Exosome Enveloped by Nano Lipid Particle a New Model for Signal Transducer and Activator of Transcription 3 Silencer Ribonucleic Acid Delivery System to a Glioblastoma Mice Model
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture Experiments
2.1.1. Intact Exosome Purification and Exosomal RNA Isolation Followed by Complementary DNA Synthesis
2.1.2. siRNAs Sequences
2.1.3. Stability of siRNA
2.1.4. Preparation of Exo-STAT3 siRNA
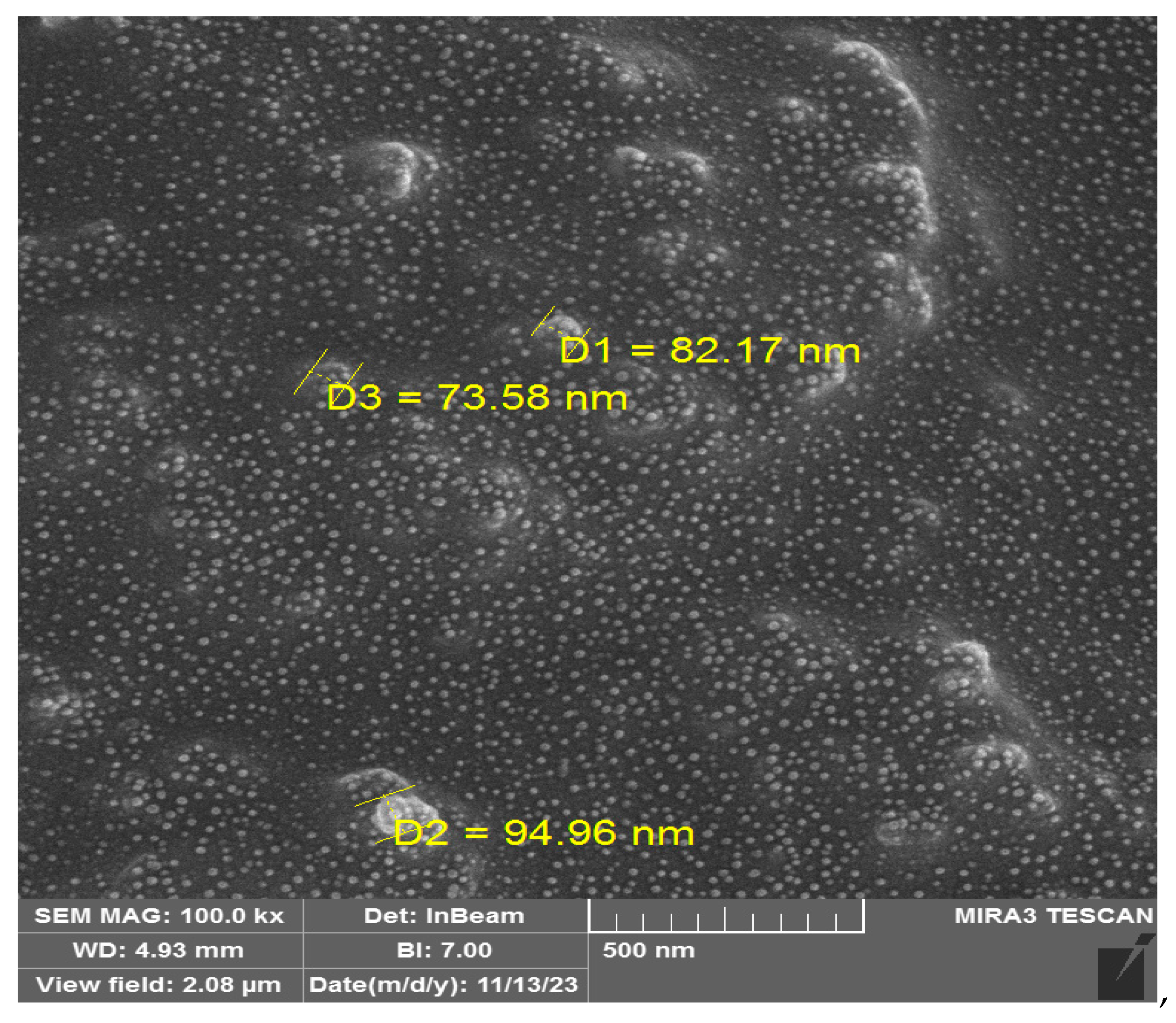
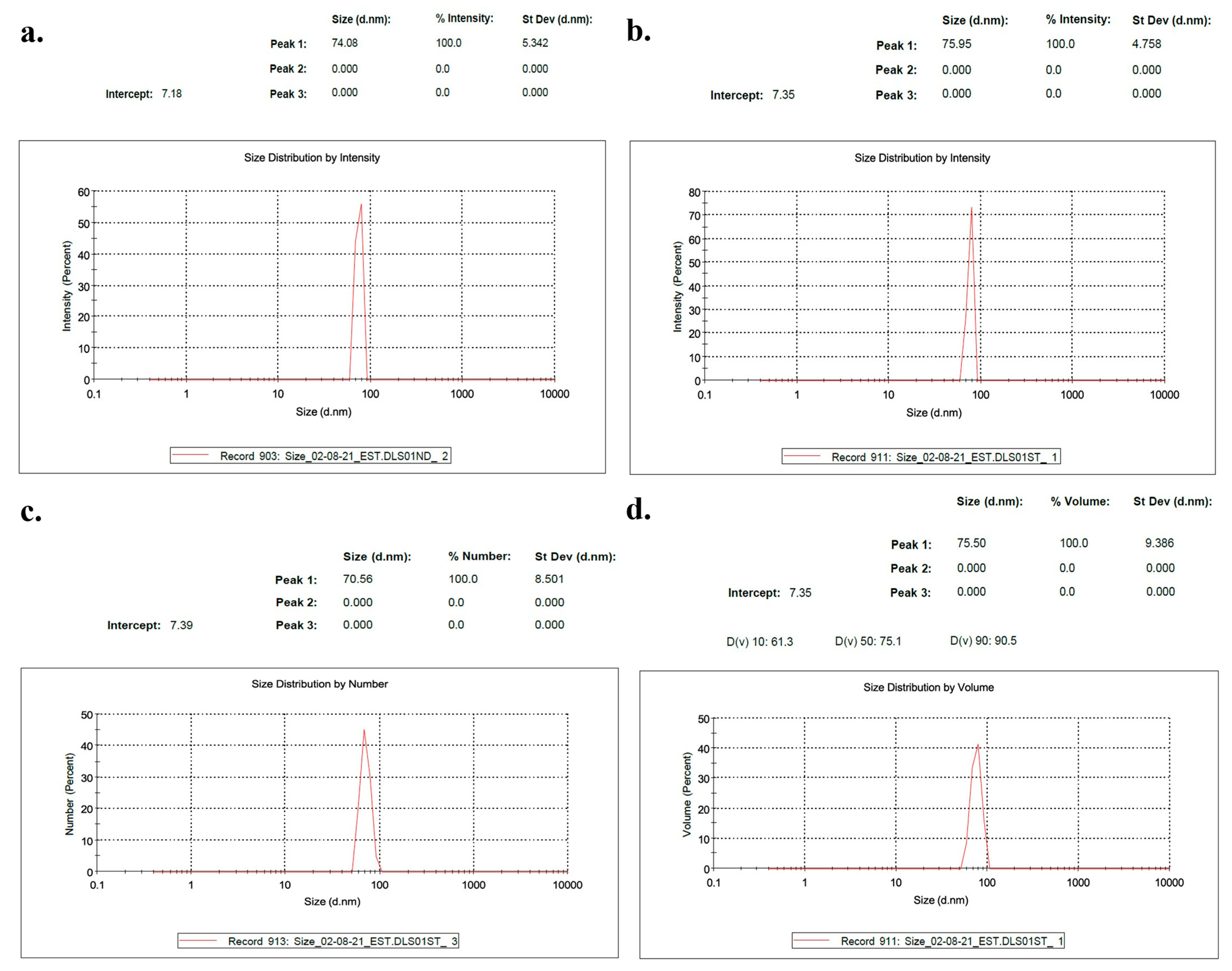
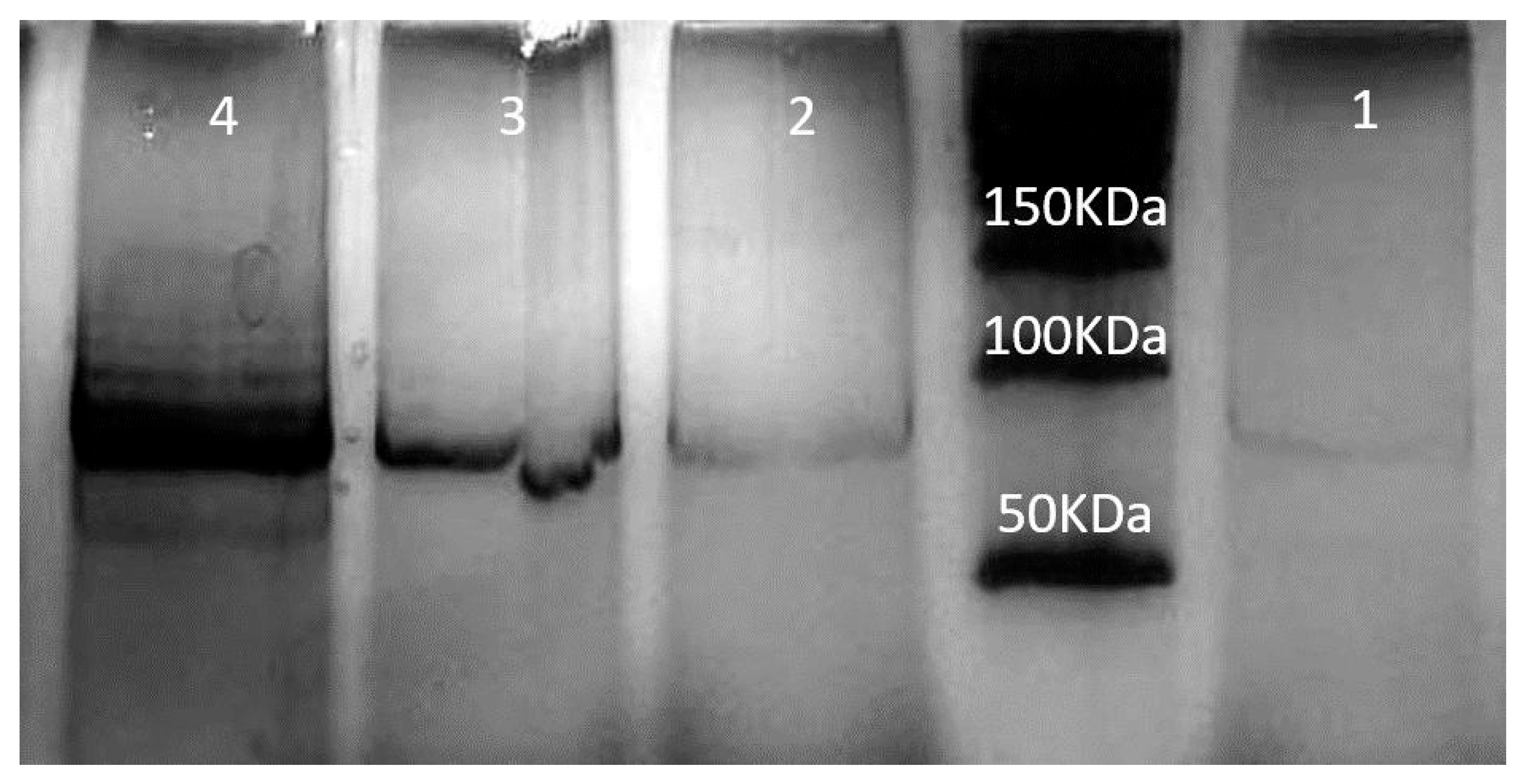
2.1.5. Characterization of Exosomes
2.1.6. Lipid Nanoparticles (NLP) Containing Exo-siRNA Using Microfluidic Technology by INSIGHT Nano Synthesis
2.1.7. Nanoparticle Characterization
2.1.8. Uptake Mechanism Studies
2.1.9. Assays for Silencing STAT3 In Vitro
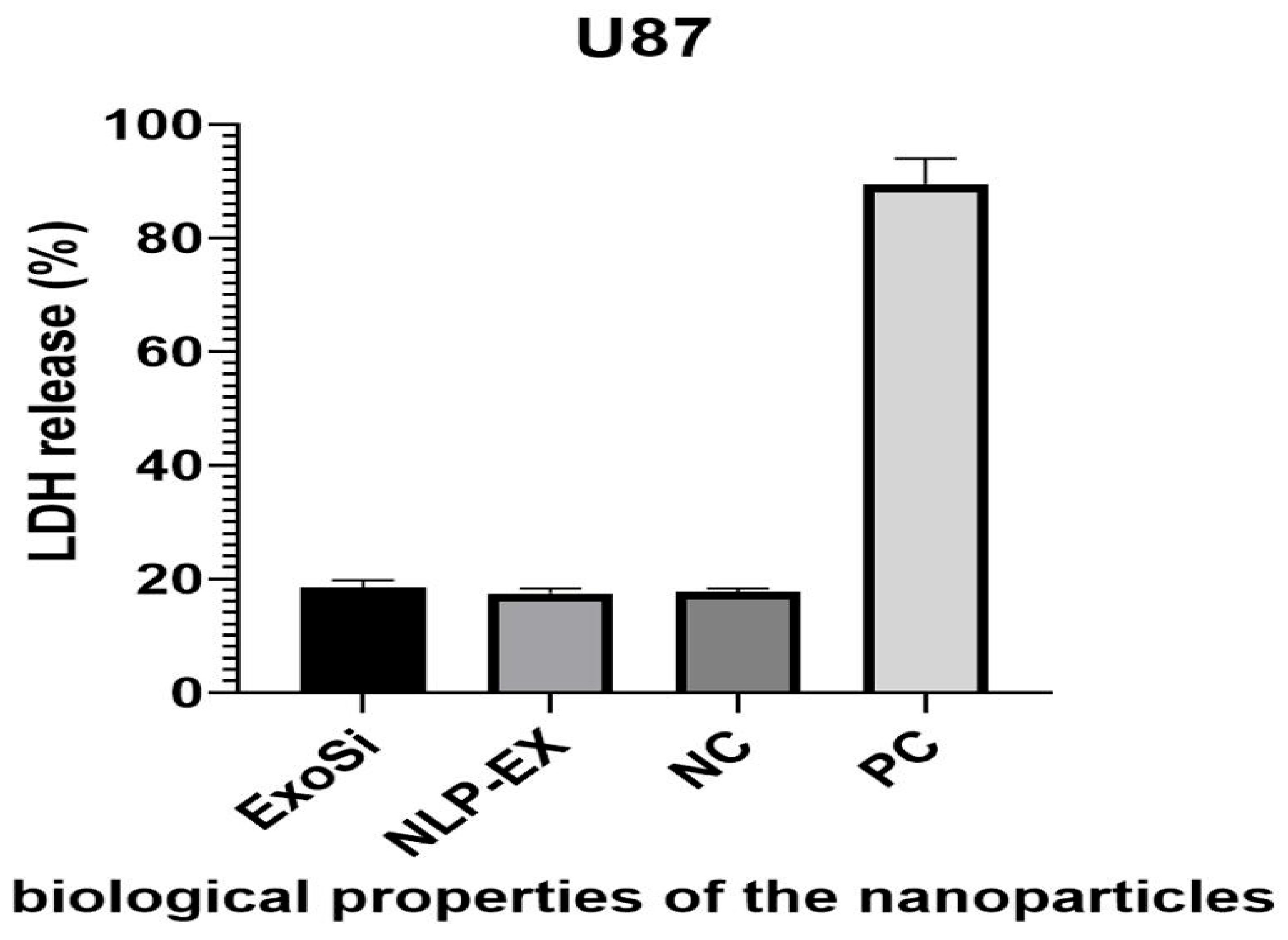
2.1.10. Cytotoxicity Assays Conducted in a Laboratory Setting
2.1.11. Apoptosis Assays
2.1.12. Western Blotting
2.2. Animal Experiments
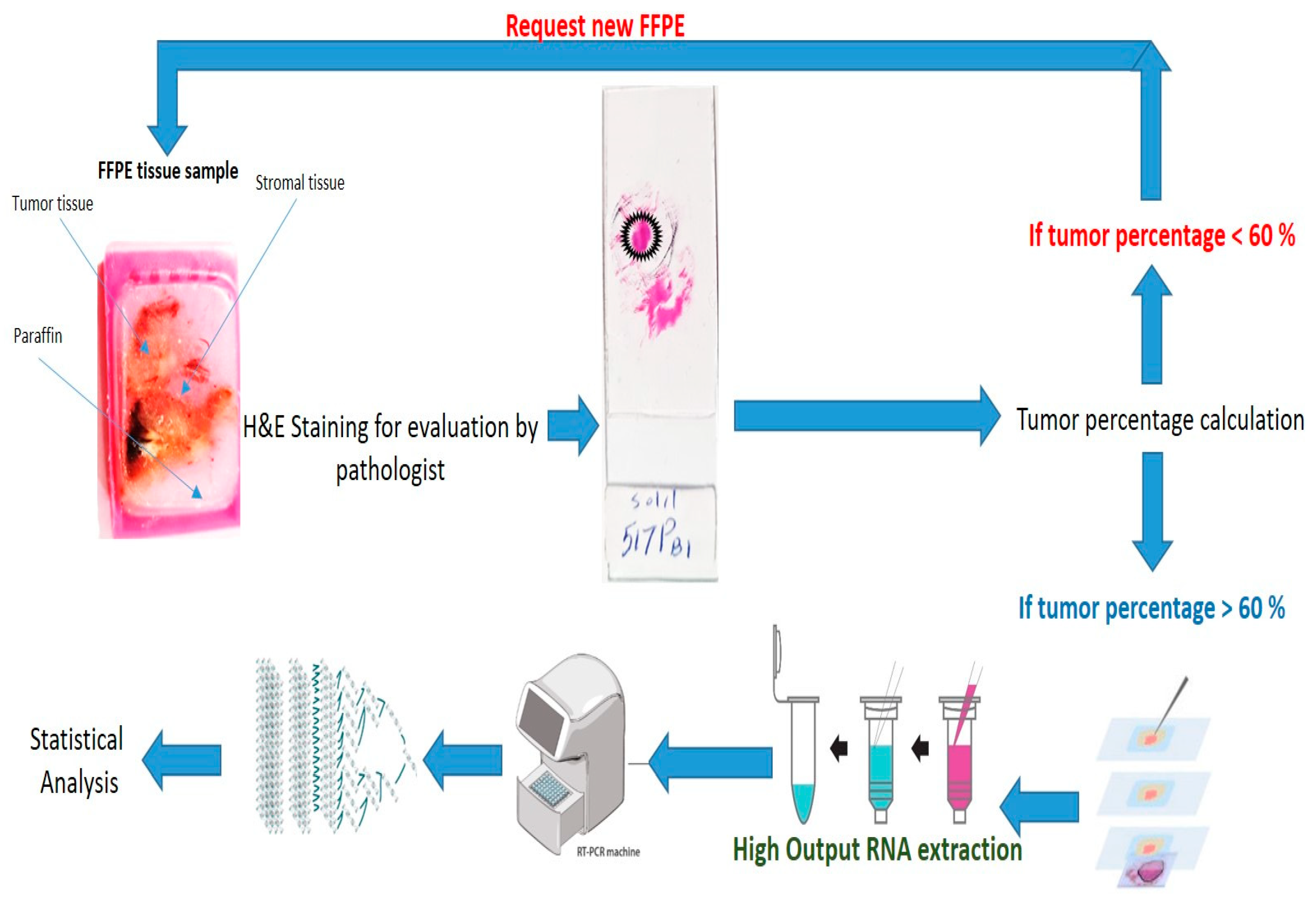
2.2.1. Laser-Capture Microdissection
2.2.2. MRI Acquisition
2.2.3. Determination of Tumor Volume
2.2.4. QRT-PCR
2.3. Statistics
3. Results
3.1. The Use of NLP-EXOSOME COMPLEX Nanoparticles Results in the Production of Reliable and Efficient Formulations for Delivering siRNA
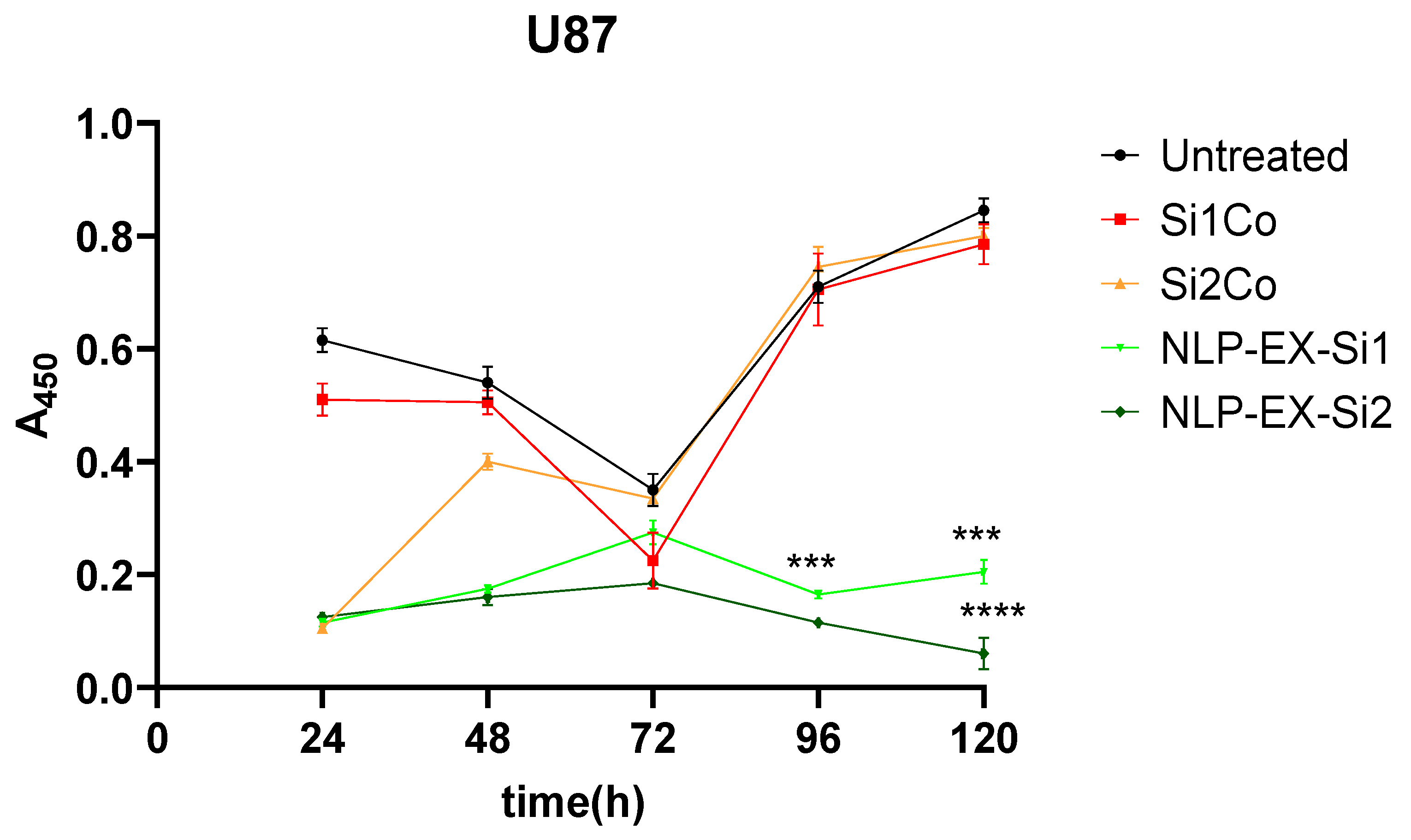
3.2. The Inhibition of STAT3 Downregulate Tumorigenesis Both In Vitro and In Vivo
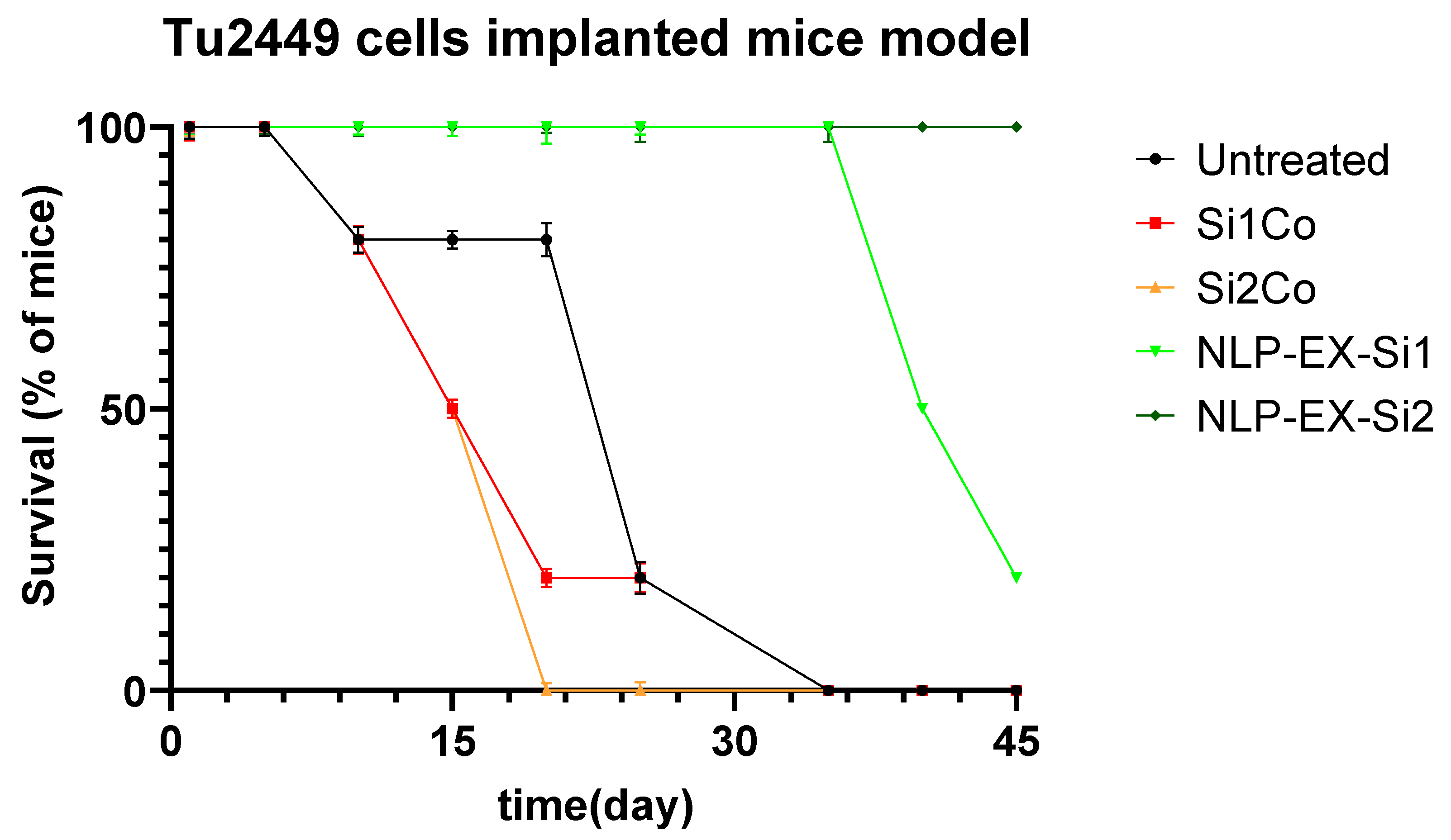
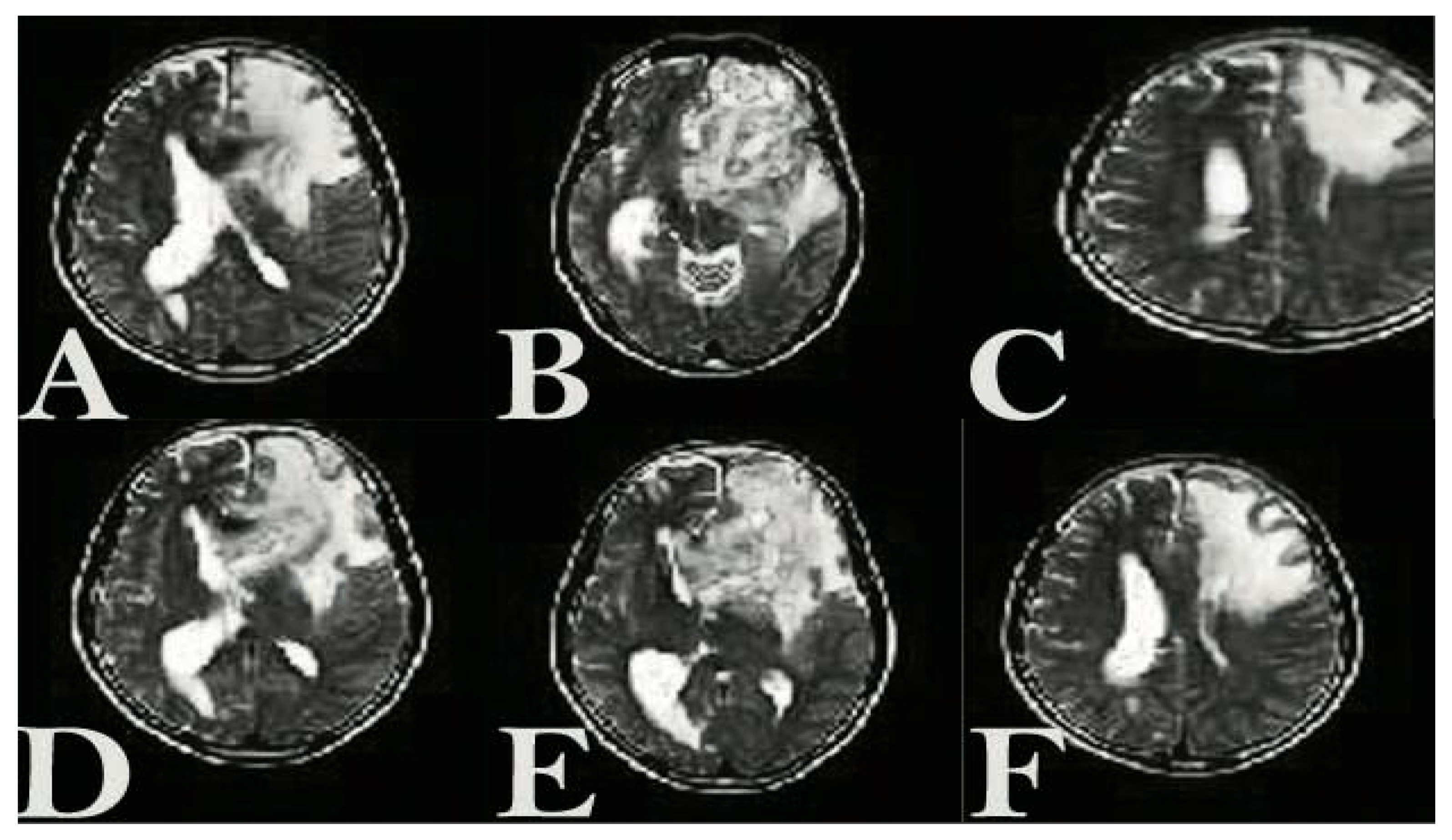
3.3. The Presence of NLP-EXOSOME COMPLEX Formulated Stat3 Silencer, Leads to an Increase in the Overall Survival Rate and a Decrease in Tumor Size in Mice with Glioma
