Establishment of Two Novel Ovarian Tumor Cell Lines with Characteristics of Mucinous Borderline Tumors or Dedifferentiated Carcinoma—Implications for Tumor Heterogeneity and the Complex Carcinogenesis of Mucinous Tumors
Simple Summary
Abstract
1. Introduction
2. Methods and Materials
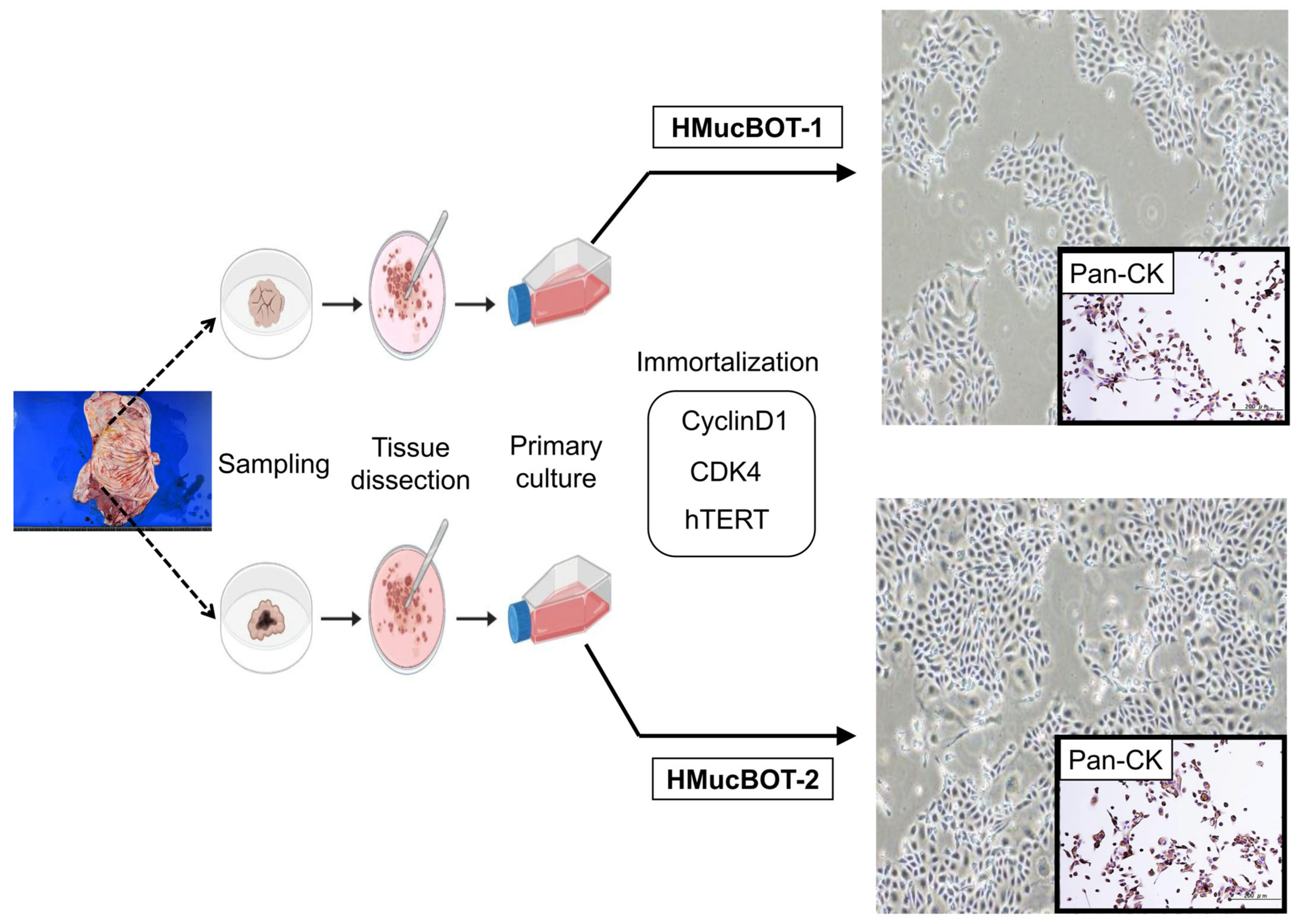
2.1. Cell Preparation and Establishment of the Cell Lines
2.2. Short Tandem Repeat (STR) Analysis
2.3. Mouse Tumorigenicity
2.4. Whole-Exome Sequencing
2.5. Immunohistochemistry Assay
2.6. Cell Proliferation Assay
2.7. Soft Agar Colony Formation Assay
2.8. Invasion Assay
2.9. Migration Assay
2.10. Responses to Chemotherapeutic Agents
2.11. Statistical Analysis
3. Results
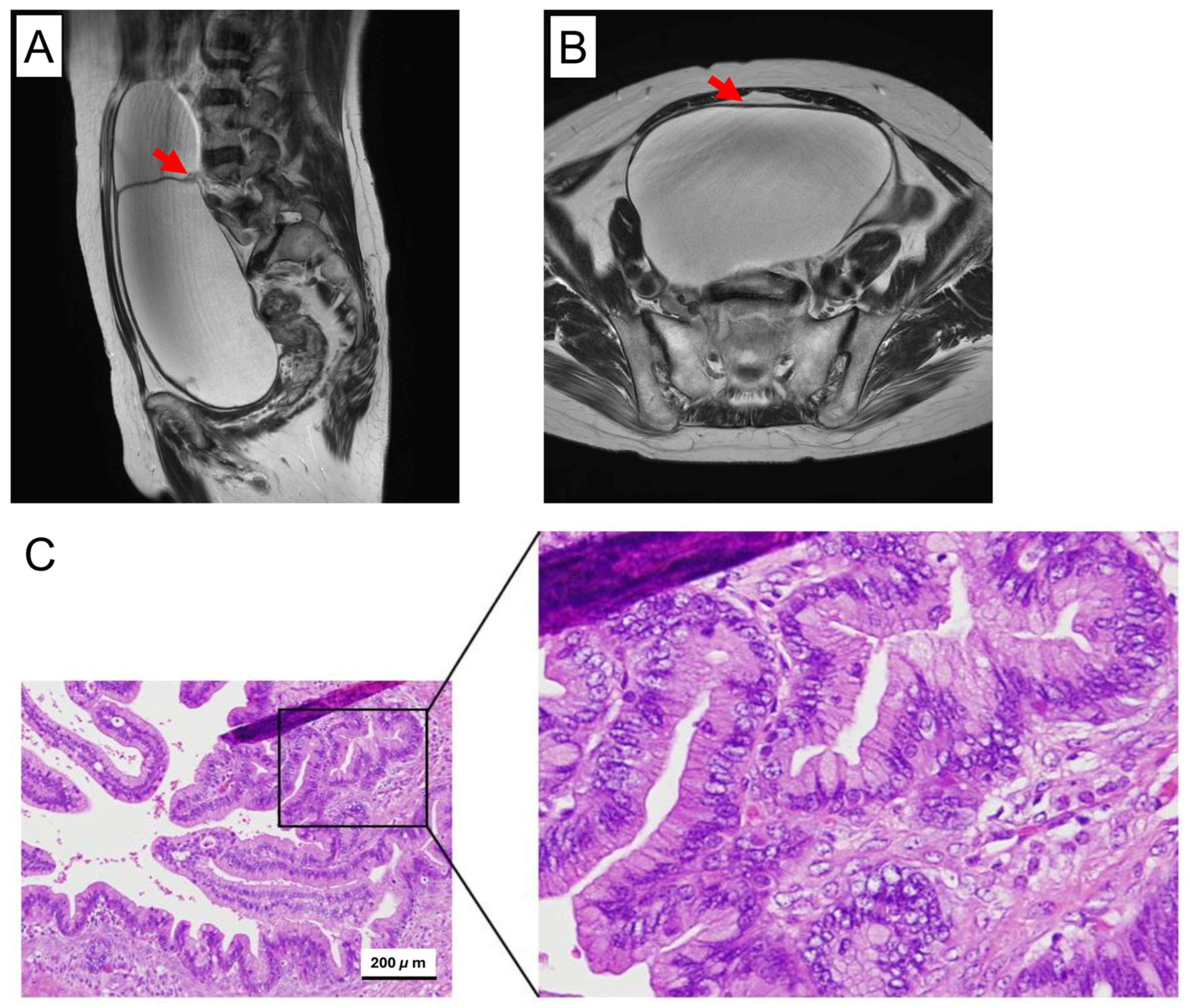
3.1. Histopathological Overview of the Original Tumor
3.2. Establishment of the MBOT Cell Lines
3.3. Histological Diversity of Mouse Tumors Derived from HMucBOT-1 and HMucBOT-2 Cells
3.4. HMucBOT-2 Cells Showed Genetic Alterations Distinct from the Original Tumor
3.5. Biological Behavior of HMucBOT-1 and HMucBOT-2 Cells
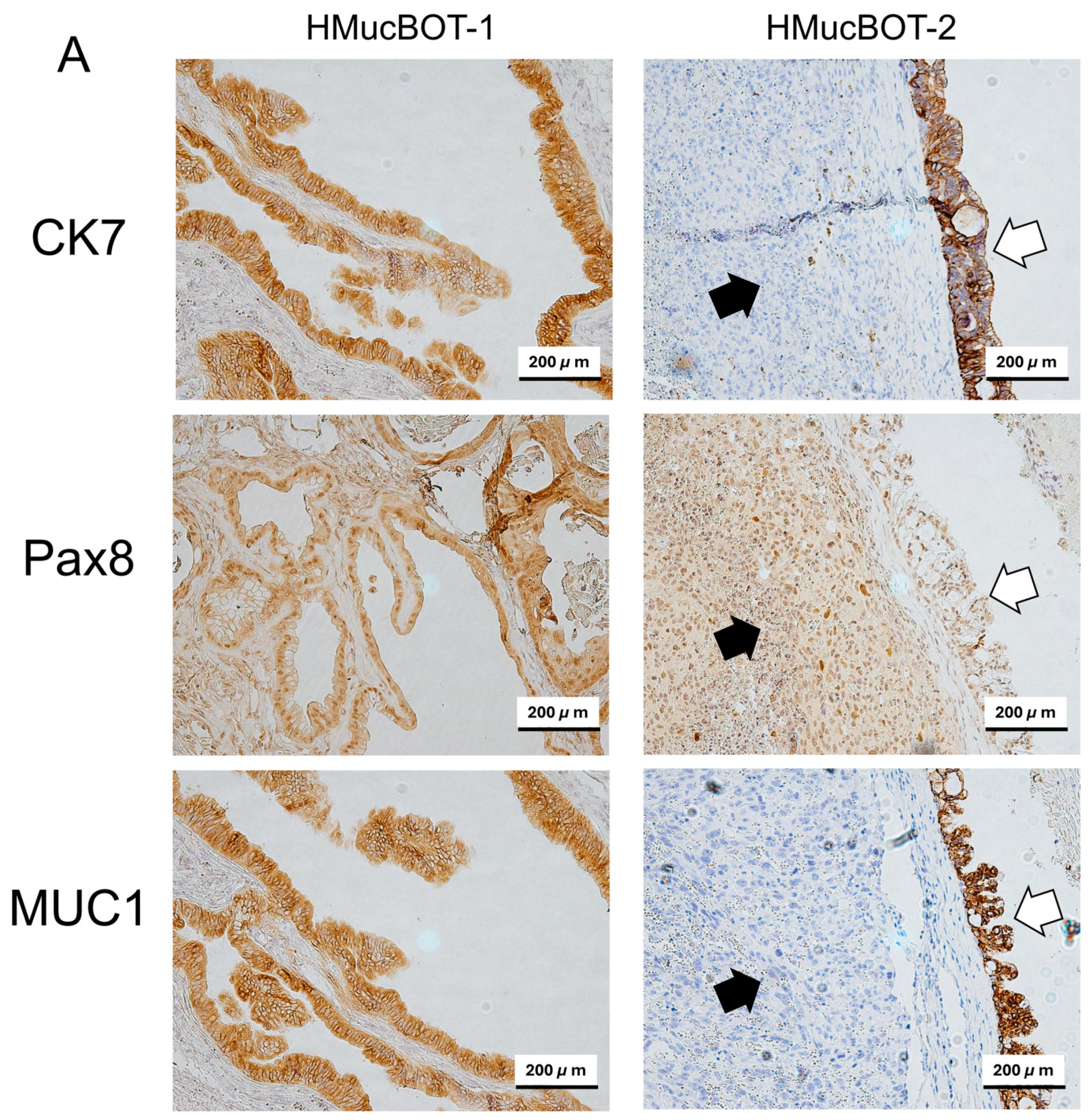
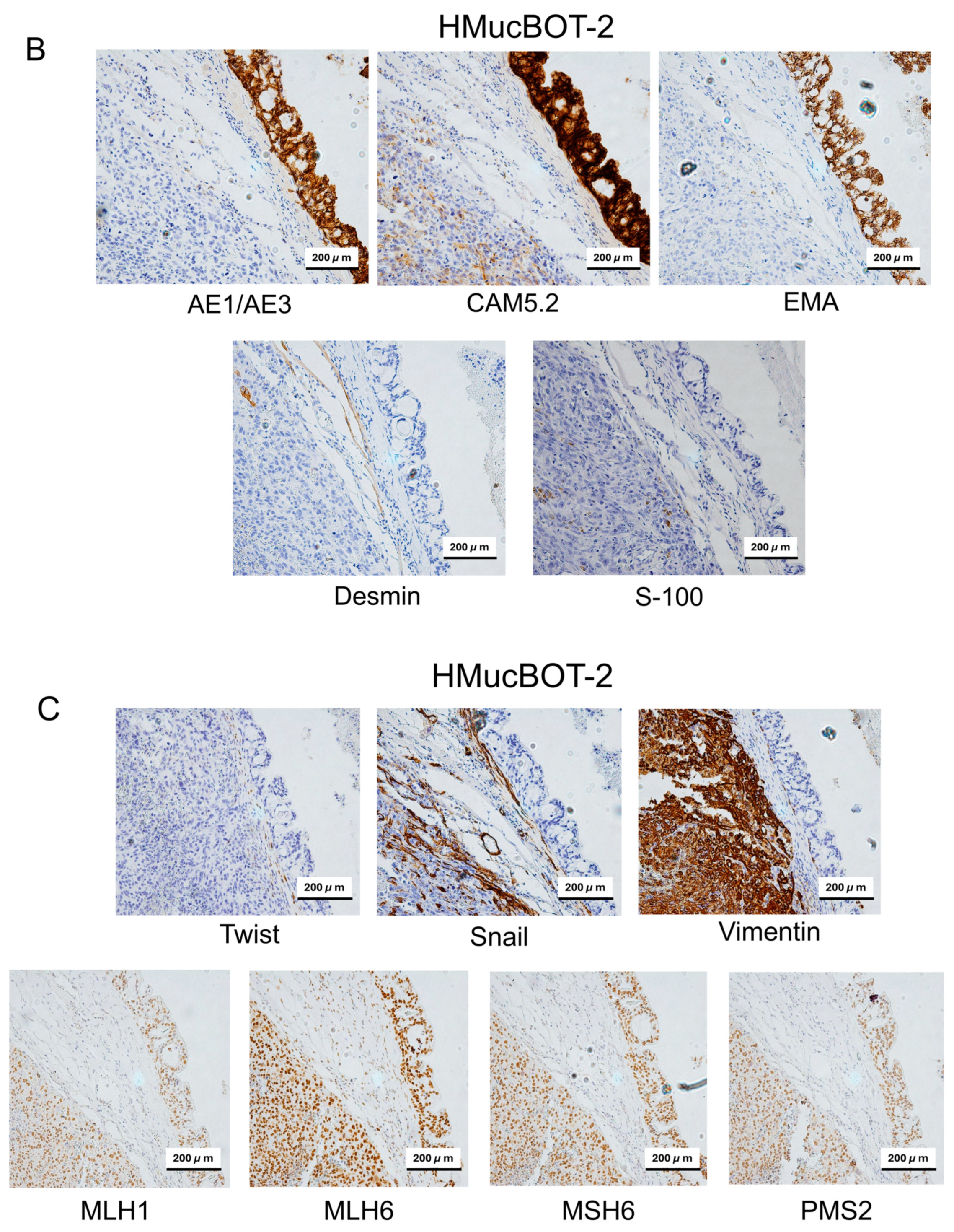
3.6. Characterization of Mouse Xenograft Tumors
3.7. The Response to Cytotoxic Drugs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Correction Statement
Abbreviations
| MBOT | Mucinous borderline ovarian tumor |
| STR | Short tandem repeat |
| BOT | Borderline ovarian tumor |
| MOC | Mucinous ovarian carcinoma |
| KRAS | Kirsten rat sarcoma virus |
| ARID1A | AT-rich interaction domain 1A |
| PIK3CA | Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha |
| TP53 | Tumor protein 53 |
| CNA | Copy number alteration |
| HMucBOT | Human mucinous borderline tumor |
| FFPE | Formalin-fixed paraffin-embedded |
| EMT | Epithelial-to-mesenchymal transition |
| MMR | Mismatch repair |
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global Cancer Statistics 2022: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Kratzer, T.B.; Giaquinto, A.N.; Sung, H.; Jemal, A. Cancer Statistics, 2025. CA Cancer J. Clin. 2025, 75, 10–45. [Google Scholar] [CrossRef]
- Alvarez, R.M.; Vazquez-Vicente, D. Fertility Sparing Treatment in Borderline Ovarian Tumours. Ecancermedicalscience 2015, 9, 507. [Google Scholar] [CrossRef] [PubMed]
- Camara, C.; Caroline Vos, M.; de Rooij, B.H.; Pijnenborg, J.M.A.; Boll, D.; van de Poll-Franse, L.V.; Ezendam, N.P.M. The Role of Positive Psychological Changes in Anxiety and Depression of Patients with Ovarian Tumors and Their Partners: An Observational Study from the Population-Based PROFILES Registry. Support. Care Cancer 2019, 27, 423–431. [Google Scholar] [CrossRef] [PubMed]
- Hauptmann, S.; Friedrich, K.; Redline, R.; Avril, S. Ovarian Borderline Tumors in the 2014 WHO Classification: Evolving Concepts and Diagnostic Criteria. Virchows Arch. 2017, 470, 125–142. [Google Scholar] [CrossRef]
- Silverberg, S.G.; Bell, D.A.; Kurman, R.J.; Seidman, J.D.; Prat, J.; Ronnett, B.M.; Copeland, L.; Silva, E.; Gorstein, F.; Young, R.H. Borderline Ovarian Tumors: Key Points and Workshop Summary. Hum. Pathol. 2004, 35, 910–917. [Google Scholar] [CrossRef]
- Văduva, C.-C.; Constantinescu, C.; Ţenovici, M.; Boldeanu, L.; Istrate-Ofiţeru, A.-M. Conservative Treatment of Borderline Ovarian Tumors: A Retrospective Study. Rom. J. Morphol. Embryol. 2023, 64, 143–150. [Google Scholar] [CrossRef]
- Seidman, J.D.; Soslow, R.A.; Vang, R.; Berman, J.J.; Stoler, M.H.; Sherman, M.E.; Oliva, E.; Kajdacsy-Balla, A.; Berman, D.M.; Copeland, L.J. Borderline Ovarian Tumors: Diverse Contemporary Viewpoints on Terminology and Diagnostic Criteria with Illustrative Images. Hum. Pathol. 2004, 35, 918–933. [Google Scholar] [CrossRef]
- Song, T.; Lee, Y.-Y.; Choi, C.H.; Kim, T.-J.; Lee, J.-W.; Bae, D.-S.; Kim, B.-G. Histologic Distribution of Borderline Ovarian Tumors Worldwide: A Systematic Review. J. Gynecol. Oncol. 2013, 24, 44–51. [Google Scholar] [CrossRef]
