Extracellular Vesicles for Childhood Cancer Liquid Biopsy
Abstract
Simple Summary
Abstract
1. Introduction
| Tissue Fragment | Description | Analytical Techniques | Advantages | Limitations | References |
|---|---|---|---|---|---|
| Circulating tumor cells (CTCs) | Cancer cells detach from the primary tumor or metastatic lesion and circulate in the bloodstream. |
|
|
| [27,28,29,31,32,33] |
| Circulating tumor DNA (ctDNA) | ctDNA is shed by primary tumor cells and subsequently enters the bloodstream. As such, ctDNA carries the genetic alterations present in the original tumor. |
|
|
| [35,36,37,41,45,53,54,55] |
| Extracellular vesicles (EVs) | Tiny membranous structures enclosed by a lipid bilayer are secreted by both normal and cancerous cells. EVs contain a diverse assortment of molecular constituents, including DNA, RNA, non-coding RNA, and proteins. |
|
|
| [51,56,57,58] |
2. EV Definition and Methods: A Brief Summary
3. EV-Based Liquid Biopsy for Childhood Cancers
3.1. Pediatric Neuroblastoma
3.2. Medulloblastoma
3.3. Pediatric Gliomas
3.4. Hepatoblastoma
3.5. Osteosarcoma
3.6. Ewing’s Sarcoma
3.7. Rhabdomyosarcoma
3.8. Pediatric Lymphoma
4. Future Prospects
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Malone, E.R.; Oliva, M.; Sabatini, P.J.B.; Stockley, T.L.; Siu, L.L. Molecular profiling for precision cancer therapies. Genome Med. 2020, 12, 8. [Google Scholar] [CrossRef] [PubMed]
- Alieva, M.; van Rheenen, J.; Broekman, M.L.D. Potential impact of invasive surgical procedures on primary tumor growth and metastasis. Clin. Exp. Metastasis 2018, 35, 319–331. [Google Scholar] [CrossRef] [PubMed]
- Gilson, P.; Merlin, J.L.; Harle, A. Deciphering Tumour Heterogeneity: From Tissue to Liquid Biopsy. Cancers 2022, 14, 1384. [Google Scholar] [CrossRef] [PubMed]
- Castro-Giner, F.; Gkountela, S.; Donato, C.; Alborelli, I.; Quagliata, L.; Ng, C.K.Y.; Piscuoglio, S.; Aceto, N. Cancer Diagnosis Using a Liquid Biopsy: Challenges and Expectations. Diagnostics 2018, 8, 31. [Google Scholar] [CrossRef]
- Lone, S.N.; Nisar, S.; Masoodi, T.; Singh, M.; Rizwan, A.; Hashem, S.; El-Rifai, W.; Bedognetti, D.; Batra, S.K.; Haris, M.; et al. Liquid biopsy: A step closer to transform diagnosis, prognosis and future of cancer treatments. Mol. Cancer 2022, 21, 79. [Google Scholar] [CrossRef] [PubMed]
- Aravanis, A.M.; Lee, M.; Klausner, R.D. Next-Generation Sequencing of Circulating Tumor DNA for Early Cancer Detection. Cell 2017, 168, 571–574. [Google Scholar] [CrossRef] [PubMed]
- Nikanjam, M.; Kato, S.; Kurzrock, R. Liquid biopsy: Current technology and clinical applications. J. Hematol. Oncol. 2022, 15, 131. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Xu, T.; Zhu, H.; Yu, J. Clinical Applications of Cerebrospinal Fluid Circulating Tumor DNA as a Liquid Biopsy for Central Nervous System Tumors. Onco Targets Ther. 2020, 13, 719–731. [Google Scholar] [CrossRef]
- Han, H.S.; Lee, K.W. Liquid Biopsy: An Emerging Diagnostic, Prognostic, and Predictive Tool in Gastric Cancer. J. Gastric Cancer 2024, 24, 4–28. [Google Scholar] [CrossRef]
- Khurshid, Z.; Zafar, M.S.; Khan, R.S.; Najeeb, S.; Slowey, P.D.; Rehman, I.U. Role of Salivary Biomarkers in Oral Cancer Detection. Adv. Clin. Chem. 2018, 86, 23–70. [Google Scholar] [CrossRef]
- Zhang, H.; Deng, D.; Li, S.; Ren, J.; Huang, W.; Liu, D.; Wang, W. Bronchoalveolar lavage fluid assessment facilitates precision medicine for lung cancer. Cancer Biol. Med. 2023, 21, 230–251. [Google Scholar] [CrossRef] [PubMed]
- Oshi, M.; Murthy, V.; Takahashi, H.; Huyser, M.; Okano, M.; Tokumaru, Y.; Rashid, O.M.; Matsuyama, R.; Endo, I.; Takabe, K. Urine as a Source of Liquid Biopsy for Cancer. Cancers 2021, 13, 2652. [Google Scholar] [CrossRef] [PubMed]
- Heitzer, E.; Perakis, S.; Geigl, J.B.; Speicher, M.R. The potential of liquid biopsies for the early detection of cancer. NPJ Precis. Oncol. 2017, 1, 36. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Li, M.; Ren, S.; Liu, Y.; Zhang, J.; Li, S.; Gao, W.; Gong, X.; Liu, J.; Wang, Y.; et al. Exploring genetic alterations in circulating tumor DNA from cerebrospinal fluid of pediatric medulloblastoma. Sci. Rep. 2021, 11, 5638. [Google Scholar] [CrossRef] [PubMed]
- Chicard, M.; Colmet-Daage, L.; Clement, N.; Danzon, A.; Bohec, M.; Bernard, V.; Baulande, S.; Bellini, A.; Deveau, P.; Pierron, G.; et al. Whole-Exome Sequencing of Cell-Free DNA Reveals Temporo-spatial Heterogeneity and Identifies Treatment-Resistant Clones in Neuroblastoma. Clin. Cancer Res. 2018, 24, 939–949. [Google Scholar] [CrossRef] [PubMed]
- Connal, S.; Cameron, J.M.; Sala, A.; Brennan, P.M.; Palmer, D.S.; Palmer, J.D.; Perlow, H.; Baker, M.J. Liquid biopsies: The future of cancer early detection. J. Transl. Med. 2023, 21, 118. [Google Scholar] [CrossRef]
- Vasseur, D.; Sassi, H.; Bayle, A.; Tagliamento, M.; Besse, B.; Marzac, C.; Arbab, A.; Auger, N.; Cotteret, S.; Aldea, M.; et al. Next-Generation Sequencing on Circulating Tumor DNA in Advanced Solid Cancer: Swiss Army Knife for the Molecular Tumor Board? A Review of the Literature Focused on FDA Approved Test. Cells 2022, 11, 1901. [Google Scholar] [CrossRef]
- Andersson, D.; Fagman, H.; Dalin, M.G.; Stahlberg, A. Circulating cell-free tumor DNA analysis in pediatric cancers. Mol. Aspects Med. 2020, 72, 100819. [Google Scholar] [CrossRef] [PubMed]
- Miguez, A.C.K.; Barros, B.D.F.; de Souza, J.E.S.; da Costa, C.M.L.; Cunha, I.W.; Barbosa, P.; Apezzato, M.L.P.; de Souza, S.J.; Carraro, D.M. Assessment of somatic mutations in urine and plasma of Wilms tumor patients. Cancer Med. 2020, 9, 5948–5959. [Google Scholar] [CrossRef]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy and Primary Brain Tumors. Cancers 2021, 13, 5429. [Google Scholar] [CrossRef]
- Huang, T.Y.; Piunti, A.; Lulla, R.R.; Qi, J.; Horbinski, C.M.; Tomita, T.; James, C.D.; Shilatifard, A.; Saratsis, A.M. Detection of Histone H3 mutations in cerebrospinal fluid-derived tumor DNA from children with diffuse midline glioma. Acta Neuropathol. Commun. 2017, 5, 28. [Google Scholar] [CrossRef] [PubMed]
- Miller, A.M.; Shah, R.H.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658. [Google Scholar] [CrossRef] [PubMed]