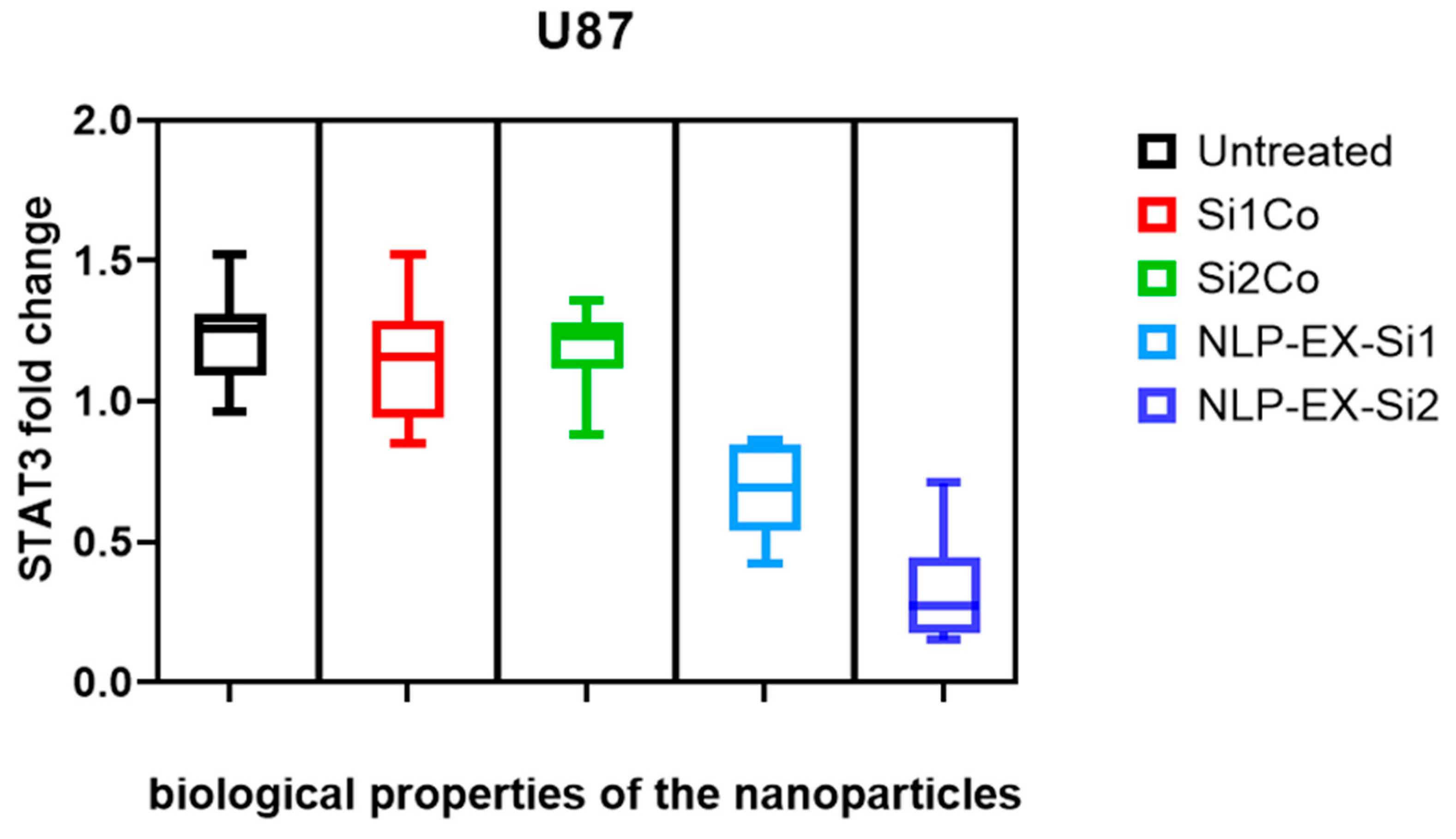
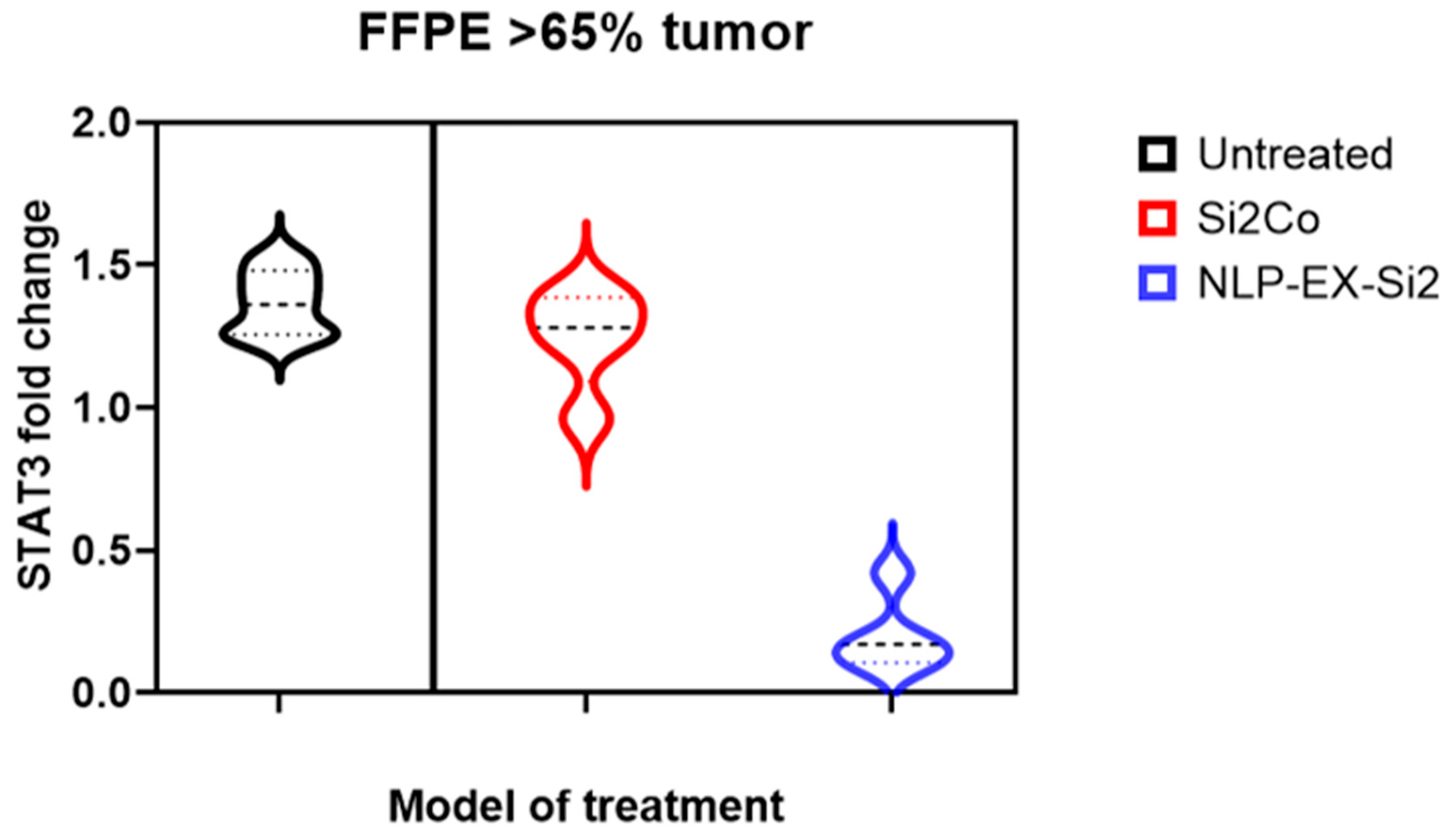
3.4. The Presence of NLP-EXOSOME COMPLEX Resulted in a Decrease in the Expression of STAT3 Within the Tumor
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Li, C.; Li, B.; Wang, H.; Qu, L.; Liu, H.; Weng, C.; Han, J.; Li, Y. Role of N6-methyladenosine methylation in glioma: Recent insights and future directions. Cell Mol. Biol. Lett. 2023, 28, 103. [Google Scholar] [CrossRef]
- Seker-Polat, F.; Pinarbasi Degirmenci, N.; Solaroglu, I.; Bagci-Onder, T. Tumor Cell Infiltration into the Brain in Glioblastoma: From Mechanisms to Clinical Perspectives. Cancers 2022, 14, 443. [Google Scholar] [CrossRef] [PubMed]
- Stoyanov, G.S.; Lyutfi, E.; Georgieva, R.; Georgiev, R.; Dzhenkov, D.L.; Petkova, L.; Ivanov, B.D.; Kaprelyan, A.; Ghenev, P. Reclassification of Glioblastoma Multiforme According to the 2021 World Health Organization Classification of Central Nervous System Tumors: A Single Institution Report and Practical Significance. Cureus 2022, 14, e21822. [Google Scholar] [CrossRef] [PubMed]
- Hanif, F.; Muzaffar, K.; Perveen, K.; Malhi, S.M.; Simjee Sh, U. Glioblastoma Multiforme: A Review of its Epidemiology and Pathogenesis through Clinical Presentation and Treatment. Asian Pac. J. Cancer Prev. 2017, 18, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Verdugo, E.; Puerto, I.; Medina, M.A. An update on the molecular biology of glioblastoma, with clinical implications and progress in its treatment. Cancer Commun. 2022, 42, 1083–1111. [Google Scholar] [CrossRef]