- Young, R. WHO Classification of Tumours of Female Reproductive Organs, 2014th ed.; IARC: Lyon, France, 2014; ISBN 978-92-832-2435-8. [Google Scholar]
- Nagase, S.; Ohta, T.; Takahashi, F.; Yaegashi, N. Board members of the 2020 Committee on Gynecologic Oncology of the Japan Society of Obstetrics and Gynecology Annual Report of the Committee on Gynecologic Oncology, the Japan Society of Obstetrics and Gynecology: Annual Patient Report for 2017 and Annual Treatment Report for 2012. J. Obstet. Gynaecol. Res. 2021, 47, 1631–1642. [Google Scholar] [CrossRef]
- WHO Classification of Tumours Editorial Board. Female Genital Tumours; WHO: Geneva, Switzerland, 2020; ISBN 978-92-832-4504-9. [Google Scholar]
- Wahab, S.A.; Tobler, J.J. MR Imaging of Epithelial Ovarian Neoplasms Part I: Benign and Borderline. Magn. Reson. Imaging Clin. North. Am. 2023, 31, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Herrington, C.S. Tumors of the Ovary, Maldeveloped Gonads, Fallopian Tube and Broad Ligament. Atlas of Tumor Pathology. Third Series, Fascicle 23 Robert E. Scully, Robert H. Young and Philip B. Clement. Armed Forces Institute of Pathology, Washington, DC, 1998. No. of Pages: 527. Price: $95.00. ISBN: 1881041433. J. Pathol. 1999, 189, 145. [Google Scholar] [CrossRef]
- Sun, L.; Li, N.; Song, Y.; Wang, G.; Zhao, Z.; Wu, L. Clinicopathologic Features and Risk Factors for Recurrence of Mucinous Borderline Ovarian Tumors: A Retrospective Study with Follow-up of More Than 10 Years. Int. J. Gynecol. Cancer 2018, 28, 1643–1649. [Google Scholar] [CrossRef]
- Karlsen, N.M.S.; Karlsen, M.A.; Høgdall, E.; Nedergaard, L.; Christensen, I.J.; Høgdall, C. Relapse and Disease Specific Survival in 1143 Danish Women Diagnosed with Borderline Ovarian Tumours (BOT). Gynecol. Oncol. 2016, 142, 50–53. [Google Scholar] [CrossRef] [PubMed]
- Song, T.; Lee, Y.-Y.; Choi, C.H.; Kim, T.-J.; Lee, J.-W.; Kim, B.-G.; Bae, D.-S. Prognosis in Patients with Serous and Mucinous Stage I Borderline Ovarian Tumors. Int. J. Gynecol. Cancer 2012, 22, 770–777. [Google Scholar] [CrossRef] [PubMed]
- Poonyakanok, V.; Warnnissorn, M.; Chaopotong, P. Oncological Outcomes and Risk Factors for Recurrence of Mucinous Borderline Ovarian Tumors: A 15-Year Experience at a Tertiary Center. J. Obstet. Gynaecol. Res. 2024, 50, 2081–2092. [Google Scholar] [CrossRef]
- Nakagawa, K.; Nakayama, K.; Nakamura, A.; Hadano, N.; Kurose, S.; Razia, S.; Aoki, S.; Kyo, S. A Novel Case of Recurrent Mucinous Borderline Ovarian Tumor: Early Relapse and Fatal Outcome. Reports 2022, 5, 15. [Google Scholar] [CrossRef]
- Wakazono, E.; Taki, M.; Watanabe, K.; Yamanoi, K.; Murakami, R.; Kakiuchi, N.; Yamaguchi, K.; Hamanishi, J.; Minamiguchi, S.; Ogawa, S.; et al. A Case Report of Mucinous Borderline Ovarian Tumor with Recurrence as Invasive Carcinoma with High Copy Number Alterations. Int. Cancer Conf. J. 2024, 13, 520–524. [Google Scholar] [CrossRef]
- Mackenzie, R.; Kommoss, S.; Winterhoff, B.J.; Kipp, B.R.; Garcia, J.J.; Voss, J.; Halling, K.; Karnezis, A.; Senz, J.; Yang, W.; et al. Targeted Deep Sequencing of Mucinous Ovarian Tumors Reveals Multiple Overlapping RAS-Pathway Activating Mutations in Borderline and Cancerous Neoplasms. BMC Cancer 2015, 15, 415. [Google Scholar] [CrossRef]
- Takahashi, M.; Nakamura, K.; Chiyoda, T.; Okada, C.; Nohara, S.; Takamatsu, R.; Yanazume, S.; Kobayashi, H.; Nishihara, H.; Yamagami, W. Transcriptome Concordance Between Borderline Tumors and Endometrioid Carcinoma: An Integrative Genomic Analysis. Cancer Med. 2025, 14, e70601. [Google Scholar] [CrossRef]
- Anglesio, M.S.; Kommoss, S.; Tolcher, M.C.; Clarke, B.; Galletta, L.; Porter, H.; Damaraju, S.; Fereday, S.; Winterhoff, B.J.; Kalloger, S.E.; et al. Molecular Characterization of Mucinous Ovarian Tumours Supports a Stratified Treatment Approach with HER2 Targeting in 19% of Carcinomas. J. Pathol. 2013, 229, 111–120. [Google Scholar] [CrossRef] [PubMed]