- Escudero, L.; Llort, A.; Arias, A.; Diaz-Navarro, A.; Martinez-Ricarte, F.; Rubio-Perez, C.; Mayor, R.; Caratu, G.; Martinez-Saez, E.; Vazquez-Mendez, E.; et al. Circulating tumour DNA from the cerebrospinal fluid allows the characterisation and monitoring of medulloblastoma. Nat. Commun. 2020, 11, 5376. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Ji, H.; Yang, W.; Zhang, M.; Guo, Y.; Li, B.; Wang, J.; Chen, R.; Chen, Y.; Wang, X. Liquid biopsy using ascitic fluid and pleural effusion supernatants for genomic profiling in gastrointestinal and lung cancers. BMC Cancer 2022, 22, 1020. [Google Scholar] [CrossRef] [PubMed]
- Lousada-Fernandez, F.; Rapado-Gonzalez, O.; Lopez-Cedrun, J.L.; Lopez-Lopez, R.; Muinelo-Romay, L.; Suarez-Cunqueiro, M.M. Liquid Biopsy in Oral Cancer. Int. J. Mol. Sci. 2018, 19, 1704. [Google Scholar] [CrossRef] [PubMed]
- De Schutter, H.; Spaepen, M.; Mc Bride, W.H.; Nuyts, S. The clinical relevance of microsatellite alterations in head and neck squamous cell carcinoma: A critical review. Eur. J. Hum. Genet. 2007, 15, 734–741. [Google Scholar] [CrossRef]
- Chiangjong, W.; Bhakdi, S.C.; Woramongkolchai, N.; Vanichapol, T.; Pongsakul, N.; Hongeng, S.; Chutipongtanate, S. Cell-Main Spectra Profile Screening Technique in Simulation of Circulating Tumour Cells Using MALDI-TOF Mass Spectrometry. Cancers 2021, 13, 3775. [Google Scholar] [CrossRef]
- Hu, X.; Zang, X.; Lv, Y. Detection of circulating tumor cells: Advances and critical concerns. Oncol. Lett. 2021, 21, 422. [Google Scholar] [CrossRef]
- Yang, R.; Zheng, S.; Dong, R. Circulating tumor cells in neuroblastoma: Current status and future perspectives. Cancer Med. 2023, 12, 7–19. [Google Scholar] [CrossRef]
- Sanders, D.G.; Wiley, F.M.; Moss, T.J. Serial immunocytologic analysis of blood for tumor cells in two patients with neuroblastoma. Cancer 1991, 67, 1423–1427. [Google Scholar] [CrossRef]
- Magbanua, M.J.; Park, J.W. Isolation of circulating tumor cells by immunomagnetic enrichment and fluorescence-activated cell sorting (IE/FACS) for molecular profiling. Methods 2013, 64, 114–118. [Google Scholar] [CrossRef] [PubMed]
- Burchill, S.A.; Bradbury, F.M.; Smith, B.; Lewis, I.J.; Selby, P. Neuroblastoma cell detection by reverse transcriptase-polymerase chain reaction (RT-PCR) for tyrosine hydroxylase mRNA. Int. J. Cancer 1994, 57, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Z.; Zhang, B.; Zheng, Y.; Zheng, C.; Liu, B.; Zheng, S.; Dong, K.; Dong, R. Circulating tumor cells detection in neuroblastoma patients by EpCAM-independent enrichment and immunostaining-fluorescence in situ hybridization. EBioMedicine 2018, 35, 244–250. [Google Scholar] [CrossRef] [PubMed]
- Haber, D.A.; Velculescu, V.E. Blood-based analyses of cancer: Circulating tumor cells and circulating tumor DNA. Cancer Discov. 2014, 4, 650–661. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Su, L.; Qian, C. Circulating tumor DNA: A promising biomarker in the liquid biopsy of cancer. Oncotarget 2016, 7, 48832–48841. [Google Scholar] [CrossRef]
- Doculara, L.; Trahair, T.N.; Bayat, N.; Lock, R.B. Circulating Tumor DNA in Pediatric Cancer. Front. Mol. Biosci. 2022, 9, 885597. [Google Scholar] [CrossRef] [PubMed]
- Marczynski, G.T.; Laus, A.C.; Dos Reis, M.B.; Reis, R.M.; Vazquez, V.L. Circulating tumor DNA (ctDNA) detection is associated with shorter progression-free survival in advanced melanoma patients. Sci. Rep. 2020, 10, 18682. [Google Scholar] [CrossRef]
- Kahana-Edwin, S.; Cain, L.E.; McCowage, G.; Darmanian, A.; Wright, D.; Mullins, A.; Saletta, F.; Karpelowsky, J. Neuroblastoma Molecular Risk-Stratification of DNA Copy Number and ALK Genotyping via Cell-Free Circulating Tumor DNA Profiling. Cancers 2021, 13, 3365. [Google Scholar] [CrossRef]
- Krumbholz, M.; Hellberg, J.; Steif, B.; Bauerle, T.; Gillmann, C.; Fritscher, T.; Agaimy, A.; Frey, B.; Juengert, J.; Wardelmann, E.; et al. Genomic EWSR1 Fusion Sequence as Highly Sensitive and Dynamic Plasma Tumor Marker in Ewing Sarcoma. Clin. Cancer Res. 2016, 22, 4356–4365. [Google Scholar] [CrossRef]
- Kjaer, E.K.R.; Vase, C.B.; Rossing, M.; Ahlborn, L.B.; Hjalgrim, L.L. Detection of circulating tumor-derived material in peripheral blood of pediatric sarcoma patients: A systematic review. Transl. Oncol. 2023, 34, 101690. [Google Scholar] [CrossRef]
- Jimenez, I.; Chicard, M.; Colmet-Daage, L.; Clement, N.; Danzon, A.; Lapouble, E.; Pierron, G.; Bohec, M.; Baulande, S.; Berrebi, D.; et al. Circulating tumor DNA analysis enables molecular characterization of pediatric renal tumors at diagnosis. Int. J. Cancer 2019, 144, 68–79. [Google Scholar] [CrossRef] [PubMed]
- Tombolan, L.; Rossi, E.; Binatti, A.; Zin, A.; Manicone, M.; Facchinetti, A.; Lucchetta, S.; Affinita, M.C.; Bonvini, P.; Bortoluzzi, S.; et al. Clinical significance of circulating tumor cells and cell-free DNA in pediatric rhabdomyosarcoma. Mol. Oncol. 2022, 16, 2071–2085. [Google Scholar] [CrossRef] [PubMed]
- Berry, J.L.; Xu, L.; Kooi, I.; Murphree, A.L.; Prabakar, R.K.; Reid, M.; Stachelek, K.; Le, B.H.A.; Welter, L.; Reiser, B.J.; et al. Genomic cfDNA Analysis of Aqueous Humor in Retinoblastoma Predicts Eye Salvage: The Surrogate Tumor Biopsy for Retinoblastoma. Mol. Cancer Res. 2018, 16, 1701–1712. [Google Scholar] [CrossRef] [PubMed]
- Leung, F.; Kulasingam, V.; Diamandis, E.P.; Hoon, D.S.; Kinzler, K.; Pantel, K.; Alix-Panabieres, C. Circulating Tumor DNA as a Cancer Biomarker: Fact or Fiction? Clin. Chem. 2016, 62, 1054–1060. [Google Scholar] [CrossRef]
- De Mattos-Arruda, L.; Cortes, J.; Santarpia, L.; Vivancos, A.; Tabernero, J.; Reis-Filho, J.S.; Seoane, J. Circulating tumour cells and cell-free DNA as tools for managing breast cancer. Nat. Rev. Clin. Oncol. 2013, 10, 377–389. [Google Scholar] [CrossRef]
- Panachan, J.; Rojsirikulchai, N.; Pongsakul, N.; Khowawisetsut, L.; Pongphitcha, P.; Siriboonpiputtana, T.; Chareonsirisuthigul, T.; Phornsarayuth, P.; Klinkulab, N.; Jinawath, N.; et al. Extracellular Vesicle-Based Method for Detecting MYCN Amplification Status of Pediatric Neuroblastoma. Cancers 2022, 14, 2627. [Google Scholar] [CrossRef]
- Chaemsaithong, P.; Luewan, S.; Taweevisit, M.; Chiangjong, W.; Pongchaikul, P.; Thorner, P.S.; Tongsong, T.; Chutipongtanate, S. Placenta-Derived Extracellular Vesicles in Pregnancy Complications and Prospects on a Liquid Biopsy for Hemoglobin Bart’s Disease. Int. J. Mol. Sci. 2023, 24, 5658. [Google Scholar] [CrossRef]
