Computational Mutagenesis of GPx7 and GPx8: Structural and Stability Insights into Rare Genetic and Somatic Missense Mutations and Their Implications for Cancer Development
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Selection and Alignment of the Sequence and Structure of GPx7 and GPx8
2.2. Phylogenetic Analysis of the Glutathione Family of Proteins
2.3. Collection of Cancer-Causing Somatic Mutations
2.4. Saturated Computational Mutagenesis
2.5. Calculations of Gibbs Free Energy Changes
2.6. Comparative and Statistical Analyses
3. Results
3.1. Phylogenetic Analysis of Gpx7 and GPx8
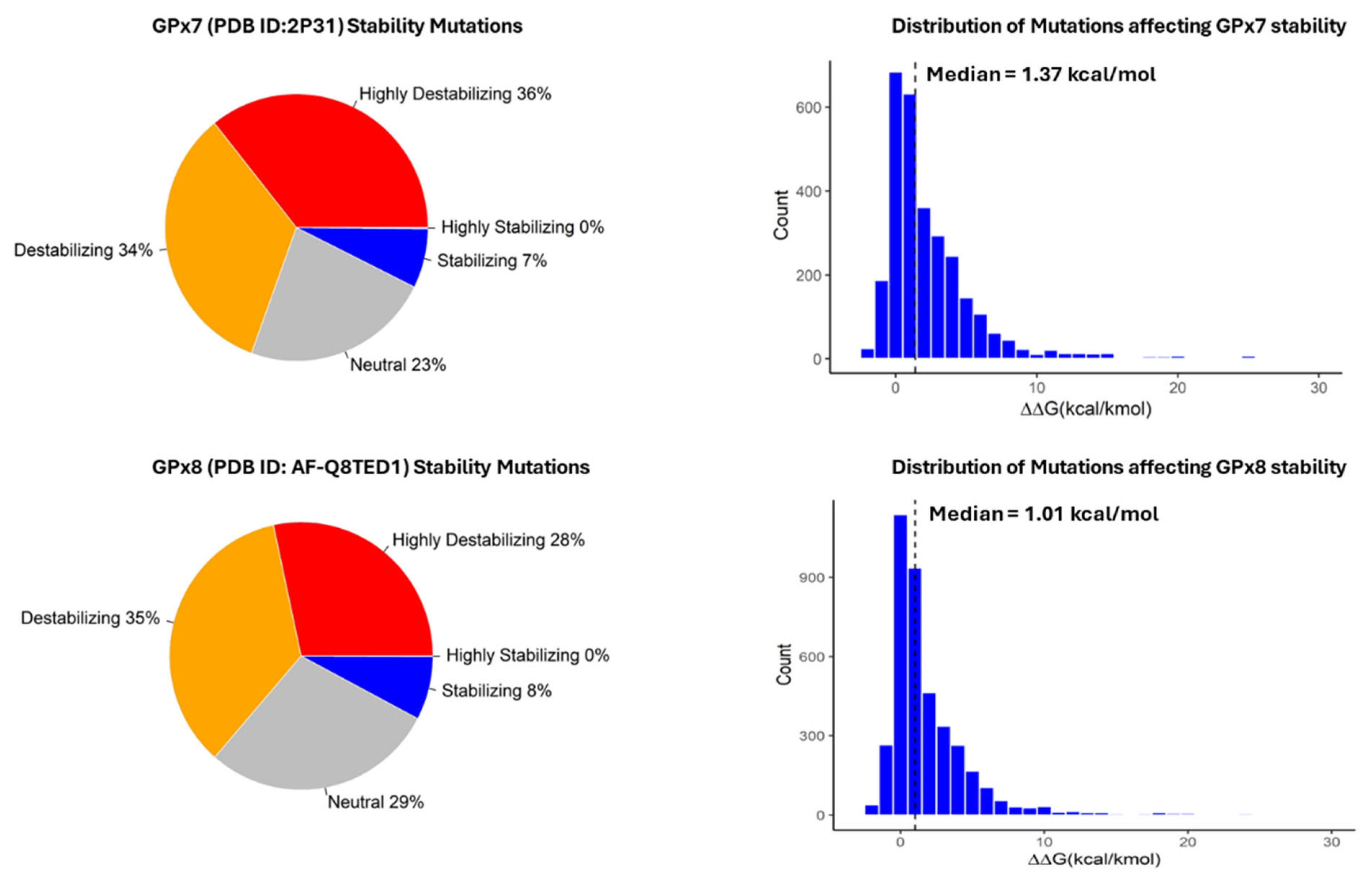
3.2. Classification and Distribution of Missense Mutations
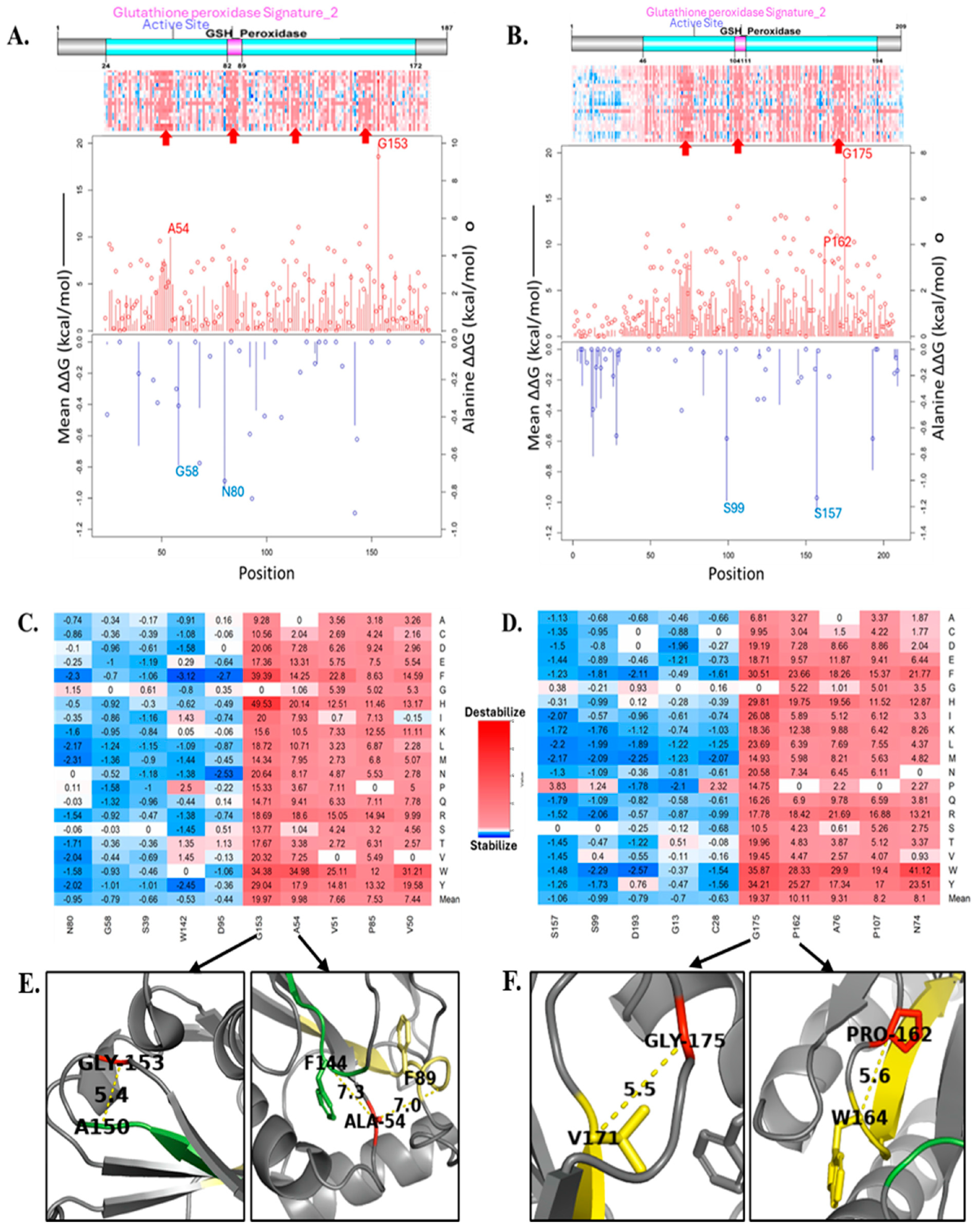
3.3. Computational Analysis of the Effect of Missense Mutations on GPx7 and GPx8
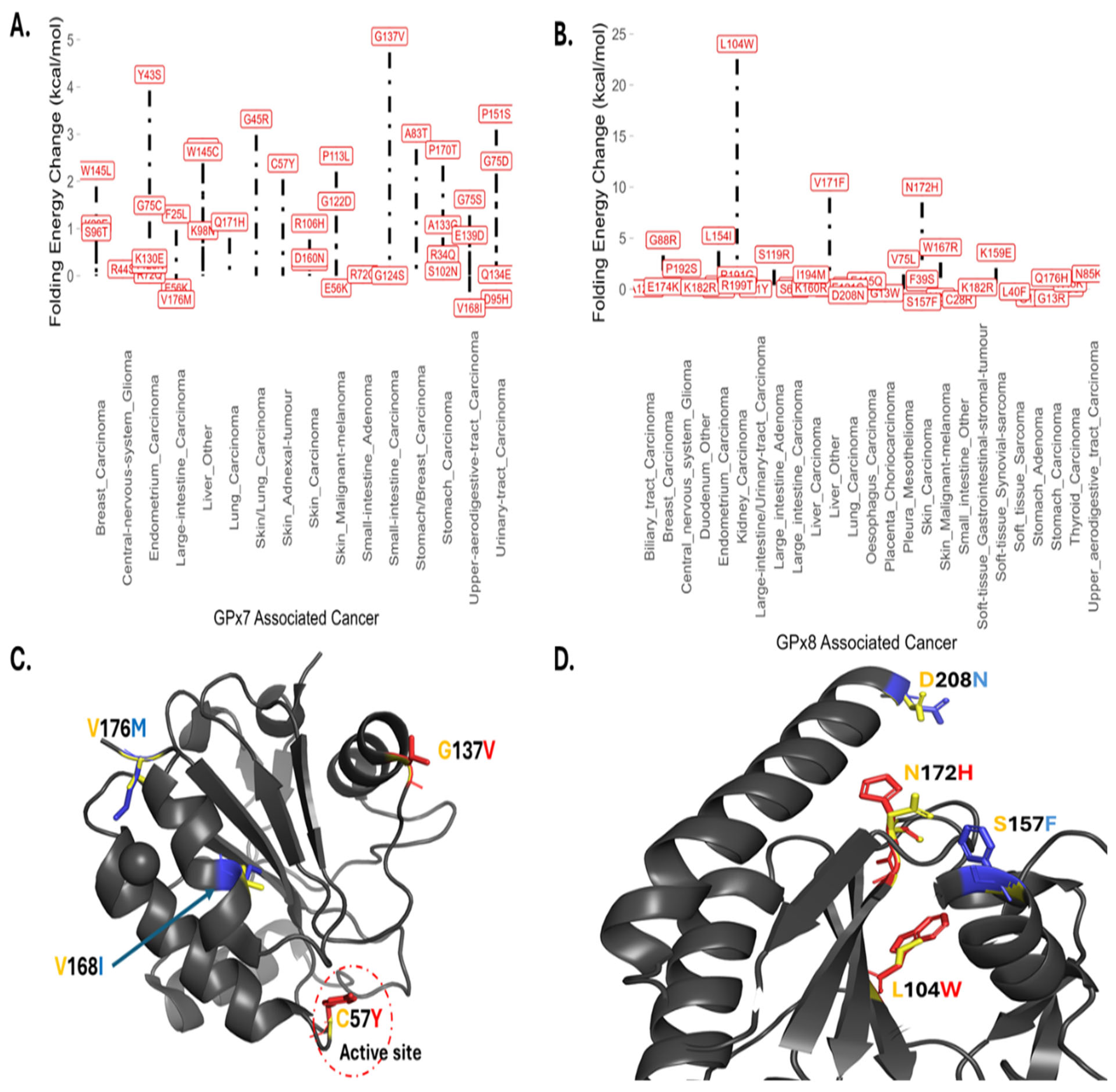
3.4. Somatic Mutations, Energy Changes, and Cancer Types
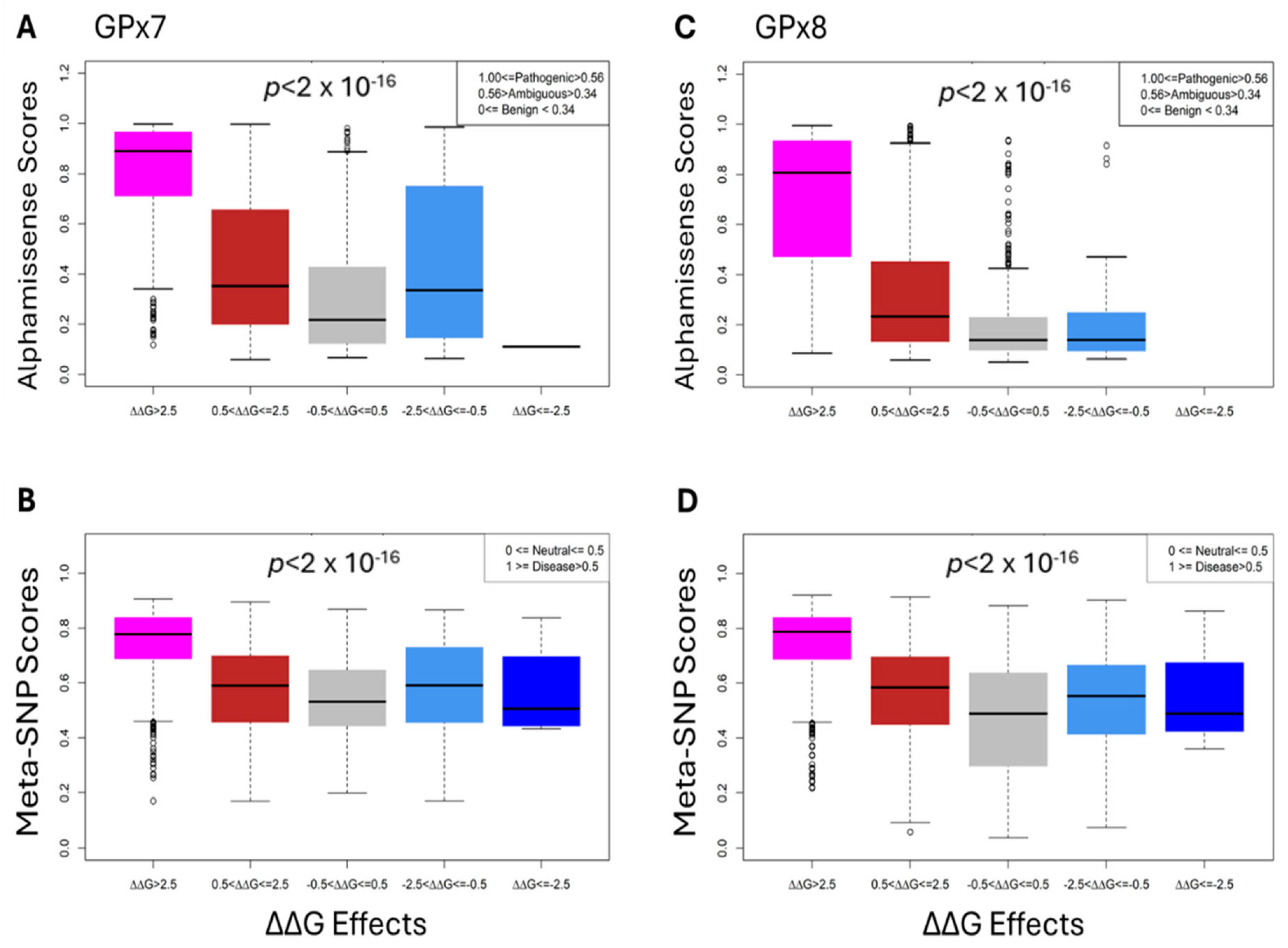
3.5. Comparative and Statistical Analyses
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pei, J.; Pan, X.; Wei, G.; Hua, Y. Research progress of glutathione peroxidase family (GPX) in redoxidation. Front. Pharmacol. 2023, 14, 1147414. [Google Scholar] [CrossRef] [PubMed]