- Therachiyil, L.; Anand, A.; Azmi, A.; Bhat, A.; Korashy, H.M.; Uddin, S. Role of RAS Signaling in Ovarian Cancer. F1000Res 2022, 11, 1253. [Google Scholar] [CrossRef]
- Lee, Y.-J.; Lee, M.-Y.; Ruan, A.; Chen, C.-K.; Liu, H.-P.; Wang, C.-J.; Chao, W.-R.; Han, C.-P. Multipoint Kras Oncogene Mutations Potentially Indicate Mucinous Carcinoma on the Entire Spectrum of Mucinous Ovarian Neoplasms. Oncotarget 2016, 7, 82097–82103. [Google Scholar] [CrossRef] [PubMed]
- Sonkin, D.; Thomas, A.; Teicher, B.A. Cancer Treatments: Past, Present, and Future. Cancer Genet. 2024, 286–287, 18–24. [Google Scholar] [CrossRef]
- Hossain, M.M.; Nakayama, K.; Shanta, K.; Razia, S.; Ishikawa, M.; Ishibashi, T.; Yamashita, H.; Sato, S.; Iida, K.; Kanno, K.; et al. Establishment of a Novel In Vitro Model of Endometriosis with Oncogenic KRAS and PIK3CA Mutations for Understanding the Underlying Biology and Molecular Pathogenesis. Cancers 2021, 13, 3174. [Google Scholar] [CrossRef] [PubMed]
- Sohel, H.I.; Kiyono, T.; Zahan, U.F.; Razia, S.; Ishikawa, M.; Yamashita, H.; Kanno, K.; Sonia, S.B.; Nakayama, K.; Kyo, S. Establishment of a Novel In Vitro and In Vivo Model to Understand Molecular Carcinogenesis of Endometriosis-Related Ovarian Neoplasms. Int. J. Mol. Sci. 2025, 26, 1995. [Google Scholar] [CrossRef]
- Bono, Y.; Kyo, S.; Takakura, M.; Maida, Y.; Mizumoto, Y.; Nakamura, M.; Nomura, K.; Kiyono, T.; Inoue, M. Creation of Immortalised Epithelial Cells from Ovarian Endometrioma. Br. J. Cancer 2012, 106, 1205–1213. [Google Scholar] [CrossRef]
- Wölfel, T.; Hauer, M.; Schneider, J.; Serrano, M.; Wölfel, C.; Klehmann-Hieb, E.; De Plaen, E.; Hankeln, T.; Büschenfelde, K.-H.M.Z.; Beach, D. A p16INK4a-Insensitive CDK4 Mutant Targeted by Cytolytic T Lymphocytes in a Human Melanoma. Science 1995, 269, 1281–1284. [Google Scholar] [CrossRef]
- Kajiwara, K.; Tanemoto, T.; Wada, S.; Karibe, J.; Ihara, N.; Ikemoto, Y.; Kawasaki, T.; Oishi, Y.; Samura, O.; Okamura, K.; et al. Fetal Therapy Model of Myelomeningocele with Three-Dimensional Skin Using Amniotic Fluid Cell-Derived Induced Pluripotent Stem Cells. Stem Cell Rep. 2017, 8, 1701–1713. [Google Scholar] [CrossRef]
- Nakayama, K.; Miyazaki, K.; Kanzaki, A.; Fukumoto, M.; Takebayashi, Y. Expression and Cisplatin Sensitivity of Copper-Transporting P-Type Adenosine Triphosphatase (ATP7B) in Human Solid Carcinoma Cell Lines. Oncol. Rep. 2001, 8, 1285–1287. [Google Scholar] [CrossRef]
- Shiomi, K.; Kiyono, T.; Okamura, K.; Uezumi, M.; Goto, Y.; Yasumoto, S.; Shimizu, S.; Hashimoto, N. CDK4 and Cyclin D1 Allow Human Myogenic Cells to Recapture Growth Property without Compromising Differentiation Potential. Gene Ther. 2011, 18, 857–866. [Google Scholar] [CrossRef]
- Karst, A.M.; Drapkin, R. Primary Culture and Immortalization of Human Fallopian Tube Secretory Epithelial Cells. Nat. Protoc. 2012, 7, 1755–1764. [Google Scholar] [CrossRef]
- Maeda, T.; Tashiro, H.; Katabuchi, H.; Begum, M.; Ohtake, H.; Kiyono, T.; Okamura, H. Establishment of an Immortalised Human Ovarian Surface Epithelial Cell Line without Chromosomal Instability. Br. J. Cancer 2005, 93, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, B.; Buddenkote, T.; Smith, H.; Shah, M.; Freeman, S.; Hulse, D.; Funingana, G.; Alcaraz, M.-L.; Ortuzar, M.-C.; Brenton, J.; et al. Growth Kinetics of High-Grade Serous Ovarian Cancer Using Longitudinal Clinical Data—Implications for Early Detection. medRxiv 2024. [Google Scholar] [CrossRef]
- Chiang, Y.-C.; Cheng, W.-F.; Chang, M.-C.; Lu, T.-P.; Kuo, K.-T.; Lin, H.-P.; Hsieh, C.-Y.; Chen, C.-A. Establishment of a New Ovarian Cancer Cell Line CA5171. Reprod. Sci. 2015, 22, 725–734. [Google Scholar] [CrossRef]
