Clinical Prediction Models for Prognosis of Colorectal Liver Metastases: A Comprehensive Review of Regression-Based and Machine Learning Models
Abstract
Simple Summary
Abstract
1. Introduction
2. Traditional Prediction Models
3. Methods
4. Outcome Types
4.1. Postoperative Complications/Mortality
4.2. Survival
4.3. Recurrence
5. Specific Patient and Treatment Characteristics
5.1. Simultaneous Resections
5.2. Upfront Surgery versus Neoadjuvant Chemotherapy
5.3. Systemic Therapies
5.4. Special Treatment Modalities (RFA, MWA, HAIP, SIRT, ALPPS)
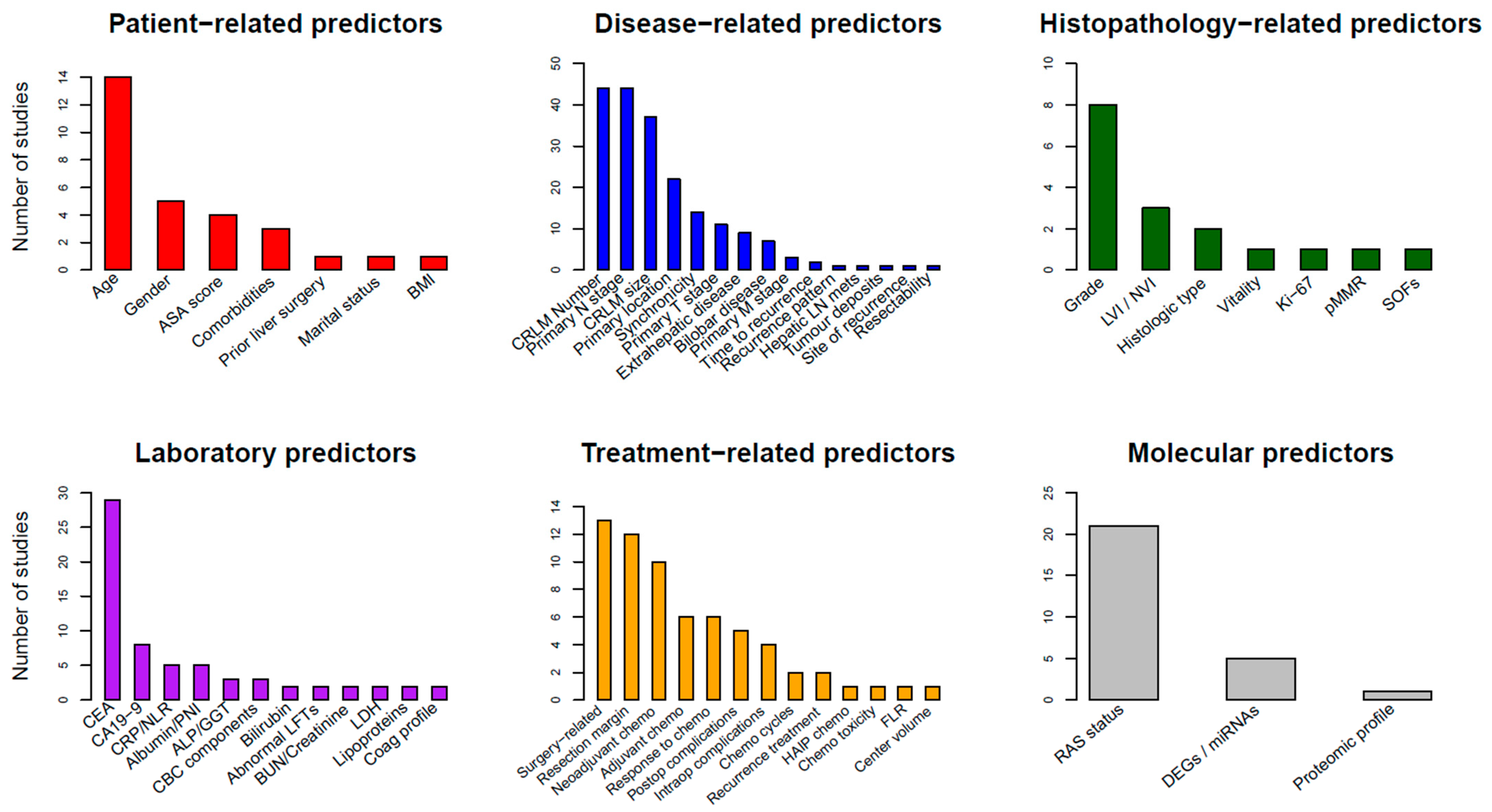
6. Predictor Types
6.1. Patient-Related Predictors
6.2. Laboratory Biomarkers
6.3. Disease-Related Predictors
6.4. Histopathological Predictors
6.5. Treatment-Related Predictors
6.6. RAS Status and Molecular Predictors
7. Development and Validation Techniques
8. Model Performance
9. Critical Appraisal of Published Models
10. Conclusions and Future Directions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Zeineddine, F.A.; Zeineddine, M.A.; Yousef, A.; Gu, Y.; Chowdhury, S.; Dasari, A.; Huey, R.W.; Johnson, B.; Kee, B.; Lee, M.S.; et al. Survival Improvement for Patients with Metastatic Colorectal Cancer over Twenty Years. NPJ Precis. Oncol. 2023, 7, 16. [Google Scholar] [CrossRef]
- Cervantes, A.; Adam, R.; Roselló, S.; Arnold, D.; Normanno, N.; Taïeb, J.; Sel09igmann, J.; De Baere, T.; Osterlund, P.; Yoshino, T.; et al. Metastatic Colorectal Cancer: ESMO Clinical Practice Guideline for Diagnosis, Treatment and Follow-Up. Ann. Oncol. 2023, 34, 10–32. [Google Scholar] [CrossRef] [PubMed]
- Martin, J.; Petrillo, A.; Smyth, E.C.; Shaida, N.; Khwaja, S.; Cheow, H.; Duckworth, A.; Heister, P.; Praseedom, R.; Jah, A.; et al. Colorectal Liver Metastases: Current Management and Future Perspectives. World J. Clin. Oncol. 2020, 11, 761–808. [Google Scholar] [CrossRef] [PubMed]
- Rompianesi, G.; Pegoraro, F.; Ceresa, C.D.; Montalti, R.; Troisi, R.I. Artificial Intelligence in the Diagnosis and Management of Colorectal Cancer Liver Metastases. World J. Gastroenterol. 2022, 28, 108–122. [Google Scholar] [CrossRef] [PubMed]