- Chutipongtanate, S.; Pattanapanyasat, K.; Meller, J.; Serrano Lopez, J. Editorial: Driving extracellular vesicles toward applications in precision medicine. Front. Med. (Lausanne) 2022, 9, 1049697. [Google Scholar] [CrossRef]
- Yu, W.; Hurley, J.; Roberts, D.; Chakrabortty, S.K.; Enderle, D.; Noerholm, M.; Breakefield, X.O.; Skog, J.K. Exosome-based liquid biopsies in cancer: Opportunities and challenges. Ann. Oncol. 2021, 32, 466–477. [Google Scholar] [CrossRef]
- Pitt, J.M.; Kroemer, G.; Zitvogel, L. Extracellular vesicles: Masters of intercellular communication and potential clinical interventions. J. Clin. Investig. 2016, 126, 1139–1143. [Google Scholar] [CrossRef]
- Yu, D.; Li, Y.; Wang, M.; Gu, J.; Xu, W.; Cai, H.; Fang, X.; Zhang, X. Exosomes as a new frontier of cancer liquid biopsy. Mol. Cancer 2022, 21, 56. [Google Scholar] [CrossRef] [PubMed]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef] [PubMed]
- Dawson, S.J.; Tsui, D.W.; Murtaza, M.; Biggs, H.; Rueda, O.M.; Chin, S.F.; Dunning, M.J.; Gale, D.; Forshew, T.; Mahler-Araujo, B.; et al. Analysis of circulating tumor DNA to monitor metastatic breast cancer. N. Engl. J. Med. 2013, 368, 1199–1209. [Google Scholar] [CrossRef] [PubMed]
- Cabus, L.; Lagarde, J.; Curado, J.; Lizano, E.; Perez-Boza, J. Current challenges and best practices for cell-free long RNA biomarker discovery. Biomark. Res. 2022, 10, 62. [Google Scholar] [CrossRef] [PubMed]
- Wan, J.C.M.; Heider, K.; Gale, D.; Murphy, S.; Fisher, E.; Mouliere, F.; Ruiz-Valdepenas, A.; Santonja, A.; Morris, J.; Chandrananda, D.; et al. ctDNA monitoring using patient-specific sequencing and integration of variant reads. Sci. Transl. Med. 2020, 12, aaz8084. [Google Scholar] [CrossRef] [PubMed]
- Malenica, M.; Vukomanovic, M.; Kurtjak, M.; Masciotti, V.; Dal Zilio, S.; Greco, S.; Lazzarino, M.; Krusic, V.; Percic, M.; Jelovica Badovinac, I.; et al. Perspectives of Microscopy Methods for Morphology Characterisation of Extracellular Vesicles from Human Biofluids. Biomedicines 2021, 9, 603. [Google Scholar] [CrossRef] [PubMed]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [PubMed]
- Welsh, J.A.; Goberdhan, D.C.I.; O’Driscoll, L.; Buzas, E.I.; Blenkiron, C.; Bussolati, B.; Cai, H.; Di Vizio, D.; Driedonks, T.A.P.; Erdbrugger, U.; et al. Minimal information for studies of extracellular vesicles (MISEV2023): From basic to advanced approaches. J. Extracell. Vesicles 2024, 13, e12404. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; LeBleu, V.S. The biology, function, and biomedical applications of exosomes. Science 2020, 367, aau6977. [Google Scholar] [CrossRef]
- Yue, B.; Yang, H.; Wang, J.; Ru, W.; Wu, J.; Huang, Y.; Lan, X.; Lei, C.; Chen, H. Exosome biogenesis, secretion and function of exosomal miRNAs in skeletal muscle myogenesis. Cell Prolif. 2020, 53, e12857. [Google Scholar] [CrossRef]
- Schwager, S.C.; Reinhart-King, C.A. Mechanobiology of microvesicle release, uptake, and microvesicle-mediated activation. Curr. Top. Membr. 2020, 86, 255–278. [Google Scholar] [CrossRef]
- Li, B.; Antonyak, M.A.; Zhang, J.; Cerione, R.A. RhoA triggers a specific signaling pathway that generates transforming microvesicles in cancer cells. Oncogene 2012, 31, 4740–4749. [Google Scholar] [CrossRef]
- Clos-Sansalvador, M.; Monguio-Tortajada, M.; Roura, S.; Franquesa, M.; Borras, F.E. Commonly used methods for extracellular vesicles’ enrichment: Implications in downstream analyses and use. Eur. J. Cell Biol. 2022, 101, 151227. [Google Scholar] [CrossRef] [PubMed]
- Oeyen, E.; Van Mol, K.; Baggerman, G.; Willems, H.; Boonen, K.; Rolfo, C.; Pauwels, P.; Jacobs, A.; Schildermans, K.; Cho, W.C.; et al. Ultrafiltration and size exclusion chromatography combined with asymmetrical-flow field-flow fractionation for the isolation and characterisation of extracellular vesicles from urine. J. Extracell. Vesicles 2018, 7, 1490143. [Google Scholar] [CrossRef]
- Stam, J.; Bartel, S.; Bischoff, R.; Wolters, J.C. Isolation of extracellular vesicles with combined enrichment methods. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2021, 1169, 122604. [Google Scholar] [CrossRef]
- Turner, N.P.; Abeysinghe, P.; Kwan Cheung, K.A.; Vaswani, K.; Logan, J.; Sadowski, P.; Mitchell, M.D. A Comparison of Blood Plasma Small Extracellular Vesicle Enrichment Strategies for Proteomic Analysis. Proteomes 2022, 10, 19. [Google Scholar] [CrossRef]
- Chutipongtanate, S.; Kongsomros, S.; Pongsakul, N.; Panachan, J.; Khowawisetsut, L.; Pattanapanyasat, K.; Hongeng, S.; Thitithanyanont, A. Anti-SARS-CoV-2 effect of extracellular vesicles released from mesenchymal stem cells. J. Extracell. Vesicles 2022, 11, e12201. [Google Scholar] [CrossRef] [PubMed]
- Willms, E.; Cabanas, C.; Mager, I.; Wood, M.J.A.; Vader, P. Extracellular Vesicle Heterogeneity: Subpopulations, Isolation Techniques, and Diverse Functions in Cancer Progression. Front. Immunol. 2018, 9, 738. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.E. Extracellular Vesicles and Metastasis. Cold Spring Harb. Perspect. Med. 2020, 10, a037275. [Google Scholar] [CrossRef]
- Haug, B.H.; Hald, O.H.; Utnes, P.; Roth, S.A.; Lokke, C.; Flaegstad, T.; Einvik, C. Exosome-like Extracellular Vesicles from MYCN-amplified Neuroblastoma Cells Contain Oncogenic miRNAs. Anticancer Res. 2015, 35, 2521–2530. [Google Scholar]
- Ma, J.; Xu, M.; Yin, M.; Hong, J.; Chen, H.; Gao, Y.; Xie, C.; Shen, N.; Gu, S.; Mo, X. Exosomal hsa-miR199a-3p Promotes Proliferation and Migration in Neuroblastoma. Front. Oncol. 2019, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Colletti, M.; Petretto, A.; Galardi, A.; Di Paolo, V.; Tomao, L.; Lavarello, C.; Inglese, E.; Bruschi, M.; Lopez, A.A.; Pascucci, L.; et al. Proteomic Analysis of Neuroblastoma-Derived Exosomes: New Insights into a Metastatic Signature. Proteomics 2017, 17, 1600430. [Google Scholar] [CrossRef] [PubMed]
- Kaid, C.; Assoni, A.; Marcola, M.; Semedo-Kuriki, P.; Bortolin, R.H.; Carvalho, V.M.; Okamoto, O.K. Proteome and miRNome profiling of microvesicles derived from medulloblastoma cell lines with stem-like properties reveals biomarkers of poor prognosis. Brain Res. 2020, 1730, 146646. [Google Scholar] [CrossRef]