- Margis, R.; Dunand, C.; Teixeira, F.K.; Margis-Pinheiro, M. Glutathione peroxidase family—An evolutionary overview. FEBS J. 2008, 275, 3959–3970. [Google Scholar] [CrossRef] [PubMed]
- Buday, K.; Conrad, M. Emerging roles for non-selenium containing ER-resident glutathione peroxidases in cell signaling and disease. Biol. Chem. 2021, 402, 271–287. [Google Scholar] [CrossRef]
- Chen, Y.I.; Wei, P.C.; Hsu, J.L.; Su, F.Y.; Lee, W.H. NPGPx (GPx7): A novel oxidative stress sensor/transmitter with multiple roles in redox homeostasis. Am. J. Transl. Res. 2016, 8, 1626–1640. [Google Scholar] [PubMed]
- Zhao, Q.; Zhang, L.; Wang, Y.; Sun, Y.; Wang, T.; Cao, J.; Qi, M.; Du, X.; Xia, Z.; Zhang, R.; et al. A Bioinformatic Analysis: The Overexpression and Prognostic Potential of GPX7 in Lower-Grade Glioma. Int. J. Gen. Med. 2022, 15, 4321–4337. [Google Scholar] [CrossRef] [PubMed]
- Khatib, A.; Solaimuthu, B.; Ben Yosef, M.; Abu Rmaileh, A.; Tanna, M.; Oren, G.; Schlesinger Frisch, M.; Axelrod, J.H.; Lichtenstein, M.; Shaul, Y.D. The glutathione peroxidase 8 (GPX8)/IL-6/STAT3 axis is essential in maintaining an aggressive breast cancer phenotype. Proc. Natl. Acad. Sci. USA 2020, 117, 21420–21431. [Google Scholar] [CrossRef]
- Madeira, F.; Pearce, M.; Tivey, A.R.N.; Basutkar, P.; Lee, J.; Edbali, O.; Madhusoodanan, N.; Kolesnikov, A.; Lopez, R. Search and sequence analysis tools services from EMBL-EBI in 2022. Nucleic Acids Res. 2022, 50, W276–W279. [Google Scholar] [CrossRef] [PubMed]