- Tang, Y.; Qazi, M.A.; Brown, K.R.; Mikolajewicz, N.; Moffat, J.; Singh, S.K.; McNicholas, P.D. Identification of five important genes to predict glioblastoma subtypes. Neurooncol. Adv. 2021, 3, vdab144. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Bellettato, C.M.; Scarpa, M. Possible strategies to cross the blood-brain barrier. Ital. J. Pediatr. 2018, 44, 131. [Google Scholar] [CrossRef]
- Luissint, A.C.; Artus, C.; Glacial, F.; Ganeshamoorthy, K.; Couraud, P.O. Tight junctions at the blood brain barrier: Physiological architecture and disease-associated dysregulation. Fluids Barriers CNS 2012, 9, 23. [Google Scholar] [CrossRef]
- Nakagawa, S.; Deli, M.A.; Kawaguchi, H.; Shimizudani, T.; Shimono, T.; Kittel, A.; Tanaka, K.; Niwa, M. A new blood-brain barrier model using primary rat brain endothelial cells, pericytes and astrocytes. Neurochem. Int. 2009, 54, 253–263. [Google Scholar] [CrossRef]
- Tsinman, O.; Tsinman, K.; Sun, N.; Avdeef, A. Physicochemical selectivity of the BBB microenvironment governing passive diffusion—matching with a porcine brain lipid extract artificial membrane permeability model. Pharm. Res. 2011, 28, 337–363. [Google Scholar] [CrossRef]
- Pardridge, W.M. Blood-brain barrier endogenous transporters as therapeutic targets: A new model for small molecule CNS drug discovery. Expert. Opin. Ther. Targets 2015, 19, 1059–1072. [Google Scholar] [CrossRef]
- Pardridge, W.M. Drug transport across the blood-brain barrier. J. Cereb. Blood Flow. Metab. 2012, 32, 1959–1972. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Chen, Q.; Chen, X.; Han, F.; Chen, Z.; Wang, Y. The blood-brain barrier: Structure, regulation, and drug delivery. Signal Transduct. Target. Ther. 2023, 8, 217. [Google Scholar] [CrossRef] [PubMed]
