Simple Summary
Pancreatic cancer remains one of the deadliest malignancies. This review summarizes preclinical mouse models in early pancreatic cancer research, focusing on genetically engineered models ranging from transgenic to inducible gene editing approaches. These models offer diverse platforms for deciphering carcinogenesis mechanisms and testing therapeutic strategies. We emphasize their applications in investigating environmental risk factors, aiming to provide researchers with a better understanding of their utility and limitations in translational pancreatic cancer research.
Abstract
Pancreatic cancer is characterized by late diagnosis, therapy resistance, and poor prognosis, necessitating the exploration of early carcinogenesis and prevention methods. Preclinical mouse models have evolved from cell line-based to human tumor tissue- or organoid-derived xenografts, now to humanized mouse models and genetically engineered mouse models (GEMMs). GEMMs, primarily driven by oncogenic Kras mutations and tumor suppressor gene alterations, offer a realistic platform for investigating pancreatic cancer initiation, progression, and metastasis. The incorporation of inducible somatic mutations and CRISPR-Cas9 screening methods has expanded their utility. To better recapitulate tumor initiation triggered by inflammatory cues, common pancreatic risk factors are being integrated into model designs. This approach aims to decipher the role of environmental factors as secondary or parallel triggers of tumor initiation alongside oncogenic burdens. Emerging models exploring pancreatitis, obesity, diabetes, and other risk factors offer significant translational potential. This review describes current mouse models for studying pancreatic carcinogenesis, their combination with inflammatory factors, and their utility in evaluating pathogenesis, providing guidance for selecting the most suitable models for pancreatic cancer research.
1. Introduction
Pancreatic cancer remains a tremendous challenge in oncology, ranking the third leading cause of cancer deaths [1]. Despite modest improvements in recent years, the 5-year relative survival rate for pancreatic cancer stands at 13%. Pancreatic ductal adenocarcinoma (PDAC), the most prevalent type of pancreatic cancer, is characterized by its aggressive nature, lack of early detection methods, and an immunosuppressive tumor microenvironment [2,3]. These factors contribute to a narrow therapeutic window, underscoring the critical need for research focused on risk factors and early carcinogenesis. Understanding the key genetic and environmental risk factors that drive pancreatic cancer development is essential for improving early detection strategies, prevention approaches, and treatment outcomes.
Epidemiological studies have identified several well-established risk factors for pancreatic cancer, including both modifiable lifestyle/environmental factors and non-modifiable factors such as age and genetic predisposition [4] (Figure 1). Among the modifiable factors, smoking is one of the most significant with the highest risk observed in heavy smokers compared to non-smokers, and the elevated risk persists for at least 10 years after cessation [5]. Alcohol consumption is also associated with increased risk, either through direct genotoxic effects on the pancreas or indirectly via the induction of pancreatitis [6]. Pancreatitis, particularly chronic pancreatitis, significantly elevates pancreatic cancer risk, with acute pancreatitis also contributing to short-term risk within the first three years after diagnosis [7,8]. Furthermore, obesity is another major modifiable risk factor, often associated with poor prognosis and treatment outcomes in pancreatic cancer patients [9,10]. Obesity-induced insulin resistance can lead to type 2 diabetes, which—along with non-obese diabetes—is a recognized risk factor. Infections with pathogens such as Hepatitis B and C viruses and Helicobacter pylori have also been implicated in pancreatic cancer development, suggesting a role for microbiome alterations in disease progression [11,12].
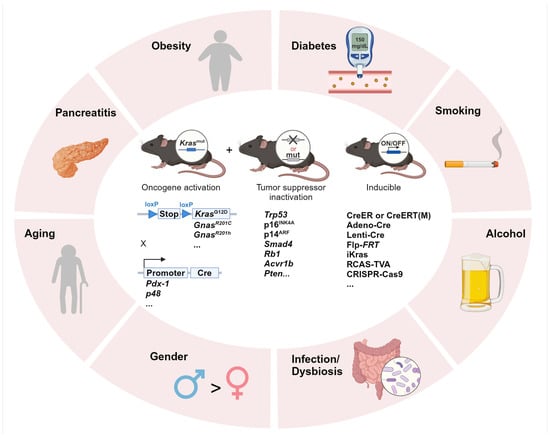
Figure 1.
Integrating environmental risk factors into pancreatic carcinogenesis research using GEMMs. Summary of the commonly investigated environmental risk factors in PDAC research and GEMMs of transgenic or inducible oncogene activation and/or tumor suppressor gene inactivation. Adeno-Cre: adenoviral-Cre; Lenti-Cre: lentiviral-Cre; Flp-FRT: flippase-FRT; iKras: doxycycline-inducible Kras; RCAS: replication-competent avian sarcoma-leukosis virus long terminal repeat with splice acceptor; TVA: tumor virus A; CRISPR: clustered regularly interspaced short palindromic repeats; Cas9: CRISPR-associated 9.
In addition to modifiable factors, non-modifiable contributors also play a significant role. Aging is strongly associated with pancreatic cancer incidence, with most PDAC diagnoses occurring between 60 and 80 years of age [13]. The accumulation of oncogenic mutations over time, along with inherited genetic predispositions, further increases susceptibility.
Understanding how these risk factors influence the molecular progression of pancreatic carcinogenesis is critical. Pancreatic carcinogenesis typically progresses through a series of sequential steps, often beginning with acinar-to-ductal metaplasia (ADM), which is initially reversible [14]. However, with persistent inflammation or injury, as induced by the risk factors described above, ADM can progress to premalignant pancreatic intraepithelial neoplasias (PanINs). These PanINs are graded from low-grade (PanIN-1 and PanIN-2) to high-grade (PanIN-3) lesions, which can ultimately develop into invasive PDAC [15]. This progression is induced or accompanied by the accumulation of genetic alterations, including activating mutations in the Kras oncogene (in >90% of PDAC cases) and inactivation of tumor suppressor genes such as CDKN2A, TP53, and SMAD4 [16]. Alternative precursor lesions, including intraductal papillary mucinous neoplasms (IPMNs) and mucinous cystic neoplasms (MCNs), represent additional routes to PDAC [17].
While existing preclinical mouse models have significantly advanced our understanding of pancreatic tumor biology and therapy, many fail to accurately replicate the early events of tumorigenesis or to include risk factors [18]. Traditional xenograft models using established cell lines typically represent advanced disease states and lack the immune components [19] (Figure 2A). Patient-derived xenografts (PDXs) and patient-derived organoid xenografts (PDOXs) have further improved the retention of parental tumor features such as heterogeneity, but they also rely on immunodeficient hosts and fail to replicate PDAC development in the context of native inflammatory and immune environment [20] (Figure 2B). To better capture tumor–immune dynamics, humanized mouse models, syngeneic models, and GEMMs have been developed. Humanized mouse models, lacking B, T, and NK cell activities, allow the engraftment of human tumor tissue and immune cells, thereby restoring some human-specific immune–tumor interactions while retaining interpatient tumor heterogeneity (Figure 2C). These models, however, exhibit mismatched stromal environments and do not recapitulate de novo tumor initiation [21,22]. Syngeneic models, involving the implantation of Panc02 [23] or KrasG12D; Trp53R172H; Pdx-1-Cre (KPC)-derived cell lines into immunocompetent mice, allow for the study of tumor-immune interactions in genetically matched hosts [24] (Figure 2D). Nevertheless, these models remain implantation-based and genetically uniform. In contrast, GEMMs support spontaneous tumor initiation and progression, capturing dynamic tumor–stroma–immune interactions. Yet, they are typically based on engineered mutations and may not incorporate the full spectrum of environmental, metabolic, or inflammatory factors that drive human disease. Constructing GEMMs can be labor-intensive and costly, requiring specialized equipment and expertise, and the success rate of generating GEMMs can also vary depending on the specific genetic alterations and the strain of mice used [25]. Despite these challenges, GEMMs remain a cornerstone of PDAC research, offering valuable insights into the molecular mechanisms of PDAC, particularly when used in conjunction with relevant risk factors.
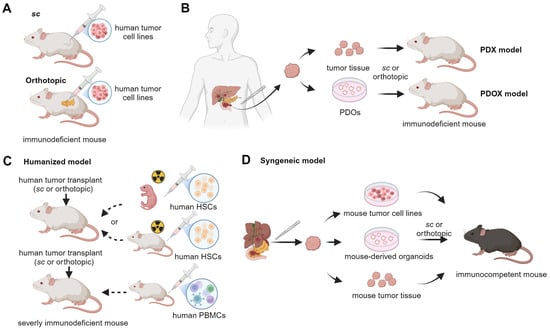
Figure 2.
Transplantation-based mouse models studying PDAC. (A) Subcutaneous (sc) and orthotopic models are generated in immunodeficient mice through the injection of tumor cells under the skin and into the pancreas, respectively. (B) Immunocompromised mice are transplanted either subcutaneously or orthotopically with human tumor tissues for PDX (patient-derived xenografts) models or with PDOs (patient-derived organoids) for PDOX (patient-derived organoid xenografts) models. (C) Humanized mouse models are developed by first injecting peripheral blood mononuclear cells (PBMCs) into an adult severely immunodeficient mouse or by first engrafting hematopoietic stem cells (HSCs) into irradiated neonatal or adult severely immunodeficient mice. Then, the mice are transplanted with human tumor tissues (cell line, PDX, PDOX, etc.). (D) Syngeneic mouse models are generated by injecting mouse tumor materials subcutaneously or orthotopically. These materials include cell lines, PDX (patient-derived xenografts), and PDOX (patient-derived orthotopic xenografts) developed from mouse tumors.
This review aims to provide an overview of the most widely used and promising GEMMs in pancreatic carcinogenesis, with a further emphasis on their applications in studying modifiable risk factors such as pancreatitis, obesity, and diabetes, which represent major modifiable risk factors for human PDAC. Additionally, we will explore the cutting-edge engineering methods including genome-wide and targeted CRISPR screening methods that have expanded the utility of animal models, enabling more precise and comprehensive studies of pancreatic cancer biology.
2. GEMMs
GEMMs have been instrumental in elucidating the mechanisms of PDAC initiation and progression. While comprehensive overviews of existing GEMMs are available elsewhere [26,27], here we briefly highlight the most relevant models that form the basis for studying genetic changes in PDAC, especially those that can later be used to explore how risk factors affect the disease.
Pioneering animal studies first utilized the elastase (EL) promoter controlled SV40 T-antigen [28], c-H-ras [29], Myc [30], and TGF-α [31] to study the roles of these genes during early cancer development. Among these, only TGF-α activation can induce ADM followed by fibrosis and eventually advanced PDAC, while the other genes primarily affect acinar cells. Moreover, these mice did not develop PDAC through well-defined PanIN. The discovery that KRAS mutations—especially the G12D isoform—occur in over 90% of PanIN-1 lesions [32] and increase with disease stage led to the development of the Lox-Stop-Lox (LSL)-KrasG12D/+ model (KC mice), controlled by Cre recombinase under pancreas-specific promoter Pdx-1 or p48 (Ptf1a) [33]. This model faithfully recapitulates human PanIN progression with the PanIN-1 lesions appearing by 8 weeks of age but rarely advances to invasive PDAC without additional genetic events or environmental stimulus.
Concomitant expression of heterozygous Trp53R172H and KrasG12D in the mouse pancreas, known as the KPC model, induce invasive and widely metastatic PDAC [34], while mice with homozygous Trp53 mutations (KPPC) develop more aggressive tumors with faster growth and shorter survival [35]. In contrast, mice with Trp53 depletion and KrasG12D/+ develop similar PanIN lesions and tumor stages as KPC mice but show significantly less metastasis [36]. Other combinations, such as Rb1 loss [37] or Ink4a/Arf deletion [38] with KrasG12D, also result in aggressive tumorigenesis due to the disruption of cell cycle regulation pathways.
Notably, concomitant deletion of Smad4 with mutant KrasG12D promotes tumor progression via IPMN/MCN lesions [39]. This SMAD4-dependent phenotype may be influenced by TGF-β superfamily ligands such as activin, as loss of activin receptor 1B (ACVR1B) accelerates IPMN-to-PDAC progression in the context of oncogenic Kras [40]. Interestingly, inactivation of TGFβR2, an upstream receptor of SMAD4, also enhances PanIN and PDAC development in a SMAD4-independent manner [41]. While TGF-β generally acts as a tumor suppressor, it may shift to a tumor-promoting role during later stages through interaction with the stroma, underscoring the complex and context-dependent role of TGF-β signaling in PDAC [42]. Similarly, Gnas gain-of-function mutation (R201C [43] or R201H [44]) induces low-grade IPMN that, when combined with oncogenic Kras, results in accelerated progression to invasive PDAC.
GEMMs also serve as valuable platforms for evaluating therapeutic strategies targeting the tumor microenvironment in PDAC. For example, stromal depletion through sonic hedgehog (SHH) loss [45] or αSMA+ myofibroblast ablation [46] enhances tumor aggressiveness in the background of oncogenic Kras, challenging classical views of the stroma as predominantly tumor-promoting [47]. Moreover, GEMMs enable preclinical testing of immunotherapies, such as checkpoint inhibitors (α-CTLA-4 and α-PD-L1) [48], as well as combination therapies (e.g., gemcitabine plus VEGFR/EGFR inhibitors [26]), which have shown encouraging outcomes. These models are thus critical for translating mechanistic findings into therapeutic advances, as reviewed in depth by Gopinathan et al. [49].
Although GEMMs may not fully capture the high mutation burden and abundant neoantigens seen in human PDAC, they offer unique advantages in modeling specific genetic alterations and their functional consequences. Importantly, these models serve as the foundation for incorporating various environmental and physiological risk factors, which will be discussed in detail in the following sections.
3. Inducible Somatic Gene Editing Models
To more closely mimic the somatic mutations that temporally occur in human pancreatic cancer, different inducible somatic mutation models prompt special tools to control gene expression. One of the most commonly applied tools is the inducible Cre system. The CreER system or the advanced version CreERT(M) utilizes a fusion of Cre recombinase with a modified estrogen receptor that allows tamoxifen-inducible Cre activity. Lee et al. used KrasLSL-G12D; Trp53flox/flox; Sox9CreERTM and KrasLSL-G12D; Trp53flox/flox; Ptf1aCreERTM to manipulate the gene expressions of adult mice in ductal and acinar cells, respectively, and found that ductal cells are more primed readily to form carcinoma upon oncogenic Kras and Trp53 deletion, while acinar cells require longer period and develop from widespread low-grade PanINs to PDAC [50]. To model PDAC-induced cachexia, KrasG12D/+; Ptf1aER-Cre/+; Ptenflox/flox (KPP) mice were generated with controlled activation of oncogenic Kras and Pten depletion post-natally. The mice developed moderate PDAC and also exhibited a progressive wasting phenotype that closely mimics human PDAC-associated cachexia [51]. The Cre recombinase can also be delivered by retrograde pancreatic ductal injection of both adenoviral-Cre (Adeno-Cre) and lentiviral-Cre (Lenti-Cre) vectors to activate the LSL-KrasG12D allele in the pancreas [52].
