Applications of Urinary Extracellular Vesicles in the Diagnosis and Active Surveillance of Prostate Cancer
Abstract
Simple Summary
Abstract
1. Introduction
- (i)
- Higher specificity and sensitivity in distinguishing clinically significant prostate cancer from indolent disease [11];
- (ii)
- Improved accuracy of diagnosis in the context of multifocal disease [12];
- (iii)
- Feasibility for non-invasive and remote testing [13];
- (iv)
- Improved cost-effectiveness [14];
- (v)
- Improved environmental sustainability [15].
2. Prostate-Derived Extracellular Vesicles
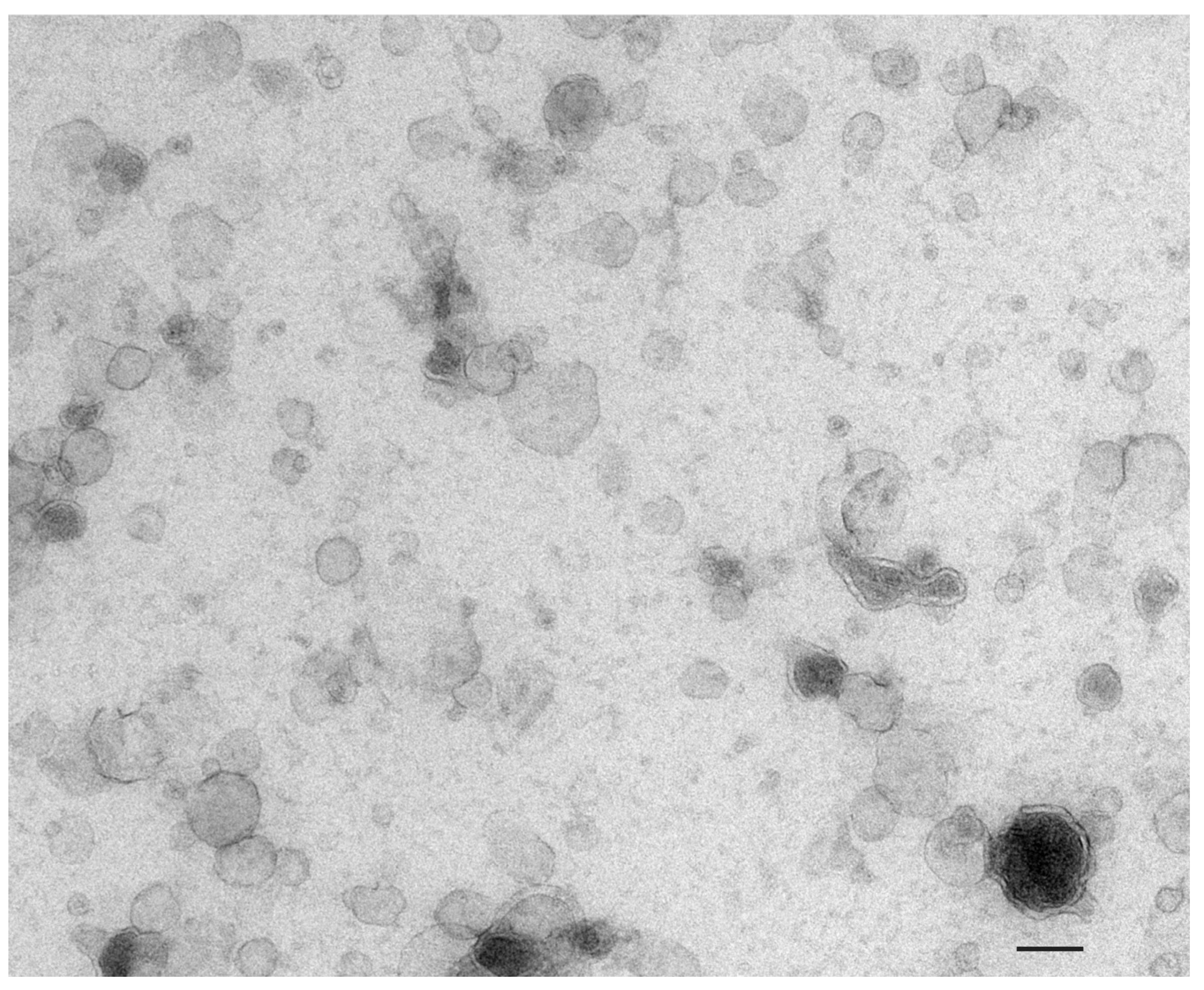
2.1. Extracellular Vesicles: Definition and Terminology
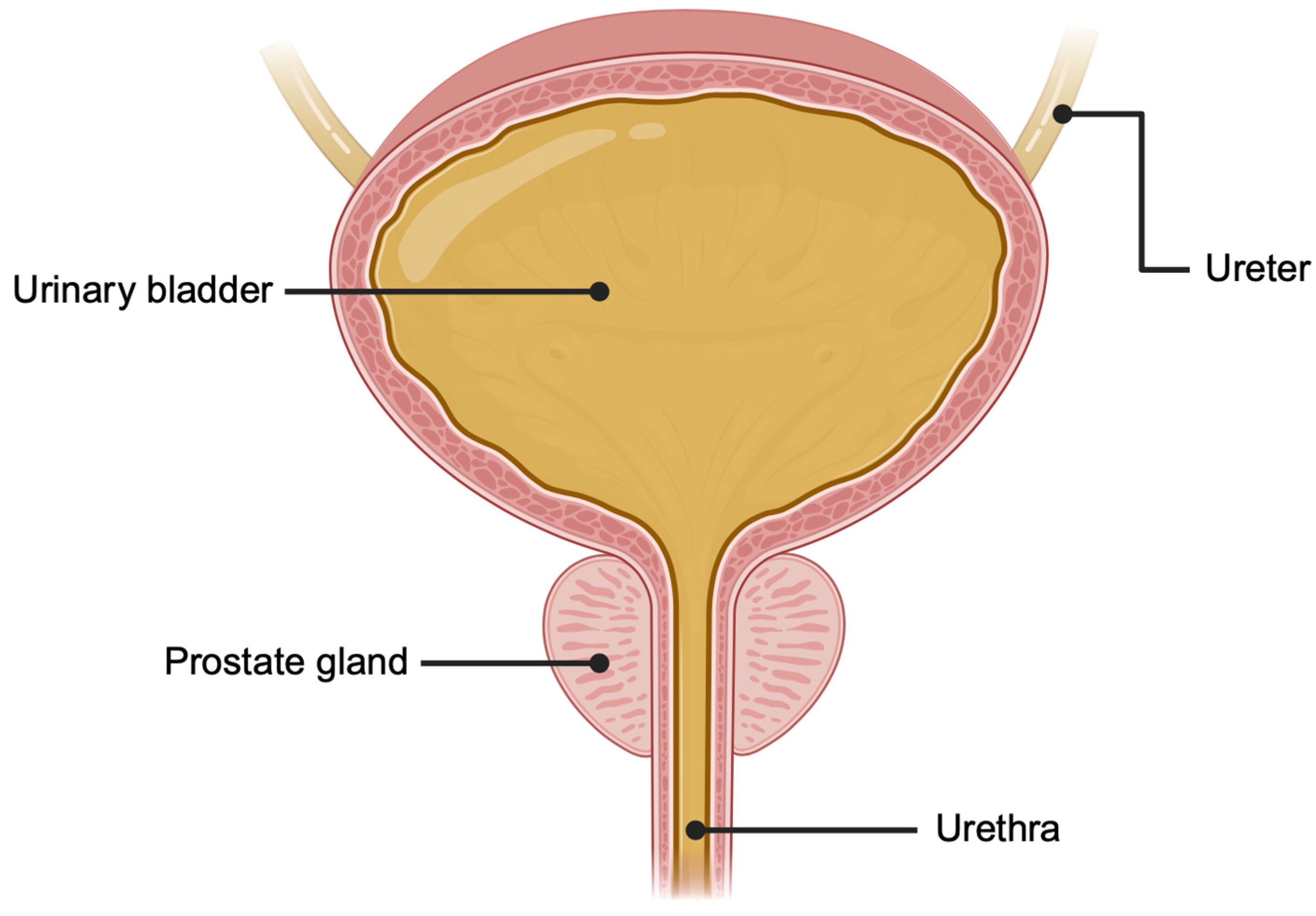
2.2. Urine as a Source of Prostate-Derived Extracellular Vesicles
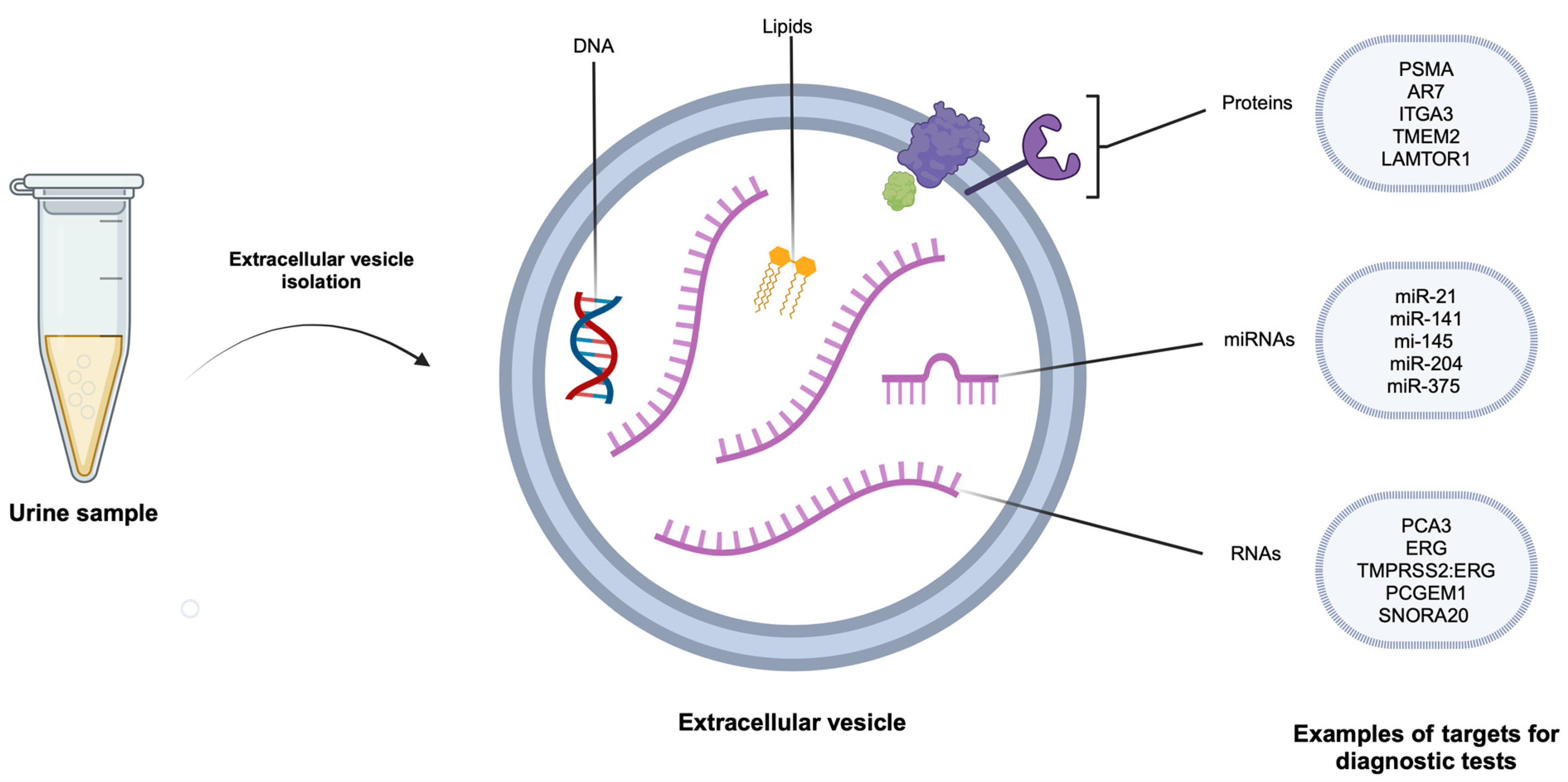
2.3. Urinary Prostate-Derived Extracellular Vesicle Contents and Their Physiological Role
3. Extracellular Vesicle Functions in Prostate Cancer
3.1. Sustaining Proliferative Signalling
3.2. Enabling Replicative Immortality
3.3. Evading Growth Suppression
3.4. Activating Invasion and Metastasis
3.5. Inducing Angiogenesis
3.6. Genomic Instability and Mutation
3.7. Resisting Cell Death
3.8. Deregulating Cellular Metabolism
3.9. Avoiding Immune Destruction
3.10. Tumour-Promoting Inflammation
3.11. Polymorphic Microbiomes as a Prostate Cancer-Enabling Characteristic
4. Urinary EV-Based Diagnostic Tests for Prostate Cancer
4.1. Overview
4.2. Urinary EV RNA-Based Diagnostic Tests
| Name of Test | Description | Diagnostic Accuracy (AUC) | Reference |
|---|---|---|---|
| Two-gene panel | PCA3 and PCGEM1 | 0.88 (95% CI 0.81–0.93) | Kohaar, 2021 [132] |
| 3-lncRNA diagnostic model (Clnc) | AC0150987.1, CTD-2589M5.4, RP11-363E6.3 | 0.776 (95%CI 0.713–0.838) | Tao, 2023 [129] |
| EPI score (ExoDx Prostate Intelliscore) | ERG and PCA3 relative to SPDEF | 0.70 (95% CI 0.65–0.75) | McKiernan, 2018 [126] |
| EXO106 (EPI ExoDx) | ERG and PCA3 relative to SPDEF | 0.764 (95% CI 0.691–0.837) | Donovan, 2015 [125] |