- Mikitsh, J.L.; Chacko, A.M. Pathways for small molecule delivery to the central nervous system across the blood-brain barrier. Perspect. Medicin Chem. 2014, 6, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Niazi, S.K. Non-Invasive Drug Delivery across the Blood-Brain Barrier: A Prospective Analysis. Pharmaceutics 2023, 15, 2599. [Google Scholar] [CrossRef]
- Yang, J.; Luly, K.M.; Green, J.J. Nonviral nanoparticle gene delivery into the CNS for neurological disorders and brain cancer applications. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2023, 15, e1853. [Google Scholar] [CrossRef]
- Sen, S.; Xavier, J.; Kumar, N.; Ahmad, M.Z.; Ranjan, O.P. Exosomes as natural nanocarrier-based drug delivery system: Recent insights and future perspectives. 3 Biotech 2023, 13, 101. [Google Scholar] [CrossRef]
- Batrakova, E.V.; Kim, M.S. Using exosomes, naturally-equipped nanocarriers, for drug delivery. J. Control. Release 2015, 219, 396–405. [Google Scholar] [CrossRef]
- Dilsiz, N. A comprehensive review on recent advances in exosome isolation and characterization: Toward clinical applications. Transl. Oncol. 2024, 50, 102121. [Google Scholar] [CrossRef]
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal 2013, 11, 88. [Google Scholar] [CrossRef] [PubMed]
- Xiong, A.; Yang, Z.; Shen, Y.; Zhou, J.; Shen, Q. Transcription Factor STAT3 as a Novel Molecular Target for Cancer Prevention. Cancers 2014, 6, 926–957. [Google Scholar] [CrossRef] [PubMed]
- Aheget, H.; Mazini, L.; Martin, F.; Belqat, B.; Marchal, J.A.; Benabdellah, K. Exosomes: Their Role in Pathogenesis, Diagnosis and Treatment of Diseases. Cancers 2020, 13, 84. [Google Scholar] [CrossRef] [PubMed]