- Tate, J.G.; Bamford, S.; Jubb, H.C.; Sondka, Z.; Beare, D.M.; Bindal, N.; Boutselakis, H.; Cole, C.G.; Creatore, C.; Dawson, E.; et al. COSMIC: The Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019, 47, D941–D947. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Chapman, M.; Evans, K.; Azevedo, L.; Hayden, M.; Heywood, S.; Millar, D.S.; Phillips, A.D.; et al. The Human Gene Mutation Database (HGMD((R))): Optimizing its use in a clinical diagnostic or research setting. Hum. Genet. 2020, 139, 1197–1207. [Google Scholar] [CrossRef] [PubMed]
- Schymkowitz, J.; Borg, J.; Stricher, F.; Nys, R.; Rousseau, F.; Serrano, L. The FoldX web server: An online force field. Nucleic Acids Res. 2005, 33, W382–W388. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, E.; Altman, R.B.; Bromberg, Y. Collective judgment predicts disease-associated single nucleotide variants. BMC Genom. 2013, 14 (Suppl. 3), S2. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Novati, G.; Pan, J.; Bycroft, C.; Zemgulyte, A.; Applebaum, T.; Pritzel, A.; Wong, L.H.; Zielinski, M.; Sargeant, T.; et al. Accurate proteome-wide missense variant effect prediction with AlphaMissense. Science 2023, 381, eadg7492. [Google Scholar] [CrossRef] [PubMed]