| ExoGrail | ERG exons 4–5, ERG exons 6–7, GJB1, HOXC6, HPN, PCA3, PPFIA2, SLC12A1, TMEM45B, TMPRSS2/ERG fusion combined with clinical parameters and EN2 levels | 0.84 (95%CI 0.78–0.89) | O’Connell, 2021 [135] |
| ExoMeth | ERG exons 4–5, ERG exons 6–7, GJB1, HOXC6, HPN, PCA3, SNORA20, TIMP4, TMPRSS2/ERG fusion combined with clinical parameters and urine cell DNA methylation data | 0.89 (95% CI 0.84–0.93) | O’Connell, 2020 [134] |
| ExoSpec | ERG exons 4–5, PCA3, SLC12A1, TMEM45B combined with clinical parameters and peptides | 0.71 (95% CI not reported) | O’Connell, 2022 [133] |
| GAPT-E score | GATA2, PCA3, TMPRSS-2 | 0.71 (95% CI not reported) | Woo, 2020 [136] |
| LBXexo score | PCA3 and PRAC | 0.736 (95% CI 0.592–0.868) | Ye, 2020 [123] |
| lncRNA-p21 | lncRNA-p21 | 0.663 (95% CI not reported) | Isin, 2015 [137] |
| Novel urine exosomal lncRNA assay | PCA3 and MALAT1 | 0.831 (95% CI not reported) | Li, 2021 [124] |
| Prostate urine risk (PUR) | AMACR, MEX3A, AMH, MEMO1, ANKRD34B, MME, APOC1, MMP11AR(exons 4–8), MMP26, DPP4, NKAIN1, ERG(exons 4–5), PALM3, GABARAPL2, PCA3, GAPDH, PPFIA2, GDF15, SIM2 (short), HOXC6, SMIM1, HPN, SSPO, IGFBP3, SULT1A1, IMPDH2, TDRD1, ITGBL1, TMPRSS2/ERG fusion, KLK4, TRPM4, MARCH5, TWIST1, MED4, UPK2 | 0.77 (95% CI 0.70–0.84) | O’Connell, 2019 [138] |
4.3. Urinary EV miRNA Based Diagnostic Tests
4.4. Urinary EV Protein-Based Diagnostic Tests
5. Urinary Prostatic EVs in Active Surveillance of Prostate Cancer
- Year 1:
- ○
- PSA every 3 to 4 months
- ○
- DRE at 12 months
- ○
- mpMRI at 12 to 18 months
- Year 2 and beyond:
- ○
- PSA every 6 months
- ○
- DRE every 12 months
- ○
- PSA kinetics monitoring, with concerning changes to be re-assessed with mpMRI and/or re-biopsy
6. Future Directions and Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- National Prostate Cancer Audit (NPCA). Annual Report 2022. Available online: https://www.npca.org.uk/content/uploads/2023/01/NPCA-Annual-Report-2022_12.01.23.pdf (accessed on 14 April 2024).
- United States Cancer Statistics (USCS). Center for Disease Control and Prevention. Prostate Cancer Stat Bite. Available online: https://www.cdc.gov/cancer/uscs/about/stat-bites/stat-bite-prostate.htm (accessed on 14 April 2024).