- Ozes, A.R.; Pulliam, N.; Ertosun, M.G.; Yilmaz, O.; Tang, J.; Copuroglu, E.; Matei, D.; Ozes, O.N.; Nephew, K.P. Protein kinase A-mediated phosphorylation regulates STAT3 activation and oncogenic EZH2 activity. Oncogene 2018, 37, 3589–3600. [Google Scholar] [CrossRef]
- Ganguly, D.; Fan, M.; Yang, C.H.; Zbytek, B.; Finkelstein, D.; Roussel, M.F.; Pfeffer, L.M. The critical role that STAT3 plays in glioma-initiating cells: STAT3 addiction in glioma. Oncotarget 2018, 9, 22095–22112. [Google Scholar] [CrossRef]
- Zhang, G.; Hou, S.; Li, S.; Wang, Y.; Cui, W. Role of STAT3 in cancer cell epithelial-mesenchymal transition (Review). Int. J. Oncol. 2024, 64, 48. [Google Scholar] [CrossRef]
- Carpenter, R.L.; Lo, H.W. STAT3 Target Genes Relevant to Human Cancers. Cancers 2014, 6, 897–925. [Google Scholar] [CrossRef] [PubMed]
- Monfaredan, A.; Rahim, F.; Tavoosidana, G.; Modarressi, M.H.; Hosseininasab, A.; Aghajani-Afrouzi, A.-A.; Sabet, M.S.; Motevaseli, E. A microchip for exosome isolation that can be impregnated with imatinib simultaneously: An in vitro analysis. Russ. Open Med. J. 2024, 13, 11. [Google Scholar] [CrossRef]
- Montano-Samaniego, M.; Bravo-Estupinan, D.M.; Mendez-Guerrero, O.; Alarcon-Hernandez, E.; Ibanez-Hernandez, M. Strategies for Targeting Gene Therapy in Cancer Cells With Tumor-Specific Promoters. Front. Oncol. 2020, 10, 605380. [Google Scholar] [CrossRef]
- Kang, H.; Ga, Y.J.; Kim, S.H.; Cho, Y.H.; Kim, J.W.; Kim, C.; Yeh, J.Y. Small interfering RNA (siRNA)-based therapeutic applications against viruses: Principles, potential, and challenges. J. Biomed. Sci. 2023, 30, 88. [Google Scholar] [CrossRef]