- Wagner, P.L.; Stiedl, A.-C.; Wilbertz, T.; Petersen, K.; Scheble, V.; Menon, R.; Reischl, M.; Mikut, R.; Rubin, M.A.; Fend, F.; et al. Frequency and Clinicopathologic Correlates of KRAS Amplification in Non-Small Cell Lung Carcinoma. Lung Cancer 2011, 74, 118–123. [Google Scholar] [CrossRef]
- Rehkaemper, J.; Korenkov, M.; Quaas, A.; Rueschoff, J.; Pamuk, A.; Zander, T.; Hillmer, A.M.; Buettner, R.; Hoelscher, A.H.; Bruns, C.J.; et al. Amplification of KRAS and Its Heterogeneity in Non-Asian Gastric Adenocarcinomas. BMC Cancer 2020, 20, 587. [Google Scholar] [CrossRef] [PubMed]
- Birkeland, E.; Wik, E.; Mjøs, S.; Hoivik, E.A.; Trovik, J.; Werner, H.M.J.; Kusonmano, K.; Petersen, K.; Raeder, M.B.; Holst, F.; et al. KRAS Gene Amplification and Overexpression but Not Mutation Associates with Aggressive and Metastatic Endometrial Cancer. Br. J. Cancer 2012, 107, 1997–2004. [Google Scholar] [CrossRef]
- Balikani, L.; Policarpio-Nicolas, M.L.C. Undifferentiated Component of Dedifferentiated Endometrial Carcinoma Presenting as a “Small Round Cell” Malignancy in a Mediastinal Lymph Node, Mimicking Small Cell Carcinoma. Diagn. Cytopathol. 2023, 51, 519–524. [Google Scholar] [CrossRef]
- Tessier-Cloutier, B.; Kommoss, F.K.F.; Kolin, D.L.; Němejcová, K.; Smith, D.; Pors, J.; Stewart, C.J.R.; McCluggage, W.G.; Foulkes, W.D.; von Deimling, A.; et al. Dedifferentiated and Undifferentiated Ovarian Carcinoma: An Aggressive and Molecularly Distinct Ovarian Tumor Characterized by Frequent SWI/SNF Complex Inactivation. Mod. Pathol. 2024, 37, 100374. [Google Scholar] [CrossRef]
- Xu, Z.; Saikia, K.; Yuan, L. Dedifferentiated Endometrial Carcinoma Arising from Serous Carcinoma: Diagnostic Challenges and Recommendations. Gynecol. Oncol. Rep. 2023, 47, 101188. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Bae, H.; Kim, H.-S. Dedifferentiated Leiomyosarcoma of the Uterine Corpus with Heterologous Component: Clinicopathological Analysis of Five Consecutive Cases from a Single Institution and Comprehensive Literature Review. Diagnostics 2024, 14, 160. [Google Scholar] [CrossRef]
- Makhdoum, S.; Quddus, M.R.; Lomme, M.M.; Hansen, K.; Lawrence, W.D. Dedifferentiated Endometrial Adenocarcinoma with Neuroendocrine Differentiation and Ballooning-Cell Features: Report of a Rare Entity with an Unusual Histology. Human. Pathol. Case Rep. 2019, 15, 92–94. [Google Scholar] [CrossRef]
- Travaglino, A.; Raffone, A.; Gencarelli, A.; Saracinelli, S.; Riccardi, C.; Mollo, A.; Zullo, F.; Insabato, L. Clinico-Pathological Features Associated with Mismatch Repair Deficiency in Endometrial Undifferentiated/Dedifferentiated Carcinoma: A Systematic Review and Meta-Analysis. Gynecol. Oncol. 2021, 160, 579–585. [Google Scholar] [CrossRef] [PubMed]
- Tafe, L.J.; Garg, K.; Chew, I.; Tornos, C.; Soslow, R.A. Endometrial and Ovarian Carcinomas with Undifferentiated Components: Clinically Aggressive and Frequently Underrecognized Neoplasms. Mod. Pathol. 2010, 23, 781–789. [Google Scholar] [CrossRef]
- Ono, R.; Nakayama, K.; Nakamura, K.; Yamashita, H.; Ishibashi, T.; Ishikawa, M.; Minamoto, T.; Razia, S.; Ishikawa, N.; Otsuki, Y.; et al. Dedifferentiated Endometrial Carcinoma Could Be A Target for Immune Checkpoint Inhibitors (Anti PD-1/PD-L1 Antibodies). Int. J. Mol. Sci. 2019, 20, 3744. [Google Scholar] [CrossRef]
- Laban, M.; Chen, X.; Guo, B. Seromucinous and Mucinous Borderline Ovarian Tumors: We Need to Know More. Reprod. Sci. 2023, 30, 1684–1685. [Google Scholar] [CrossRef]
- Shylasree, T.S.; Mahajan, D.; Chaturvedi, A.; Menon, S.; Gupta, S.; Thakur, M.; Poddar, P.; Maheshwari, A. Clinicopathological and Oncological Outcomes of Borderline Mucinous Tumours of Ovary: A Large Case Series. Indian. J. Surg. Oncol. 2024, 15, 88–94. [Google Scholar] [CrossRef]
- Sharma, S.V.; Haber, D.A.; Settleman, J. Cell Line-Based Platforms to Evaluate the Therapeutic Efficacy of Candidate Anticancer Agents. Nat. Rev. Cancer 2010, 10, 241–253. [Google Scholar] [CrossRef]