- Tolkach, Y.; Kristiansen, G. The Heterogeneity of Prostate Cancer: A Practical Approach. Pathobiology 2018, 85, 108–116. [Google Scholar] [CrossRef]
- Sakr, W.A.; Grignon, D.J.; Crissman, J.D.; Heilbrun, L.K.; Cassin, B.J.; Pontes, J.J.; Haas, G.P. High grade prostatic intraepithelial neoplasia (HGPIN) and prostatic adenocarcinoma between the ages of 20–69: An autopsy study of 249 cases. In Vivo 1994, 8, 439–443. [Google Scholar] [PubMed]
- Hamdy, F.C.; Donovan, J.L.; Lane, J.A.; Mason, M.; Metcalfe, C.; Holding, P.; Davis, M.; Peters, T.J.; Turner, E.L.; Martin, R.M.; et al. 10-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Localized Prostate Cancer. N. Engl. J. Med. 2016, 375, 1415–1424. [Google Scholar] [CrossRef] [PubMed]
- Lardas, M.; Liew, M.; van den Bergh, R.C.; De Santis, M.; Bellmunt, J.; Van den Broeck, T.; Cornford, P.; Cumberbatch, M.G.; Fossati, N.; Gross, T.; et al. Quality of Life Outcomes after Primary Treatment for Clinically Localised Prostate Cancer: A Systematic Review. Eur. Urol. 2017, 72, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Loeb, S.; Bjurlin, M.A.; Nicholson, J.; Tammela, T.L.; Penson, D.F.; Carter, H.B.; Carroll, P.; Etzioni, R. Overdiagnosis and overtreatment of prostate cancer. Eur. Urol. 2014, 65, 1046–1055. [Google Scholar] [CrossRef] [PubMed]
- Hamdy, F.C.; Donovan, J.L.; Lane, J.A.; Metcalfe, C.; Davis, M.; Turner, E.L.; Martin, R.M.; Young, G.J.; Walsh, E.I.; Bryant, R.J.; et al. Fifteen-Year Outcomes after Monitoring, Surgery, or Radiotherapy for Prostate Cancer. N. Engl. J. Med. 2023, 388, 1547–1558. [Google Scholar] [CrossRef] [PubMed]
- NICE. Prostate Cancer Diagnosis and Management; National Instititute for Health and Care Excellence: Manchester, UK, 2021. [Google Scholar]
- Walker, C.H.; Marchetti, K.A.; Singhal, U.; Morgan, T.M. Active surveillance for prostate cancer: Selection criteria, guidelines, and outcomes. World J. Urol. 2022, 40, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Kohaar, I.; Petrovics, G.; Srivastava, S. A Rich Array of Prostate Cancer Molecular Biomarkers: Opportunities and Challenges. Int. J. Mol. Sci. 2019, 20, 1813. [Google Scholar] [CrossRef] [PubMed]
- Haffner, M.C.; Zwart, W.; Roudier, M.P.; True, L.D.; Nelson, W.G.; Epstein, J.I.; De Marzo, A.M.; Nelson, P.S.; Yegnasubramanian, S. Genomic and phenotypic heterogeneity in prostate cancer. Nat. Rev. Urol. 2021, 18, 79–92. [Google Scholar] [CrossRef]
- Pinsky, P.F.; Parnes, H.L.; Andriole, G. Mortality and complications after prostate biopsy in the Prostate, Lung, Colorectal and Ovarian Cancer Screening (PLCO) trial. BJU Int. 2014, 113, 254–259. [Google Scholar] [CrossRef]
- Thorn, J.C.; Turner, E.L.; Walsh, E.I.; Donovan, J.L.; Neal, D.E.; Hamdy, F.C.; Martin, R.M.; Noble, S.M. Impact of PSA testing on secondary care costs in England and Wales: Estimates from the Cluster randomised triAl of PSA testing for Prostate cancer (CAP). BMC Health Serv. Res. 2023, 23, 610. [Google Scholar] [CrossRef] [PubMed]
- Chaban, Y.V.; Vosshenrich, J.; McKee, H.; Gunasekaran, S.; Brown, M.J.; Atalay, M.K.; Heye, T.; Markl, M.; Woolen, S.A.; Simonetti, O.P.; et al. Environmental Sustainability and MRI: Challenges, Opportunities, and a Call for Action. J. Magn. Reson. Imaging 2024, 59, 1149–1167. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed]
- Jeppesen, D.K.; Zhang, Q.; Franklin, J.L.; Coffey, R.J. Extracellular vesicles and nanoparticles: Emerging complexities. Trends Cell Biol. 2023, 33, 667–681. [Google Scholar] [CrossRef] [PubMed]
- Erdbrügger, U.; Blijdorp, C.J.; Bijnsdorp, I.V.; Borràs, F.E.; Burger, D.; Bussolati, B.; Byrd, J.B.; Clayton, A.; Dear, J.W.; Falcón-Pérez, J.M.; et al. Urinary extracellular vesicles: A position paper by the Urine Task Force of the International Society for Extracellular Vesicles. J. Extracell. Vesicles 2021, 10, e12093. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrügger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Chitti, S.V.; Gummadi, S.; Kang, T.; Shahi, S.; Marzan, A.L.; Nedeva, C.; Sanwlani, R.; Bramich, K.; Stewart, S.; Petrovska, M.; et al. Vesiclepedia 2024: An extracellular vesicles and extracellular particles repository. Nucleic Acids Res. 2024, 52, D1694–D1698. [Google Scholar] [CrossRef] [PubMed]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef] [PubMed]
- Wiggins, R.C.; Glatfelter, A.; Kshirsagar, B.; Brukman, J. Procoagulant activity in normal human urine associated with subcellular particles. Kidney Int. 1986, 29, 591–597. [Google Scholar] [CrossRef]
- Pisitkun, T.; Shen, R.F.; Knepper, M.A. Identification and proteomic profiling of exosomes in human urine. Proc. Natl. Acad. Sci. USA 2004, 101, 13368–13373. [Google Scholar] [CrossRef]
- Hurst, R.; Meader, E.; Gihawi, A.; Rallapalli, G.; Clark, J.; Kay, G.L.; Webb, M.; Manley, K.; Curley, H.; Walker, H.; et al. Microbiomes of Urine and the Prostate Are Linked to Human Prostate Cancer Risk Groups. Eur. Urol. Oncol. 2022, 5, 412–419. [Google Scholar] [CrossRef]
- Park, J.Y.; Kang, C.S.; Seo, H.C.; Shin, J.C.; Kym, S.M.; Park, Y.S.; Shin, T.S.; Kim, J.G.; Kim, Y.K. Bacteria-Derived Extracellular Vesicles in Urine as a Novel Biomarker for Gastric Cancer: Integration of Liquid Biopsy and Metagenome Analysis. Cancers 2021, 13, 4687. [Google Scholar] [CrossRef]
- Drake, R.R.; White, K.Y.; Fuller, T.W.; Igwe, E.; Clements, M.A.; Nyalwidhe, J.O.; Given, R.W.; Lance, R.S.; Semmes, O.J. Clinical collection and protein properties of expressed prostatic secretions as a source for biomarkers of prostatic disease. J. Proteom. 2009, 72, 907–917. [Google Scholar] [CrossRef]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef]
- Webb, M.; Manley, K.; Olivan, M.; Guldvik, I.; Palczynska, M.; Hurst, R.; Connell, S.P.; Mills, I.G.; Brewer, D.S.; Mills, R.; et al. Methodology for the at-home collection of urine samples for prostate cancer detection. Biotechniques 2020, 68, 65–71. [Google Scholar] [CrossRef]
- Wu, Z.; Zhang, Z.; Xia, W.; Cai, J.; Li, Y.; Wu, S. Extracellular vesicles in urologic malignancies-Implementations for future cancer care. Cell Prolif. 2019, 52, e12659. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; O’Neill, V.; Bentink, S.; Noerholm, M.; Belzer, S.; Skog, J.; Kattan, M.W.; Partin, A.; Andriole, G.; et al. A Novel Urine Exosome Gene Expression Assay to Predict High-grade Prostate Cancer at Initial Biopsy. JAMA Oncol. 2016, 2, 882–889. [Google Scholar] [CrossRef]
- Brody, I.; Ronquist, G.; Gottfries, A. Ultrastructural localization of the prostasome—An organelle in human seminal plasma. Ups. J. Med. Sci. 1983, 88, 63–80. [Google Scholar] [CrossRef]
- Nilsson, B.O.; Jin, M.; Einarsson, B.; Persson, B.-E.; Ronquist, G. Monoclonal antibodies against human prostasomes. Prostate 1998, 35, 178–184. [Google Scholar] [CrossRef]
- Aalberts, M.; Stout, T.A.E.; Stoorvogel, W. Prostasomes: Extracellular vesicles from the prostate. Reproduction 2014, 147, R1–R14. [Google Scholar] [CrossRef]
- Ronquist, G.; Brody, I. The prostasome: Its secretion and function in man. Biochim. Biophys. Acta 1985, 822, 203–218. [Google Scholar] [CrossRef]
- van Doormaal, F.F.; Kleinjan, A.; Di Nisio, M.; Büller, H.R.; Nieuwland, R. Cell-derived microvesicles and cancer. Neth. J. Med. 2009, 67, 266–273. [Google Scholar]
- Kim, J.H.; Lee, J.; Park, J.; Gho, Y.S. Gram-negative and Gram-positive bacterial extracellular vesicles. Semin. Cell Dev. Biol. 2015, 40, 97–104. [Google Scholar] [CrossRef]
- Bose, S.; Aggarwal, S.; Singh, D.V.; Acharya, N. Extracellular vesicles: An emerging platform in gram-positive bacteria. Microb. Cell 2020, 7, 312–322. [Google Scholar] [CrossRef]