- Brachet-Botineau, M.; Polomski, M.; Neubauer, H.A.; Juen, L.; Hedou, D.; Viaud-Massuard, M.C.; Prie, G.; Gouilleux, F. Pharmacological Inhibition of Oncogenic STAT3 and STAT5 Signaling in Hematopoietic Cancers. Cancers 2020, 12, 240. [Google Scholar] [CrossRef]
- Linder, B.; Weirauch, U.; Ewe, A.; Uhmann, A.; Seifert, V.; Mittelbronn, M.; Harter, P.N.; Aigner, A.; Kogel, D. Therapeutic Targeting of Stat3 Using Lipopolyplex Nanoparticle-Formulated siRNA in a Syngeneic Orthotopic Mouse Glioma Model. Cancers 2019, 11, 333. [Google Scholar] [CrossRef]
- Fatima, S.; Qaiser, A.; Andleeb, S.; Hashmi, A.H.; Manzoor, S. Navigating the brain: The role of exosomal shuttles in precision therapeutics. Front. Neurol. 2023, 14, 1324216. [Google Scholar] [CrossRef]
- Ezike, T.C.; Okpala, U.S.; Onoja, U.L.; Nwike, C.P.; Ezeako, E.C.; Okpara, O.J.; Okoroafor, C.C.; Eze, S.C.; Kalu, O.L.; Odoh, E.C.; et al. Advances in drug delivery systems, challenges and future directions. Heliyon 2023, 9, e17488. [Google Scholar] [CrossRef]
- Koh, H.B.; Kim, H.J.; Kang, S.W.; Yoo, T.H. Exosome-Based Drug Delivery: Translation from Bench to Clinic. Pharmaceutics 2023, 15, 2042. [Google Scholar] [CrossRef]
- Zeng, H.; Guo, S.; Ren, X.; Wu, Z.; Liu, S.; Yao, X. Current Strategies for Exosome Cargo Loading and Targeting Delivery. Cells 2023, 12, 1416. [Google Scholar] [CrossRef]
- Ohka, F.; Natsume, A.; Wakabayashi, T. Current trends in targeted therapies for glioblastoma multiforme. Neurol. Res. Int. 2012, 2012, 878425. [Google Scholar] [CrossRef]
