2021, 9, 619330. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Jiménez, F.; Muiños, F.; Sentís, I.; Deu-Pons, J.; Reyes-Salazar, I.; Arnedo-Pac, C.; Mularoni, L.; Pich, O.; Bonet, J.; Kranas, H.; et al. A Compendium of Mutational Cancer Driver Genes. Nat. Rev. Cancer 2020, 20, 555–572. [Google Scholar] [CrossRef]
- Futreal, P.A.; Coin, L.; Marshall, M.; Down, T.; Hubbard, T.; Wooster, R.; Rahman, N.; Stratton, M.R. A Census of Human Cancer Genes. Nat. Rev. Cancer 2004, 4, 177–183. [Google Scholar] [CrossRef]
- Tischoff, I. Pathologische Diagnostik Beim CUP-Syndrom. Der Onkol. 2021, 27, 642–650. [Google Scholar] [CrossRef]
- Moran, S.; Martínez-Cardús, A.; Sayols, S.; Musulén, E.; Balañá, C.; Estival-Gonzalez, A.; Moutinho, C.; Heyn, H.; Diaz-Lagares, A.; de Moura, M.C.; et al. Epigenetic Profiling to Classify Cancer of Unknown Primary: A Multicentre, Retrospective Analysis. Lancet Oncol. 2016, 17, 1386–1395. [Google Scholar] [CrossRef]
- Vural, S.; Wang, X.; Guda, C. Classification of Breast Cancer Patients Using Somatic Mutation Profiles and Machine Learning Approaches. BMC Syst. Biol. 2016, 10, 62. [Google Scholar] [CrossRef]
- He, B.; Dai, C.; Lang, J.; Bing, P.; Tian, G.; Wang, B.; Yang, J. A Machine Learning Framework to Trace Tumor Tissue-of-Origin of 13 Types of Cancer Based on DNA Somatic Mutation. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165916. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Li, L.; Peng, L.; Wang, B.; Lang, J.; Lu, Q.; Zhang, X.; Sun, Y.; Tian, G.; Zhang, H.; et al. Predicting Cancer Tissue-of-Origin by a Machine Learning Method Using DNA Somatic Mutation Data. Front. Genet. 2020, 11, 674. [Google Scholar] [CrossRef] [PubMed]
- Hauser, K.; Kurz, A.; Haggenmüller, S.; Maron, R.C.; von Kalle, C.; Utikal, J.S.; Meier, F.; Hobelsberger, S.; Gellrich, F.F.; Sergon, M.; et al. Explainable Artificial Intelligence in Skin Cancer Recognition: A Systematic Review. Eur. J. Cancer 2022, 167, 54–69. [Google Scholar] [CrossRef] [PubMed]
- Shaban-Nejad, A.; Michalowski, M.; Brownstein, J.; Buckeridge, D. Guest Editorial Explainable AI: Towards Fairness, Accountability, Transparency and Trust in Healthcare. IEEE J. Biomed. Health Inform. 2021, 25, 2374–2375. [Google Scholar] [CrossRef]
- Holzinger, A.; Biemann, C.; Pattichis, C.S.; Kell, D.B. What Do We Need to Build Explainable AI Systems for the Medical Domain? arXiv 2017, arXiv:1712.09923. [Google Scholar]


| Author | Year | Title | AI Method | Input Data Source |
|---|---|---|---|---|
| Khan et al. | 2001 | 10.1038/89044 [27] | ANN/DNN | Expression (RNA-Seq) |
| Peng et al. | 2006 | 10.1016/j.compbiomed.2005.04.001 [36] | SVM | Expression (Microarray) |
| Chen et al. | 2015 | 10.1155/2015/491502 [32] | SVM | Somatic alterations |
| Yuan et al. | 2016 | 10.1186/s12859-016-1334-9 [34] | ANN/DNN | Somatic alterations |
| Liu et al. | 2019 | 10.3390/genes10100778 [37] | ANN/DNN | Expression (Methyl-Seq) |
| Jiao et al. | 2020 | 10.1038/s41467-019-13825-8 [35] | ANN/DNN, RF | Somatic alterations |
| Mostavi et al. | 2020 | 10.1186/s12920-020-0677-2 [28] | CNN | Expression (RNA-Seq) |
| Kim et al. | 2020 | 10.1093/bioinformatics/btz772 [38] | SVM, RF, ANN/DNN | Expression (Single cell RNA-Seq) |