- Zhu, L.Y.; Wu, X.Y.; Liu, X.D.; Zheng, D.F.; Li, H.S.; Yang, B.; Zhang, J.; Chang, Q. Aggressive Medulloblastoma-Derived Exosomal miRNAs Promote In Vitro Invasion and Migration of Tumor Cells Via Ras/MAPK Pathway. J. Neuropathol. Exp. Neurol. 2020, 79, 734–745. [Google Scholar] [CrossRef]
- Jackson, H.K.; Mitoko, C.; Linke, F.; Macarthur, D.; Kerr, I.D.; Coyle, B. Extracellular Vesicles Potentiate Medulloblastoma Metastasis in an EMMPRIN and MMP-2 Dependent Manner. Cancers 2023, 15, 2601. [Google Scholar] [CrossRef]
- Chen, W.W.; Balaj, L.; Liau, L.M.; Samuels, M.L.; Kotsopoulos, S.K.; Maguire, C.A.; Loguidice, L.; Soto, H.; Garrett, M.; Zhu, L.D.; et al. BEAMing and Droplet Digital PCR Analysis of Mutant IDH1 mRNA in Glioma Patient Serum and Cerebrospinal Fluid Extracellular Vesicles. Mol. Ther. Nucleic Acids 2013, 2, e109. [Google Scholar] [CrossRef]
- Yue, X.; Lan, F.; Xia, T. Hypoxic Glioma Cell-Secreted Exosomal miR-301a Activates Wnt/beta-catenin Signaling and Promotes Radiation Resistance by Targeting TCEAL7. Mol. Ther. 2019, 27, 1939–1949. [Google Scholar] [CrossRef]
- Zeng, A.; Wei, Z.; Yan, W.; Yin, J.; Huang, X.; Zhou, X.; Li, R.; Shen, F.; Wu, W.; Wang, X.; et al. Exosomal transfer of miR-151a enhances chemosensitivity to temozolomide in drug-resistant glioblastoma. Cancer Lett. 2018, 436, 10–21. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Zeng, A.; Zhang, Z.; Shi, Z.; Yan, W.; You, Y. Exosomal transfer of miR-1238 contributes to temozolomide-resistance in glioblastoma. EBioMedicine 2019, 42, 238–251. [Google Scholar] [CrossRef]
- Jiao, C.; Jiao, X.; Zhu, A.; Ge, J.; Xu, X. Exosomal miR-34s panel as potential novel diagnostic and prognostic biomarker in patients with hepatoblastoma. J. Pediatr. Surg. 2017, 52, 618–624. [Google Scholar] [CrossRef]
- Liu, W.; Chen, S.; Liu, B. Diagnostic and prognostic values of serum exosomal microRNA-21 in children with hepatoblastoma: A Chinese population-based study. Pediatr. Surg. Int. 2016, 32, 1059–1065. [Google Scholar] [CrossRef]
- Hu, Y.; Zai, H.; Jiang, W.; Yao, Y.; Ou, Z.; Zhu, Q. miR-126 in Extracellular Vesicles Derived from Hepatoblastoma Cells Promotes the Tumorigenesis of Hepatoblastoma through Inducing the Differentiation of BMSCs into Cancer Stem Cells. J. Immunol. Res. 2021, 2021, 6744715. [Google Scholar] [CrossRef]
- Miller, I.V.; Raposo, G.; Welsch, U.; Prazeres da Costa, O.; Thiel, U.; Lebar, M.; Maurer, M.; Bender, H.U.; von Luettichau, I.; Richter, G.H.; et al. First identification of Ewing’s sarcoma-derived extracellular vesicles and exploration of their biological and potential diagnostic implications. Biol. Cell 2013, 105, 289–303. [Google Scholar] [CrossRef] [PubMed]
- Tsugita, M.; Yamada, N.; Noguchi, S.; Yamada, K.; Moritake, H.; Shimizu, K.; Akao, Y.; Ohno, T. Ewing sarcoma cells secrete EWS/Fli-1 fusion mRNA via microvesicles. PLoS ONE 2013, 8, e77416. [Google Scholar] [CrossRef]
- Ventura, S.; Aryee, D.N.; Felicetti, F.; De Feo, A.; Mancarella, C.; Manara, M.C.; Picci, P.; Colombo, M.P.; Kovar, H.; Care, A.; et al. CD99 regulates neural differentiation of Ewing sarcoma cells through miR-34a-Notch-mediated control of NF-kappaB signaling. Oncogene 2016, 35, 3944–3954. [Google Scholar] [CrossRef] [PubMed]
- Araki, Y.; Aiba, H.; Yoshida, T.; Yamamoto, N.; Hayashi, K.; Takeuchi, A.; Miwa, S.; Igarashi, K.; Nguyen, T.D.; Ishii, K.A.; et al. Osteosarcoma-Derived Small Extracellular Vesicles Enhance Tumor Metastasis and Suppress Osteoclastogenesis by miR-146a-5p. Front. Oncol. 2021, 11, 667109. [Google Scholar] [CrossRef] [PubMed]
- Jerez, S.; Araya, H.; Hevia, D.; Irarrazaval, C.E.; Thaler, R.; van Wijnen, A.J.; Galindo, M. Extracellular vesicles from osteosarcoma cell lines contain miRNAs associated with cell adhesion and apoptosis. Gene 2019, 710, 246–257. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, T.; Uotani, K.; Yoshida, A.; Morita, T.; Nezu, Y.; Kobayashi, E.; Yoshida, A.; Uehara, T.; Omori, T.; Sugiu, K.; et al. Clinical significance of circulating miR-25-3p as a novel diagnostic and prognostic biomarker in osteosarcoma. Oncotarget 2017, 8, 33375–33392. [Google Scholar] [CrossRef]
- Zhong, L.; Liao, D.; Li, J.; Liu, W.; Wang, J.; Zeng, C.; Wang, X.; Cao, Z.; Zhang, R.; Li, M.; et al. Rab22a-NeoF1 fusion protein promotes osteosarcoma lung metastasis through its secretion into exosomes. Signal Transduct. Target. Ther. 2021, 6, 59. [Google Scholar] [CrossRef]
- Endo-Munoz, L.; Cai, N.; Cumming, A.; Macklin, R.; Merida de Long, L.; Topkas, E.; Mukhopadhyay, P.; Hill, M.; Saunders, N.A. Progression of Osteosarcoma from a Non-Metastatic to a Metastatic Phenotype Is Causally Associated with Activation of an Autocrine and Paracrine uPA Axis. PLoS ONE 2015, 10, e0133592. [Google Scholar] [CrossRef]
- Ghayad, S.E.; Rammal, G.; Ghamloush, F.; Basma, H.; Nasr, R.; Diab-Assaf, M.; Chelala, C.; Saab, R. Exosomes derived from embryonal and alveolar rhabdomyosarcoma carry differential miRNA cargo and promote invasion of recipient fibroblasts. Sci. Rep. 2016, 6, 37088. [Google Scholar] [CrossRef] [PubMed]
- Fahs, A.; Hussein, N.; Zalzali, H.; Ramadan, F.; Ghamloush, F.; Tamim, H.; El Homsi, M.; Badran, B.; Boulos, F.; Tawil, A.; et al. CD147 Promotes Tumorigenesis via Exosome-Mediated Signaling in Rhabdomyosarcoma. Cells 2022, 11, 2267. [Google Scholar] [CrossRef] [PubMed]
- Rammal, G.; Fahs, A.; Kobeissy, F.; Mechref, Y.; Zhao, J.; Zhu, R.; Diab-Assaf, M.; Saab, R.; Ghayad, S.E. Proteomic Profiling of Rhabdomyosarcoma-Derived Exosomes Yield Insights into Their Functional Role in Paracrine Signaling. J. Proteome Res. 2019, 18, 3567–3579. [Google Scholar] [CrossRef] [PubMed]
- Damanti, C.C.; Ferrone, L.; Gaffo, E.; Garbin, A.; Tosato, A.; Contarini, G.; Gallingani, I.; Angioni, R.; Molon, B.; Borile, G.; et al. Plasma small-extracellular vesicles enriched in miR-122-5p promote disease aggressiveness in pediatric anaplastic large-cell lymphoma. Cancer Commun. 2023, 43, 630–634. [Google Scholar] [CrossRef] [PubMed]
- Garbin, A.; Contarini, G.; Damanti, C.C.; Tosato, A.; Bortoluzzi, S.; Gaffo, E.; Pizzi, M.; Carraro, E.; Lo Nigro, L.; Vinti, L.; et al. MiR-146a-5p enrichment in small-extracellular vesicles of relapsed pediatric ALCL patients promotes macrophages infiltration and differentiation. Biochem. Pharmacol. 2023, 215, 115747. [Google Scholar] [CrossRef] [PubMed]