- He, J.; Zhang, C.; Ozkan, A.; Feng, T.; Duan, P.; Wang, S.; Yang, X.; Xie, J.; Liu, X. Patient-Derived Tumor Models and Their Distinctive Applications in Personalized Drug Therapy. Mechanobiol. Med. 2023, 1, 100014. [Google Scholar] [CrossRef]
- Lakshmi Haridas, K.; Menon, A.; Deepthi, B. Borderline Ovarian Mucinous Tumor with Anaplastic Carcinomatous Mural Nodule: A Case Report. Gynecol. Oncol. Rep. 2021, 37, 100809. [Google Scholar] [CrossRef]
- Dundr, P.; Singh, N.; Nožičková, B.; Němejcová, K.; Bártů, M.; Stružinská, I. Primary Mucinous Ovarian Tumors vs. Ovarian Metastases from Gastrointestinal Tract, Pancreas and Biliary Tree: A Review of Current Problematics. Diagn. Pathol. 2021, 16, 20. [Google Scholar] [CrossRef] [PubMed]
- Hou, R.; Jiang, L.; Liu, D.; Lin, B.; Hu, Z.; Gao, J.; Zhang, D.; Zhang, S.; Iwamori, M. Lewis(y) Antigen Promotes the Progression of Epithelial Ovarian Cancer by Stimulating MUC1 Expression. Int. J. Mol. Med. 2017, 40, 293–302. [Google Scholar] [CrossRef]
- Lan, Y.; Ni, W.; Tai, G. Expression of MUC1 in Different Tumours and Its Clinical Significance (Review). Mol. Clin. Oncol. 2022, 17, 161. [Google Scholar] [CrossRef]
- Liu, H.; Li, Y.; Karsidag, M.; Tu, T.; Wang, P. Technical and Biological Biases in Bulk Transcriptomic Data Mining for Cancer Research. J. Cancer 2025, 16, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.H.; Mao, T.-L.; Vang, R.; Ayhan, A.; Wang, T.-L.; Kurman, R.J.; Shih, I.-M. Endocervical-Type Mucinous Borderline Tumors Are Related to Endometrioid Tumors Based on Mutation and Loss of Expression of ARID1A. Int. J. Gynecol. Pathol. 2012, 31, 297–303. [Google Scholar] [CrossRef] [PubMed]
- Fontana, B.; Gallerani, G.; Salamon, I.; Pace, I.; Roncarati, R.; Ferracin, M. ARID1A in Cancer: Friend or Foe? Front. Oncol. 2023, 13, 1136248. [Google Scholar] [CrossRef]
- Shi, Y.; Zheng, H.; Wang, T.; Zhou, S.; Zhao, S.; Li, M.; Cao, B. Targeting KRAS: From Metabolic Regulation to Cancer Treatment. Mol. Cancer 2025, 24, 9. [Google Scholar] [CrossRef]
- Krasinskas, A.M.; Moser, A.J.; Saka, B.; Adsay, N.V.; Chiosea, S.I. KRAS Mutant Allele-Specific Imbalance Is Associated with Worse Prognosis in Pancreatic Cancer and Progression to Undifferentiated Carcinoma of the Pancreas. Mod. Pathol. 2013, 26, 1346–1354. [Google Scholar] [CrossRef]
- Kandoth, C.; McLellan, M.D.; Vandin, F.; Ye, K.; Niu, B.; Lu, C.; Xie, M.; Zhang, Q.; McMichael, J.F.; Wyczalkowski, M.A.; et al. Mutational Landscape and Significance across 12 Major Cancer Types. Nature 2013, 502, 333–339. [Google Scholar] [CrossRef]
- Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; Murray, B.A.; et al. Distinct Patterns of Somatic Genome Alterations in Lung Adenocarcinomas and Squamous Cell Carcinomas. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Grasso, C.S.; Wu, Y.-M.; Robinson, D.R.; Cao, X.; Dhanasekaran, S.M.; Khan, A.P.; Quist, M.J.; Jing, X.; Lonigro, R.J.; Brenner, J.C.; et al. The Mutational Landscape of Lethal Castration-Resistant Prostate Cancer. Nature 2012, 487, 239–243. [Google Scholar] [CrossRef]
- Northcott, P.A.; Shih, D.J.H.; Peacock, J.; Garzia, L.; Sorana Morrissy, A.; Zichner, T.; Stütz, A.M.; Korshunov, A.; Reimand, J.; Schumacher, S.E.; et al. Subgroup-Specific Structural Variation across 1000 Medulloblastoma Genomes. Nature 2012, 488, 49–56. [Google Scholar] [CrossRef]
- Parsons, D.W.; Li, M.; Zhang, X.; Jones, S.; Leary, R.J.; Lin, J.C.-H.; Boca, S.M.; Carter, H.; Samayoa, J.; Bettegowda, C.; et al. The Genetic Landscape of the Childhood Cancer Medulloblastoma. Science 2011, 331, 435–439. [Google Scholar] [CrossRef] [PubMed]
- Pugh, T.J.; Weeraratne, S.D.; Archer, T.C.; Pomeranz Krummel, D.A.; Auclair, D.; Bochicchio, J.; Carneiro, M.O.; Carter, S.L.; Cibulskis, K.; Erlich, R.L.; et al. Medulloblastoma Exome Sequencing Uncovers Subtype-Specific Somatic Mutations. Nature 2012, 488, 106–110. [Google Scholar] [CrossRef]