- Tolomeo, M.; Cascio, A. The Multifaced Role of STAT3 in Cancer and Its Implication for Anticancer Therapy. Int. J. Mol. Sci. 2021, 22, 603. [Google Scholar] [CrossRef]
- Zhuang, X.; Xiang, X.; Grizzle, W.; Sun, D.; Zhang, S.; Axtell, R.C.; Ju, S.; Mu, J.; Zhang, L.; Steinman, L.; et al. Treatment of brain inflammatory diseases by delivering exosome encapsulated anti-inflammatory drugs from the nasal region to the brain. Mol. Ther. 2011, 19, 1769–1779. [Google Scholar] [CrossRef]
- Li, Z.; Yang, L.; Wang, H.; Binzel, D.W.; Williams, T.M.; Guo, P. Non-Small-Cell Lung Cancer Regression by siRNA Delivered Through Exosomes That Display EGFR RNA Aptamer. Nucleic Acid. Ther. 2021, 31, 364–374. [Google Scholar] [CrossRef]
- Esposito, C.L.; Nuzzo, S.; Catuogno, S.; Romano, S.; de Nigris, F.; de Franciscis, V. STAT3 Gene Silencing by Aptamer-siRNA Chimera as Selective Therapeutic for Glioblastoma. Mol. Ther. Nucleic Acids 2018, 10, 398–411. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Gerson, S.L. MGMT: Its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer 2004, 4, 296–307. [Google Scholar] [CrossRef]
- Hegi, M.E.; Diserens, A.C.; Gorlia, T.; Hamou, M.F.; de Tribolet, N.; Weller, M.; Kros, J.M.; Hainfellner, J.A.; Mason, W.; Mariani, L.; et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N. Engl. J. Med. 2005, 352, 997–1003. [Google Scholar] [CrossRef]
- Yu, H.; Pardoll, D.; Jove, R. STATs in cancer inflammation and immunity: A leading role for STAT3. Nat. Rev. Cancer 2009, 9, 798–809. [Google Scholar] [CrossRef]