An alternative time- and host-specific recombination system that can be used alongside or instead of Cre-lox is flippase-FRT (Flp-FRT) [53]. In the FSF-KrasG12D/++; FSF-R26CAG−CreERT2/++; Trp53flox/flox; Pdx1-Flp mice, under the control of Pdx1-Flip, oncogenic Kras and CreERTM can be induced in the pancreas. After Tamoxifen administration, Cre functions to deplete Trp53 by the Cre-loxP system. This dual-recombinase system can also be applied to study the tumor microenvironment. For example, in FSF-KrasG12D/+; Trp53frt/frt; Pdx1-Flp; αSMA-Cre; Col1a1flox/flox mice, oncogenic Kras is controlled by Flp-FRT in the pancreas and the Col1a1 is depleted in the myofibroblasts, allowing for more sophisticated spatial genetic manipulations [54].
Additionally, the doxycycline-inducible Kras system (iKras) allows for temporary control of the gene expression. Collins et al. generated triple transgenic R26-rtTa-IRES-EGFP; TetO-KrasG12D; p48-Cre mice [55]. Upon doxycycline administration, the reverse tetracycline transactivator (rtTa)-doxycycline complex, which is specifically expressed in the pancreas controlled by p48-Cre, binds to the tetracycline-responsive element (TRE), allowing downstream KrasG12D expression. They found that inactivation of KrasG12D in established precursor lesions resulted in regression of these lesions, demonstrating that sustained KrasG12D expression is essential for tumor maintenance.
The avian sarcoma-leukosis virus-A-derived vector, namely RCAS (replication-competent avian sarcoma-leukosis virus long terminal repeat with splice acceptor), can deliver cDNA, shRNAs, non-coding RNAs, or CRISPR components to tumor virus A (TVA)-expressing cells under the control of promoters like elastase [56]. The RCAS-TVA system allows for versatile gene delivery in transgenic mice expressing the TVA receptor. In LSL-KrasG12D; Ptf1a-cre; elastase-tva mice, injected DF1 chicken fibroblasts producing RCAS-Wnt1, -GFP, or -β-cateninS37A selectively control Wnt signaling in pancreatic tissues already primed for oncogenic Kras expression [57]. This approach provides the flexibility to combine multiple genetic alterations for investigating various molecular mechanisms.
CRISPR-based somatic genome engineering has emerged as a powerful method for modeling PDAC with the advantage of increased speed, flexibility, and cost-effectiveness. Two main delivery methods have been developed for in vivo CRISPR editing in PDAC models: viral and non-viral approaches [58]. Viral delivery systems, particularly adeno-associated viruses (AAVs), have been widely adopted. In one common approach, KrasLSL-G12D; Rosa26CAG-LSL-Cas9; Ptf1aCre mice express oncogenic KrasG12D and Cas9 specifically in pancreatic cells. Researchers can then deliver vectors encoding sgRNAs by using self-complementary AAV targeting genes of interest, such as Trp53, via retrograde pancreatic ductal injection. Non-viral delivery methods, such as pancreas electroporation, offer an alternative approach with potentially reduced immunogenicity and larger cargo capacity [58]. In this technique, plasmid DNA encoding Cas9 and sgRNAs is directly introduced into pancreatic cells through electrical pulses. The choice of plasmids often depends on the specific experimental design. In mice that already express Cas9 (e.g., Rosa26CAG-LSL-Cas9 strains), only sgRNAs need to be delivered. Conversely, in wild-type mice, vectors containing both Cas9 and sgRNAs should be used. These CRISPR-based approaches have significantly accelerated PDAC research, allowing for rapid screening of potential tumor suppressors and oncogenes. The in vivo CRISPR screening method will be further reviewed in the following sections.
4. CRISPR-Cas9 Screening Models
CRISPR-Cas9-based genetic screening is a powerful tool for high-throughput functional genomics. Typically, sgRNA libraries are delivered via lentiviral vectors alongside Cas9 to induce gene knockouts in cell populations, with proliferation-based drop-out assays used to identify fitness or drug-sensitivity genes (Figure 3A) [59,60,61]. While informative, such systems lack the complexity of in vivo tumor biology. To address this limitation, CRISPR screening has been adapted to in vivo models, where transplantation-based approaches introduce perturbed cell pools derived from xenografts or established lines into host animals, offering a level of robustness comparable to in vitro systems [62,63,64] (Figure 3B). This enables assessment of gene function under physiological conditions and has revealed both conserved and context-specific metabolic dependencies. For example, in vivo screens, compared with in vitro screens, have identified genes regulating heme metabolism, oxidative phosphorylation, nucleotide synthesis, and antigen presentation as crucial for tumor formation [65]. Additionally, immune profiling using immunocompetent versus immunodeficient mice identified interferon-γ (IFNγ) as a key mediator of immune evasion and tumor suppression in pancreatic cancer [66]. Drug-assisted in vivo CRISPR screens using this platform have also uncovered targets that enhance the efficacy of gemcitabine or MEK inhibition, highlighting its therapeutic relevance [67].
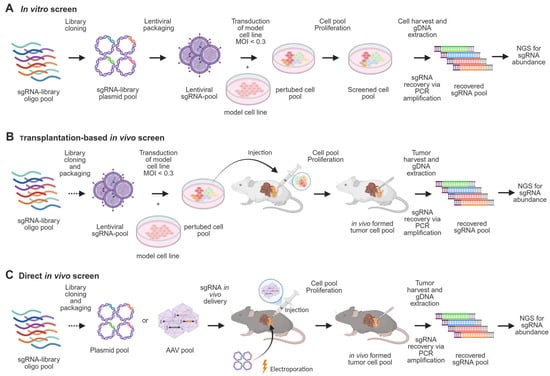
Figure 3.
Screening methods in PDAC research. (A) CRISPR in vitro screening begins with cloning an sgRNA pool into a library, which is used to produce a lentiviral pool. The model cell line is transduced at a multiplicity of infection (MOI) < 0.3 to ensure only one sgRNA integrates per cell. The perturbed cell pool is then expanded for the required number of doublings, followed by genomic DNA extraction and PCR amplification of sgRNA sequences to determine their abundance. (B) For in vivo screens using transplantation-based methods, the perturbed cell pool is injected intravenously, subcutaneously, or orthotopically into the host model. (C) Alternatively, somatic in vivo screening methods deliver sgRNA libraries directly into pancreatic tissue through electroporation or AAV injection. In both cases, tumor formation is monitored, and sgRNA abundance is analyzed post-harvest.
More recently, somatic CRISPR screening using GEMMs has enabled direct analysis of gene function during tumor initiation (Figure 3C). In these models, sgRNA libraries are delivered via electroporation or AAV into pancreatic tissue, allowing in situ gene editing within a native microenvironment [68]. For instance, a recent study used an in vivo screening method with an adeno-associated virus-sgRNA library targeting 125 recurrently mutated PDAC genes [69]. Using KC mice with inducible Kras and Cas9-GFP, the study identified accelerated tumor formation upon Cas9-mediated knockout of suppressor genes such as Cdkn2a, Rnf43, and Fbxw7, validating the somatic screen’s effectiveness in phenotyping relevant PanIN development hits. Notably, novel tumor progression regulators, SCAF1 and USP15, were discovered, shown to be suppressive in pancreatic tumorigenesis. These findings significantly contribute to understanding early PDAC carcinogenesis mechanisms, providing more insights into key regulators and pathways like TGF-β regulation, MAPK signaling, and NF-κB signaling. Although this method is pivotal for addressing tumor suppressors in early PDAC development, the library size was limited due to constraints such as viral titer and transduction efficiency. To ensure each cell received only one sgRNA and to maintain statistical robustness, infection efficiencies were optimized at low levels, which inherently restricted the number of sgRNAs that could be screened.
Emerging systems like organoids, either directly cultured from patient tissue such as IPMNs [70] or derived from induced pluripotent stem cells (iPSCs) [71], offer advantages over traditional 2D cell culture by better mimicking cancer heterogeneity, making them clinically relevant for transplantation-based screening methods. However, challenges such as sgRNA efficiency, clonal heterogeneity [72], and variability in transduction efficiency and proliferation among clones [73] lead to high false-positive rates. While such approaches hold promise for functional studies, they are not yet established for pancreatic or iPSC-derived organoids. Furthermore, most in vivo screens rely on CRISPR/Cas9 or RNAi, but the rapidly expanding CRISPR toolbox offers even greater potential. Emerging technologies such as CRISPR interference (CRISPRi), CRISPR activation (CRISPRa) [74], and prime editing [75], which allows precise base conversion without double-strand breaks, enable refined modulation of gene function. Additionally, CRISPR base editing has been applied in PDAC models to correct oncogenic KRAS mutations in organoids and cell lines, offering a potential tool for early intervention [76]. TALENs (transcription activator-like effector nucleases), though less widely used in PDAC research, remain a valuable alternative for precise gene editing, particularly when off-target minimization is critical [77]. Alternative CRISPR systems such as Cas12 [78] and Cas13 [79] also hold promise for refined gene regulation studies in vivo. Additionally, applying CRISPR screening to in vivo models that incorporate risk factors such as pancreatitis, obesity, or diabetes as discussed in the flowing sections could provide valuable insights into early PDAC carcinogenesis.
5. Risk Factor: Pancreatitis
Pancreatitis is a well-established risk factor for pancreatic cancer [80,81]. While evidence regarding acute pancreatitis (AP) as a risk factor remains inconsistent [8,82], numerous studies have shown that chronic pancreatitis (CP) is particularly significant in increasing the risk of PDAC [83,84,85]. CP is a complex disease influenced by factors such as smoking [86], alcohol abuse [87], pancreatic duct obstruction [88], autoimmunity [89], and genetic mutations [90,91,92]. Hereditary CP typically begins with recurrent episodes of AP that progress to CP over 10–15 years, significantly increasing the risk of PDAC in affected individuals by creating a chronic inflammatory microenvironment. This hereditary form, often caused by mutations in genes encoding protease serine 1 (PRSS1), chymotrypsin C (CTRC), serine protease inhibitor Kazal type 1 (SPINK1), and/or carboxypeptidase A1 (CPA1), serves as a genetic background for studying the mechanisms underlying pancreatitis and its progression to PDAC.
The p.R122H mutation in the PRSS1 gene that encodes the human cationic trypsinogen is the first reported mutation that was identified in hereditary CP [90]. In the subsequent years, several mutations like the p.N21I [93] and p.A16V [94,95] in the PRSS1 gene were also linked to hereditary CP. All these mutations lead to elevated levels of intrapancreatic trypsin that causes damage to the organ. Trypsin is produced and secreted by the pancreas in an inactive form called trypsinogen. Trypsinogen is converted to the active form by proteolysis of its activation peptide. This proteolysis occurs by enterokinase or trypsin itself. However, mutations in trypsinogen or the effector proteins of the protective mechanisms can disturb the homeostasis and lead to pancreatitis. Several studies also reported that increased autoactivation of trypsinogen is the underlying mechanism of action for other cationic trypsinogen mutations [96,97]. The first transgenic mouse model expressing the PRSS1 mutant p.R122H was generated by Archer et al., mimicking key features of human pancreatitis, including acinar cell damage, fibrosis, and inflammation [98]. However, the penetrance of the p.R122H mutation in this model was only 40%, compared to 80% in hereditary pancreatitis patients. Another model system, developed by Athwal et al. [99], expressed human PRSS1 wild-type protein or mutants (p.R122H and p.N29I), predisposing mice to pancreatitis. Upon stimulation with low-dose cerulein, these mice progressed to PDAC. Additionally, mutations that increase trypsinogen autoactivation, such as p.D23A [100] and p.D22N/K24R [101], have been used to generate mouse models that replicate hallmarks of pancreatitis, including inflammatory cell infiltration and acinar cell necrosis. These preclinical models provide valuable tools for studying pancreatitis and early steps of PDAC.
In humans, mutations in the CTRC gene are strongly associated with chronic pancreatitis [102,103]. Under normal physiological conditions, CTRC protects against pancreatitis by degrading intrapancreatic trypsinogen. Loss-of-function mutations in CTRC impair this protective mechanism, increasing susceptibility to pancreatitis. Interestingly, C57BL/6N mice naturally lack CTRC expression, with chymotrypsin B1 (CTRB1) serving as the major chymotrypsin isoform in these mice. Using CRISPR-Cas9 technology, Jancsó et al. [104] generated a Ctrb1-deficient mouse model, which exhibited more severe cerulein-induced pancreatitis compared to controls due to elevated intrapancreatic trypsin activation. These findings highlight the critical role of chymotrypsin in maintaining protease homeostasis in the pancreas.
Another defense strategy of the pancreas against trypsinogen autoactivation is the enzyme SPINK1, which inactivates prematurely activated trypsin and protects against pancreatitis [105]. Over 20 SPINK1 variants have been reported, with the p.N34S mutation being the most frequent [91]. Given SPINK1’s critical role in the human pancreas, several mouse models have been developed to elucidate its mechanisms and explore its potential as a therapeutic target. It is important to note that the mouse ortholog of human SPINK1 shares 63% amino acid sequence identity with the human protein, while rats possess homologous proteins known as pancreatic secretory trypsin inhibitors I and II (PSTI-I and -II) [106]. While Spink1-null mice (previously referred to as Spink3-null) did not survive beyond two weeks [107], heterozygous Spink1-deficient mice exhibited reduced SPINK1 expression without altered susceptibility to transient cerulein-induced pancreatitis. However, recent studies demonstrated that prolonged cerulein hyperstimulation in Spink1-KOhet mice led to hallmarks of acute pancreatitis, underscoring SPINK1’s protective role in trypsin-dependent pancreatitis [108]. Furthermore, when crossed with trypsinogen mutant strains (T7D23A or T7D22N/K24R), which spontaneously develop CP, Spink1-KOhet mice exhibited accelerated CP progression, suggesting that even slightly reduced SPINK1 levels can exacerbate CP development [108]. These findings underscore SPINK1’s significance as a disease modifier in pancreatitis and its potential relevance to therapeutic strategies.