- Brown, L.; Wolf, J.M.; Prados-Rosales, R.; Casadevall, A. Through the wall: Extracellular vesicles in Gram-positive bacteria, mycobacteria and fungi. Nat. Rev. Microbiol. 2015, 13, 620–630. [Google Scholar] [CrossRef]
- Yates, A.G.; Pink, R.C.; Erdbrügger, U.; Siljander, P.R.-M.; Dellar, E.R.; Pantazi, P.; Akbar, N.; Cooke, W.R.; Vatish, M.; Dias-Neto, E.; et al. In sickness and in health: The functional role of extracellular vesicles in physiology and pathology in vivo. J. Extracell. Vesicles 2022, 11, e12151. [Google Scholar] [CrossRef]
- Ronquist, G.; Nilsson, B.O.; Hjertën, S. Interaction between prostasomes and spermatozoa from human semen. Arch. Androl. 1990, 24, 147–157. [Google Scholar] [CrossRef]
- Park, K.H.; Kim, B.J.; Kang, J.; Nam, T.S.; Lim, J.M.; Kim, H.T.; Park, J.K.; Kim, Y.G.; Chae, S.W.; Kim, U.H. Ca2+ signaling tools acquired from prostasomes are required for progesterone-induced sperm motility. Sci. Signal 2011, 4, ra31. [Google Scholar] [CrossRef]
- Chisholm, J.; Haas-Neill, S.; Margetts, P.; Al-Nedawi, K. Characterization of proteins, mRNAs, and miRNAs of circulating extracellular vesicles from prostate cancer patients compared to healthy subjects. Front. Oncol. 2022, 12, 895555. [Google Scholar] [CrossRef]
- Kharmate, G.; Hosseini-Beheshti, E.; Caradec, J.; Chin, M.Y.; Tomlinson Guns, E.S. Epidermal Growth Factor Receptor in Prostate Cancer Derived Exosomes. PLoS ONE 2016, 11, e0154967. [Google Scholar] [CrossRef]
- Logozzi, M.; Angelini, D.F.; Iessi, E.; Mizzoni, D.; Di Raimo, R.; Federici, C.; Lugini, L.; Borsellino, G.; Gentilucci, A.; Pierella, F.; et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett. 2017, 403, 318–329. [Google Scholar] [CrossRef]
- Bertokova, A.; Svecova, N.; Kozics, K.; Gabelova, A.; Vikartovska, A.; Jane, E.; Hires, M.; Bertok, T.; Tkac, J. Exosomes from prostate cancer cell lines: Isolation optimisation and characterisation. Biomed. Pharmacother. 2022, 151, 113093. [Google Scholar] [CrossRef]
- Logozzi, M.; Mizzoni, D.; Di Raimo, R.; Giuliani, A.; Maggi, M.; Sciarra, A.; Fais, S. Plasmatic Exosome Number and Size Distinguish Prostate Cancer Patients From Healthy Individuals: A Prospective Clinical Study. Front. Oncol. 2021, 11, 727317. [Google Scholar] [CrossRef]
- Choi, W.W.Y.; Sánchez, C.; Li, J.J.; Dinarvand, M.; Adomat, H.; Ghaffari, M.; Khoja, L.; Vafaee, F.; Joshua, A.M.; Chi, K.N.; et al. Extracellular vesicles from biological fluids as potential markers in castration resistant prostate cancer. J. Cancer Res. Clin. Oncol. 2023, 149, 4701–4717. [Google Scholar] [CrossRef]
- Johnstone, R.M.; Mathew, A.; Mason, A.B.; Teng, K. Exosome formation during maturation of mammalian and avian reticulocytes: Evidence that exosome release is a major route for externalization of obsolete membrane proteins. J. Cell. Physiol. 1991, 147, 27–36. [Google Scholar] [CrossRef]
- Lai, C.P.; Kim, E.Y.; Badr, C.E.; Weissleder, R.; Mempel, T.R.; Tannous, B.A.; Breakefield, X.O. Visualization and tracking of tumour extracellular vesicle delivery and RNA translation using multiplexed reporters. Nat. Commun. 2015, 6, 7029. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46. [Google Scholar] [CrossRef]
- Jorissen, R.N.; Walker, F.; Pouliot, N.; Garrett, T.P.; Ward, C.W.; Burgess, A.W. Epidermal growth factor receptor: Mechanisms of activation and signalling. Exp. Cell Res. 2003, 284, 31–53. [Google Scholar] [CrossRef] [PubMed]
- Frawley, T.; Piskareva, O. Extracellular Vesicle Dissemination of Epidermal Growth Factor Receptor and Ligands and Its Role in Cancer Progression. Cancers 2020, 12, 3200. [Google Scholar] [CrossRef]
- Di Lorenzo, G.; Tortora, G.; D’Armiento, F.P.; De Rosa, G.; Staibano, S.; Autorino, R.; D’Armiento, M.; De Laurentiis, M.; De Placido, S.; Catalano, G.; et al. Expression of Epidermal Growth Factor Receptor Correlates with Disease Relapse and Progression to Androgen-independence in Human Prostate Cancer1. Clin. Cancer Res. 2002, 8, 3438–3444. [Google Scholar]
- DeRita, R.M.; Zerlanko, B.; Singh, A.; Lu, H.; Iozzo, R.V.; Benovic, J.L.; Languino, L.R. c-Src, Insulin-Like Growth Factor I Receptor, G-Protein-Coupled Receptor Kinases and Focal Adhesion Kinase are Enriched Into Prostate Cancer Cell Exosomes. J. Cell. Biochem. 2017, 118, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Mills, I.G. Maintaining and reprogramming genomic androgen receptor activity in prostate cancer. Nat. Rev. Cancer 2014, 14, 187–198. [Google Scholar] [CrossRef]
- Read, J.; Ingram, A.; Al Saleh, H.A.; Platko, K.; Gabriel, K.; Kapoor, A.; Pinthus, J.; Majeed, F.; Qureshi, T.; Al-Nedawi, K. Nuclear transportation of exogenous epidermal growth factor receptor and androgen receptor via extracellular vesicles. Eur. J. Cancer 2017, 70, 62–74. [Google Scholar] [CrossRef]
- Zheng, Y.; Li, J.X.; Chen, C.J.; Lin, Z.Y.; Liu, J.X.; Lin, F.J. Extracellular vesicle-derived circ_SLC19A1 promotes prostate cancer cell growth and invasion through the miR-497/septin 2 pathway. Cell Biol. Int. 2020, 44, 1037–1045. [Google Scholar] [CrossRef]
- Huang, B.; Zhou, D.; Huang, X.; Xu, X.; Xu, Z. Silencing circSLC19A1 Inhibits Prostate Cancer Cell Proliferation, Migration and Invasion Through Regulating miR-326/MAPK1 Axis. Cancer Manag. Res. 2020, 12, 11883–11895. [Google Scholar] [CrossRef]
- Yardy, G.W.; Brewster, S.F. Wnt signalling and prostate cancer. Prostate Cancer Prostatic Dis. 2005, 8, 119–126. [Google Scholar] [CrossRef]
- March-Villalba, J.A.; Martínez-Jabaloyas, J.M.; Herrero, M.J.; Santamaria, J.; Aliño, S.F.; Dasí, F. Cell-Free Circulating Plasma hTERT mRNA Is a Useful Marker for Prostate Cancer Diagnosis and Is Associated with Poor Prognosis Tumor Characteristics. PLoS ONE 2012, 7, e43470. [Google Scholar] [CrossRef]
- Goldvaser, H.; Gutkin, A.; Beery, E.; Edel, Y.; Nordenberg, J.; Wolach, O.; Rabizadeh, E.; Uziel, O.; Lahav, M. Characterisation of blood-derived exosomal hTERT mRNA secretion in cancer patients: A potential pan-cancer marker. Br. J. Cancer 2017, 117, 353–357. [Google Scholar] [CrossRef]
- Taylor, B.S.; Schultz, N.; Hieronymus, H.; Gopalan, A.; Xiao, Y.; Carver, B.S.; Arora, V.K.; Kaushik, P.; Cerami, E.; Reva, B.; et al. Integrative genomic profiling of human prostate cancer. Cancer Cell 2010, 18, 11–22. [Google Scholar] [CrossRef] [PubMed]
- Di Cristofano, A.; Pandolfi, P.P. The multiple roles of PTEN in tumor suppression. Cell 2000, 100, 387–390. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.A.; Xu, Y.; Tao, W.; Ubellacker, J.M.; Lim, M.; Aum, D.; Lee, G.Y.; Zhou, K.; Zope, H.; Yu, M.; et al. Restoration of tumour-growth suppression in vivo via systemic nanoparticle-mediated delivery of PTEN mRNA. Nat. Biomed. Eng. 2018, 2, 850–864. [Google Scholar] [CrossRef] [PubMed]
- Gabriel, K.; Ingram, A.; Austin, R.; Kapoor, A.; Tang, D.; Majeed, F.; Qureshi, T.; Al-Nedawi, K. Regulation of the tumor suppressor PTEN through exosomes: A diagnostic potential for prostate cancer. PLoS ONE 2013, 8, e70047. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.G.; Ray, R.; Bhattacharya, D.; Karmakar, P. DNA damage induced cellular senescence and it’s PTEN-armed exosomes—The warriors against prostate carcinoma cells. Med. Oncol. 2022, 39, 34. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Mu, X.; Yin, Q.; Hu, K. miR-106a contributes to prostate carcinoma progression through PTEN. Oncol. Lett. 2019, 17, 1327–1332. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hu, C.; Chao, H.; Zhang, Y.; Li, Y.; Hou, J.; Huang, L. Exosomal transfer of miR-106a-5p contributes to cisplatin resistance and tumorigenesis in nasopharyngeal carcinoma. J. Cell. Mol. Med. 2021, 25, 9183–9198. [Google Scholar] [CrossRef] [PubMed]