| Zelli et al. | 2023 | 10.1186/s12967-023-04720-4 [39] | XGBoost | Somatic alterations |
| Darmofal et al. | 2024 | 10.1158/2159-8290.CD-23-0996 [40] | RF, ANN/DNN | Somatic alterations |
| Alanazi et al. | 2024 | 10.1016/j.sjbs.2023.103918 [29] | SVM, RF, ANN/DNN etc. | Expression (RNA-Seq) |
| Dataset Name | Input |
|---|---|
| WGS_MS | Mutation counts in 3-nucleotide-, 2-nucleotide and 1-nucleotide context (mutation context) on all mutations across the genome |
| WGS_MS + Bins | Mutation context on all mutations across the genome + number of mutations in 1 Mbp-bins |
| WES_MS | Mutation context on all mutations in exonic regions |
| WES_MS + Bins | Mutation context on all mutations in exonic regions + number of mutations in 1 Mbp-bins |
| WIIS_MS | Mutation context on all mutations in intronic and intergenic regions |
| WIIS_MS + Bins | Mutation context on all mutations in intronic and intergenic regions + number of mutations in 1 Mbp-bins |
| WGS_GeneM | Mutation counts in 3-nucleotide-context on all mutations separated by genes |
| Dataset | Crossfold Iteration | Precision | Recall | F1 Score | MCC Score |
|---|---|---|---|---|---|
| WGS_MS + Bins | 1 | 0.63 | 0.65 | 0.62 | 0.72 |
| 2 | 0.61 | 0.61 | 0.60 | 0.67 | |
| 3 | 0.64 | 0.63 | 0.63 | 0.72 | |
| 4 | 0.61 | 0.61 | 0.59 | 0.67 | |
| 5 | 0.67 | 0.63 | 0.61 | 0.68 | |
| 6 | 0.68 | 0.64 | 0.65 | 0.72 | |
| 7 | 0.60 | 0.58 | 0.58 | 0.68 | |
| 8 | 0.57 | 0.59 | 0.56 | 0.66 | |
| 9 | 0.60 | 0.59 | 0.57 | 0.66 | |
| 10 | 0.61 | 0.61 | 0.58 | 0.67 | |
| WES_MS + Bins | 1 | 0.47 | 0.43 | 0.44 | 0.48 |
| 2 | 0.40 | 0.41 | 0.40 | 0.44 | |
| 3 | 0.40 | 0.41 | 0.39 | 0.43 | |
| 4 | 0.48 | 0.45 | 0.45 | 0.50 | |
| 5 | 0.49 | 0.43 | 0.44 | 0.46 | |
| 6 | 0.40 | 0.39 | 0.39 | 0.46 | |
| 7 | 0.52 | 0.42 | 0.44 | 0.46 | |
| 8 | 0.44 | 0.43 | 0.42 | 0.44 | |
| 9 | 0.47 | 0.45 | 0.44 | 0.47 | |
| 10 | 0.42 | 0.39 | 0.39 | 0.43 | |
| WIIS_MS + Bins | 1 | 0.58 | 0.54 | 0.54 | 0.64 |
| 2 | 0.54 | 0.54 | 0.52 | 0.61 | |
| 3 | 0.56 | 0.56 | 0.54 | 0.64 | |
| 4 | 0.56 | 0.54 | 0.53 | 0.63 | |
| 5 | 0.55 | 0.54 | 0.53 | 0.65 | |
| 6 | 0.53 | 0.54 | 0.52 | 0.63 | |
| 7 | 0.54 | 0.57 | 0.54 | 0.65 | |
| 8 | 0.55 | 0.55 | 0.53 | 0.64 | |
| 9 | 0.57 | 0.56 | 0.54 | 0.66 | |
| 10 | 0.60 | 0.55 | 0.55 | 0.66 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wagner, J.; Oldenburg, J.; Nath, N.; Simm, S. Explainable AI Model Reveals Informative Mutational Signatures for Cancer-Type Classification. Cancers 2025, 17, 1731. https://doi.org/10.3390/cancers17111731
Wagner J, Oldenburg J, Nath N, Simm S. Explainable AI Model Reveals Informative Mutational Signatures for Cancer-Type Classification. Cancers. 2025; 17(11):1731. https://doi.org/10.3390/cancers17111731
Chicago/Turabian StyleWagner, Jonas, Jan Oldenburg, Neetika Nath, and Stefan Simm. 2025. "Explainable AI Model Reveals Informative Mutational Signatures for Cancer-Type Classification" Cancers 17, no. 11: 1731. https://doi.org/10.3390/cancers17111731
APA StyleWagner, J., Oldenburg, J., Nath, N., & Simm, S. (2025). Explainable AI Model Reveals Informative Mutational Signatures for Cancer-Type Classification. Cancers, 17(11), 1731. https://doi.org/10.3390/cancers17111731