Recent studies have shown that CP can also occur through mechanisms independent of trypsin-dependent pathways. The second most abundant enzyme in pancreatic juice, pro-carboxypeptidase A1 (proCPA1) [109], is activated to its functional form, CPA1, by trypsin and CTRC [110]. DNA sequencing of 944 individuals with CP showed that loss-of-function CPA1 variants play a role in the development of CP. Forty-one percent of individuals with functionally impaired CPA1 variants carried the c.768C > G (p.Asn256Lys) mutation [92]. In 2019, a mouse model that recapitulated CPA1-associated CP was developed [111]. Experiments using this model demonstrated that misfolding of the mutant CPA1 protein induces endoplasmic reticulum (ER) stress, leading to CP. Importantly, this model differs from trypsin-dependent models by highlighting ER stress as a distinct mechanism in CP pathogenesis, underscoring the complexity of the disease and the need to investigate multiple molecular pathways.
Cerulein, a cholecystokinin analogue, is the most commonly used agent to induce acute or chronic pancreatitis in rodents. The severity of pancreatic damage in cerulein-induced models is directly proportional to the dose and duration of administration, making it a reproducible and widely used experimental system. Studies by Guerra et al. indicate that activation of oncogenic KrasG12V in adult acinar cells alone does not lead to PDAC; however, pretreatment with cerulein to induce CP results in PDAC development, suggesting that cerulein sensitizes the pancreas for tumorigenesis [112]. Nonetheless, a major drawback of cerulein-induced mouse models is that they do not represent the clinical situation, where various factors such as alcohol abuse, gallstones, or genetic predisposition contribute to pancreatitis.
Several important factors must be considered when selecting a mouse model to study the role of CP in PDAC. The model should faithfully replicate the stages of human disease and allow disease induction with minimal artificial intervention. For instance, mouse models that develop pancreatitis while harboring genetic predispositions to PDAC provide a more patient-relevant platform. It is also important to note that early steps of PDAC and CP share similar histopathological features such as ADM and inflammatory cell infiltration, and hence these mouse models offer the opportunity to understand the different stages of the diseases in a context-dependent manner. Furthermore, CP and PDAC develop over years in humans, involving mutation accumulation, DNA damage, and neoplastic lesion formation. Mouse models offer the advantage of recapitulating these processes under controlled conditions, enabling comprehensive studies of genetic and environmental influences on disease progression. Combining the described genetic or chemically induced pancreatitis models with well-established cancer models, such as inducible KrasG12D mice, can enhance our understanding of CP as a risk factor for PDAC and provide insights into the molecular mechanisms driving this progression.
6. Risk Factor: Obesity
Obesity is a chronic disease characterized by abnormal and excessive fat accumulation, posing significant health risks worldwide. Since 1990, adult obesity rates have more than doubled, while adolescent obesity has quadrupled globally, contributing to a growing public health crisis [113]. A hallmark of obesity is hypertrophic and dysfunctional adipose tissue, which acts not only as an energy storage depot but also as an endocrine and immunologically active organ [114]. By releasing adipokines, it influences both local microenvironments and distant organs systemically [115]. Consequently, obesity is associated with systemic and chronic low-grade inflammation [116].
Over recent decades, obesity has been identified as a significant risk factor for numerous chronic diseases, including cardiovascular conditions, type 2 diabetes, and various cancers such as PDAC [9,117,118]. Additionally, obesity elevates the risk of developing pancreatic precancerous lesions [119]. Preclinical studies suggest several mechanisms by which obesity promotes pancreatic tumorigenesis, including hyperinsulinaemia and insulin resistance, hyperglycaemia, inflammation, altered cellular metabolism, hormone dysregulation, cellular stress, microbial dysbiosis, as well as activation of oncogenic drivers [120,121,122] (Figure 4A).
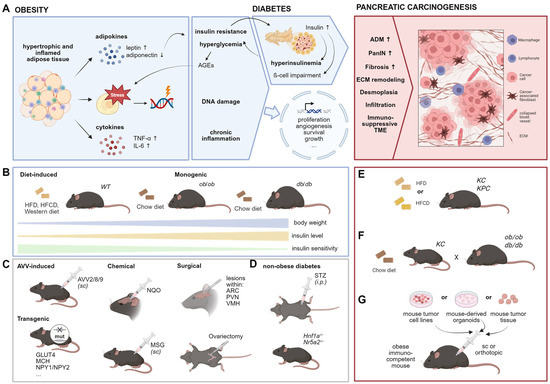
Figure 4.
Animal models to investigate the impact of obesity on PDAC. (A) Selected hypothesized links between obesity and early pancreatic carcinogenesis. Dysfunctional adipose tissue results in cellular stress and altered hormone and cytokine secretion causing DNA damage and a status of chronic inflammation, respectively (left). Obesity can further result in diabetes (middle) in which insulin resistance causes increased insulin production within pancreatic islets, which in turn leads to hyperinsulinaemia and β-cell impairment. The resulting hyperglycaemia promotes the generation of advanced glycation end products (AGEs) which further fuel cellular stress. The mentioned links may promote altered gene expression of processes associated with carcinogenesis, promoting features of early pancreatic carcinogenesis (right). ADM: acinar to ductal metaplasia, PanIN: pancreatic intraepithelial neoplasia, ECM: extracellular matrix, TME: tumor microenvironment. (B) Common obesity mouse models. HFD: high-fat diet, HFCD: high-fat, high-calorie diet. (C) Additional models to induce obesity in rodents. AVV: adeno-associated virus vector, sc: subcutaneous, NQO: 4-nitroquinoline 1-oxide, MSG: monosodium glutamate, ARC: arcuate nucleus, PVN: paraventricular nucleus, VMH: ventromedial hypothalamus. (D) Mouse models to induce non-obese diabetes. STZ: streptozotocin, i.p.: intraperitoneal (E) Combination of genetic obesity and PDAC models. (F) Diet-induced obesity in PDAC mouse models. (G) Syngeneic models to investigate tumor progression in the context of obesity.
Numerous murine obesity models have been studied in this context [123] and reviewed in detail by Kfoury et al., among others [123,124,125,126]. The most commonly used models are genetically engineered or diet-induced obesity (DIO) models (Figure 4B). For example, the ob/ob and db/db mice are monogenic models that represent mutations in the leptin gene or its receptor, respectively. These mutations lead to hyperphagia, resulting in severe early-onset obesity and, in the case of db/db mice, diabetes [127]. The ob/ob mice exhibit impaired glucose tolerance and insulin sensitivity reflecting a more pre-diabetic stage, whereas db/db mice are insulin resistant and display hyperinsulinemia as well as hyperleptinaemia. Obesity can also be modeled using other approaches, including nutritional, surgical, chemical, and AAV methods (Figure 4C), which differ from non-obese diabetes models (Figure 4D) that will be discussed in the following section.
To investigate the effect of obesity on pancreatic carcinogenesis, recent studies have combined obesity models with PDAC models. For instance, crossing ob/ob mice with KC mice generates KCO mice (Figure 4E) [128], which develop early-onset obesity and show increased pancreatic tumor burden. Chung et al. have detailed the impact of obesity on early pancreatic carcinogenesis and demonstrated that weight loss—induced either by caloric restriction or exogenous AAV-leptin administration—can reverse these effects [128]. They also identified an endocrine–exocrine signaling pathway involving cholecystokinin derived from stressed β-cells in pancreatic islets, highlighting local obesity-associated changes as pivotal in early pancreatic carcinogenesis. However, several studies have highlighted leptin as an adipokine with both systemic and obesity-independent effects, acting as a potent anti-apoptotic, proliferative, and inflammatory agent [129,130], which significantly influences the immune response [131,132] and thus may directly impact tumorigenesis. In vitro studies further showed that leptin increases the migratory capacity of pancreatic cancer cells [133,134,135]. Given these findings, models based on modifications in leptin signaling, such as ob/ob mice, may not accurately reflect the microenvironment of pre-neoplastic lesions or pancreatic cancer. Additionally, db/db mice are less suitable for studying obesity-related pancreatic carcinogenesis due to confounding effects from diabetes. Non-diabetic obesity models are therefore recommended to ensure more accurate results.
Another commonly used model to investigate obesity-related effects on pancreatic carcinogenesis is the DIO model (Figure 4F). These models involve feeding wild-type or transgenic mice high-caloric diets, resulting in a slower onset of obesity compared to ob/ob and db/db mice, which more closely mimics the human pathogenesis of obesity. Chronic high-fat diet (HFD) consumption in mice leads to glucose intolerance, impaired insulin sensitivity, and enhanced β-cell mass and proliferation [136,137,138,139], reflecting a pre-diabetic stage that progressively worsens over time. Several studies have demonstrated that HFD-induced obesity promotes pancreatic cancer development in KC and KPC mice [122,128,140,141,142,143,144]. HFD has been shown to increase inflammation, fibrosis, and PanIN lesions while accelerating progression into more aggressive PDAC [122,140,144,145]. Notably, it has been suggested that HFD contributes to pancreatic cancer not only through obesity and pre-diabetes but also by directly affecting carcinogenic processes [146]. However, the HFD of these studies varied in composition, fatty acid (FA) ratio, and duration and onset of intervention. Ead et al. reported that feeding KC mice an HFD with 60% of calories from fat (based on lard; 9:1 n-6:n-3 FA ratio) resulted in only mild effects on early pancreatic ADM without cancer development [142]. In contrast, a high-fat, high-calorie diet with 40% of calories from corn oil (50:1 n-6:n-3 FA ratio) in KC mice led to more advanced PanIN lesions [144] and tumor development already at an age of 3 months [141]. Furthermore, KPC mice fed a HFD (60% calories from fat; source: beef tallow and safflower oil) developed larger primary tumors and exhibited higher metastatic rates [143]. Compared to HFD, a high-carbohydrate diet displayed lower tumorigenic potential, while a high-protein diet was comparable to a normal diet [147]. These findings emphasize the significance of factors such as calorie percentage from fat, fat source, and n-6:n-3 FA ratio in influencing the obese phenotype and metabolic signaling linked to pancreatic carcinogenesis [121]. Additionally, when interpreting results from DIO models, researchers must consider confounding variables such as housing conditions, microbiome composition, age, gender, and feeding duration.
An alternative experimental design involves combining KC mice with gene therapy, where AAV vectors (AVV2/9/8) [140,148,149,150] are injected into the adipose tissue to induce obesity. Using this strategy, the precise timing of obesity induction based on the experimental design can be controlled, thereby avoiding undesired effects on embryonic development and hormone-related processes [126].
Since in humans, obesity develops gradually over time and pancreatic cancer-driving mutations typically occur in adulthood, GEMMs for PDAC that present mutations from birth may not accurately replicate human disease pathogenesis, especially when combining two GEMMs (e.g., KCO mice). Therefore, inducing pancreatic carcinogenesis in pre-existing obesity models by inducible oncogene mutations [151] or orthotopic implantation models—using PDAC cells, organoids [152] or tumor xeno-/allografts—offer promising tools to study the impact of obesity onset on pancreatic cancer progression (Figure 4G).
7. Risk Factor: Diabetes
Diabetes and pancreatic cancer share some concurrent hereditary predispositions, particularly in subgroups of patients with family history and younger age at diagnosis, and these concomitant genetic susceptibilities have been comprehensively reviewed recently by Popovic et al. [153]. Mutations in HNF1A, PDX1, and HNF1B, which affect pancreatic development, have been associated with increased risk of monogenic forms of maturity onset diabetes of the young (MODY types 3, 4, and 5) and pancreatic cancer [154,155]. For instance, by crossing KC mice with Hnf1aflox/flox mice, Kalisz et al. found that Hnf1a loss cooperates with the KrasG12D mutation to promote pancreatic carcinogenesis [156]. The nuclear receptor NR5A2 plays a dual role in pancreatic health and disease. Agonistic activation of NR5A2 promotes beta cell regeneration, potentially reversing type 1 diabetes [157]. Conversely, Nr5a2 heterozygosity sensitizes the pancreas to pancreatitis-induced inflammation, delays the ADM regeneration, and accelerates oncogenic Kras-driven carcinogenesis in mice [158,159]. Additionally, UCP2 gene polymorphisms are associated with both diabetes and PDAC, with UCP2 loss significantly reducing oncogenic Kras-induced PDAC growth [160]. These findings collectively indicate that certain genetic factors contribute to both diabetes and pancreatic cancer susceptibility, and designing mouse models for understanding these shared genetic pathways may provide new insights into prevention strategies and targeted therapies for both conditions.
Type 2 diabetes has been associated with an approximately two-fold overall increased risk of pancreatic cancer, with the highest risk observed in the first year following diabetes diagnosis gradually diminishing over time [161,162]. Interestingly, recent studies have revealed that type 1 diabetes patients also exhibit modestly elevated hazard ratios for pancreatic cancer, with men showing a slightly higher risk than women [163]. This similarity in risk profile between type 1 and type 2 diabetes suggests that the underlying mechanism may be related to shared metabolic pathology, likely chronic hyperglycaemia, rather than factors specific to type 2 diabetes. The strong association between hyperglycaemia and pancreatic cancer risk is further supported by the finding that each 0.56 mmol/L (10 mg/dL) increment in fasting glucose is associated with a 14% increase in the incidence rate of pancreatic cancer [164]. Experimental evidence from mouse models has provided mechanistic insights into this relationship. Non-obese-associated diabetes induced by streptozotocin accelerates the PDAC development in KC mice (Figure 4D), an effect that can be prevented by scavenger of reactive carbonyl species (RCS) and advanced glycation end-product (AGE) inhibitor, indicating that hyperglycaemia-derived carbonyl stress plays a crucial role in pancreatic carcinogenesis, particularly in high-risk diabetic individuals [165]. These findings also highlight the importance of glycaemic control and cancer surveillance in all diabetic patients, regardless of diabetes classification.