- Moons, K.G.M.; Altman, D.G.; Reitsma, J.B.; Ioannidis, J.P.A.; Macaskill, P.; Steyerberg, E.W.; Vickers, A.J.; Ransohoff, D.F.; Collins, G.S. Transparent Reporting of a Multivariable Prediction Model for Individual Prognosis or Diagnosis (TRIPOD): Explanation and Elaboration. Ann. Intern. Med. 2015, 162, W1–W73. [Google Scholar] [CrossRef] [PubMed]
- Nordlinger, B.; Guiguet, M.; Vaillant, J.-C.; Balladur, P.; Boudjema, K.; Bachellier, P.; Jaeck, D.; de Chirurgie, A.F. Surgical Resection of Colorectal Carcinoma Metastases to the Liver: A Prognostic Scoring System to Improve Case Selection, Based on 1568 Patients. Cancer 1996, 77, 1254–1262. [Google Scholar] [CrossRef]
- Fong, Y.; Fortner, J.; Sun, R.L.; Brennan, M.F.; Blumgart, L.H. Clinical Score for Predicting Recurrence after Hepatic Resection for Metastatic Colorectal Cancer: Analysis of 1001 Consecutive Cases. Ann. Surg. 1999, 230, 309–321. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, K.; Shimada, H.; Fujii, Y.; Endo, I.; Sekido, H.; Togo, S.; Ike, H. Pre-Hepatectomy Prognostic Staging to Determine Treatment Strategy for Colorectal Cancer Metastases to the Liver. Langenbeck’s Arch. Surg. 2004, 389, 371–379. [Google Scholar] [CrossRef] [PubMed]
- Minagawa, M. Simplified Staging System for Predicting the Prognosis of Patients with Resectable Liver Metastasis. Arch. Surg. 2007, 142, 269. [Google Scholar] [CrossRef]
- Yamaguchi, T.; Mori, T.; Takahashi, K.; Matsumoto, H.; Miyamoto, H.; Kato, T. A New Classification System for Liver Metastases from Colorectal Cancer in Japanese Multicenter Analysis. Hepatogastroenterology 2008, 55, 173–178. [Google Scholar]
- Adam, R.; de Haas, R.J.; Wicherts, D.A.; Vibert, E.; Salloum, C.; Azoulay, D.; Castaing, D. Concomitant Extrahepatic Disease in Patients with Colorectal Liver Metastases. Ann. Surg. 2011, 253, 349–359. [Google Scholar] [CrossRef] [PubMed]