- Damanti, C.C.; Gaffo, E.; Lovisa, F.; Garbin, A.; Di Battista, P.; Gallingani, I.; Tosato, A.; Pillon, M.; Carraro, E.; Mascarin, M.; et al. MiR-26a-5p as a Reference to Normalize MicroRNA qRT-PCR Levels in Plasma Exosomes of Pediatric Hematological Malignancies. Cells 2021, 10, 101. [Google Scholar] [CrossRef] [PubMed]
- Lovisa, F.; Garbin, A.; Crotti, S.; Di Battista, P.; Gallingani, I.; Damanti, C.C.; Tosato, A.; Carraro, E.; Pillon, M.; Mafakheri, E.; et al. Increased Tenascin C, Osteopontin and HSP90 Levels in Plasmatic Small Extracellular Vesicles of Pediatric ALK-Positive Anaplastic Large Cell Lymphoma: New Prognostic Biomarkers? Diagnostics 2021, 11, 253. [Google Scholar] [CrossRef]
- Repetto, O.; Lovisa, F.; Elia, C.; Enderle, D.; Romanato, F.; Buffardi, S.; Sala, A.; Pillon, M.; Steffan, A.; Burnelli, R.; et al. Proteomic Exploration of Plasma Exosomes and Other Small Extracellular Vesicles in Pediatric Hodgkin Lymphoma: A Potential Source of Biomarkers for Relapse Occurrence. Diagnostics 2021, 11, 917. [Google Scholar] [CrossRef] [PubMed]
- Johnsen, J.I.; Dyberg, C.; Wickstrom, M. Neuroblastoma-A Neural Crest Derived Embryonal Malignancy. Front. Mol. Neurosci. 2019, 12, 9. [Google Scholar] [CrossRef]
- Maris, J.M. Recent advances in neuroblastoma. N. Engl. J. Med. 2010, 362, 2202–2211. [Google Scholar] [CrossRef]
- Honore, N.; Galot, R.; van Marcke, C.; Limaye, N.; Machiels, J.P. Liquid Biopsy to Detect Minimal Residual Disease: Methodology and Impact. Cancers 2021, 13, 5364. [Google Scholar] [CrossRef] [PubMed]
- Orr, B.A. Pathology, diagnostics, and classification of medulloblastoma. Brain Pathol. 2020, 30, 664–678. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Ronvaux, L.; Riva, M.; Coosemans, A.; Herzog, M.; Rommelaere, G.; Donis, N.; D’Hondt, L.; Douxfils, J. Liquid Biopsy in Glioblastoma. Cancers 2022, 14, 3394. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Romero, N.; Carrion-Navarro, J.; Areal-Hidalgo, P.; Ortiz de Mendivil, A.; Asensi-Puig, A.; Madurga, R.; Nunez-Torres, R.; Gonzalez-Neira, A.; Belda-Iniesta, C.; Gonzalez-Rumayor, V.; et al. BRAF V600E Detection in Liquid Biopsies from Pediatric Central Nervous System Tumors. Cancers 2019, 12, 66. [Google Scholar] [CrossRef]
- Wong, H.A.; Fatimy, R.E.; Onodera, C.; Wei, Z.; Yi, M.; Mohan, A.; Gowrisankaran, S.; Karmali, P.; Marcusson, E.; Wakimoto, H.; et al. The Cancer Genome Atlas Analysis Predicts MicroRNA for Targeting Cancer Growth and Vascularization in Glioblastoma. Mol. Ther. 2015, 23, 1234–1247. [Google Scholar] [CrossRef]
- Ranganathan, S.; Lopez-Terrada, D.; Alaggio, R. Hepatoblastoma and Pediatric Hepatocellular Carcinoma: An Update. Pediatr. Dev. Pathol. 2020, 23, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M.; Munding, J.; Gruner, M.; Liffers, S.T.; Verdoodt, B.; Hauk, J.; Steinstraesser, L.; Tannapfel, A.; Hermeking, H. Frequent concomitant inactivation of miR-34a and miR-34b/c by CpG methylation in colorectal, pancreatic, mammary, ovarian, urothelial, and renal cell carcinomas and soft tissue sarcomas. Virchows Arch. 2011, 458, 313–322. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Wu, B.; Wang, X.; Jiang, Y.; Zhu, D. Role of extracellular vesicles in osteosarcoma. Int. J. Med. Sci. 2022, 19, 1216–1226. [Google Scholar] [CrossRef]
- Biazzo, A.; De Paolis, M. Multidisciplinary approach to osteosarcoma. Acta Orthop. Belg. 2016, 82, 690–698. [Google Scholar]
- Chou, A.J.; Gorlick, R. Chemotherapy resistance in osteosarcoma: Current challenges and future directions. Expert Rev. Anticancer Ther. 2006, 6, 1075–1085. [Google Scholar] [CrossRef] [PubMed]
- Ucci, A.; Cappariello, A.; Ponzetti, M.; Tennant, F.; Loftus, A.E.P.; Shefferd, K.; Maurizi, A.; Delle Monache, S.; Teti, A.; Rucci, N. Anti-osteoblastogenic, pro-inflammatory and pro-angiogenic effect of extracellular vesicles isolated from the human osteosarcoma cell line MNNG/HOS. Bone 2021, 153, 116130. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Samuel, G.; Crow, J.; Godwin, A.K.; Zeng, Y. Molecular assessment of circulating exosomes toward liquid biopsy diagnosis of Ewing sarcoma family of tumors. Transl. Res. 2018, 201, 136–153. [Google Scholar] [CrossRef]
- Skapek, S.X.; Ferrari, A.; Gupta, A.A.; Lupo, P.J.; Butler, E.; Shipley, J.; Barr, F.G.; Hawkins, D.S. Rhabdomyosarcoma. Nat. Rev. Dis. Primers 2019, 5, 1. [Google Scholar] [CrossRef]
- Satou, A.; Bennani, N.N.; Feldman, A.L. Update on the classification of T-cell lymphomas, Hodgkin lymphomas, and histiocytic/dendritic cell neoplasms. Expert Rev. Hematol. 2019, 12, 833–843. [Google Scholar] [CrossRef]
- Sandlund, J.T. Non-Hodgkin Lymphoma in Children. Curr. Hematol. Malig. Rep. 2015, 10, 237–243. [Google Scholar] [CrossRef]
- Becker, A.; Thakur, B.K.; Weiss, J.M.; Kim, H.S.; Peinado, H.; Lyden, D. Extracellular Vesicles in Cancer: Cell-to-Cell Mediators of Metastasis. Cancer Cell 2016, 30, 836–848. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Wills, C.A.; Chen, L.; Zhang, J.; Zhao, Y.; Zhou, M.; Sundstrom, J.M.; Schell, T.; Spiegelman, V.S.; Young, M.M.; et al. Small extracellular vesicles induce resistance to anti-GD2 immunotherapy unveiling tipifarnib as an adjunct to neuroblastoma immunotherapy. J. Immunother. Cancer 2022, 10, e004399. [Google Scholar] [CrossRef]
- Chen, W.; Hao, X.; Yang, B.; Zhang, Y.; Sun, L.; Hua, Y.; Yang, L.; Yu, J.; Zhao, J.; Hou, L.; et al. MYCN-amplified neuroblastoma cell-derived exosomal miR-17-5p promotes proliferation and migration of non-MYCN amplified cells. Mol. Med. Rep. 2021, 23, 245. [Google Scholar] [CrossRef]
- Challagundla, K.B.; Wise, P.M.; Neviani, P.; Chava, H.; Murtadha, M.; Xu, T.; Kennedy, R.; Ivan, C.; Zhang, X.; Vannini, I.; et al. Exosome-mediated transfer of microRNAs within the tumor microenvironment and neuroblastoma resistance to chemotherapy. J. Natl. Cancer Inst. 2015, 107, djv135. [Google Scholar] [CrossRef]