Diabetes and pancreatic cancer share some common risk factors, such as obesity—as earlier mentioned—highlighting the complexity and overlap of underlying mechanisms [166]. Obesity-induced insulin resistance is a critical link in this relationship, contributing to both diabetes and carcinogenesis [167]. Experimental evidence supports this connection, as obese-associated diabetic db/db mice demonstrate accelerated PDAC progression and metastasis [168]. To elucidate the specific role of insulin in PDAC initiation, Zhang et al. generated LSL-KrasG12D; Ins1+/−; Ins2−/−; Ptf1aCreER mice with genetic manipulations of Ins1 and Ins2 resulting in a sustained reduction in fasting insulin while maintaining unimpaired glucose homeostasis in female mice. Their findings revealed that HFD-mediated increases in insulin, in the absence of changes in fasting glucose, promoted PanIN development [169]. In a case-control study involving 973 patients with PDAC, Li et al. found that insulin or insulin secretagogues increased the risk of PDAC, while metformin administration in diabetic patients lowered the risk [170]. Preclinical studies have further demonstrated that metformin suppresses PDAC initiation and progression in KC and KPC mice [171]. These findings underscore the importance of further investigating the causal relationship between pharmacological therapies for type 2 diabetes and PDAC development.
New-onset diabetes is significantly associated with an increased incidence of pancreatic cancer compared to long-term diabetes (more than 2 years) prior to pancreatic cancer diagnosis, suggesting a potential reverse causation, where diabetes occurs as a consequence of PDAC development and may present as a subclinical manifestation [172]. Supporting this hypothesis, Parajuli et al. demonstrated in a KC mouse model that TGF-β signaling-activated apoptosis during PDAC progression caused depletion of β-cell mass, resulting in diabetic susceptibility [173]. This evidence that pancreatic cancer can trigger diabetes by damaging islet β-cells further contributes to the complexity.
In summary, it is crucial to recognize that chronic inflammation, hyperglycaemia, and insulin resistance can contribute to pancreatic carcinogenesis, consistent with the observation that long-term type 2 diabetes patients have an increased pancreatic cancer incidence compared to those with short-term type 2 diabetes (2–5 years) [172]. Besides, diabetes is often associated with comorbidities such as obesity and pancreatitis, which are themselves risk factors for pancreatic cancer. This complexity underscores the importance of precisely defining mouse models to avoid confounding factors and clearly delineate specific causal relationships. Conversely, pancreatic cancer development can also cause the onset of diabetes. These bidirectional relationships highlight the importance of carefully designing relevant mouse models to decipher the underlying mechanisms.
8. Other Risk Factors
Other than the previously mentioned risk factors that have been extensively investigated, additional cancer risk factors such as smoking, alcohol consumption, and infections [174] are also explored in GEMMs concerning the epidemiology of pancreatic cancer.
Patients with a history of pancreatitis, especially heavy smokers, exhibit a significantly elevated risk of developing pancreatic cancer [162]. Data regarding the impact of smoking on PDAC carcinogenesis is inconsistent. For instance, KC mice exposed to cigarette smoke for two weeks experienced significant weight loss but showed decreased cell proliferation in pancreatic ductal and acinar cells [175]. However, a six-week exposure to tobacco smoke promoted the formation of PanINs in KC mice, which was further exacerbated by the induction of chronic pancreatitis [176]. This discrepancy may arise from variations in the source, duration, and concentration of cigarette smoke used in different studies. Another investigation exposed KC mice to cigarette smoke for up to 20 weeks and observed overexpression of stem cell features such as Paf1 and Sox9; this stemness may contribute to the expansion of PanINs [177]. Nicotine, the primary addictive substance in cigarette smoke, has been shown to accelerate ADM and tumor formation by activating AKT/ERK/MYC signaling in both KC and KPC mice [178]. When interpreting these results, it is essential to consider confounding factors such as smoking-induced genotoxicity [179] and its interplay with other risk factors like diabetes, obesity, and chronic pancreatitis. These interactions have been extensively reviewed elsewhere [180].
Excessive alcohol consumption is associated with an increased risk of pancreatic cancer [6]. Moderate alcohol intake has been shown to lead to the development of advanced PanIN and subsequent PDAC in KC mice [181]. However, it is important to note that alcohol is also a leading cause of pancreatitis, which can act as a confounding factor in this context.
Infection with pathogens such as Hepatitis B virus and Helicobacter pylori has been identified as potential risk factors for PDAC, highlighting the role of microbiome alterations and dysfunction in the disease’s development. Studies have investigated the gut microbiome in KC mice [182] and the fecal and tumoral microbiome in KPC mice [183] to better understand the relationship between microbiome composition and PDAC development. Additionally, the oral microbiome, particularly Porphyromonas gingivalis, has been linked to an increased risk of pancreatic cancer [184], prompting further investigation using GEMMs. Administration of Porphyromonas gingivalis induced pancreatic ADM in wild-type mice and accelerated PDAC progression from PanIN lesions in KrasG12D/+; Ptf1aER-Cre/+ mice, while also altering the intrapancreatic microbiome composition [185]. Furthermore, ablation of the gut microbiome in KrasG12D/+; PTENflox/+; Pdx1-Cre mice resulted in decreased tumor growth and a reduction in the suppressive immune microenvironment [186]. Beyond these examples, recent studies have manipulated the gut microbiome through methods such as fecal microbial transplants and systemic antibiotics or antifungals in mouse models to explore specific microbiome relationships with pancreatic carcinogenesis. This area of research has been extensively reviewed, emphasizing the potential for microbial modification as a therapeutic strategy against pancreatic cancer [187,188].
9. Conclusions and Perspectives
Preclinical mouse models offer diverse approaches for investigating PDAC tumor development and identifying potential therapeutic targets. While xenograft models derived from PDAC cell lines, tumor tissues, or PDOs allow for genetic manipulation and drug testing in a relatively high-throughput manner, they have limitations in monitoring early carcinogenesis and testing early triggers. Humanized models add another dimension by better mimicking the human immune system but still fall short in replicating the complete process of early tumor development. GEMMs, particularly those manipulating somatic gene expression, are invaluable for exploring genetic alterations in early pancreatic carcinogenesis within the context of a competent immune system. Although often time consuming, these models provide crucial insights into tumor initiation and progression. The new introduction of large-scale CRISPR screening has further extended the utility of GEMMs, enabling unbiased identification of key genetic players in PDAC development.
Recent research has increasingly focused on the role of inflammatory triggers in PDAC initiation. Applying environmental risk factors or targeting underlying mechanisms in GEMMs can broaden our understanding of carcinogenesis, potentially leading to early prevention strategies and the development of druggable targets to halt cancer progression at its earliest stages.
However, caution is necessary when designing and interpreting results from mouse models. While GEMMs like the KPC model display good tumor heterogeneity, they may not consistently correlate with all clinical features seen in PDAC patients, such as cachexia. Off-target effects should also be considered, particularly when using Cre-lox systems. For instance, drivers like PDX-1 and Ptf1a can be expressed in organs other than the pancreas during embryonic development. Notably, the development of thymic tumors in KC mice has been reported and should be accounted for to avoid misinterpretation of results [189]. Furthermore, inflammatory factors are often interconnected and can act as confounders for each other, necessitating the careful interpretation of results. Finally, it is crucial to verify the translational relevance of findings from mouse models in human systems before drawing conclusions.
Author Contributions
Conceptualization, B.Y., A.-K.F., E.H., T.V.I., H.L., M.B., M.G., P.M. and J.R.; writing—original draft preparation, B.Y., A.-K.F., T.V.I. and E.H.; visualization, B.Y., A.-K.F. and E.H.; writing—review and editing, B.Y., A.-K.F., T.V.I., E.H., H.L., M.B., M.G., P.M. and J.R.; funding acquisition, H.L., M.B., M.G., P.M. and J.R. All authors have read and agreed to the published version of the manuscript.
Funding
The work was conducted within the Research Training Group InCuPanc (GRK 2751/1-2022 Project number: 449501615) funded by the DFG.
Acknowledgments
The figures were created with BioRender.com.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zheng, Y.; Yang, F.; Zhu, L.; Zhu, X.-Q.; Wang, Z.-F.; Wu, X.-L.; Zhou, C.-H.; Yan, J.-Y.; Hu, B.-Y.; et al. The molecular biology of pancreatic adenocarcinoma: Translational challenges and clinical perspectives. Signal Transduct. Target. Ther. 2021, 6, 249. [Google Scholar] [CrossRef] [PubMed]
- Mizrahi, J.D.; Surana, R.; Valle, J.W.; Shroff, R.T. Pancreatic cancer. Lancet 2020, 395, 2008–2020. [Google Scholar] [CrossRef] [PubMed]
- Capasso, M.; Franceschi, M.; Rodriguez-Castro, K.I.; Crafa, P.; Cambiè, G.; Miraglia, C.; Barchi, A.; Nouvenne, A.; Leandro, G.; Meschi, T.; et al. Epidemiology and risk factors of pancreatic cancer. Acta Biomed. 2018, 89, 141–146. [Google Scholar] [CrossRef]
- Iodice, S.; Gandini, S.; Maisonneuve, P.; Lowenfels, A.B. Tobacco and the risk of pancreatic cancer: A review and meta-analysis. Langenbeck’s Arch. Surg. 2008, 393, 535–545. [Google Scholar] [CrossRef]
- Wang, Y.-T.; Gou, Y.-W.; Jin, W.-W.; Xiao, M.; Fang, H.-Y. Association between alcohol intake and the risk of pancreatic cancer: A dose–response meta-analysis of cohort studies. BMC Cancer 2016, 16, 212. [Google Scholar] [CrossRef]
- Gandhi, S.; de la Fuente, J.; Murad, M.H.; Majumder, S. Chronic Pancreatitis Is a Risk Factor for Pancreatic Cancer, and Incidence Increases with Duration of Disease: A Systematic Review and Meta-analysis. Clin. Transl. Gastroenterol. 2022, 13, e00463. [Google Scholar] [CrossRef]
- Ma, D.-M.; Dong, X.-W.; Han, X.; Ling, Z.; Lu, G.-T.; Sun, Y.-Y.; Yin, X.-D. Pancreatitis and Pancreatic Cancer Risk. Technol. Cancer Res. Treat. 2023, 22, 15330338231164875. [Google Scholar] [CrossRef]
- Calle, E.E.; Rodriguez, C.; Walker-Thurmond, K.; Thun, M.J. Overweight, obesity, and mortality from cancer in a prospectively studied cohort of U.S. adults. N. Engl. J. Med. 2003, 348, 1625–1638. [Google Scholar] [CrossRef]
- Berrington de Gonzalez, A.; Sweetland, S.; Spencer, E. A meta-analysis of obesity and the risk of pancreatic cancer. Br. J. Cancer 2003, 89, 519–523. [Google Scholar] [CrossRef]