- Jones, D.T.W.; Jäger, N.; Kool, M.; Zichner, T.; Hutter, B.; Sultan, M.; Cho, Y.-J.; Pugh, T.J.; Hovestadt, V.; Stütz, A.M.; et al. Dissecting the Genomic Complexity Underlying Medulloblastoma. Nature 2012, 488, 100–105. [Google Scholar] [CrossRef]
- Robinson, G.; Parker, M.; Kranenburg, T.A.; Lu, C.; Chen, X.; Ding, L.; Phoenix, T.N.; Hedlund, E.; Wei, L.; Zhu, X.; et al. Novel Mutations Target Distinct Subgroups of Medulloblastoma. Nature 2012, 488, 43–48. [Google Scholar] [CrossRef] [PubMed]
- Morin, R.D.; Mendez-Lago, M.; Mungall, A.J.; Goya, R.; Mungall, K.L.; Corbett, R.D.; Johnson, N.A.; Severson, T.M.; Chiu, R.; Field, M.; et al. Frequent Mutation of Histone-Modifying Genes in Non-Hodgkin Lymphoma. Nature 2011, 476, 298–303. [Google Scholar] [CrossRef]
- Dalgliesh, G.L.; Furge, K.; Greenman, C.; Chen, L.; Bignell, G.; Butler, A.; Davies, H.; Edkins, S.; Hardy, C.; Latimer, C.; et al. Systematic Sequencing of Renal Carcinoma Reveals Inactivation of Histone Modifying Genes. Nature 2010, 463, 360–363. [Google Scholar] [CrossRef]
- Gui, Y.; Guo, G.; Huang, Y.; Hu, X.; Tang, A.; Gao, S.; Wu, R.; Chen, C.; Li, X.; Zhou, L.; et al. Frequent Mutations of Chromatin Remodeling Genes in Transitional Cell Carcinoma of the Bladder. Nat. Genet. 2011, 43, 875–878. [Google Scholar] [CrossRef]
- Hillman, R.T.; Celestino, J.; Terranova, C.; Beird, H.C.; Gumbs, C.; Little, L.; Nguyen, T.; Thornton, R.; Tippen, S.; Zhang, J.; et al. KMT2D/MLL2 Inactivation Is Associated with Recurrence in Adult-Type Granulosa Cell Tumors of the Ovary. Nat. Commun. 2018, 9, 2496. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Chang, D.K.; Nones, K.; Johns, A.L.; Patch, A.-M.; Gingras, M.-C.; Miller, D.K.; Christ, A.N.; Bruxner, T.J.C.; Quinn, M.C.; et al. Genomic Analyses Identify Molecular Subtypes of Pancreatic Cancer. Nature 2016, 531, 47–52. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Kim, J.; Bowlby, R.; Mungall, A.J.; Robertson, A.G.; Odze, R.D.; Cherniack, A.D.; Shih, J.; Pedamallu, C.S.; Cibulskis, C.; et al. Integrated Genomic Characterization of Oesophageal Carcinoma. Nature 2017, 541, 169–175. [Google Scholar] [CrossRef]
- The Cancer Genome Atlas Research Network; Bass, A.J.; Thorsson, V.; Shmulevich, I.; Reynolds, S.M.; Miller, M.; Bernard, B.; Hinoue, T.; Laird, P.W.; Curtis, C.; et al. Comprehensive Molecular Characterization of Gastric Adenocarcinoma. Nature 2014, 513, 202–209. [Google Scholar] [CrossRef]
- Lee, Y.; Moon, S.; Seok, J.Y.; Lee, J.-H.; Nam, S.; Chung, Y.S. Characterization of the Genomic Alterations in Poorly Differentiated Thyroid Cancer. Sci. Rep. 2023, 13, 19154. [Google Scholar] [CrossRef]
- Linehan, A.; O’Reilly, M.; McDermott, R.; O’Kane, G.M. Targeting KRAS Mutations in Pancreatic Cancer: Opportunities for Future Strategies. Front Med. 2024, 11, 1369136. [Google Scholar] [CrossRef] [PubMed]
- Shahrouzi, P.; Forouz, F.; Mathelier, A.; Kristensen, V.N.; Duijf, P.H.G. Copy Number Alterations: A Catastrophic Orchestration of the Breast Cancer Genome. Trends Mol. Med. 2024, 30, 750–764. [Google Scholar] [CrossRef]
- Grist, E.; Friedrich, S.; Brawley, C.; Mendes, L.; Parry, M.; Ali, A.; Haran, A.; Hoyle, A.; Gilson, C.; Lall, S.; et al. Accumulation of Copy Number Alterations and Clinical Progression across Advanced Prostate Cancer. Genome Med. 2022, 14, 102. [Google Scholar] [CrossRef]
- Bassaganyas, L.; Pinyol, R.; Esteban-Fabró, R.; Torrens, L.; Torrecilla, S.; Willoughby, C.E.; Franch-Expósito, S.; Vila-Casadesús, M.; Salaverria, I.; Montal, R.; et al. Copy Number Alteration Burden Differentially Impacts Immune Profiles and Molecular Features of Hepatocellular Carcinoma. Clin. Cancer Res. 2020, 26, 6350–6361. [Google Scholar] [CrossRef]
- Perren, T.J. Mucinous Epithelial Ovarian Carcinoma. Ann. Oncol. 2016, 27 (Suppl. S1), i53–i57. [Google Scholar] [CrossRef]