- Gassmann, H.; Schneider, K.; Evdokimova, V.; Ruzanov, P.; Schober, S.J.; Xue, B.; von Heyking, K.; Thiede, M.; Richter, G.H.S.; Pfaffl, M.W.; et al. Ewing Sarcoma-Derived Extracellular Vesicles Impair Dendritic Cell Maturation and Function. Cells 2021, 10, 2081. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.A.; Koh, E.J.; Kim, R.N.; Byun, J.W.; Phi, J.H.; Yang, J.; Wang, K.C.; Park, A.K.; Hwang, D.W.; Lee, J.Y.; et al. Extracellular vesicle-associated miR-135b and -135a regulate stemness in Group 4 medulloblastoma cells by targeting angiomotin-like 2. Cancer Cell Int. 2020, 20, 558. [Google Scholar] [CrossRef] [PubMed]
- Epple, L.M.; Griffiths, S.G.; Dechkovskaia, A.M.; Dusto, N.L.; White, J.; Ouellette, R.J.; Anchordoquy, T.J.; Bemis, L.T.; Graner, M.W. Medulloblastoma exosome proteomics yield functional roles for extracellular vesicles. PLoS ONE 2012, 7, e42064. [Google Scholar] [CrossRef] [PubMed]
- Purvis, I.J.; Velpula, K.K.; Guda, M.R.; Nguyen, D.; Tsung, A.J.; Asuthkar, S. B7-H3 in Medulloblastoma-Derived Exosomes; A Novel Tumorigenic Role. Int. J. Mol. Sci. 2020, 21, 7050. [Google Scholar] [CrossRef] [PubMed]
- Bisaro, B.; Mandili, G.; Poli, A.; Piolatto, A.; Papa, V.; Novelli, F.; Cenacchi, G.; Forni, M.; Zanini, C. Proteomic analysis of extracellular vesicles from medullospheres reveals a role for iron in the cancer progression of medulloblastoma. Mol. Cell Ther. 2015, 3, 8. [Google Scholar] [CrossRef] [PubMed]
- Dhamdhere, M.R.; Gowda, C.P.; Singh, V.; Liu, Z.; Carruthers, N.; Grant, C.N.; Sharma, A.; Dovat, S.; Sundstrom, J.M.; Wang, H.G.; et al. IGF2BP1 regulates the cargo of extracellular vesicles and promotes neuroblastoma metastasis. Oncogene 2023, 42, 1558–1571. [Google Scholar] [CrossRef]
- Mancarella, C.; Giusti, V.; Caldoni, G.; Laginestra, M.A.; Parra, A.; Toracchio, L.; Giordano, G.; Roncuzzi, L.; Piazzi, M.; Blalock, W.; et al. Extracellular vesicle-associated IGF2BP3 tunes Ewing sarcoma cell migration and affects PI3K/Akt pathway in neighboring cells. Cancer Gene Ther. 2023, 30, 1285–1295. [Google Scholar] [CrossRef] [PubMed]
- Macklin, R.; Wang, H.; Loo, D.; Martin, S.; Cumming, A.; Cai, N.; Lane, R.; Ponce, N.S.; Topkas, E.; Inder, K.; et al. Extracellular vesicles secreted by highly metastatic clonal variants of osteosarcoma preferentially localize to the lungs and induce metastatic behaviour in poorly metastatic clones. Oncotarget 2016, 7, 43570–43587. [Google Scholar] [CrossRef] [PubMed]
- Bell, J.L.; Turlapati, R.; Liu, T.; Schulte, J.H.; Huttelmaier, S. IGF2BP1 harbors prognostic significance by gene gain and diverse expression in neuroblastoma. J. Clin. Oncol. 2015, 33, 1285–1293. [Google Scholar] [CrossRef]
- Xue, P.; Huang, S.; Han, X.; Zhang, C.; Yang, L.; Xiao, W.; Fu, J.; Li, H.; Zhou, Y. Exosomal miR-101-3p and miR-423-5p inhibit medulloblastoma tumorigenesis through targeting FOXP4 and EZH2. Cell Death Differ. 2022, 29, 82–95. [Google Scholar] [CrossRef]
- Huang, S.; Xue, P.; Han, X.; Zhang, C.; Yang, L.; Liu, L.; Wang, X.; Li, H.; Fu, J.; Zhou, Y. Exosomal miR-130b-3p targets SIK1 to inhibit medulloblastoma tumorigenesis. Cell Death Dis. 2020, 11, 408. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Romero, N.; Esteban-Rubio, S.; Rackov, G.; Carrion-Navarro, J.; Belda-Iniesta, C.; Ayuso-Sacido, A. Extracellular vesicles compartment in liquid biopsies: Clinical application. Mol. Aspects Med. 2018, 60, 27–37. [Google Scholar] [CrossRef] [PubMed]
- Irmer, B.; Chandrabalan, S.; Maas, L.; Bleckmann, A.; Menck, K. Extracellular Vesicles in Liquid Biopsies as Biomarkers for Solid Tumors. Cancers 2023, 15, 1307. [Google Scholar] [CrossRef] [PubMed]

| Cancer Type | Vesicle Type | Transcript | Protein | Main Finding | References |
|---|---|---|---|---|---|
| Neuroblastoma | LEVs derived from MYCN-amplified neuroblastoma cells and patient-derived bone marrow plasma | X | MYCN mRNA was detectable in LEVs, but not sEVs, of MYCN-amplified neuroblastoma cell lines and patient-derived bone marrow plasma, where the presence and absence of MYCN-LEVs were associated with MYCN amplification status and treatment–relapse disease states. | [46] | |
| sEVs derived from MYCN-amplified neuroblastoma cell lines | X | miR-92a was the highest expressed in EV derived from the neuroblastoma cell line. | [70] | ||
| sEVs derived from plasma of healthy and neuroblastoma patients | X | RT-qPCR validation discovered that sEVs (exosome) containing miR-199a-3p were significantly higher expressed in neuroblastoma patients compared with the healthy donors. | [71] | ||
| sEVs isolated from neuroblastoma cells derived from abdominal primary tumors and bone marrow metastasis | X | Six proteins uniquely presented in metastatic neuroblastoma sEVs, e.g., signal peptidase complex catalytic subunit SEC11, cell division cycle-associated protein 3, nuclear pore complex protein Nup107, calcium, and integrin-binding protein 1. EV proteins of primary tumors are involved in neuronal development and function, while proteins exclusively present in EVs derived from neuroblastoma–bone marrow metastatic models are associated with cell survival, proliferation, and progression. | [72] | ||
| Medulloblastoma | LEVs derived from highly aggressive stem-like medulloblastoma cells overexpressing the pluripotent factor OCT4A | X | X | The interactome analysis of distinct proteins and miRNA suggested that ERK, PI3K/AKT/mTOR, EGF/EFGR, and stem cell self-renewal are the main oncogenic signaling pathways altered in these aggressive medulloblastoma cells. LEVs carried four proteins (UBE2M, HNRNPCL2, HNRNPCL3, and HNRNPCL4) and five miRNAs (miR-4449, miR-500b, miR-3648, miR-1291, and miR-3607). | [73] |
| sEVs derived from group 3 medulloblastoma cell lines | X | sEVs (Exosome) derived from the group 3 medulloblastoma cell line with increased levels of miR-181a-5p, miR-125b-5p, and let-7b-5p could promote in vitro invasion and migratory abilities of a less invasive medulloblastoma cell line through the activation of ERK in the Ras/MAPK pathway. | [74] | ||
| sEVs derived from metastatic medulloblastoma cell lines compared with non-metastatic medulloblastoma cell lines | X | The sEVs derived from the metastatic medulloblastoma cell line had significantly increased in MMP-2 localized on their external surface. This study found a high level of MMP-2 activity in CSF from three of four patients associated with tumor progression. | [75] | ||
| Glioma | sEVs derived from CSF and serum of glioma patients | X | A combination of biofluid EV-derived RNA and BEAMing RT-PCR could detect and quantify mutant and wild-type IDH1 RNA transcripts in CSF of patients with gliomas. | [76] | |