- Zanini, S.; Renzi, S.; Limongi, A.R.; Bellavite, P.; Giovinazzo, F.; Bermano, G. A review of lifestyle and environment risk factors for pancreatic cancer. Eur. J. Cancer 2021, 145, 53–70. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.H.; Fu, J.J.; Wang, X.L.; Zhu, J.Y.; Ye, X.H.; Chen, S.D. Hepatitis B or C viral infection and risk of pancreatic cancer: A meta-analysis of observational studies. World J. Gastroenterol. 2013, 19, 4234–4241. [Google Scholar] [CrossRef]
- Grigorescu, R.R.; Husar-Sburlan, I.A.; Gheorghe, C. Pancreatic Cancer: A Review of Risk Factors. Life 2024, 14, 980. [Google Scholar] [CrossRef] [PubMed]
- Marstrand-Daucé, L.; Lorenzo, D.; Chassac, A.; Nicole, P.; Couvelard, A.; Haumaitre, C. Acinar-to-Ductal Metaplasia (ADM): On the Road to Pancreatic Intraepithelial Neoplasia (PanIN) and Pancreatic Cancer. Int. J. Mol. Sci. 2023, 24, 9946. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Xie, K.; Zheng, S. Molecular Biomarkers of Pancreatic Intraepithelial Neoplasia and Their Implications in Early Diagnosis and Therapeutic Intervention of Pancreatic Cancer. Int. J. Biol. Sci. 2016, 12, 292–301. [Google Scholar] [CrossRef]
- Hu, H.-F.; Ye, Z.; Qin, Y.; Xu, X.-W.; Yu, X.-J.; Zhuo, Q.-F.; Ji, S.-R. Mutations in key driver genes of pancreatic cancer: Molecularly targeted therapies and other clinical implications. Acta Pharmacol. Sin. 2021, 42, 1725–1741. [Google Scholar] [CrossRef]
- de Wilde, R.F.; Hruban, R.H.; Maitra, A.; Offerhaus, G.J.A. Reporting precursors to invasive pancreatic cancer: Pancreatic intraepithelial neoplasia, intraductal neoplasms and mucinous cystic neoplasm. Diagn. Histopathol. 2012, 18, 17–30. [Google Scholar] [CrossRef]
- Mallya, K.; Gautam, S.K.; Aithal, A.; Batra, S.K.; Jain, M. Modeling pancreatic cancer in mice for experimental therapeutics. Biochim. Biophys. Acta Rev. Cancer 2021, 1876, 188554. [Google Scholar] [CrossRef]
- Fernandez, J.L.; Årbogen, S.; Sadeghinia, M.J.; Haram, M.; Snipstad, S.; Torp, S.H.; Einen, C.; Mühlenpfordt, M.; Maardalen, M.; Vikedal, K.; et al. A Comparative Analysis of Orthotopic and Subcutaneous Pancreatic Tumour Models: Tumour Microenvironment and Drug Delivery. Cancers 2023, 15, 5415. [Google Scholar] [CrossRef]
- Wang, E.; Xiang, K.; Zhang, Y.; Wang, X.-F. Patient-derived organoids (PDOs) and PDO-derived xenografts (PDOXs): New opportunities in establishing faithful pre-clinical cancer models. J. Natl. Cancer Cent. 2022, 2, 263–276. [Google Scholar] [CrossRef]
- He, M.; Henderson, M.; Muth, S.; Murphy, A.; Zheng, L. Preclinical mouse models for immunotherapeutic and non-immunotherapeutic drug development for pancreatic ductal adenocarcinoma. Ann. Pancreat. Cancer 2020, 3, 7. [Google Scholar] [CrossRef] [PubMed]
- Guil-Luna, S.; Sedlik, C.; Piaggio, E. Humanized Mouse Models to Evaluate Cancer Immunotherapeutics. Annu. Rev. Cancer Biol. 2021, 5, 119–136. [Google Scholar] [CrossRef]
- Corbett, T.H.; Roberts, B.J.; Leopold, W.R.; Peckham, J.C.; Wilkoff, L.J.; Griswold, D.P., Jr.; Schabel, F.M., Jr. Induction and chemotherapeutic response of two transplantable ductal adenocarcinomas of the pancreas in C57BL/6 mice. Cancer Res. 1984, 44, 717–726. [Google Scholar]
- Torres, M.P.; Rachagani, S.; Souchek, J.J.; Mallya, K.; Johansson, S.L.; Batra, S.K. Novel Pancreatic Cancer Cell Lines Derived from Genetically Engineered Mouse Models of Spontaneous Pancreatic Adenocarcinoma: Applications in Diagnosis and Therapy. PLoS ONE 2013, 8, e80580. [Google Scholar] [CrossRef] [PubMed]
- Kersten, K.; de Visser, K.E.; van Miltenburg, M.H.; Jonkers, J. Genetically engineered mouse models in oncology research and cancer medicine. EMBO Mol. Med. 2017, 9, 137–153. [Google Scholar] [CrossRef]
- Westphalen, C.B.; Olive, K.P. Genetically engineered mouse models of pancreatic cancer. Cancer J. 2012, 18, 502–510. [Google Scholar] [CrossRef]
- Vudatha, V.; Herremans, K.M.; Freudenberger, D.C.; Liu, C.; Trevino, J.G. Chapter Three—In vivo models of pancreatic ductal adenocarcinoma. In Advances in Cancer Research; Emdad, L., Atfi, A., Gogna, R., Trevino, J.G., Fisher, P.B., Eds.; Academic Press: New York, NY, USA, 2023; Volume 159, pp. 75–112. [Google Scholar]
- Ornitz, D.M.; Hammer, R.E.; Messing, A.; Palmiter, R.D.; Brinster, R.L. Pancreatic Neoplasia Induced by SV40 T-Antigen Expression in Acinar Cells of Transgenic Mice. Science 1987, 238, 188–193. [Google Scholar] [CrossRef]
- Quaife, C.J.; Pinkert, C.A.; Ornitz, D.M.; Palmiter, R.D.; Brinster, R.L. Pancreatic neoplasia induced by ras expression in acinar cells of transgenic mice. Cell 1987, 48, 1023–1034. [Google Scholar] [CrossRef]
- Sandgren, E.P.; Quaife, C.J.; Paulovich, A.G.; Palmiter, R.D.; Brinster, R.L. Pancreatic tumor pathogenesis reflects the causative genetic lesion. Proc. Natl. Acad. Sci. USA 1991, 88, 93–97. [Google Scholar] [CrossRef]
- Sandgren, E.P.; Luetteke, N.C.; Palmiter, R.D.; Brinster, R.L.; Lee, D.C. Overexpression of TGFα in transgenic mice: Induction of epithelial hyperplasia, pancreatic metaplasia, and carcinoma of the breast. Cell 1990, 61, 1121–1135. [Google Scholar] [CrossRef]
- Kanda, M.; Matthaei, H.; Wu, J.; Hong, S.M.; Yu, J.; Borges, M.; Hruban, R.H.; Maitra, A.; Kinzler, K.; Vogelstein, B.; et al. Presence of somatic mutations in most early-stage pancreatic intraepithelial neoplasia. Gastroenterology 2012, 142, 730–733.e739. [Google Scholar] [CrossRef] [PubMed]
- Hingorani, S.R.; Petricoin, E.F.; Maitra, A.; Rajapakse, V.; King, C.; Jacobetz, M.A.; Ross, S.; Conrads, T.P.; Veenstra, T.D.; Hitt, B.A.; et al. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell 2003, 4, 437–450. [Google Scholar] [CrossRef]
- Hingorani, S.R.; Wang, L.; Multani, A.S.; Combs, C.; Deramaudt, T.B.; Hruban, R.H.; Rustgi, A.K.; Chang, S.; Tuveson, D.A. Trp53R172H and KrasG12D cooperate to promote chromosomal instability and widely metastatic pancreatic ductal adenocarcinoma in mice. Cancer Cell 2005, 7, 469–483. [Google Scholar] [CrossRef] [PubMed]
- Hirth, M.; Xie, Y.; Höper, C.; Prats, A.; Hackert, T.; Ebert, M.P.; Kuner, R. Genetic Mouse Models to Study Pancreatic Cancer-Induced Pain and Reduction in Well-Being. Cells 2022, 11, 2634. [Google Scholar] [CrossRef]
- Morton, J.P.; Timpson, P.; Karim, S.A.; Ridgway, R.A.; Athineos, D.; Doyle, B.; Jamieson, N.B.; Oien, K.A.; Lowy, A.M.; Brunton, V.G.; et al. Mutant p53 drives metastasis and overcomes growth arrest/senescence in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2010, 107, 246–251. [Google Scholar] [CrossRef]
- Carrière, C.; Gore, A.J.; Norris, A.M.; Gunn, J.R.; Young, A.L.; Longnecker, D.S.; Korc, M. Deletion of Rb Accelerates Pancreatic Carcinogenesis by Oncogenic Kras and Impairs Senescence in Premalignant Lesions. Gastroenterology 2011, 141, 1091–1101. [Google Scholar] [CrossRef] [PubMed]
- Aguirre, A.J.; Bardeesy, N.; Sinha, M.; Lopez, L.; Tuveson, D.A.; Horner, J.; Redston, M.S.; DePinho, R.A. Activated Kras and Ink4a/Arf deficiency cooperate to produce metastatic pancreatic ductal adenocarcinoma. Genes Dev. 2003, 17, 3112–3126. [Google Scholar] [CrossRef]
- Izeradjene, K.; Combs, C.; Best, M.; Gopinathan, A.; Wagner, A.; Grady, W.M.; Deng, C.-X.; Hruban, R.H.; Adsay, N.V.; Tuveson, D.A.; et al. KrasG12D and Smad4/Dpc4 Haploinsufficiency Cooperate to Induce Mucinous Cystic Neoplasms and Invasive Adenocarcinoma of the Pancreas. Cancer Cell 2007, 11, 229–243. [Google Scholar] [CrossRef]
- Qiu, W.; Tang, S.M.; Lee, S.; Turk, A.T.; Sireci, A.N.; Qiu, A.; Rose, C.; Xie, C.; Kitajewski, J.; Wen, H.-J.; et al. Loss of Activin Receptor Type 1B Accelerates Development of Intraductal Papillary Mucinous Neoplasms in Mice with Activated KRAS. Gastroenterology 2016, 150, 218–228.e212. [Google Scholar] [CrossRef]
- Ijichi, H.; Chytil, A.; Gorska, A.E.; Aakre, M.E.; Fujitani, Y.; Fujitani, S.; Wright, C.V.; Moses, H.L. Aggressive pancreatic ductal adenocarcinoma in mice caused by pancreas-specific blockade of transforming growth factor-beta signaling in cooperation with active Kras expression. Genes Dev. 2006, 20, 3147–3160. [Google Scholar] [CrossRef] [PubMed]
- Costanza, B.; Umelo, I.A.; Bellier, J.; Castronovo, V.; Turtoi, A. Stromal Modulators of TGF-β in Cancer. J. Clin. Med. 2017, 6, 7. [Google Scholar] [CrossRef] [PubMed]
- Ideno, N.; Yamaguchi, H.; Ghosh, B.; Gupta, S.; Okumura, T.; Steffen, D.J.; Fisher, C.G.; Wood, L.D.; Singhi, A.D.; Nakamura, M.; et al. GNAS(R201C) Induces Pancreatic Cystic Neoplasms in Mice That Express Activated KRAS by Inhibiting YAP1 Signaling. Gastroenterology 2018, 155, 1593–1607.e1512. [Google Scholar] [CrossRef] [PubMed]
- Taki, K.; Ohmuraya, M.; Tanji, E.; Komatsu, H.; Hashimoto, D.; Semba, K.; Araki, K.; Kawaguchi, Y.; Baba, H.; Furukawa, T. GNAS(R201H) and Kras(G12D) cooperate to promote murine pancreatic tumorigenesis recapitulating human intraductal papillary mucinous neoplasm. Oncogene 2016, 35, 2407–2412. [Google Scholar] [CrossRef]
- Rhim, A.D.; Oberstein, P.E.; Thomas, D.H.; Mirek, E.T.; Palermo, C.F.; Sastra, S.A.; Dekleva, E.N.; Saunders, T.; Becerra, C.P.; Tattersall, I.W.; et al. Stromal Elements Act to Restrain, Rather Than Support, Pancreatic Ductal Adenocarcinoma. Cancer Cell 2014, 25, 735–747. [Google Scholar] [CrossRef]
- Özdemir, B.C.; Pentcheva-Hoang, T.; Carstens, J.L.; Zheng, X.; Wu, C.-C.; Simpson, T.R.; Laklai, H.; Sugimoto, H.; Kahlert, C.; Novitskiy, S.V.; et al. Depletion of Carcinoma-Associated Fibroblasts and Fibrosis Induces Immunosuppression and Accelerates Pancreas Cancer with Reduced Survival. Cancer Cell 2014, 25, 719–734. [Google Scholar] [CrossRef] [PubMed]
- Polani, F.; Grierson, P.M.; Lim, K.H. Stroma-targeting strategies in pancreatic cancer: Past lessons, challenges and prospects. World J. Gastroenterol. 2021, 27, 2105–2121. [Google Scholar] [CrossRef] [PubMed]
- Feig, C.; Jones, J.O.; Kraman, M.; Wells, R.J.B.; Deonarine, A.; Chan, D.S.; Connell, C.M.; Roberts, E.W.; Zhao, Q.; Caballero, O.L.; et al. Targeting CXCL12 from FAP-expressing carcinoma-associated fibroblasts synergizes with anti–PD-L1 immunotherapy in pancreatic cancer. Proc. Natl. Acad. Sci. USA 2013, 110, 20212–20217. [Google Scholar] [CrossRef]
- Gopinathan, A.; Morton, J.P.; Jodrell, D.I.; Sansom, O.J. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis. Model. Mech. 2015, 8, 1185–1200. [Google Scholar] [CrossRef]
- Lee, A.Y.L.; Dubois, C.L.; Sarai, K.; Zarei, S.; Schaeffer, D.F.; Sander, M.; Kopp, J.L. Cell of origin affects tumour development and phenotype in pancreatic ductal adenocarcinoma. Gut 2019, 68, 487–498. [Google Scholar] [CrossRef]
- Talbert, E.E.; Cuitiño, M.C.; Ladner, K.J.; Rajasekerea, P.V.; Siebert, M.; Shakya, R.; Leone, G.W.; Ostrowski, M.C.; Paleo, B.; Weisleder, N.; et al. Modeling Human Cancer-induced Cachexia. Cell Rep. 2019, 28, 1612–1622.e1614. [Google Scholar] [CrossRef]
- Chiou, S.H.; Winters, I.P.; Wang, J.; Naranjo, S.; Dudgeon, C.; Tamburini, F.B.; Brady, J.J.; Yang, D.; Grüner, B.M.; Chuang, C.H.; et al. Pancreatic cancer modeling using retrograde viral vector delivery and in vivo CRISPR/Cas9-mediated somatic genome editing. Genes Dev. 2015, 29, 1576–1585. [Google Scholar] [CrossRef] [PubMed]
- Schönhuber, N.; Seidler, B.; Schuck, K.; Veltkamp, C.; Schachtler, C.; Zukowska, M.; Eser, S.; Feyerabend, T.B.; Paul, M.C.; Eser, P.; et al. A next-generation dual-recombinase system for time- and host-specific targeting of pancreatic cancer. Nat. Med. 2014, 20, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Kim, J.; Yang, S.; Wang, H.; Wu, C.-J.; Sugimoto, H.; LeBleu, V.S.; Kalluri, R. Type I collagen deletion in αSMA+ myofibroblasts augments immune suppression and accelerates progression of pancreatic cancer. Cancer Cell 2021, 39, 548–565.e546. [Google Scholar] [CrossRef]