- Denisova, E.; Westphal, D.; Surowy, H.M.; Meier, F.; Hutter, B.; Reifenberger, J.; Rutten, A.; Schulz, A.; Sergon, M.; Ziemer, M.; et al. Whole-exome sequencing in eccrine porocarcinoma indicates promising therapeutic strategies. Cancer Gene Ther. 2022, 29, 697–708. [Google Scholar] [CrossRef] [PubMed]
- Sobitan, A.; Edwards, W.; Jalal, M.S.; Kolawole, A.; Ullah, H.; Duttaroy, A.; Li, J.; Teng, S. Prediction of the Effects of Missense Mutations on Human Myeloperoxidase Protein Stability Using In Silico Saturation Mutagenesis. Genes 2022, 13, 1412. [Google Scholar] [CrossRef] [PubMed]
- Sobitan, A.; Gebremedhin, B.; Yao, Q.; Xie, G.; Gu, X.; Li, J.; Teng, S. A Computational Approach: The Functional Effects of Thyroid Peroxidase Variants in Thyroid Cancer and Genetic Disorders. JCO Clin. Cancer Inform. 2024, 8, e2300140. [Google Scholar] [CrossRef] [PubMed]
- Kvarnung, M.; Taylan, F.; Nilsson, D.; Anderlid, B.M.; Malmgren, H.; Lagerstedt-Robinson, K.; Holmberg, E.; Burstedt, M.; Nordenskjold, M.; Nordgren, A.; et al. Genomic screening in rare disorders: New mutations and phenotypes, highlighting ALG14 as a novel cause of severe intellectual disability. Clin. Genet. 2018, 94, 528–537. [Google Scholar] [CrossRef]