- Cao, Z.; Xu, L.; Zhao, S. Exosome-derived miR-27a produced by PSC-27 cells contributes to prostate cancer chemoresistance through p53. Biochem. Biophys. Res. Commun. 2019, 515, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Antin, P.; Berx, G.; Blanpain, C.; Brabletz, T.; Bronner, M.; Campbell, K.; Cano, A.; Casanova, J.; Christofori, G.; et al. Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020, 21, 341–352. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, C.; Rani, S.; O’Brien, K.; O’Neill, A.; Prencipe, M.; Sheikh, R.; Webb, G.; McDermott, R.; Watson, W.; Crown, J.; et al. Docetaxel-resistance in prostate cancer: Evaluating associated phenotypic changes and potential for resistance transfer via exosomes. PLoS ONE 2012, 7, e50999. [Google Scholar] [CrossRef]
- Brzozowski, J.S.; Bond, D.R.; Jankowski, H.; Goldie, B.J.; Burchell, R.; Naudin, C.; Smith, N.D.; Scarlett, C.J.; Larsen, M.R.; Dun, M.D.; et al. Extracellular vesicles with altered tetraspanin CD9 and CD151 levels confer increased prostate cell motility and invasion. Sci. Rep. 2018, 8, 8822. [Google Scholar] [CrossRef]
- Cao, Z.; Kyprianou, N. Mechanisms navigating the TGF-β pathway in prostate cancer. Asian J. Urol. 2015, 2, 11–18. [Google Scholar] [CrossRef] [PubMed]
- El-Sayed, I.Y.; Daher, A.; Destouches, D.; Firlej, V.; Kostallari, E.; Maillé, P.; Huet, E.; Haidar-Ahmad, N.; Jenster, G.; de la Taille, A.; et al. Extracellular vesicles released by mesenchymal-like prostate carcinoma cells modulate EMT state of recipient epithelial-like carcinoma cells through regulation of AR signaling. Cancer Lett. 2017, 410, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Oregel-Cortez, M.I.; Frayde-Gómez, H.; Quintana-González, G.; García-González, V.; Vazquez-Jimenez, J.G.; Galindo-Hernández, O. Resistin Induces Migration and Invasion in PC3 Prostate Cancer Cells: Role of Extracellular Vesicles. Life 2023, 13, 2321. [Google Scholar] [CrossRef] [PubMed]
- Tapial Martínez, P.; López Navajas, P.; Lietha, D. FAK Structure and Regulation by Membrane Interactions and Force in Focal Adhesions. Biomolecules 2020, 10, 179. [Google Scholar] [CrossRef] [PubMed]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef]
- Deep, G.; Jain, A.; Kumar, A.; Agarwal, C.; Kim, S.; Leevy, W.M.; Agarwal, R. Exosomes secreted by prostate cancer cells under hypoxia promote matrix metalloproteinases activity at pre-metastatic niches. Mol. Carcinog. 2020, 59, 323–332. [Google Scholar] [CrossRef] [PubMed]
- Motamedinia, P.; Scott, A.N.; Bate, K.L.; Sadeghi, N.; Salazar, G.; Shapiro, E.; Ahn, J.; Lipsky, M.; Lin, J.; Hruby, G.W.; et al. Urine Exosomes for Non-Invasive Assessment of Gene Expression and Mutations of Prostate Cancer. PLoS ONE 2016, 11, e0154507. [Google Scholar] [CrossRef] [PubMed]
- Perner, S.; Mosquera, J.M.; Demichelis, F.; Hofer, M.D.; Paris, P.L.; Simko, J.; Collins, C.; Bismar, T.A.; Chinnaiyan, A.M.; De Marzo, A.M.; et al. TMPRSS2-ERG fusion prostate cancer: An early molecular event associated with invasion. Am. J. Surg. Pathol. 2007, 31, 882–888. [Google Scholar] [CrossRef] [PubMed]
- Soller, M.J.; Isaksson, M.; Elfving, P.; Soller, W.; Lundgren, R.; Panagopoulos, I. Confirmation of the high frequency of the TMPRSS2/ERG fusion gene in prostate cancer. Genes Chromosomes Cancer 2006, 45, 717–719. [Google Scholar] [CrossRef]
- Leshem, O.; Madar, S.; Kogan-Sakin, I.; Kamer, I.; Goldstein, I.; Brosh, R.; Cohen, Y.; Jacob-Hirsch, J.; Ehrlich, M.; Ben-Sasson, S.; et al. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS ONE 2011, 6, e21650. [Google Scholar] [CrossRef]
- Shin, S.; Park, Y.H.; Jung, S.H.; Jang, S.H.; Kim, M.Y.; Lee, J.Y.; Chung, Y.J. Urinary exosome microRNA signatures as a noninvasive prognostic biomarker for prostate cancer. NPJ Genom. Med. 2021, 6, 45. [Google Scholar] [CrossRef] [PubMed]
- Tan, Y.F.; Chen, Z.Y.; Wang, L.; Wang, M.; Liu, X.H. MiR-142-3p functions as an oncogene in prostate cancer by targeting FOXO1. J. Cancer 2020, 11, 1614–1624. [Google Scholar] [CrossRef] [PubMed]
- Reis, S.T.; Pontes-Junior, J.; Antunes, A.A.; Dall’Oglio, M.F.; Dip, N.; Passerotti, C.C.; Rossini, G.A.; Morais, D.R.; Nesrallah, A.J.; Piantino, C.; et al. miR-21 may acts as an oncomir by targeting RECK, a matrix metalloproteinase regulator, in prostate cancer. BMC Urol. 2012, 12, 14. [Google Scholar] [CrossRef] [PubMed]
- Oh-Hohenhorst, S.J.; Lange, T. Role of Metastasis-Related microRNAs in Prostate Cancer Progression and Treatment. Cancers 2021, 13, 4492. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Schöpfer, A.; Wagner, U.; Sauter, G.; Moch, H.; Willi, N.; Gasser, T.C.; Mihatsch, M.J. Metastatic patterns of prostate cancer: An autopsy study of 1,589 patients. Hum. Pathol. 2000, 31, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Akoto, T.; Saini, S. Role of Exosomes in Prostate Cancer Metastasis. Int. J. Mol. Sci. 2021, 22, 3528. [Google Scholar] [CrossRef]
- Henrich, S.E.; McMahon, K.M.; Plebanek, M.P.; Calvert, A.E.; Feliciano, T.J.; Parrish, S.; Tavora, F.; Mega, A.; De Souza, A.; Carneiro, B.A.; et al. Prostate cancer extracellular vesicles mediate intercellular communication with bone marrow cells and promote metastasis in a cholesterol-dependent manner. J. Extracell. Vesicles 2020, 10, e12042. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, K.; Ochi, H.; Sunamura, S.; Kosaka, N.; Mabuchi, Y.; Fukuda, T.; Yao, K.; Kanda, H.; Ae, K.; Okawa, A.; et al. Cancer-secreted hsa-miR-940 induces an osteoblastic phenotype in the bone metastatic microenvironment via targeting ARHGAP1 and FAM134A. Proc. Natl. Acad. Sci. USA 2018, 115, 2204–2209. [Google Scholar] [CrossRef] [PubMed]
- Luo, J.-q.; Yang, T.-w.; Wu, J.; Lai, H.-h.; Zou, L.-b.; Chen, W.-b.; Zhou, X.-m.; Lv, D.-j.; Cen, S.-r.; Long, Z.-n.; et al. Exosomal PGAM1 promotes prostate cancer angiogenesis and metastasis by interacting with ACTG1. Cell Death Dis. 2023, 14, 502. [Google Scholar] [CrossRef]
- Varkaris, A.; Katsiampoura, A.D.; Araujo, J.C.; Gallick, G.E.; Corn, P.G. Src signaling pathways in prostate cancer. Cancer Metastasis Rev. 2014, 33, 595–606. [Google Scholar] [CrossRef] [PubMed]
- Chennakrishnaiah, S.; Tsering, T.; Gregory, C.; Tawil, N.; Spinelli, C.; Montermini, L.; Karatzas, N.; Aprikian, S.; Choi, D.; Klewes, L.; et al. Extracellular vesicles from genetically unstable, oncogene-driven cancer cells trigger micronuclei formation in endothelial cells. Sci. Rep. 2020, 10, 8532. [Google Scholar] [CrossRef] [PubMed]
- Elbakrawy, E.M.; Mayah, A.; Hill, M.A.; Kadhim, M. Induction of Genomic Instability in a Primary Human Fibroblast Cell Line Following Low-Dose Alpha-Particle Exposure and the Potential Role of Exosomes. Biology 2020, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Boysen, G.; Barbieri, C.E.; Prandi, D.; Blattner, M.; Chae, S.S.; Dahija, A.; Nataraj, S.; Huang, D.; Marotz, C.; Xu, L.; et al. SPOP mutation leads to genomic instability in prostate cancer. eLife 2015, 4, e09207. [Google Scholar] [CrossRef] [PubMed]
- Lyu, F.; Shang, S.Y.; Gao, X.S.; Ma, M.W.; Xie, M.; Ren, X.Y.; Liu, M.Z.; Chen, J.Y.; Li, S.S.; Huang, L. Uncovering the Secrets of Prostate Cancer’s Radiotherapy Resistance: Advances in Mechanism Research. Biomedicines 2023, 11, 1628. [Google Scholar] [CrossRef] [PubMed]
- Lane, D.P. Cancer. p53, guardian of the genome. Nature 1992, 358, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Ozaki, T.; Nakagawara, A. Role of p53 in Cell Death and Human Cancers. Cancers 2011, 3, 994–1013. [Google Scholar] [CrossRef] [PubMed]