| sEVs derived from serum or conditioned media of glioma patients | X | Hypoxic GBM cells secrete sEVs containing miR-301a, which can promote radiation resistance in normoxia-cultured cells. Hypoxic sEVs containing miR-301a directly targeted GBM tumor suppressor TCEAL7 genes and actively suppressed their expression in normoxic glioma cells. | [77] | ||
| sEVs derived from temozolomide (TMZ)-resistant glioblastoma multiforme cells | X | The lower expression of miR-151 in sEVs was related to an increased resistance to temozolomide (TMZ), in which the restoration of miR-151a expression sensitized TMZ-resistant glioblastoma multiforme cells. | [78] | ||
| sEVs derived from TMZ-resistant glioblastoma cells | X | The high expression of sEVs containing miR-1238 led to the acquired resistance against temozolomide in glioblastoma-sensitive cells. | [79] | ||
| Hepatoblastoma | sEVs derived from the serum of hepatoblastoma patients | X | The level of sEV-derived miR-34a/b/c was significantly lower in the serum of patients with hepatoblastoma compared to healthy control groups. | [80] | |
| sEVs derived from plasma of pediatric hepatoblastoma patients | X | The elevated level of miR-21 containing sEVs in hepatoblastoma patients might be another biomarker for hepatoblastoma. | [81] | ||
| sEVs derived from the hepatoblastoma cell line | X | sEV-derived miR-126 was upregulated in hepatoblastoma cells, which suggested that this microRNA promoted the tumorigenesis of liver cancer. | [82] | ||
| Ewing’s sarcoma | sEVs derived from Ewing’s sarcoma cell line | X | RT-qPCR detected ES-specific transcripts such as EWSR1-FLI1 from Ewing’s sarcoma-derived EVs. | [83] | |
| LEVs derived from Ewing’s sarcoma cell of xenografted mice | X | EWS/FLI1 fusion mRNA (resulting from the t(11;22) (q24;q12) translocation) could be identified in MVs derived from Ewing sarcoma cells and was also detectable in MVs from plasma samples of ES cell-xenografted animals. | [84] | ||
| sEVs isolated from silenced CD99 expression of patients derived Ewing’s sarcoma cell line | X | Ewing sarcoma cells with silenced CD99 expression released EVs containing high levels of miR-34a. | [85] | ||
| Osteosarcoma | sEVs derived from malignant human osteosarcoma cell lines | X | Small extracellular vesicles derived from malignant human osteosarcomas contain miR-146a-5p. | [86] | |
| sEVs derived from six different human osteosarcoma or osteoblast cell lines | X | Next-generation miRNA sequencing revealed miRNAs in cell lines with different degrees of metastatic potential and found that mi-21-5p, miR-143-3p, miR-148a-3p, and 181a-5p are highly expressed in sEV-derived metastatic SAOS2 cells. | [87] | ||
| sEVs derived from the osteosarcoma cell line | X | Circulating miR-17-5p and miR-25-3p could be identified in osteosarcoma cells, which are used as a novel diagnostic and prognostic biomarker and also reflect tumor burden in the osteosarcoma mouse model. | [88] | ||
| sEVs derived from the Rab22a-NeoF1 fusion protein cell line | X | The osteosarcoma Rab22a-NeoF1 fusion protein was secreted via EV by binding to the KFERQ-like motif of HSP90, which was taken up by macrophages and other cancer cells. EV derived from osteosarcoma contained programmed death-ligand 1 (PD-L1) and N-cadherin. | [89] | ||
| sEVs derived from the metastatic osteosarcoma cell line | X | The elevation of uPA and its receptor (uPAR) was cargo via sEVs secreted from metastatic osteosarcoma cells. | [90] | ||
| Rhabdomyosarcoma | sEVs derived from rhabdomyosarcoma cell lines | X | Ten miRNAs were common among the two rhabdomyosarcoma cell lines (JR1 and RD), while only two miRNAs (miR-1246 and miR-1268) were present in EVs of all cell lines. | [91] | |
| sEV derived from the rhabdomyosarcoma cell lines | X | CD147 was exclusively expressed in metastatic tumors of human rhabdomyosarcoma tissue, which was involved in modulating the microenvironment through rhabdomyosarcoma-secreted sEVs. | [92] | ||
| sEV derived from alveolar and embryonal rhabdomyosarcoma cell lines | X | A proteomic study revealed 122 common proteins in alveolar rhabdomyosarcoma-derived EVs and 161 common proteins in embryonal rhabdomyosarcoma-derived EVs. The biological process analysis suggested that 81 proteins were common to both subtypes, which involved cell signaling, cell movement, and cancer. | [93] | ||
| Pediatric lymphoma | sEVs derived from plasma of ALCL patients | X | miR-122-5p was elevated in plasma sEVs derived from ALCL patients, which are critical in promoting tumor cell dissemination and aggressiveness. | [94] | |
| sEVs derived from plasma of 20 pediatric anaplastic lymphoma kinase-positive ALCL patients | X | Small RNA-sequencing analysis in plasma sEVs from 20 pediatric ALCA suggested that miR-146a-5p and miR-378a-3p showed a negative prognostic impact in both univariate and multivariate analysis. | [95] | ||
| sEVs derived from Burkitt lymphoma, Hodgkin lymphoma, and mature B-cell acute lymphoblastic leukemia | X | Burkitt lymphoma, Hodgkin lymphoma, and mature B-cell acute lymphoblastic leukemia most stably expressed miR-26a-5p and miR-486-5p in sEVs. | [96] | ||
| sEVs derived from plasma pediatric ALCL patients and healthy donors | X | The Reactome database and KEGG networks highlighted a dramatic increase in proteins of the PI3K/AKT pathway in ALCL-sEVs, which included heat shock protein 90-kDa isoform alpha-1, osteopontin, and tenascin plasma EV derived from pediatric ALCL patients. | [97] | ||
| sEVs derived from plasma of non-relapsed and relapsed nodular sclerosis Hodgkin lymphoma | X | LC-MS/MS identified these 11 unique protein spots, including five more abundant in non-relapsed HL (e.g., isoform 2 preproprotein of complement C4-A, complement C4-B, fibrinogen γ chain, inter-α-trypsin inhibitor heavy chain H2, and immunoglobulin heavy chain constant region mu) and six more abundant in relapsed HL (e.g., apolipoprotein A-I, apolipoprotein A-IV, clusterin, haptoglobin, α-1-acid glycoprotein 1, and transthyretin). | [98] |
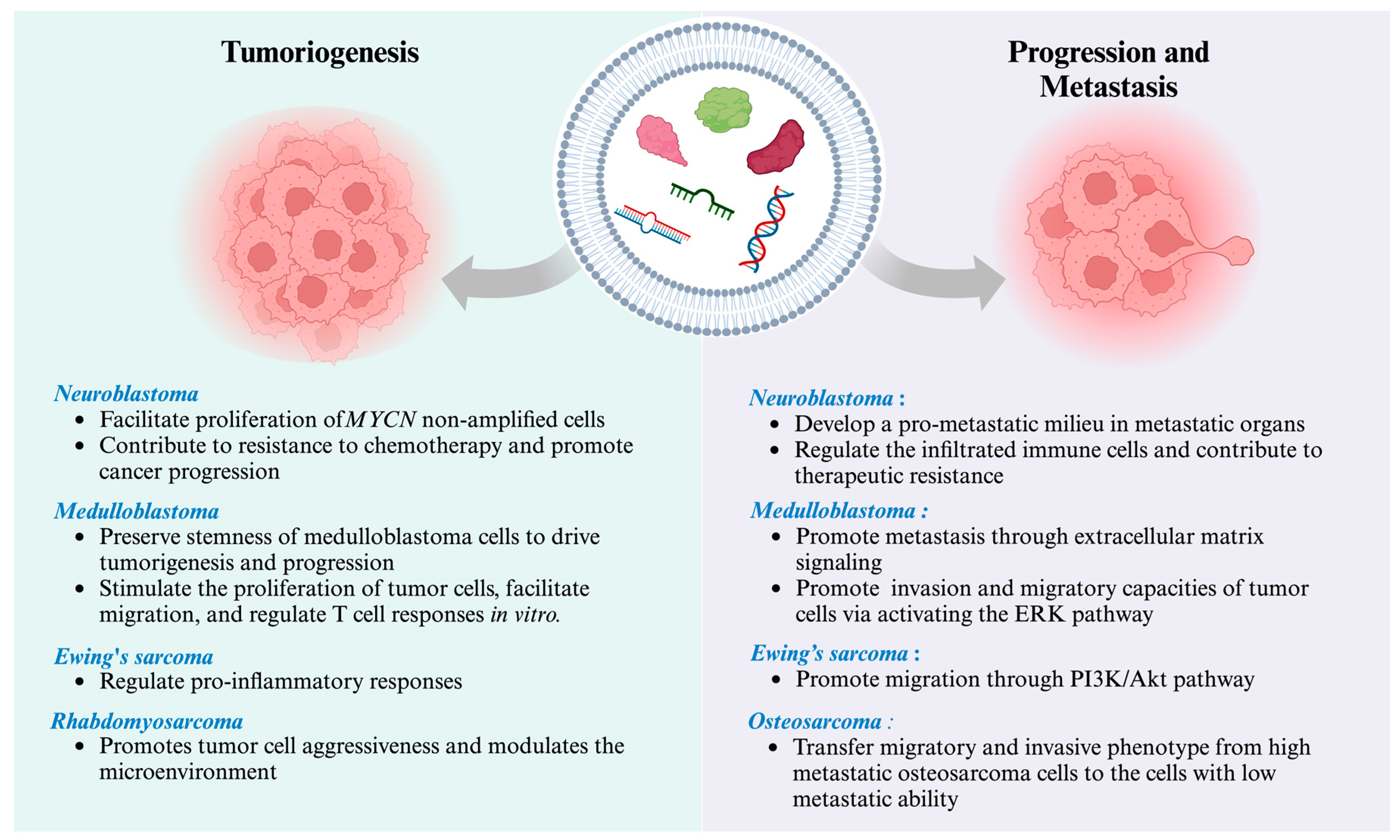
| Disease Stage | Disease | Significant Biology Process | Vesicle Type | EV Molecular Cargo | Main Finding | References |