- Permuth, J.B.; Pirie, A.; Ann Chen, Y.; Lin, H.Y.; Reid, B.M.; Chen, Z.; Monteiro, A.; Dennis, J.; Mendoza-Fandino, G.; AOCS Study Group; et al. Exome genotyping arrays to identify rare and low frequency variants associated with epithelial ovarian cancer risk. Hum. Mol. Genet. 2016, 25, 3600–3612. [Google Scholar] [CrossRef]
- Li, L.; Jiang, D.; Liu, H.; Guo, C.; Zhao, R.; Zhang, Q.; Xu, C.; Qin, Z.; Feng, J.; Liu, Y.; et al. Comprehensive proteogenomic characterization of early duodenal cancer reveals the carcinogenesis tracks of different subtypes. Nat. Commun. 2023, 14, 1751. [Google Scholar] [CrossRef]
- Pang, Y.; Xie, F.; Cao, H.; Wang, C.; Zhu, M.; Liu, X.; Lu, X.; Huang, T.; Shen, Y.; Li, K.; et al. Mutational inactivation of mTORC1 repressor gene DEPDC5 in human gastrointestinal stromal tumors. Proc. Natl. Acad. Sci. USA 2019, 116, 22746–22753. [Google Scholar] [CrossRef] [PubMed]
- Klomsiri, C.; Karplus, P.A.; Poole, L.B. Cysteine-based redox switches in enzymes. Antioxid. Redox Signal. 2011, 14, 1065–1077. [Google Scholar] [CrossRef]
- Zhang, H.; Huang, Y.; Bei, Z.; Ju, Z.; Meng, J.; Hao, M.; Zhang, J.; Zhang, H.; Xi, W. Inter-Residue Distance Prediction From Duet Deep Learning Models. Front. Genet. 2022, 13, 887491. [Google Scholar] [CrossRef] [PubMed]