- Reifenberger, G.; Wirsching, H.G.; Knobbe-Thomsen, C.B.; Weller, M. Advances in the molecular genetics of gliomas—Implications for classification and therapy. Nat. Rev. Clin. Oncol. 2017, 14, 434–452. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Monfaredan, A.; Şen, S.; Hosseininasab, A.; Taştekin, D.; Fazli, G.; Bozbey, H.U.; Yousefi, N.; Hocaoğlu, M.; Öncül, M.O.; Özen, R.S. Exosome Enveloped by Nano Lipid Particle a New Model for Signal Transducer and Activator of Transcription 3 Silencer Ribonucleic Acid Delivery System to a Glioblastoma Mice Model. Cancers 2025, 17, 1648. https://doi.org/10.3390/cancers17101648
Monfaredan A, Şen S, Hosseininasab A, Taştekin D, Fazli G, Bozbey HU, Yousefi N, Hocaoğlu M, Öncül MO, Özen RS. Exosome Enveloped by Nano Lipid Particle a New Model for Signal Transducer and Activator of Transcription 3 Silencer Ribonucleic Acid Delivery System to a Glioblastoma Mice Model. Cancers. 2025; 17(10):1648. https://doi.org/10.3390/cancers17101648
Chicago/Turabian StyleMonfaredan, Amir, Sena Şen, Alaviyehsadat Hosseininasab, Didem Taştekin, Ghazaleh Fazli, Hamza Uğur Bozbey, Nasrin Yousefi, Merve Hocaoğlu, Mustafa Oral Öncül, and Rıdvan Seçkin Özen. 2025. "Exosome Enveloped by Nano Lipid Particle a New Model for Signal Transducer and Activator of Transcription 3 Silencer Ribonucleic Acid Delivery System to a Glioblastoma Mice Model" Cancers 17, no. 10: 1648. https://doi.org/10.3390/cancers17101648
APA StyleMonfaredan, A., Şen, S., Hosseininasab, A., Taştekin, D., Fazli, G., Bozbey, H. U., Yousefi, N., Hocaoğlu, M., Öncül, M. O., & Özen, R. S. (2025). Exosome Enveloped by Nano Lipid Particle a New Model for Signal Transducer and Activator of Transcription 3 Silencer Ribonucleic Acid Delivery System to a Glioblastoma Mice Model. Cancers, 17(10), 1648. https://doi.org/10.3390/cancers17101648