- Prat, J. Pathology of Borderline and Invasive Cancers. Best. Pract. Res. Clin. Obstet. Gynaecol. 2017, 41, 15–30. [Google Scholar] [CrossRef]
- Cuatrecasas, M.; Villanueva, A.; Matias-Guiu, X.; Prat, J. K-Ras Mutations in Mucinous Ovarian Tumors: A Clinicopathologic and Molecular Study of 95 Cases. Cancer 1997, 79, 1581–1586. [Google Scholar] [CrossRef]
- Enomoto, T.; Weghorst, C.M.; Inoue, M.; Tanizawa, O.; Rice, J.M. K-Ras Activation Occurs Frequently in Mucinous Adenocarcinomas and Rarely in Other Common Epithelial Tumors of the Human Ovary. Am. J. Pathol. 1991, 139, 777–785. [Google Scholar]
- Xue, W.; Wu, K.; Guo, X.; Chen, C.; Huang, T.; Li, L.; Liu, B.; Chang, H.; Zhao, J. The Pan-Cancer Landscape of Glutamate and Glutamine Metabolism: A Comprehensive Bioinformatic Analysis across 32 Solid Cancer Types. Biochim. Biophys. Acta Mol. Basis Dis. 2024, 1870, 166982. [Google Scholar] [CrossRef] [PubMed]
- Jahangiri, L. Updates on Liquid Biopsies in Neuroblastoma for Treatment Response, Relapse and Recurrence Assessment. Cancer Genet. 2024, 288–289, 32–39. [Google Scholar] [CrossRef]
- Ohyama, H.; Hirotsu, Y.; Amemiya, K.; Mikata, R.; Amano, H.; Hirose, S.; Oyama, T.; Iimuro, Y.; Kojima, Y.; Mochizuki, H.; et al. Development of a Molecular Barcode Detection System for Pancreaticobiliary Malignancies and Comparison with Next-Generation Sequencing. Cancer Genet. 2024, 280–281, 6–12. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, T.; Nie, Q.; Chaudhary, L.N.; Basel, D.; Reddi, H.V. Methylation Signatures as Biomarkers for Non-Invasive Early Detection of Breast Cancer: A Systematic Review of the Literature. Cancer Genet. 2024, 282–283, 1–8. [Google Scholar] [CrossRef]
- He, S.; Xiao, X.; Lei, R.; Chen, J.; Huang, H.; Yilihamu, A.; Guo, M.; Tan, C.; Li, X.; Zhuang, Z.; et al. Establishment of Breast Phyllodes Tumor Cell Lines Preserving the Features of Phyllodes Tumors. BIO Integr. 2023, 4, 7. [Google Scholar] [CrossRef]





| Cell Line Name | Results (Tumor Formation in SCID Mice) | Remarks |
|---|---|---|
| HMucBOT-1 | 5/5 | Mucinous borderline tumor |
| HMucBOT-2 | 5/5 | Dedifferentiated carcinoma |
| Gene Type | Gene | Mutation (VAF%) | Plession Score | Cos. | CV. | CNA | eCN |
|---|---|---|---|---|---|---|---|
| HRD | ARID1A | p.N106 * (21.6) | 2 | Neutral | 2.5 | ||
| HRD | ARID1A | p.L2270Ifs * 8 (23.0) | 2 | Neutral | 2.5 | ||
| OG | KRAS | p.G12V (22.3) | 3 | 10,787 | P | Neutral | 2.8 |
| TSG | CDKN2A | Wild type | 3 | HD | 0.1 | ||
| TSG | ARID1B | p.W1937 * (19.3) | 2 | P | Neutral | 2.1 | |
| TSG | NF1 | p.K354N (31.1) | 2 | P | Neutral | 1.8 | |
| Other | GAA | p.S332L (12.1) | 0.5 | Neutral | …… |
| Gene Type | Gene | Mutation (VAF%) | Plession Score | Cos. | CV. | CNA | eCN |
|---|---|---|---|---|---|---|---|
| HRD | ARID1A | p.N106 * (32.2) | 2 | 0 | Neutral | 2.1 | |
| HRD | ARID1A | p.L2270Ifs * 8 (41.7) | 2 | 0 | Neutral | 2.1 | |
| HRD | BRIP1 | p.D153H (5.0) | 1 | 0 | Neutral | 2.0 | |
| OG | KRAS | p.G12V (31.6) | 3 | 10,787 | P | Amp | 3.1 |
| OG | ACSL3 | p.L186F (51.5) | 1.5 | 2 | Neutral | 2 | |
| OG | SUZ12 | p.V68A (6.2) | 1 | 0 | Neutral | 2 | |
| OG | TERT | p.I587V (32.3) | 1 | 0 | Amp | 3.3 | |
| OG | GRM3 | p.R716M (6.9) | 1 | 0 | Neutral | 2 | |
| OG | EWSR1 | p.R511L (8.3) | 1 | 0 | Neutral | 2 | |
| OG | HMGA1 | p.K46R (5.9) | 1 | 0 | Neutral | 2 | |
| OG | IRF4 | p.A16T (54.0) | 1 | 0 | Neutral | 2 | |
| OG | GNAS | p.S340A (50.1) | 1 | 0 | Neutral | 2 | |
| OG | EZH2 | p.C329S (6.6) | 1 | 0 | Neutral | 2 | |
| OG | BIRC6 | p.N2640K (5.4) | 0.5 | 0 | Neutral | 2 | |
| OG | MPL | p.S162T (5.8) | 0.5 | 0 | Neutral | 2.1 | |
| OG | CLIP1 | p.K1156I (6.6) | 0.5 | 0 | Amp | 3.1 | |
| OG | PTK6 | p.G10V (6.7) | 0.5 | 0 | Neutral | 2 | |
| OG | SPECC1 | p.K950R (5.1) | 0.5 | 0 | Neutral | 2 | |
| OG | CCND1 | c.415-2A>T (22.6) | 0.5 | 1 | Neutral | 2 | |
| OG | PDE4DIP | p.E1577G (6.7) | 0.5 | 0 | Neutral | 2.1 | |