- Mateo, J.; Seed, G.; Bertan, C.; Rescigno, P.; Dolling, D.; Figueiredo, I.; Miranda, S.; Nava Rodrigues, D.; Gurel, B.; Clarke, M.; et al. Genomics of lethal prostate cancer at diagnosis and castration resistance. J. Clin. Investig. 2020, 130, 1743–1751. [Google Scholar] [CrossRef] [PubMed]
- Beier, A.K.; Puhr, M.; Stope, M.B.; Thomas, C.; Erb, H.H.H. Metabolic changes during prostate cancer development and progression. J. Cancer Res. Clin. Oncol. 2023, 149, 2259–2270. [Google Scholar] [CrossRef] [PubMed]
- Singh, K.K.; Desouki, M.M.; Franklin, R.B.; Costello, L.C. Mitochondrial aconitase and citrate metabolism in malignant and nonmalignant human prostate tissues. Mol. Cancer 2006, 5, 14. [Google Scholar] [CrossRef]
- Scaglia, N.; Frontini-López, Y.R.; Zadra, G. Prostate Cancer Progression: As a Matter of Fats. Front. Oncol. 2021, 11, 719865. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 2016, 5, e10250. [Google Scholar] [CrossRef] [PubMed]
- Sena, L.A.; Denmeade, S.R. Fatty Acid Synthesis in Prostate Cancer: Vulnerability or Epiphenomenon? Cancer Res. 2021, 81, 4385–4393. [Google Scholar] [CrossRef] [PubMed]
- Abusamra, A.J.; Zhong, Z.; Zheng, X.; Li, M.; Ichim, T.E.; Chin, J.L.; Min, W.-P. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood Cells Mol. Dis. 2005, 35, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Miyazaki, T.; Ikeda, K.; Sato, W.; Horie-Inoue, K.; Inoue, S. Extracellular vesicle-mediated EBAG9 transfer from cancer cells to tumor microenvironment promotes immune escape and tumor progression. Oncogenesis 2018, 7, 7. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Morley, S.; Le, M.; Bedoret, D.; Umetsu, D.T.; Di Vizio, D.; Freeman, M.R. Enhanced shedding of extracellular vesicles from amoeboid prostate cancer cells: Potential effects on the tumor microenvironment. Cancer Biol. Ther. 2014, 15, 409–418. [Google Scholar] [CrossRef] [PubMed]
- Archer, M.; Dogra, N.; Kyprianou, N. Inflammation as a Driver of Prostate Cancer Metastasis and Therapeutic Resistance. Cancers 2020, 12, 2984. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Allavena, P.; Sica, A.; Balkwill, F. Cancer-related inflammation. Nature 2008, 454, 436–444. [Google Scholar] [CrossRef]
- Voronov, E.; Shouval, D.S.; Krelin, Y.; Cagnano, E.; Benharroch, D.; Iwakura, Y.; Dinarello, C.A.; Apte, R.N. IL-1 is required for tumor invasiveness and angiogenesis. Proc. Natl. Acad. Sci. USA 2003, 100, 2645–2650. [Google Scholar] [CrossRef]
- Nguyen, D.P.; Li, J.; Tewari, A.K. Inflammation and prostate cancer: The role of interleukin 6 (IL-6). BJU Int. 2014, 113, 986–992. [Google Scholar] [CrossRef]
- Yuan, A.; Chen, J.J.; Yao, P.L.; Yang, P.C. The role of interleukin-8 in cancer cells and microenvironment interaction. Front. Biosci. 2005, 10, 853–865. [Google Scholar] [CrossRef]
- Fitzgerald, W.; Freeman, M.L.; Lederman, M.M.; Vasilieva, E.; Romero, R.; Margolis, L. A System of Cytokines Encapsulated in ExtraCellular Vesicles. Sci. Rep. 2018, 8, 8973. [Google Scholar] [CrossRef]
- Alaimo, A.; Genovesi, S.; Annesi, N.; De Felice, D.; Subedi, S.; Macchia, A.; La Manna, F.; Ciani, Y.; Vannuccini, F.; Mugoni, V.; et al. Sterile inflammation via TRPM8 RNA-dependent TLR3-NF-kB/IRF3 activation promotes antitumor immunity in prostate cancer. EMBO j. 2024, 43, 780–805. [Google Scholar] [CrossRef]
- Sass, D.; Fitzgerald, W.; Barb, J.J.; Kupzyk, K.; Margolis, L.; Saligan, L. An exploratory analysis of extracellular vesicle-associated and soluble cytokines in cancer-related fatigue in men with prostate cancer. Brain Behav. Immun. Health 2020, 9, 100140. [Google Scholar] [CrossRef]
- Park, J.; Kim, N.E.; Yoon, H.; Shin, C.M.; Kim, N.; Lee, D.H.; Park, J.Y.; Choi, C.H.; Kim, J.G.; Kim, Y.K.; et al. Fecal Microbiota and Gut Microbe-Derived Extracellular Vesicles in Colorectal Cancer. Front. Oncol. 2021, 11, 650026. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.R.; Han, K.; Han, Y.; Kang, N.; Shin, T.S.; Park, H.J.; Kim, H.; Kwon, W.; Lee, S.; Kim, Y.K.; et al. Microbiome Markers of Pancreatic Cancer Based on Bacteria-Derived Extracellular Vesicles Acquired from Blood Samples: A Retrospective Propensity Score Matching Analysis. Biology 2021, 10, 219. [Google Scholar] [CrossRef] [PubMed]
- Mishra, S.; Tejesvi, M.V.; Hekkala, J.; Turunen, J.; Kandikanti, N.; Kaisanlahti, A.; Suokas, M.; Leppä, S.; Vihinen, P.; Kuitunen, H.; et al. Gut microbiome-derived bacterial extracellular vesicles in patients with solid tumours. J. Adv. Res. 2024; online ahead of print. [Google Scholar] [CrossRef]
- Khoo, A.; Govindarajan, M.; Qiu, Z.; Liu, L.Y.; Ignatchenko, V.; Waas, M.; Macklin, A.; Keszei, A.; Main, B.P.; Yang, L.; et al. Prostate Cancer Reshapes the Secreted and Extracellular Vesicle Urinary Proteomes. bioRxiv 2023. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Bajo-Santos, C.; Line, A.; Martens-Uzunova, E.S.; de la Fuente, J.M.; Moros, M.; Soekmadji, C.; Tasken, K.A.; Llorente, A. Extracellular vesicles as a source of prostate cancer biomarkers in liquid biopsies: A decade of research. Br. J. Cancer 2022, 126, 331–350. [Google Scholar] [CrossRef] [PubMed]
- Miranda, K.C.; Bond, D.T.; McKee, M.; Skog, J.; Păunescu, T.G.; Da Silva, N.; Brown, D.; Russo, L.M. Nucleic acids within urinary exosomes/microvesicles are potential biomarkers for renal disease. Kidney Int. 2010, 78, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.F.; He, S.; Wu, X.; Jiang, S.; Zhang, R.C.; Yang, Z.S.; Chen, F.W.; Pan, D.L.; Li, D.; Li, G. Detection of Prostate Cancer Antigen 3 and Prostate Cancer Susceptibility Candidate in Non-DRE Urine Improves Diagnosis of Prostate Cancer in Chinese Population. Prostate Cancer 2020, 2020, 3964615. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Ji, J.; Lyu, J.; Jin, X.; He, X.; Mo, S.; Xu, H.; He, J.; Cao, Z.; Chen, X.; et al. A novel urine exosomal lncRNA assay to improve the detection of prostate cancer at initial biopsy: A retrospective multicenter diagnostic feasibility study. Cancers 2021, 13, 4075. [Google Scholar] [CrossRef]
- Donovan, M.J.; Noerholm, M.; Bentink, S.; Belzer, S.; Skog, J.; O’Neill, V.; Cochran, J.S.; Brown, G.A. A molecular signature of PCA3 and ERG exosomal RNA from non-DRE urine is predictive of initial prostate biopsy result. Prostate Cancer Prostatic Dis. 2015, 18, 370–375. [Google Scholar] [CrossRef]
- McKiernan, J.; Donovan, M.J.; Margolis, E.; Partin, A.; Carter, B.; Brown, G.; Torkler, P.; Noerholm, M.; Skog, J.; Shore, N.; et al. A Prospective Adaptive Utility Trial to Validate Performance of a Novel Urine Exosome Gene Expression Assay to Predict High-Grade Prostate Cancer in Patients with Prostate-Specific Antigen 2-10 ng/mL at Initial Biopsy. Eur. Urol. 2018, 74, 731–738. [Google Scholar] [CrossRef] [PubMed]
- Margolis, E.; Brown, G.; Partin, A.; Carter, B.; McKiernan, J.; Tutrone, R.; Torkler, P.; Fischer, C.; Tadigotla, V.; Noerholm, M.; et al. Predicting high-grade prostate cancer at initial biopsy: Clinical performance of the ExoDx (EPI) Prostate Intelliscore test in three independent prospective studies. Prostate Cancer Prostatic Dis. 2022, 25, 296–301. [Google Scholar] [CrossRef] [PubMed]
- EAU. European Association of Urology Guidelines. Edn. Presented at the EAU Annual Congress Paris 2024. Available online: https://d56bochluxqnz.cloudfront.net/documents/full-guideline/EAU-EANM-ESTRO-ESUR-ISUP-SIOG-Guidelines-on-Prostate-Cancer-2024_2024-04-09-132035_ypmy.pdf (accessed on 14 April 2024).