- Collins, M.A.; Bednar, F.; Zhang, Y.; Brisset, J.-C.; Galbán, S.; Galbán, C.J.; Rakshit, S.; Flannagan, K.S.; Adsay, N.V.; Pasca di Magliano, M. Oncogenic Kras is required for both the initiation and maintenance of pancreatic cancer in mice. J. Clin. Investig. 2012, 122, 639–653. [Google Scholar] [CrossRef]
- Wang, W.; Dong, B.; Ittmann, M.M.; Yang, F. A Versatile Gene Delivery System for Efficient and Tumor Specific Gene Manipulation in vivo. Discoveries 2016, 4, e58. [Google Scholar] [CrossRef][Green Version]
- Sano, M.; Driscoll, D.R.; De Jesus-Monge, W.E.; Klimstra, D.S.; Lewis, B.C. Activated Wnt Signaling in Stroma Contributes to Development of Pancreatic Mucinous Cystic Neoplasms. Gastroenterology 2014, 146, 257–267. [Google Scholar] [CrossRef]
- Kaltenbacher, T.; Löprich, J.; Maresch, R.; Weber, J.; Müller, S.; Oellinger, R.; Groß, N.; Griger, J.; de Andrade Krätzig, N.; Avramopoulos, P.; et al. CRISPR somatic genome engineering and cancer modeling in the mouse pancreas and liver. Nat. Protoc. 2022, 17, 1142–1188. [Google Scholar] [CrossRef] [PubMed]
- Hanna, R.E.; Doench, J.G. Design and analysis of CRISPR–Cas experiments. Nat. Biotechnol. 2020, 38, 813–823. [Google Scholar] [CrossRef]
- Doench, J.G.; Fusi, N.; Sullender, M.; Hegde, M.; Vaimberg, E.W.; Donovan, K.F.; Smith, I.; Tothova, Z.; Wilen, C.; Orchard, R.; et al. Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 2016, 34, 184–191. [Google Scholar] [CrossRef]
- Oktriani, R.; Pirona, A.C.; Kalmár, L.; Rahadian, A.S.; Miao, B.; Bauer, A.S.; Hoheisel, J.D.; Boettcher, M.; Du, H. Genome-Wide CRISPR Screen Identifies Genes Involved in Metastasis of Pancreatic Ductal Adenocarcinoma. Cancers 2024, 16, 3684. [Google Scholar] [CrossRef]
- Chen, S.; Sanjana, N.E.; Zheng, K.; Shalem, O.; Lee, K.; Shi, X.; Scott, D.A.; Song, J.; Pan, J.Q.; Weissleder, R.; et al. Genome-wide CRISPR Screen in a Mouse Model of Tumor Growth and Metastasis. Cell 2015, 160, 1246–1260. [Google Scholar] [CrossRef]
- Braun, C.J.; Adames, A.C.; Saur, D.; Rad, R. Tutorial: Design and execution of CRISPR in vivo screens. Nat. Protoc. 2022, 17, 1903–1925. [Google Scholar] [CrossRef] [PubMed]
- Hulton, C.H.; Costa, E.A.; Shah, N.S.; Quintanal-Villalonga, A.; Heller, G.; de Stanchina, E.; Rudin, C.M.; Poirier, J.T. Direct genome editing of patient-derived xenografts using CRISPR-Cas9 enables rapid in vivo functional genomics. Nat. Cancer 2020, 1, 359–369. [Google Scholar] [CrossRef]
- Zhu, X.G.; Chudnovskiy, A.; Baudrier, L.; Prizer, B.; Liu, Y.; Ostendorf, B.N.; Yamaguchi, N.; Arab, A.; Tavora, B.; Timson, R.; et al. Functional Genomics In Vivo Reveal Metabolic Dependencies of Pancreatic Cancer Cells. Cell Metab. 2021, 33, 211–221.e216. [Google Scholar] [CrossRef] [PubMed]
- Dubrot, J.; Du, P.P.; Lane-Reticker, S.K.; Kessler, E.A.; Muscato, A.J.; Mehta, A.; Freeman, S.S.; Allen, P.M.; Olander, K.E.; Ockerman, K.M.; et al. In vivo CRISPR screens reveal the landscape of immune evasion pathways across cancer. Nat. Immunol. 2022, 23, 1495–1506. [Google Scholar] [CrossRef] [PubMed]
- Szlachta, K.; Kuscu, C.; Tufan, T.; Adair, S.J.; Shang, S.; Michaels, A.D.; Mullen, M.G.; Fischer, N.L.; Yang, J.; Liu, L.; et al. CRISPR knockout screening identifies combinatorial drug targets in pancreatic cancer and models cellular drug response. Nat. Commun. 2018, 9, 4275. [Google Scholar] [CrossRef]
- Maresch, R.; Mueller, S.; Veltkamp, C.; Öllinger, R.; Friedrich, M.; Heid, I.; Steiger, K.; Weber, J.; Engleitner, T.; Barenboim, M.; et al. Multiplexed pancreatic genome engineering and cancer induction by transfection-based CRISPR/Cas9 delivery in mice. Nat. Commun. 2016, 7, 10770. [Google Scholar] [CrossRef]
- Martinez, S.; Wu, S.; Geuenich, M.; Malik, A.; Weber, R.; Woo, T.; Zhang, A.; Jang, G.H.; Dervovic, D.; Al-Zahrani, K.N.; et al. In vivo CRISPR screens reveal SCAF1 and USP15 as drivers of pancreatic cancer. Nat. Commun. 2024, 15, 5266. [Google Scholar] [CrossRef] [PubMed]
- Beato, F.; Reverón, D.; Dezsi, K.B.; Ortiz, A.; Johnson, J.O.; Chen, D.-T.; Ali, K.; Yoder, S.J.; Jeong, D.; Malafa, M.; et al. Establishing a living biobank of patient-derived organoids of intraductal papillary mucinous neoplasms of the pancreas. Lab. Investig. 2021, 101, 204–217. [Google Scholar] [CrossRef]
- Ungricht, R.; Guibbal, L.; Lasbennes, M.-C.; Orsini, V.; Beibel, M.; Waldt, A.; Cuttat, R.; Carbone, W.; Basler, A.; Roma, G.; et al. Genome-wide screening in human kidney organoids identifies developmental and disease-related aspects of nephrogenesis. Cell Stem Cell 2022, 29, 160–175.e167. [Google Scholar] [CrossRef]
- Michels, B.E.; Mosa, M.H.; Streibl, B.I.; Zhan, T.; Menche, C.; Abou-El-Ardat, K.; Darvishi, T.; Członka, E.; Wagner, S.; Winter, J.; et al. Pooled In Vitro and In Vivo CRISPR-Cas9 Screening Identifies Tumor Suppressors in Human Colon Organoids. Cell Stem Cell 2020, 26, 782–792.e787. [Google Scholar] [CrossRef] [PubMed]
- Ringel, T.; Frey, N.; Ringnalda, F.; Janjuha, S.; Cherkaoui, S.; Butz, S.; Srivatsa, S.; Pirkl, M.; Russo, G.; Villiger, L.; et al. Genome-Scale CRISPR Screening in Human Intestinal Organoids Identifies Drivers of TGF-β Resistance. Cell Stem Cell 2020, 26, 431–440.e438. [Google Scholar] [CrossRef]
- Gilbert, L.A.; Horlbeck, M.A.; Adamson, B.; Villalta, J.E.; Chen, Y.; Whitehead, E.H.; Guimaraes, C.; Panning, B.; Ploegh, H.L.; Bassik, M.C.; et al. Genome-Scale CRISPR-Mediated Control of Gene Repression and Activation. Cell 2014, 159, 647–661. [Google Scholar] [CrossRef] [PubMed]
- Anzalone, A.V.; Randolph, P.B.; Davis, J.R.; Sousa, A.A.; Koblan, L.W.; Levy, J.M.; Chen, P.J.; Wilson, C.; Newby, G.A.; Raguram, A.; et al. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 2019, 576, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Sayed, S.; Sidorova, O.A.; Hennig, A.; Augsburg, M.; Cortés Vesga, C.P.; Abohawya, M.; Schmitt, L.T.; Sürün, D.; Stange, D.E.; Mircetic, J.; et al. Efficient Correction of Oncogenic KRAS and TP53 Mutations through CRISPR Base Editing. Cancer Res. 2022, 82, 3002–3015. [Google Scholar] [CrossRef]
- Joung, J.K.; Sander, J.D. TALENs: A widely applicable technology for targeted genome editing. Nat. Rev. Mol. Cell Biol. 2013, 14, 49–55. [Google Scholar] [CrossRef]
- DeWeirdt, P.C.; Sanson, K.R.; Sangree, A.K.; Hegde, M.; Hanna, R.E.; Feeley, M.N.; Griffith, A.L.; Teng, T.; Borys, S.M.; Strand, C.; et al. Optimization of AsCas12a for combinatorial genetic screens in human cells. Nat. Biotechnol. 2021, 39, 94–104. [Google Scholar] [CrossRef]
- Konermann, S.; Lotfy, P.; Brideau, N.J.; Oki, J.; Shokhirev, M.N.; Hsu, P.D. Transcriptome Engineering with RNA-Targeting Type VI-D CRISPR Effectors. Cell 2018, 173, 665–676.e614. [Google Scholar] [CrossRef]
- Lowenfels, A.B.; Maisonneuve, P.; Cavallini, G.; Ammann, R.W.; Lankisch, P.G.; Andersen, J.R.; Dimagno, E.P.; Andrén-Sandberg, A.; Domellöf, L. Pancreatitis and the risk of pancreatic cancer. International Pancreatitis Study Group. N. Engl. J. Med. 1993, 328, 1433–1437. [Google Scholar] [CrossRef]
- Kim, H.S.; Gweon, T.-G.; Park, S.H.; Kim, T.H.; Kim, C.W.; Chang, J.H. Incidence and risk of pancreatic cancer in patients with chronic pancreatitis: Defining the optimal subgroup for surveillance. Sci. Rep. 2023, 13, 106. [Google Scholar] [CrossRef]
- Kirkegård, J.; Cronin-Fenton, D.; Heide-Jørgensen, U.; Mortensen, F.V. Acute Pancreatitis and Pancreatic Cancer Risk: A Nationwide Matched-Cohort Study in Denmark. Gastroenterology 2018, 154, 1729–1736. [Google Scholar] [CrossRef]
- Park, S.M.; Kim, K.B.; Han, J.-H.; Kim, N.; Kang, T.U.; Swan, H.; Kim, H.J. Incidence and risk of pancreatic cancer in patients with acute or chronic pancreatitis: A population-based cohort study. Sci. Rep. 2023, 13, 18930. [Google Scholar] [CrossRef] [PubMed]
- Malka, D.; Hammel, P.; Maire, F.; Rufat, P.; Madeira, I.; Pessione, F.; Lévy, P.; Ruszniewski, P. Risk of pancreatic adenocarcinoma in chronic pancreatitis. Gut 2002, 51, 849–852. [Google Scholar] [CrossRef] [PubMed]
- Ueda, J.; Tanaka, M.; Ohtsuka, T.; Tokunaga, S.; Shimosegawa, T.; Research Committee of Intractable Diseases of the Pancreas. Surgery for chronic pancreatitis decreases the risk for pancreatic cancer: A multicenter retrospective analysis. Surgery 2013, 153, 357–364. [Google Scholar] [CrossRef]
- Yadav, D.; Hawes, R.H.; Brand, R.E.; Anderson, M.A.; Money, M.E.; Banks, P.A.; Bishop, M.D.; Baillie, J.; Sherman, S.; DiSario, J.; et al. Alcohol consumption, cigarette smoking, and the risk of recurrent acute and chronic pancreatitis. Arch. Intern. Med. 2009, 169, 1035–1045. [Google Scholar] [CrossRef]
- Pandol, S.J.; Gorelick, F.S.; Gerloff, A.; Lugea, A. Alcohol abuse, endoplasmic reticulum stress and pancreatitis. Dig. Dis. 2010, 28, 776–782. [Google Scholar] [CrossRef]
- Tanaka, T.; Miura, Y.; Matsugu, Y.; Ichiba, Y.; Ito, H.; Dohi, K. Pancreatic duct obstruction is an aggravating factor in the canine model of chronic alcoholic pancreatitis. Gastroenterology 1998, 115, 1248–1253. [Google Scholar] [CrossRef]
- Okazaki, K.; Chiba, T. Autoimmune related pancreatitis. Gut 2002, 51, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Whitcomb, D.C.; Gorry, M.C.; Preston, R.A.; Furey, W.; Sossenheimer, M.J.; Ulrich, C.D.; Martin, S.P.; Gates, L.K.; Amann, S.T.; Toskes, P.P.; et al. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene. Nat. Genet. 1996, 14, 141–145. [Google Scholar] [CrossRef] [PubMed]
- Witt, H.; Luck, W.; Hennies, H.C.; Classen, M.; Kage, A.; Lass, U.; Landt, O.; Becker, M. Mutations in the gene encoding the serine protease inhibitor, Kazal type 1 are associated with chronic pancreatitis. Nat. Genet. 2000, 25, 213–216. [Google Scholar] [CrossRef]
- Witt, H.; Beer, S.; Rosendahl, J.; Chen, J.-M.; Chandak, G.R.; Masamune, A.; Bence, M.; Szmola, R.; Oracz, G.; Macek, M.; et al. Variants in CPA1 are strongly associated with early onset chronic pancreatitis. Nat. Genet. 2013, 45, 1216–1220. [Google Scholar] [CrossRef]
- Gorry, M.C.; Gabbaizedeh, D.; Furey, W.; Gates, L.K.; Preston, R.A.; Aston, C.E.; Zhang, Y.; Ulrich, C.; Ehrlich, G.D.; Whitcomb, D.C. Mutations in the cationic trypsinogen gene are associated with recurrent acute and chronic pancreatitis. Gastroenterology 1997, 113, 1063–1068. [Google Scholar] [CrossRef] [PubMed]
- Grocock, C.J.; Rebours, V.; Delhaye, M.N.; Andrén-Sandberg, A.; Weiss, F.U.; Mountford, R.; Harcus, M.J.; Niemczyck, E.; Vitone, L.J.; Dodd, S.; et al. The variable phenotype of the p.A16V mutation of cationic trypsinogen (PRSS1) in pancreatitis families. Gut 2010, 59, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Witt, H.; Luck, W.; Becker, M. A signal peptide cleavage site mutation in the cationic trypsinogen gene is strongly associated with chronic pancreatitis. Gastroenterology 1999, 117, 7–10. [Google Scholar] [CrossRef] [PubMed]
- Teich, N.; Nemoda, Z.; Köhler, H.; Heinritz, W.; Mössner, J.; Keim, V.; Sahin-Tóth, M. Gene conversion between functional trypsinogen genes PRSS1 and PRSS2 associated with chronic pancreatitis in a six-year-old girl. Hum. Mutat. 2005, 25, 343–347. [Google Scholar] [CrossRef]
- Sahin-Tóth, M.; Tóth, M. Gain-of-function mutations associated with hereditary pancreatitis enhance autoactivation of human cationic trypsinogen. Biochem. Biophys. Res. Commun. 2000, 278, 286–289. [Google Scholar] [CrossRef]