|---|---|---|---|---|---|---|
| Tumorigenesis | Neuroblastoma | Regulation of infiltrated immune cells after chemotherapy treatment | sEVs | Not specified | Small EVs suppressed splenic NK cell maturation in vivo and dinutuximab-induced NK cell-mediated antibody-dependent cellular cytotoxicity in vitro upon dinutuximab treatment. | [118] |
| Facilitating the migration and proliferation of non-MYCN amplified cells | sEVs released by MYCN-amplified neuroblastoma cells | miR-17-5p | miR-17-5p is crucial in facilitating the migration and proliferation of non-MYCN amplified cells. | [119] | ||
| Contribute to resistance to chemotherapy and promote cancer progression | sEVs derived from neuroblastoma cell line | miR-21, miR-155 | The data presented in this study demonstrate the distinct function of EV-derived miR-21 and miR-155 in intercellular communication between neuroblastoma cells and human monocytes, contributing to developing resistance to chemotherapy and cancer progression. | [120] | ||
| Ewing’s sarcoma | Regulation of pro-inflammatory response | sEVs derived from Ewing’s sarcoma cell lines | Not specified | Exposure to Ewing’s sarcoma EVs inhibited the process of cellular development towards moDCs, as indicated by the decreased expression levels of co-stimulatory molecules (e.g., CD80, CD86, and HLA-DR). Ewing’s sarcoma EVs exhibited the ability to suppress the proliferation of CD4+ and CD8+ T cells, as well as the release of IFNγ, while simultaneously increasing the secretion of IL-10 and IL-6. | [121] | |
| Rhabdosarcoma | Promotes tumor cell aggressiveness and modulates the microenvironment | sEV derived from rhabdomyosarcoma cell lines | CD147 | Treatment of normal fibroblasts with rhabdomyosarcoma-derived EVs increased proliferation, migration, and invasion, whereas CD147-downregulated rhabdomyosarcoma cells block these effects. | [92] | |
| Medulloblastoma | Preserve stem cell characteristics or communicate with neighboring cells to promote the progression of group 4 of medulloblastoma | sEVs derived from bulk tumor cells and brain tumor sheroid-forming cells | miR-135b, miR-135a | The suppression of miR-135b and miR-135a leads to a decrease in the stemness of medulloblastoma brain tumor spheroid-forming cells, in which AMOTL2 was targeted by miR-135b and miR-135a. | [122] | |
| Promote in vitro invasion and migratory capacities of tumor cells via activating the ERK pathway in the Ras/MAPK signaling cascade | sEVs derived from Group 3 medulloblastoma cell lines | miR-181a-5p, miR-125b-5p, let-7b-5p | The upregulation of these miRNAs led to more significant in vitro invasion and migratory capacities of tumor cells via activating the ERK pathway in the Ras/MAPK signaling cascade. | [74] | ||
| Stimulate the proliferation of tumor cells, facilitate migration, and regulate T cell responses in vitro | sEVs derived from medulloblastoma cell lines | Not specified | The examination of the functional properties of EVs can stimulate the proliferation of tumor cells, facilitate migration, and regulate T cell responses, which might play a crucial role in the progression of medulloblastoma. | [123] | ||
| Play a role in the progression of medulloblastoma | sEVs derived from control and overexpressed B7-H3 cells | B7-H3 (immunosuppressive immune check point) | This study revealed a novel role in EV production and packaging for B7-H3 that may contribute to medulloblastoma progression. | [124] | ||
| Involvement in the advancement and invasion of medulloblastoma. | sEVs derived from medulloblastoma cell line | Iron carrier proteins | This study establishes the relationship between iron metabolism and the advancement and invasion of medulloblastoma. The reduction in iron induces cell cycle arrest in the G1/S phases, resulting in the suppression of cell proliferation and the initiation of apoptosis. | [125] | ||
| Cancer EVs promote metastasis | Neuroblastoma | Pro-metastatic | sEVs derived from neuroblastoma cell line | IGF2BP1 | IGF2BP1 affects the levels of SEMA3A and SHMT2 in EVs, and this regulation plays a role in developing a pro-metastatic milieu in metastatic organs. | [126] |
| Ewing’s sarcoma | Promote tumor cell migration | sEVs derived from Ewing’s sarcoma cell line | IGF2BP3 | This data indicated that IGF2BP3 containing EVs may have a role in regulating phenotypic heterogeneity and promoting cell migration of Ewing’s sarcoma. | [127] | |
| Osteosarcoma | Promote tumor cell migration and invasive phenotype | sEVs derived from high metastatic potential cell line | Several proteins relating to signaling pathways mediated by G-protein coupled receptors | The uptake of sEVs derived from the high metastatic osteosarcoma cell line by the same cell line with low metastatic ability leads to the induction of migratory and invasive phenotypes. Proteins relating to signaling pathways mediated by G-protein coupled receptors may play a crucial role in driving metastasis of osteosarcoma. | [128] | |
| Medulloblastoma | Promote medulloblastoma metastasis through extracellular matrix signaling | sEVs derived from metastatic medulloblastoma cell line | EMMPRIN, MMP-2 | This study provides evidence for the significance of EMMPRIN and MMP-2-associated sEVs in facilitating a conducive environment that promotes medulloblastoma metastasis. | [75] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Singhto, N.; Pongphitcha, P.; Jinawath, N.; Hongeng, S.; Chutipongtanate, S. Extracellular Vesicles for Childhood Cancer Liquid Biopsy. Cancers 2024, 16, 1681. https://doi.org/10.3390/cancers16091681
Singhto N, Pongphitcha P, Jinawath N, Hongeng S, Chutipongtanate S. Extracellular Vesicles for Childhood Cancer Liquid Biopsy. Cancers. 2024; 16(9):1681. https://doi.org/10.3390/cancers16091681
Chicago/Turabian StyleSinghto, Nilubon, Pongpak Pongphitcha, Natini Jinawath, Suradej Hongeng, and Somchai Chutipongtanate. 2024. "Extracellular Vesicles for Childhood Cancer Liquid Biopsy" Cancers 16, no. 9: 1681. https://doi.org/10.3390/cancers16091681
APA StyleSinghto, N., Pongphitcha, P., Jinawath, N., Hongeng, S., & Chutipongtanate, S. (2024). Extracellular Vesicles for Childhood Cancer Liquid Biopsy. Cancers, 16(9), 1681. https://doi.org/10.3390/cancers16091681