- Tao, W.; Wang, B.Y.; Luo, L.; Li, Q.; Meng, Z.A.; Xia, T.L.; Deng, W.M.; Yang, M.; Zhou, J.; Zhang, X.; et al. A urine extracellular vesicle lncRNA classifier for high-grade prostate cancer and increased risk of progression: A multi-center study. Cell Rep. Med. 2023, 4, 101240. [Google Scholar] [CrossRef]
- Gan, J.; Zeng, X.; Wang, X.; Wu, Y.; Lei, P.; Wang, Z.; Yang, C.; Hu, Z. Effective Diagnosis of Prostate Cancer Based on mRNAs From Urinary Exosomes. Front. Med. 2022, 9, 736110. [Google Scholar] [CrossRef] [PubMed]
- Lillard, J.W., Jr.; Moses, K.A.; Mahal, B.A.; George, D.J. Racial disparities in Black men with prostate cancer: A literature review. Cancer 2022, 128, 3787–3795. [Google Scholar] [CrossRef]
- Kohaar, I.; Chen, Y.; Banerjee, S.; Borbiev, T.; Kuo, H.-C.; Ali, A.; Ravindranath, L.; Kagan, J.; Srivastava, S.; Dobi, A.; et al. A Urine Exosome Gene Expression Panel Distinguishes between Indolent and Aggressive Prostate Cancers at Biopsy. J. Urol. 2021, 205, 420–425. [Google Scholar] [CrossRef]
- O’Connell, S.P.; Frantzi, M.; Latosinska, A.; Webb, M.; Mullen, W.; Pejchinovski, M.; Salji, M.; Mischak, H.; Cooper, C.S.; Clark, J.; et al. A Model to Detect Significant Prostate Cancer Integrating Urinary Peptide and Extracellular Vesicle RNA Data. Cancers 2022, 14, 1995. [Google Scholar] [CrossRef] [PubMed]
- Connell, S.P.; O’Reilly, E.; Tuzova, A.; Webb, M.; Hurst, R.; Mills, R.; Zhao, F.; Bapat, B.; Cooper, C.S.; Perry, A.S.; et al. Development of a multivariable risk model integrating urinary cell DNA methylation and cell-free RNA data for the detection of significant prostate cancer. Prostate 2020, 80, 547–558. [Google Scholar] [CrossRef]
- Connell, S.P.; Mills, R.; Pandha, H.; Morgan, R.; Cooper, C.S.; Clark, J.; Brewer, D.S. Integration of urinary EN2 protein & cell-free RNA data in the development of a multivariable risk model for the detection of prostate cancer prior to biopsy. Cancers 2021, 13, 2102. [Google Scholar] [CrossRef]
- Woo, J.; Santasusagna, S.; Banks, J.; Pastor-Lopez, S.; Yadav, K.; Carceles-Cordon, M.; Dominguez-Andres, A.; Den, R.B.; Languino, L.R.; Pippa, R.; et al. Urine Extracellular Vesicle GATA2 mRNA Discriminates Biopsy Result in Men with Suspicion of Prostate Cancer. J. Urol. 2020, 204, 691–700. [Google Scholar] [CrossRef] [PubMed]
- Işın, M.; Uysaler, E.; Özgür, E.; Köseoğlu, H.; Şanlı, Ö.; Yücel, Ö.B.; Gezer, U.; Dalay, N. Exosomal lncRNA-p21 levels may help to distinguish prostate cancer from benign disease. Front. Genet. 2015, 6, 168. [Google Scholar] [CrossRef] [PubMed]
- Connell, S.P.; Yazbek-Hanna, M.; McCarthy, F.; Hurst, R.; Webb, M.; Curley, H.; Walker, H.; Mills, R.; Ball, R.Y.; Sanda, M.G.; et al. A four-group urine risk classifier for predicting outcomes in patients with prostate cancer. BJU Int. 2019, 124, 609–620. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, M.; Bajo-Santos, C.; Hessvik, N.P.; Lorenz, S.; Fromm, B.; Berge, V.; Sandvig, K.; Linē, A.; Llorente, A. Identification of non-invasive miRNAs biomarkers for prostate cancer by deep sequencing analysis of urinary exosomes. Mol. Cancer 2017, 16, 156. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Qin, S.; An, T.; Tang, Y.; Huang, Y.; Zheng, L. MiR-145 detection in urinary extracellular vesicles increase diagnostic efficiency of prostate cancer based on hydrostatic filtration dialysis method. Prostate 2017, 77, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Foj, L.; Ferrer, F.; Serra, M.; Arevalo, A.; Gavagnach, M.; Gimenez, N.; Filella, X. Exosomal and Non-Exosomal Urinary miRNAs in Prostate Cancer Detection and Prognosis. Prostate 2017, 77, 573–583. [Google Scholar] [CrossRef]
- Wang, W.W.; Sorokin, I.; Aleksic, I.; Fisher, H.; Kaufman, R.P., Jr.; Winer, A.; McNeill, B.; Gupta, R.; Tilki, D.; Fleshner, N.; et al. Expression of Small Noncoding RNAs in Urinary Exosomes Classifies Prostate Cancer into Indolent and Aggressive Disease. J. Urol. 2020, 204, 466–475. [Google Scholar] [CrossRef]
- Wang, C.B.; Chen, S.H.; Zhao, L.; Jin, X.; Chen, X.; Ji, J.; Mo, Z.N.; Wang, F.B. Urine-derived exosomal PSMA is a promising diagnostic biomarker for the detection of prostate cancer on initial biopsy. Clin. Transl. Oncol. 2023, 25, 758–767. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, X.; Li, H.; Zhao, L.; Kong, G.; Chen, J.; Li, Z.; Qi, J.; Tian, Y.; Zhang, F. Urinary exosome-based androgen receptor-variant 7 detection in metastatic castration-resistant prostate cancer patients. Transl. Androl. Urol. 2022, 11, 202–212. [Google Scholar] [CrossRef]
- Bijnsdorp, I.V.; Geldof, A.A.; Lavaei, M.; Piersma, S.R.; van Moorselaar, R.J.; Jimenez, C.R. Exosomal ITGA3 interferes with non-cancerous prostate cell functions and is increased in urine exosomes of metastatic prostate cancer patients. J. Extracell. Vesicles 2013, 2, 22097. [Google Scholar] [CrossRef]
- Overbye, A.; Skotland, T.; Koehler, C.J.; Thiede, B.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Identification of prostate cancer biomarkers in urinary exosomes. Oncotarget 2015, 6, 30357–30376. [Google Scholar] [CrossRef] [PubMed]
- Folkmanis, K.; Junk, E.; Merdane, E.; Folkmane, I.; Folkmanis, V.; Ivanovs, I.; Eglitis, J.; Jakubovskis, M.; Laabs, S.; Isajevs, S.; et al. Clinicopathological Significance of Exosomal Proteins CD9 and CD63 and DNA Mismatch Repair Proteins in Prostate Adenocarcinoma and Benign Hyperplasia. Diagnostics 2022, 12, 287. [Google Scholar] [CrossRef] [PubMed]
- Fujita, K.; Kume, H.; Matsuzaki, K.; Kawashima, A.; Ujike, T.; Nagahara, A.; Uemura, M.; Miyagawa, Y.; Tomonaga, T.; Nonomura, N. Proteomic analysis of urinary extracellular vesicles from high Gleason score prostate cancer. Sci. Rep. 2017, 7, 42961. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Skotland, T.; Berge, V.; Sandvig, K.; Llorente, A. Exosomal proteins as prostate cancer biomarkers in urine: From mass spectrometry discovery to immunoassay-based validation. Eur. J. Pharm. Sci. 2017, 98, 80–85. [Google Scholar] [CrossRef] [PubMed]
- Royo, F.; Zuniga-Garcia, P.; Torrano, V.; Loizaga, A.; Sanchez-Mosquera, P.; Ugalde-Olano, A.; Gonzalez, E.; Cortazar, A.R.; Palomo, L.; Fernandez-Ruiz, S.; et al. Transcriptomic profiling of urine extracellular vesicles reveals alterations of CDH3 in prostate cancer. Oncotarget 2016, 7, 6835–6846. [Google Scholar] [CrossRef] [PubMed]
- Koppers-Lalic, D.; Hackenberg, M.; de Menezes, R.; Misovic, B.; Wachalska, M.; Geldof, A.; Zini, N.; de Reijke, T.; Wurdinger, T.; Vis, A.; et al. Non-invasive prostate cancer detection by measuring miRNA variants (isomiRs) in urine extracellular vesicles. Oncotarget 2016, 7, 22566–22578. [Google Scholar] [CrossRef] [PubMed]