- Archer, H.; Jura, N.; Keller, J.; Jacobson, M.; Bar-Sagi, D. A mouse model of hereditary pancreatitis generated by transgenic expression of R122H trypsinogen. Gastroenterology 2006, 131, 1844–1855. [Google Scholar] [CrossRef]
- Athwal, T.; Huang, W.; Mukherjee, R.; Latawiec, D.; Chvanov, M.; Clarke, R.; Smith, K.; Campbell, F.; Merriman, C.; Criddle, D.; et al. Expression of human cationic trypsinogen (PRSS1) in murine acinar cells promotes pancreatitis and apoptotic cell death. Cell Death Dis. 2014, 5, e1165. [Google Scholar] [CrossRef][Green Version]
- Geisz, A.; Sahin-Tóth, M. A preclinical model of chronic pancreatitis driven by trypsinogen autoactivation. Nat. Commun. 2018, 9, 5033. [Google Scholar] [CrossRef]
- Demcsák, A.; Sahin-Tóth, M. Rate of Autoactivation Determines Pancreatitis Phenotype in Trypsinogen Mutant Mice. Gastroenterology 2022, 163, 761–763. [Google Scholar] [CrossRef]
- Rosendahl, J.; Witt, H.; Szmola, R.; Bhatia, E.; Ozsvári, B.; Landt, O.; Schulz, H.-U.; Gress, T.M.; Pfützer, R.; Löhr, M.; et al. Chymotrypsin C (CTRC) variants that diminish activity or secretion are associated with chronic pancreatitis. Nat. Genet. 2008, 40, 78–82. [Google Scholar] [CrossRef] [PubMed]
- Masson, E.; Chen, J.-M.; Scotet, V.; Le Maréchal, C.; Férec, C. Association of rare chymotrypsinogen C (CTRC) gene variations in patients with idiopathic chronic pancreatitis. Hum. Genet. 2008, 123, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Jancsó, Z.; Hegyi, E.; Sahin-Tóth, M. Chymotrypsin Reduces the Severity of Secretagogue-Induced Pancreatitis in Mice. Gastroenterology 2018, 155, 1017–1021. [Google Scholar] [CrossRef]
- Laskowski, M.; Kato, I. Protein inhibitors of proteinases. Annu. Rev. Biochem. 1980, 49, 593–626. [Google Scholar] [CrossRef]
- Romac, J.M.J.; Ohmuraya, M.; Bittner, C.; Majeed, M.F.; Vigna, S.R.; Que, J.; Fee, B.E.; Wartmann, T.; Yamamura, K.-I.; Liddle, R.A. Transgenic expression of pancreatic secretory trypsin inhibitor-1 rescues SPINK3-deficient mice and restores a normal pancreatic phenotype. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 298, G518–G524. [Google Scholar] [CrossRef]
- Ohmuraya, M.; Hirota, M.; Araki, M.; Mizushima, N.; Matsui, M.; Mizumoto, T.; Haruna, K.; Kume, S.; Takeya, M.; Ogawa, M.; et al. Autophagic cell death of pancreatic acinar cells in serine protease inhibitor Kazal type 3-deficient mice. Gastroenterology 2005, 129, 696–705. [Google Scholar] [CrossRef]
- Demcsák, A.; Sahin-Tóth, M. Heterozygous Spink1 Deficiency Promotes Trypsin-dependent Chronic Pancreatitis in Mice. Cell. Mol. Gastroenterol. Hepatol. 2024, 18, 101361. [Google Scholar] [CrossRef] [PubMed]
- Scheele, G.; Bartelt, D.; Bieger, W. Characterization of human exocrine pancreatic proteins by two-dimensional isoelectric focusing/sodium dodecyl sulfate gel electrophoresis. Gastroenterology 1981, 80, 461–473. [Google Scholar] [CrossRef]
- Szmola, R.; Bence, M.; Carpentieri, A.; Szabó, A.; Costello, C.E.; Samuelson, J.; Sahin-Tóth, M. Chymotrypsin C is a co-activator of human pancreatic procarboxypeptidases A1 and A2. J. Biol. Chem. 2011, 286, 1819–1827. [Google Scholar] [CrossRef]
- Hegyi, E.; Sahin-Tóth, M. Human CPA1 mutation causes digestive enzyme misfolding and chronic pancreatitis in mice. Gut 2019, 68, 301–312. [Google Scholar] [CrossRef]
- Guerra, C.; Schuhmacher, A.J.; Cañamero, M.; Grippo, P.J.; Verdaguer, L.; Pérez-Gallego, L.; Dubus, P.; Sandgren, E.P.; Barbacid, M. Chronic pancreatitis is essential for induction of pancreatic ductal adenocarcinoma by K-Ras oncogenes in adult mice. Cancer Cell 2007, 11, 291–302. [Google Scholar] [CrossRef] [PubMed]
- Murray, C.J.L.; Aravkin, A.Y.; Zheng, P.; Abbafati, C.; Abbas, K.M.; Abbasi-Kangevari, M.; Abd-Allah, F.; Abdelalim, A.; Abdollahi, M.; Abdollahpour, I.; et al. Global burden of 87 risk factors in 204 countries and territories, 1990–2019: A systematic analysis for the Global Burden of Disease Study 2019. Lancet 2020, 396, 1223–1249. [Google Scholar] [CrossRef] [PubMed]
- Rosen, E.D.; Spiegelman, B.M. What We Talk About When We Talk About Fat. Cell 2014, 156, 20–44. [Google Scholar] [CrossRef]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Zatterale, F.; Longo, M.; Naderi, J.; Raciti, G.A.; Desiderio, A.; Miele, C.; Beguinot, F. Chronic Adipose Tissue Inflammation Linking Obesity to Insulin Resistance and Type 2 Diabetes. Front. Physiol. 2020, 10, 1607. [Google Scholar] [CrossRef]
- Quail, D.F.; Dannenberg, A.J. The obese adipose tissue microenvironment in cancer development and progression. Nat. Rev. Endocrinol. 2019, 15, 139–154. [Google Scholar] [CrossRef]
- Bhaskaran, K.; Douglas, I.; Forbes, H.; dos-Santos-Silva, I.; Leon, D.A.; Smeeth, L. Body-mass index and risk of 22 specific cancers: A population-based cohort study of 5·24 million UK adults. Lancet 2014, 384, 755–765. [Google Scholar] [CrossRef]
- Rebours, V.; Gaujoux, S.; d’Assignies, G.; Sauvanet, A.; Ruszniewski, P.; Lévy, P.; Paradis, V.; Bedossa, P.; Couvelard, A. Obesity and Fatty Pancreatic Infiltration Are Risk Factors for Pancreatic Precancerous Lesions (PanIN). Clin. Cancer Res. 2015, 21, 3522–3528. [Google Scholar] [CrossRef]
- El-Tanani, M.; Rabbani, S.A.; Aljabali, A.A.; Matalka, I.I.; El-Tanani, Y.; Rizzo, M.; Tambuwala, M.M. The Complex Connection between Obesity and Cancer: Signaling Pathways and Therapeutic Implications. Nutr. Cancer 2024, 76, 683–706. [Google Scholar] [CrossRef]
- Ruiz, C.F.; Garcia, C.; Jacox, J.B.; Lawres, L.; Muzumdar, M.D. Decoding the obesity–cancer connection: Lessons from preclinical models of pancreatic adenocarcinoma. Life Sci. Alliance 2023, 6, e202302228. [Google Scholar] [CrossRef]
- Philip, B.; Roland, C.L.; Daniluk, J.; Liu, Y.; Chatterjee, D.; Gomez, S.B.; Ji, B.; Huang, H.; Wang, H.; Fleming, J.B.; et al. A high-fat diet activates oncogenic Kras and COX2 to induce development of pancreatic ductal adenocarcinoma in mice. Gastroenterology 2013, 145, 1449–1458. [Google Scholar] [CrossRef] [PubMed]
- Suleiman, J.B.; Mohamed, M.; Bakar, A.B.A. A systematic review on different models of inducing obesity in animals: Advantages and limitations. J. Adv. Vet. Anim. Res. 2020, 7, 103–114. [Google Scholar] [CrossRef] [PubMed]
- Kfoury, S.; Michl, P.; Roth, L. Modeling Obesity-Driven Pancreatic Carcinogenesis-A Review of Current In Vivo and In Vitro Models of Obesity and Pancreatic Carcinogenesis. Cells 2022, 11, 3170. [Google Scholar] [CrossRef]
- Lutz, T.A.; Woods, S.C. Overview of animal models of obesity. Curr. Protoc. Pharmacol. 2012, 58, 5–61. [Google Scholar] [CrossRef]
- Wu, S.-C.; Lin, C.-H. Direct Adeno-associated Viruses Injection of Murine Adipose Tissue. Bio-Protocol 2023, 13, e4674. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.; Gholipourmalekabadi, M.; Shafikhani, S.H. Animal models for type 1 and type 2 diabetes: Advantages and limitations. Front. Endocrinol. 2024, 15, 1359685. [Google Scholar] [CrossRef]
- Chung, K.M.; Singh, J.; Lawres, L.; Dorans, K.J.; Garcia, C.; Burkhardt, D.B.; Robbins, R.; Bhutkar, A.; Cardone, R.; Zhao, X.; et al. Endocrine-Exocrine Signaling Drives Obesity-Associated Pancreatic Ductal Adenocarcinoma. Cell 2020, 181, 832–847.e818. [Google Scholar] [CrossRef]
- Renehan, A.G.; Zwahlen, M.; Egger, M. Adiposity and cancer risk: New mechanistic insights from epidemiology. Nat. Rev. Cancer 2015, 15, 484–498. [Google Scholar] [CrossRef]
- Iyengar, N.M.; Gucalp, A.; Dannenberg, A.J.; Hudis, C.A. Obesity and Cancer Mechanisms: Tumor Microenvironment and Inflammation. J. Clin. Oncol. 2016, 34, 4270–4276. [Google Scholar] [CrossRef]
- Vansaun, M.N. Molecular pathways: Adiponectin and leptin signaling in cancer. Clin. Cancer Res. 2013, 19, 1926–1932. [Google Scholar] [CrossRef]
- Myers, M.G.; Cowley, M.A.; Münzberg, H. Mechanisms of leptin action and leptin resistance. Annu. Rev. Physiol. 2008, 70, 537–556. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Tan, M.; Tian, X.; Zhang, J.; Zhang, J.; Chen, J.; Xu, W.; Sheng, H. Leptin receptor mediates the proliferation and glucose metabolism of pancreatic cancer cells via AKT pathway activation. Mol. Med. Rep. 2020, 21, 945–952. [Google Scholar] [CrossRef]
- Mendonsa, A.M.; Chalfant, M.C.; Gorden, L.D.; Vansaun, M.N. Modulation of the leptin receptor mediates tumor growth and migration of pancreatic cancer cells. PLoS ONE 2015, 10, e0126686. [Google Scholar] [CrossRef]
- Fan, Y.; Gan, Y.; Shen, Y.; Cai, X.; Song, Y.; Zhao, F.; Yao, M.; Gu, J.; Tu, H. Leptin signaling enhances cell invasion and promotes the metastasis of human pancreatic cancer via increasing MMP-13 production. Oncotarget 2015, 6, 16120–16134. [Google Scholar] [CrossRef] [PubMed]
- Mosser, R.E.; Maulis, M.F.; Moullé, V.S.; Dunn, J.C.; Carboneau, B.A.; Arasi, K.; Pappan, K.; Poitout, V.; Gannon, M. High-fat diet-induced β-cell proliferation occurs prior to insulin resistance in C57Bl/6J male mice. Am. J. Physiol. Endocrinol. Metab. 2015, 308, E573–E582. [Google Scholar] [CrossRef]
- Ahren, B.; Pacini, G. Insufficient islet compensation to insulin resistance vs. reduced glucose effectiveness in glucose-intolerant mice. Am. J. Physiol. Endocrinol. Metab. 2002, 283, E738–E744. [Google Scholar] [CrossRef] [PubMed]
- Berglund, E.D.; Li, C.Y.; Poffenberger, G.; Ayala, J.E.; Fueger, P.T.; Willis, S.E.; Jewell, M.M.; Powers, A.C.; Wasserman, D.H. Glucose metabolism in vivo in four commonly used inbred mouse strains. Diabetes 2008, 57, 1790–1799. [Google Scholar] [CrossRef]
- Turner, N.; Kowalski, G.M.; Leslie, S.J.; Risis, S.; Yang, C.; Lee-Young, R.S.; Babb, J.R.; Meikle, P.J.; Lancaster, G.I.; Henstridge, D.C.; et al. Distinct patterns of tissue-specific lipid accumulation during the induction of insulin resistance in mice by high-fat feeding. Diabetologia 2013, 56, 1638–1648. [Google Scholar] [CrossRef]
- Wang, D.; Tai, P.W.L.; Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 2019, 18, 358–378. [Google Scholar] [CrossRef]
- Chang, H.-H.; Moro, A.; Takakura, K.; Su, H.-Y.; Mo, A.; Nakanishi, M.; Waldron, R.T.; French, S.W.; Dawson, D.W.; Hines, O.J.; et al. Incidence of pancreatic cancer is dramatically increased by a high fat, high calorie diet in KrasG12D mice. PLoS ONE 2017, 12, e0184455. [Google Scholar] [CrossRef]
- Ead, A.S.; Wirkus, J.; Matsukuma, K.; Mackenzie, G.G. A high-fat diet induces changes in mesenteric adipose tissue accelerating early-stage pancreatic carcinogenesis in mice. J. Nutr. Biochem. 2024, 131, 109690. [Google Scholar] [CrossRef]
- Okumura, T.; Ohuchida, K.; Sada, M.; Abe, T.; Endo, S.; Koikawa, K.; Iwamoto, C.; Miura, D.; Mizuuchi, Y.; Moriyama, T.; et al. Extra-pancreatic invasion induces lipolytic and fibrotic changes in the adipose microenvironment, with released fatty acids enhancing the invasiveness of pancreatic cancer cells. Oncotarget 2017, 8, 18280–18295. [Google Scholar] [CrossRef]
- Dawson, D.W.; Hertzer, K.; Moro, A.; Donald, G.; Chang, H.-H.; Go, V.L.; Pandol, S.J.; Lugea, A.; Gukovskaya, A.S.; Li, G.; et al. High-fat, high-calorie diet promotes early pancreatic neoplasia in the conditional KrasG12D mouse model. Cancer Prev. Res. 2013, 6, 1064–1073. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Yang, Y.; Liu, M.; Wang, D.; Wang, F.; Bi, Y.; Ji, J.; Li, S.; Liu, Y.; Chen, R.; et al. Oncogenic KRAS Reduces Expression of FGF21 in Acinar Cells to Promote Pancreatic Tumorigenesis in Mice on a High-Fat Diet. Gastroenterology 2019, 157, 1413–1428.e1411. [Google Scholar] [CrossRef] [PubMed]