| Top 5 Destabilizing Mutations | Top 5 Stabilizing Mutations | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| GPx8 | Foldx | Meta-SNP | GPx8 | Foldx | Meta-SNP | ||||
| Mutations | ∆∆G | Effect | Score | Pathogenicity | Mutations | ∆∆G | Effect | Score | Pathogenicity |
| N74W | 41.12 | Highly_Des | 0.89 | Disease | S70I | −2.82 | Highly_Sta | 0.36 | Neutral |
| G175W | 35.87 | Highly_Des | 0.88 | Disease | S119P | −2.74 | Highly_Sta | 0.49 | Neutral |
| G175Y | 34.21 | Highly_Des | 0.88 | Disease | D193W | −2.57 | Highly_Sta | 0.86 | Disease |
| A126W | 33.09 | Highly_Des | 0.82 | Disease | P118D | −2.39 | Stabilizing | 0.30 | Neutral |
| G175F | 30.51 | Highly_Des | 0.87 | Disease | S99W | −2.29 | Stabilizing | 0.72 | Disease |
| GPx7 | GPx7 | ||||||||
| Mutations | ∆∆G | Effect | Score | Pathogenicity | Mutations | ∆∆G | Effect | Score | Pathogenicity |
| G153H | 49.53 | Highly_Des | 0.89 | Disease | E93M | −3.61 | Highly_Sta | 0.56 | Disease |
| G153F | 39.39 | Highly_Des | 0.87 | Disease | W142F | −3.12 | Highly_Sta | 0.84 | Disease |
| A54W | 34.98 | Highly_Des | 0.88 | Disease | D95F | −2.70 | Highly_Sta | 0.45 | Neutral |
| G153W | 34.38 | Highly_Des | 0.89 | Disease | D95N | −2.53 | Highly_Sta | 0.43 | Neutral |