- Samsonov, R.; Shtam, T.; Burdakov, V.; Glotov, A.; Tsyrlina, E.; Berstein, L.; Nosov, A.; Evtushenko, V.; Filatov, M.; Malek, A. Lectin-induced agglutination method of urinary exosomes isolation followed by mi-RNA analysis: Application for prostate cancer diagnostic. Prostate 2016, 76, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, K.; Fujita, K.; Tomiyama, E.; Hatano, K.; Hayashi, Y.; Wang, C.; Ishizuya, Y.; Yamamoto, Y.; Hayashi, T.; Kato, T.; et al. MiR-30b-3p and miR-126-3p of urinary extracellular vesicles could be new biomarkers for prostate cancer. Transl. Androl. Urol. 2021, 10, 1918–1927. [Google Scholar] [CrossRef] [PubMed]
- Van Hemelrijck, M.; Ji, X.; Helleman, J.; Roobol, M.J.; van der Linden, W.; Nieboer, D.; Bangma, C.H.; Frydenberg, M.; Rannikko, A.; Lee, L.S.; et al. Reasons for Discontinuing Active Surveillance: Assessment of 21 Centres in 12 Countries in the Movember GAP3 Consortium. Eur. Urol. 2019, 75, 523–531. [Google Scholar] [CrossRef]
- Ramirez-Garrastacho, M.; Berge, V.; Line, A.; Llorente, A. Potential of miRNAs in urinary extracellular vesicles for management of active surveillance in prostate cancer patients. Br. J. Cancer 2022, 126, 492–501. [Google Scholar] [CrossRef]
- Ling, A.S.Y.; Hakansson, A.K.; Ong, E.H.W.; Lau, A.S.Y.; Low, K.P.; Wong, T.R.; Tan, N.; Tan, J.S.H.; Tuan, J.K.L.; Tan, T.W.K.; et al. Comparative genomic analyses between Asian and Caucasian prostate cancers in an 80,829 patient cohort. J. Clin. Oncol. 2022, 40, 273. [Google Scholar] [CrossRef]
- Miah, S.; Dunford, C.; Edison, M.; Eldred-Evans, D.; Gan, C.; Shah, T.T.; Lunn, P.; Winkler, M.; Ahmed, H.U.; Gibbons, N.; et al. A prospective clinical, cost and environmental analysis of a clinician-led virtual urology clinic. Ann. R. Coll. Surg. Engl. 2019, 101, 30–34. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, B.D.; Thompson, M.; Price, C.P.; Heneghan, C.; Plüddemann, A. Home-use faecal immunochemical testing: Primary care diagnostic technology update. Br. J. Gen. Pract. 2015, 65, 156–158. [Google Scholar] [CrossRef] [PubMed]
- Yoon, H.; Kim, N.E.; Park, J.; Shin, C.M.; Kim, N.; Lee, D.H.; Park, J.Y.; Choi, C.H.; Kim, J.G.; Park, Y.S. Analysis of the gut microbiome using extracellular vesicles in the urine of patients with colorectal cancer. Korean J. Intern. Med. 2023, 38, 27–38. [Google Scholar] [CrossRef] [PubMed]
- Matuszczak, M.; Schalken, J.A.; Salagierski, M. Prostate Cancer Liquid Biopsy Biomarkers’ Clinical Utility in Diagnosis and Prognosis. Cancers 2021, 13, 3373. [Google Scholar] [CrossRef]
- Byun, Y.J.; Piao, X.M.; Jeong, P.; Kang, H.W.; Seo, S.P.; Moon, S.K.; Lee, J.Y.; Choi, Y.H.; Lee, H.Y.; Kim, W.T.; et al. Urinary microRNA-1913 to microRNA-3659 expression ratio as a non-invasive diagnostic biomarker for prostate cancer. Investig. Clin. Urol. 2021, 62, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Bryzgunova, O.E.; Zaripov, M.M.; Skvortsova, T.E.; Lekchnov, E.A.; Grigor’eva, A.E.; Zaporozhchenko, I.A.; Morozkin, E.S.; Ryabchikova, E.I.; Yurchenko, Y.B.; Voitsitskiy, V.E.; et al. Comparative Study of Extracellular Vesicles from the Urine of Healthy Individuals and Prostate Cancer Patients. PLoS ONE 2016, 11, e0157566. [Google Scholar] [CrossRef]
- Danarto, R.; Astuti, I.; Umbas, R.; Haryana, S.M. Urine miR-21-5p and miR-200c-3p as potential non-invasive biomarkers in patients with prostate cancer. Turk. J. Urol. 2020, 46, 26–30. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.Y.; Shin, H.; Moon, H.W.; Park, Y.H.; Park, J.; Lee, J.Y. Urinary exosomal microRNA profiling in intermediate-risk prostate cancer. Sci. Rep. 2021, 11, 7355. [Google Scholar] [CrossRef]
- Wani, S.; Kaul, D.; Mavuduru, R.S.; Kakkar, N.; Bhatia, A. Urinary-exosomal miR-2909: A novel pathognomonic trait of prostate cancer severity. J. Biotechnol. 2017, 259, 135–139. [Google Scholar] [CrossRef]
- Hendriks, R.; Dijkstra, S.; Cornel, E.B.; Jannink, S.; De Jong, H.; Hessels, D.; Melchers, W.; Smit, F.; Leyten, G.; De Reijke, T.; et al. Elevated HOXC6/DLX1 mRNA biomarker levels in urine to help select patients at increased risk for high-grade prostate cancer detection upon prostate biopsy. J. Clin. Oncol. 2016, 34. [Google Scholar] [CrossRef]
| Operational Term | Description | Example |
|---|---|---|
| Physical characteristics | Size with defined ranges | Small EVs (<200 nm) |
| Biochemical composition | Antigen positivity or biochemical staining | CD63+ EVs |
| Description of conditions | Cellular conditions | Hypoxic EVs |
| Cell of origin | Type of cell that EV originates from | Podocyte EVs |
| Name of Test | Diagnostic Accuracy | Reference |
|---|---|---|
| miR-21-5p | 0.65 (95% CI 0.477–0.814) | Samsonov, 2016 [152] |
| miR-30b-3p | 0.663 (95% CI 0.011–0.805) | Matsuzaki, 2021 [153] |
| miR-126-3p | 0.664 (95% CI 0.016–5.39) | Matsuzaki, 2021 [153] |
| miR-501-3p | 0.69 (95% CI 0.52–0.85) | Rodriguez, 2017 [139] |
| miR-196a-5p | 0.73 (95% CI 0.56–0.89) | Rodriguez, 2017 [139] |
| miR-574-3p | 0.85 (95% CI 0.736–0.964) | Samsonov, 2016 [152] |
| miR-141-5p | 0.86 (95% CI 0.732–0.994) | Samsonov, 2016 [152] |
| mi-145 (in combination with serum PSA) | 0.863 (95% CI 0.791–0.934) | Xu, 2017 [140] |
| miR-21, miR-204 and miR-375 (in combination with serum PSA) | 0.866 (95% CI not reported) | Koppers-Lalic, 2016 [151] |
| miR-21, miR-141, miR-375, miR-214 and let-7c | 0.872 (95% CI 0.786–0.958) | Foj, 2017 [141] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Smith, S.F.; Brewer, D.S.; Hurst, R.; Cooper, C.S. Applications of Urinary Extracellular Vesicles in the Diagnosis and Active Surveillance of Prostate Cancer. Cancers 2024, 16, 1717. https://doi.org/10.3390/cancers16091717
Smith SF, Brewer DS, Hurst R, Cooper CS. Applications of Urinary Extracellular Vesicles in the Diagnosis and Active Surveillance of Prostate Cancer. Cancers. 2024; 16(9):1717. https://doi.org/10.3390/cancers16091717
Chicago/Turabian StyleSmith, Stephanie F., Daniel S. Brewer, Rachel Hurst, and Colin S. Cooper. 2024. "Applications of Urinary Extracellular Vesicles in the Diagnosis and Active Surveillance of Prostate Cancer" Cancers 16, no. 9: 1717. https://doi.org/10.3390/cancers16091717
APA StyleSmith, S. F., Brewer, D. S., Hurst, R., & Cooper, C. S. (2024). Applications of Urinary Extracellular Vesicles in the Diagnosis and Active Surveillance of Prostate Cancer. Cancers, 16(9), 1717. https://doi.org/10.3390/cancers16091717