- Ruze, R.; Song, J.; Yin, X.; Chen, Y.; Xu, R.; Wang, C.; Zhao, Y. Mechanisms of obesity- and diabetes mellitus-related pancreatic carcinogenesis: A comprehensive and systematic review. Signal Transduct. Target. Ther. 2023, 8, 139. [Google Scholar] [CrossRef]
- Zhu, L.; Ji, J.; Ma, J.; Wang, D.; Liu, M.; Du, J.X.; Chen, R.; Hou, W.; Abbruzzese, J.L.; Logsdon, C.D.; et al. Differential Effects of Dietary Macronutrients on the Development of Oncogenic KRAS-Mediated Pancreatic Ductal Adenocarcinoma. Cancers 2022, 14, 2723. [Google Scholar] [CrossRef]
- O’Neill, S.M.; Hinkle, C.; Chen, S.-J.; Sandhu, A.; Hovhannisyan, R.; Stephan, S.; Lagor, W.R.; Ahima, R.S.; Johnston, J.C.; Reilly, M.P. Targeting adipose tissue via systemic gene therapy. Gene Ther. 2014, 21, 653–661. [Google Scholar] [CrossRef]
- Uhrig-Schmidt, S.; Geiger, M.; Luippold, G.; Birk, G.; Mennerich, D.; Neubauer, H.; Grimm, D.; Wolfrum, C.; Kreuz, S. Gene delivery to adipose tissue using transcriptionally targeted rAAV8 vectors. PLoS ONE 2014, 9, e116288. [Google Scholar] [CrossRef]
- Bates, R.; Huang, W.; Cao, L. Adipose Tissue: An Emerging Target for Adeno-associated Viral Vectors. Mol. Ther. Methods Clin. Dev. 2020, 19, 236–249. [Google Scholar] [CrossRef]
- Mallya, K.; Haridas, D.; Seshacharyulu, P.; Pothuraju, R.; Junker, W.M.; Krishn, S.R.; Muniyan, S.; Vengoji, R.; Batra, S.K.; Rachagani, S. Acinar transformed ductal cells exhibit differential mucin expression in a tamoxifen-induced pancreatic ductal adenocarcinoma mouse model. Biol. Open 2020, 9, bio052878. [Google Scholar] [CrossRef] [PubMed]
- Lupo, F.; Piro, G.; Torroni, L.; Delfino, P.; Trovato, R.; Rusev, B.; Fiore, A.; Filippini, D.; Sanctis, F.d.; Manfredi, M.; et al. Organoid-Transplant Model Systems to Study the Effects of Obesity on the Pancreatic Carcinogenesis in vivo. Front. Cell Dev. Biology 2020, 8, 308. [Google Scholar] [CrossRef] [PubMed]
- Popovic, K.; Smolović, B.; Martinović, M.; Vučković, L. The Relationship between Diabetes Mellitus and Pancreatic Cancer—Diabetes Mellitus as a Red Flag for Pancreatic Cancer. Cancer Epidemiol. Biomark. Prev. 2023, 32, 298–305. [Google Scholar] [CrossRef]
- Duvillié, B.; Kourdoughli, R.; Druillennec, S.; Eychène, A.; Pouponnot, C. Interplay Between Diabetes and Pancreatic Ductal Adenocarcinoma and Insulinoma: The Role of Aging, Genetic Factors, and Obesity. Front. Endocrinol. 2020, 11, 563267. [Google Scholar] [CrossRef] [PubMed]
- Amundadottir, L.T. Pancreatic Cancer Genetics. Int. J. Biol. Sci. 2016, 12, 314–325. [Google Scholar] [CrossRef]
- Kalisz, M.; Bernardo, E.; Beucher, A.; Maestro, M.A.; del Pozo, N.; Millán, I.; Haeberle, L.; Schlensog, M.; Safi, S.A.; Knoefel, W.T.; et al. HNF1A recruits KDM6A to activate differentiated acinar cell programs that suppress pancreatic cancer. EMBO J. 2020, 39, e102808. [Google Scholar] [CrossRef]
- Cobo-Vuilleumier, N.; Lorenzo, P.I.; Rodríguez, N.G.; Herrera Gómez, I.G.; Fuente-Martin, E.; López-Noriega, L.; Mellado-Gil, J.M.; Romero-Zerbo, S.Y.; Baquié, M.; Lachaud, C.C.; et al. LRH-1 agonism favours an immune-islet dialogue which protects against diabetes mellitus. Nat. Commun. 2018, 9, 1488. [Google Scholar] [CrossRef]
- Flandez, M.; Cendrowski, J.; Cañamero, M.; Salas, A.; del Pozo, N.; Schoonjans, K.; Real, F.X. Nr5a2 heterozygosity sensitises to, and cooperates with, inflammation in KRas(G12V)-driven pancreatic tumourigenesis. Gut 2014, 63, 647–655. [Google Scholar] [CrossRef] [PubMed]
- Cobo, I.; Martinelli, P.; Flández, M.; Bakiri, L.; Zhang, M.; Carrillo-de-Santa-Pau, E.; Jia, J.; Sánchez-Arévalo Lobo, V.J.; Megías, D.; Felipe, I.; et al. Transcriptional regulation by NR5A2 links differentiation and inflammation in the pancreas. Nature 2018, 554, 533–537. [Google Scholar] [CrossRef]
- Raho, S.; Capobianco, L.; Malivindi, R.; Vozza, A.; Piazzolla, C.; De Leonardis, F.; Gorgoglione, R.; Scarcia, P.; Pezzuto, F.; Agrimi, G.; et al. KRAS-regulated glutamine metabolism requires UCP2-mediated aspartate transport to support pancreatic cancer growth. Nat. Metab. 2020, 2, 1373–1381. [Google Scholar] [CrossRef]
- Li, D.; Tang, H.; Hassan, M.M.; Holly, E.A.; Bracci, P.M.; Silverman, D.T. Diabetes and risk of pancreatic cancer: A pooled analysis of three large case–control studies. Cancer Causes Control 2011, 22, 189–197. [Google Scholar] [CrossRef]
- Maisonneuve, P.; Lowenfels, A.B.; Bueno-de-Mesquita, H.B.; Ghadirian, P.; Baghurst, P.A.; Zatonski, W.A.; Miller, A.B.; Duell, E.J.; Boffetta, P.; Boyle, P. Past Medical History and Pancreatic Cancer Risk: Results from a Multicenter Case-Control Study. Ann. Epidemiol. 2010, 20, 92–98. [Google Scholar] [CrossRef]
- Carstensen, B.; Read, S.H.; Friis, S.; Sund, R.; Keskimäki, I.; Svensson, A.M.; Ljung, R.; Wild, S.H.; Kerssens, J.J.; Harding, J.L.; et al. Cancer incidence in persons with type 1 diabetes: A five-country study of 9,000 cancers in type 1 diabetic individuals. Diabetologia 2016, 59, 980–988. [Google Scholar] [CrossRef] [PubMed]
- Liao, W.-C.; Tu, Y.-K.; Wu, M.-S.; Lin, J.-T.; Wang, H.-P.; Chien, K.-L. Blood glucose concentration and risk of pancreatic cancer: Systematic review and dose-response meta-analysis. BMJ 2015, 349, g7371. [Google Scholar] [CrossRef] [PubMed]
- Menini, S.; Iacobini, C.; de Latouliere, L.; Manni, I.; Vitale, M.; Pilozzi, E.; Pesce, C.; Cappello, P.; Novelli, F.; Piaggio, G.; et al. Diabetes promotes invasive pancreatic cancer by increasing systemic and tumour carbonyl stress in Kras(G12D/+) mice. J. Exp. Clin. Cancer Res. 2020, 39, 152. [Google Scholar] [CrossRef]
- van Kruijsdijk, R.C.M.; van der Wall, E.; Visseren, F.L.J. Obesity and Cancer: The Role of Dysfunctional Adipose Tissue. Cancer Epidemiol. Biomark. Prev. 2009, 18, 2569–2578. [Google Scholar] [CrossRef]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Xu, X.; Liu, P.-C.; Mao, H.; Ju, S. Transcriptomic Analyses and Potential Therapeutic Targets of Pancreatic Cancer With Concomitant Diabetes. Front. Oncol. 2020, 10, 563527. [Google Scholar] [CrossRef]
- Zhang, A.M.Y.; Magrill, J.; de Winter, T.J.J.; Hu, X.; Skovsø, S.; Schaeffer, D.F.; Kopp, J.L.; Johnson, J.D. Endogenous Hyperinsulinemia Contributes to Pancreatic Cancer Development. Cell Metab. 2019, 30, 403–404. [Google Scholar] [CrossRef]
- Li, D.; Yeung, S.C.J.; Hassan, M.M.; Konopleva, M.; Abbruzzese, J.L. Antidiabetic Therapies Affect Risk of Pancreatic Cancer. Gastroenterology 2009, 137, 482–488. [Google Scholar] [CrossRef]
- Chen, K.; Qian, W.; Jiang, Z.; Cheng, L.; Li, J.; Sun, L.; Zhou, C.; Gao, L.; Lei, M.; Yan, B.; et al. Metformin suppresses cancer initiation and progression in genetic mouse models of pancreatic cancer. Mol. Cancer 2017, 16, 131. [Google Scholar] [CrossRef]
- Tan, J.; You, Y.; Guo, F.; Xu, J.; Dai, H.; Bie, P. Association of elevated risk of pancreatic cancer in diabetic patients: A systematic review and meta-analysis. Oncol. Lett. 2017, 13, 1247–1255. [Google Scholar] [CrossRef] [PubMed]
- Parajuli, P.; Nguyen, T.L.; Prunier, C.; Razzaque, M.S.; Xu, K.; Atfi, A. Pancreatic cancer triggers diabetes through TGF-β–mediated selective depletion of islet β-cells. Life Sci. Alliance 2020, 3, e201900573. [Google Scholar] [CrossRef] [PubMed]
- McGuigan, A.; Kelly, P.; Turkington, R.C.; Jones, C.; Coleman, H.G.; McCain, R.S. Pancreatic cancer: A review of clinical diagnosis, epidemiology, treatment and outcomes. World J. Gastroenterol. 2018, 24, 4846–4861. [Google Scholar] [CrossRef] [PubMed]
- Glauert, H.P.; Elliott, R.S.; Han, S.G.; Athey, M.; Lee, E.Y.; Gairola, C.G. Effect of cigarette smoke exposure and mutant Kras overexpression on pancreatic cell proliferation. Oncol. Lett. 2017, 13, 1939–1943. [Google Scholar] [CrossRef]
- Edderkaoui, M.; Grippo, P.J.; Ouhaddi, Y.; Benhaddou, H.; Xu, S.; Pinkerton, K.; Tsukamoto, H.; Knudsen, B.; Gukovskaya, A.S.; Pandol, S. Mouse models of pancreatic cancer induced by chronic pancreatitis and smoking. J. Clin. Oncol. 2014, 32, 229. [Google Scholar] [CrossRef]
- Nimmakayala, R.K.; Seshacharyulu, P.; Lakshmanan, I.; Rachagani, S.; Chugh, S.; Karmakar, S.; Rauth, S.; Vengoji, R.; Atri, P.; Talmon, G.A.; et al. Cigarette Smoke Induces Stem Cell Features of Pancreatic Cancer Cells via PAF1. Gastroenterology 2018, 155, 892–908.e896. [Google Scholar] [CrossRef]
- Hermann, P.C.; Sancho, P.; Cañamero, M.; Martinelli, P.; Madriles, F.; Michl, P.; Gress, T.; de Pascual, R.; Gandia, L.; Guerra, C.; et al. Nicotine promotes initiation and progression of KRAS-induced pancreatic cancer via Gata6-dependent dedifferentiation of acinar cells in mice. Gastroenterology 2014, 147, 1119–1133.e1114. [Google Scholar] [CrossRef]
- Momi, N.; Kaur, S.; Ponnusamy, M.P.; Kumar, S.; Wittel, U.A.; Batra, S.K. Interplay between Smoking-induced Genotoxicity and Altered Signaling in Pancreatic Carcinogenesis. Carcinogenesis 2012, 33, 1617–1628. [Google Scholar] [CrossRef]
- Weissman, S.; Takakura, K.; Eibl, G.; Pandol, S.J.; Saruta, M. The Diverse Involvement of Cigarette Smoking in Pancreatic Cancer Development and Prognosis. Pancreas 2020, 49, 612–620. [Google Scholar] [CrossRef]
- Asahina, K.; Balog, S.; Hwang, E.; Moon, E.; Wan, E.; Skrypek, K.; Chen, Y.; Fernandez, J.; Romo, J.; Yang, Q.; et al. Moderate alcohol intake promotes pancreatic ductal adenocarcinoma development in mice expressing oncogenic Kras. Am. J. Physiol. Gastrointest. Liver Physiol. 2020, 318, G265–G276. [Google Scholar] [CrossRef]
- Kaune, T.; Griesmann, H.; Theuerkorn, K.; Hämmerle, M.; Laumen, H.; Krug, S.; Plumeier, I.; Kahl, S.; Junca, H.; Gustavo Dos Anjos Borges, L.; et al. Gender-specific changes of the gut microbiome correlate with tumor development in murine models of pancreatic cancer. iScience 2023, 26, 106841. [Google Scholar] [CrossRef] [PubMed]
- Pfisterer, N.; Ammer-Herrmenau, C.; Antweiler, K.; Küffer, S.; Ellenrieder, V.; Neesse, A. Dynamics of intestinal and intratumoral microbiome signatures in genetically engineered mice and human pancreatic ductal adenocarcinoma. Pancreatology 2023, 23, 663–673. [Google Scholar] [CrossRef] [PubMed]
- Michaud, D.S.; Izard, J.; Wilhelm-Benartzi, C.S.; You, D.H.; Grote, V.A.; Tjønneland, A.; Dahm, C.C.; Overvad, K.; Jenab, M.; Fedirko, V.; et al. Plasma antibodies to oral bacteria and risk of pancreatic cancer in a large European prospective cohort study. Gut 2013, 62, 1764–1770. [Google Scholar] [CrossRef]
- Saba, E.; Farhat, M.; Daoud, A.; Khashan, A.; Forkush, E.; Menahem, N.H.; Makkawi, H.; Pandi, K.; Angabo, S.; Kawasaki, H.; et al. Oral bacteria accelerate pancreatic cancer development in mice. Gut 2024, 73, 770–786. [Google Scholar] [CrossRef] [PubMed]
- Thomas, R.M.; Gharaibeh, R.Z.; Gauthier, J.; Beveridge, M.; Pope, J.L.; Guijarro, M.V.; Yu, Q.; He, Z.; Ohland, C.; Newsome, R.; et al. Intestinal microbiota enhances pancreatic carcinogenesis in preclinical models. Carcinogenesis 2018, 39, 1068–1078. [Google Scholar] [CrossRef]
- Sexton, R.E.; Uddin, M.H.; Bannoura, S.; Khan, H.Y.; Mzannar, Y.; Li, Y.; Aboukameel, A.; Al-Hallak, M.N.; Al-Share, B.; Mohamed, A.; et al. Connecting the Human Microbiome and Pancreatic Cancer. Cancer Metastasis Rev. 2022, 41, 317–331. [Google Scholar] [CrossRef]
- Attebury, H.; Daley, D. The Gut Microbiome and Pancreatic Cancer Development and Treatment. Cancer J. 2023, 29, 49–56. [Google Scholar] [CrossRef]
- Liot, S.; El Kholti, N.; Balas, J.; Genestier, L.; Verrier, B.; Valcourt, U.; Lambert, E. Development of thymic tumor in [LSL:KrasG12D.; Pdx1-CRE] mice, an adverse effect associated with accelerated pancreatic carcinogenesis. Sci. Rep. 2021, 11, 15075. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).