| OG | CDK4 | c.633-1G>A (21.7) | 0.5 | 0 | Amp | 3.1 | |
| OG | CDK4 | c.219-2A>C (27.1) | 0.5 | 0 | Amp | 3.1 | |
| OG | CCND1 | c.414+1G>C (10.2) | 0 | 0 | Neutral | 2 | |
| OG | CCND1 | c.574+1G>A (8.6) | 0 | 0 | Neutral | 2 | |
| OG | CCND1 | c.724-1G>A (8.2) | 0 | 0 | Neutral | 2 | |
| OG | CDK4 | c.820-2A>T (9.6) | 0 | 0 | Amp | 3.1 | |
| OG | CDK4 | c.819+2T>A (10.7) | 0 | 0 | Amp | 3.1 | |
| OG | CDK4 | c.684-1G>A (6.4) | 0 | 0 | Amp | 3.1 | |
| OG | CDK4 | c.683+1G>C (8.9) | 0 | 1 | Amp | 3.1 | |
| OG | CDK4 | c.632+2T>C (13.9) | 0 | 0 | Amp | 3.1 | |
| OG | CDK4 | c.218+2T>C (10.4) | 0 | 0 | Amp | 3.1 | |
| TSG | ARID1B | p.W1937(41.3) | 2 | 0 | P | Neutral | 2 |
| TSG | NF1 | p.K354N (44.2) | 2 | 0 | P | Neutral | 2 |
| TSG | ARID1B | p.E1340(5.4) | 1.5 | 0 | Neutral | 2 | |
| TSG | ATP2B3 | c.2626-2A>G (5.2) | 1.5 | 0 | Neutral | 1.7 | |
| TSG | ARHGEF12 | p.Q1266L (6.5) | 1 | 0 | Neutral | 2 | |
| TSG | RAD21 | p.I17F (5.0) | 1 | 1 | Neutral | 1.9 | |
| TSG | PTPN13 | p.G2262V (5.8) | 1 | 0 | Neutral | 1.9 | |
| TSG | TPM3 | p.K6M (5.1) | 1 | 0 | Neutral | 2.1 | |
| TSG | ZFHX3 | p.M1294V (5.9) | 1 | 0 | Neutral | 2 | |
| TSG | SPEN | p.Y2819N (6.5) | 1 | 0 | Neutral | 2.1 | |
| TSG | UBR5 | p.P2036H (6.1) | 1 | 0 | Neutral | 1.9 | |
| TSG | UBR5 | p.C2034G (5.0) | 1 | 0 | Neutral | 1.9 | |
| TSG | KMT2D | p.A963T (5.5) | 0.5 | 0 | Amp | 3.1 | |
| TSG | CDKN2A | Wild type | 3 | HD | −0.7 | ||
| Other | APOB | p.E1601K (43.5) | 1 | 0 | - | ||
| Other | GAA | p.S332L (44.3) | 1 | 0 | - | ||
| Other | POLQ | p.L2352F (5.7) | 0.5 | 0 | Neutral | 2 | |
| Other | PRKCB | p.R415S (6.0) | 0.5 | 0 | Neutral | 2 | |
| Other | NBEA | c.6585+2958G>C (6.4) | 0.5 | 0 | Neutral | 1.9 | |
| Other | TTN | p.T25595P (5.0) | 0.5 | 0 | - | ||
| Other | TTN | p.V21151L (5.0) | 0.5 | 0 | - | ||
| Other | CTNND1 | p.P88T (5.2) | 0.5 | 0 | Neutral | 2 | |
| Other | PCBP1 | p.N48H (6.1) | 0.5 | 0 | Neutral | 2 | |
| Other | TMEM43 | p.G237E (6.9) | 0.5 | 0 | - | ||
| Other | AFDN | p.S1469R (6.2) | 0.5 | 0 | - | ||
| Other | TTN | p.I21153F (5.2) | 0.5 | 0 | - | ||
| Other | TTN | c.11311+3989T>G (5.3) | 0.5 | 0 | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sohel, H.I.; Zahan, U.F.; Kiyono, T.; Ishikawa, M.; Razia, S.; Kanno, K.; Yamashita, H.; Sonia, S.B.; Nakayama, K.; Kyo, S. Establishment of Two Novel Ovarian Tumor Cell Lines with Characteristics of Mucinous Borderline Tumors or Dedifferentiated Carcinoma—Implications for Tumor Heterogeneity and the Complex Carcinogenesis of Mucinous Tumors. Cancers 2025, 17, 1716. https://doi.org/10.3390/cancers17101716
Sohel HI, Zahan UF, Kiyono T, Ishikawa M, Razia S, Kanno K, Yamashita H, Sonia SB, Nakayama K, Kyo S. Establishment of Two Novel Ovarian Tumor Cell Lines with Characteristics of Mucinous Borderline Tumors or Dedifferentiated Carcinoma—Implications for Tumor Heterogeneity and the Complex Carcinogenesis of Mucinous Tumors. Cancers. 2025; 17(10):1716. https://doi.org/10.3390/cancers17101716
Chicago/Turabian StyleSohel, Hasibul Islam, Umme Farzana Zahan, Tohru Kiyono, Masako Ishikawa, Sultana Razia, Kosuke Kanno, Hitomi Yamashita, Shahataj Begum Sonia, Kentaro Nakayama, and Satoru Kyo. 2025. "Establishment of Two Novel Ovarian Tumor Cell Lines with Characteristics of Mucinous Borderline Tumors or Dedifferentiated Carcinoma—Implications for Tumor Heterogeneity and the Complex Carcinogenesis of Mucinous Tumors" Cancers 17, no. 10: 1716. https://doi.org/10.3390/cancers17101716
APA StyleSohel, H. I., Zahan, U. F., Kiyono, T., Ishikawa, M., Razia, S., Kanno, K., Yamashita, H., Sonia, S. B., Nakayama, K., & Kyo, S. (2025). Establishment of Two Novel Ovarian Tumor Cell Lines with Characteristics of Mucinous Borderline Tumors or Dedifferentiated Carcinoma—Implications for Tumor Heterogeneity and the Complex Carcinogenesis of Mucinous Tumors. Cancers, 17(10), 1716. https://doi.org/10.3390/cancers17101716