| V50W | 31.21 | Highly_Des | 0.86 | Disease | S48I | −2.49 | Stabilizing | 0.40 | Neutral |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sobitan, A.; Buhari, N.; Youssri, Z.; Wen, F.; Kidane, D.; Teng, S. Computational Mutagenesis of GPx7 and GPx8: Structural and Stability Insights into Rare Genetic and Somatic Missense Mutations and Their Implications for Cancer Development. Cancers 2025, 17, 105. https://doi.org/10.3390/cancers17010105
Sobitan A, Buhari N, Youssri Z, Wen F, Kidane D, Teng S. Computational Mutagenesis of GPx7 and GPx8: Structural and Stability Insights into Rare Genetic and Somatic Missense Mutations and Their Implications for Cancer Development. Cancers. 2025; 17(1):105. https://doi.org/10.3390/cancers17010105
Chicago/Turabian StyleSobitan, Adebiyi, Nosimot Buhari, Zainab Youssri, Fayuan Wen, Dawit Kidane, and Shaolei Teng. 2025. "Computational Mutagenesis of GPx7 and GPx8: Structural and Stability Insights into Rare Genetic and Somatic Missense Mutations and Their Implications for Cancer Development" Cancers 17, no. 1: 105. https://doi.org/10.3390/cancers17010105
APA StyleSobitan, A., Buhari, N., Youssri, Z., Wen, F., Kidane, D., & Teng, S. (2025). Computational Mutagenesis of GPx7 and GPx8: Structural and Stability Insights into Rare Genetic and Somatic Missense Mutations and Their Implications for Cancer Development. Cancers, 17(1), 105. https://doi.org/10.3390/cancers17010105