- Iwatsuki, S.; Dvorchik, I.; Madariaga, J.R.; Marsh, J.W.; Dodson, F.; Bonham, A.C.; Geller, D.A.; Gayowski, T.J.; Fung, J.J.; Starzl, T.E. Hepatic Resection for Metastatic Colorectal Adenocarcinoma: A Proposal of a Prognostic Scoring System11No Competing Interests Declared. J. Am. Coll. Surg. 1999, 189, 291–299. [Google Scholar] [CrossRef]
- Lise, M.; Bacchetti, S.; Da Pian, P.; Nitti, D.; Pilati, P. Patterns of Recurrence after Resection of Colorectal Liver Metastases: Prediction by Models of Outcome Analysis. World J. Surg. 2001, 25, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, I.; Takada, T.; Matsuda, K.; Adachi, M.; Nagawa, H.; Muto, T.; Okinaga, K. A New Scoring System to Classify Patients with Colorectal Liver Metastases: Proposal of Criteria to Select Candidates for Hepatic Resection. J. Hepatobiliary Pancreat. Surg. 2004, 11, 79–83. [Google Scholar] [CrossRef] [PubMed]
- Schindl, M. Prognostic Scoring in Colorectal Cancer Liver Metastases. Arch. Surg. 2005, 140, 183. [Google Scholar] [CrossRef] [PubMed]
- Malik, H.Z.; Prasad, K.R.; Halazun, K.J.; Aldoori, A.; Al-Mukhtar, A.; Gomez, D.; Lodge, J.P.A.; Toogood, G.J. Preoperative Prognostic Score for Predicting Survival After Hepatic Resection for Colorectal Liver Metastases. Ann. Surg. 2007, 246, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Ueno, H.; Mochizuki, H.; Hatsuse, K.; Hase, K.; Yamamoto, T. Indicators for Treatment Strategies of Colorectal Liver Metastases. Ann. Surg. 2000, 231, 59. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.S.; Kim, M.J.; Yun, S.H.; Chun, H.K.; Lee, W.Y.; Kim, S.J.; Choi, S.H.; Heo, J.S.; Joh, J.W.; Kim, Y. Il Risk Factor Stratification after Simultaneous Liver and Colorectal Resection for Synchronous Colorectal Metastasis. Langenbeck’s Arch. Surg. 2008, 393, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Konopke, R.; Kersting, S.; Distler, M.; Dietrich, J.; Gastmeier, J.; Heller, A.; Kulisch, E.; Saeger, H. Prognostic Factors and Evaluation of a Clinical Score for Predicting Survival after Resection of Colorectal Liver Metastases. Liver Int. 2009, 29, 89–102. [Google Scholar] [CrossRef]
- Buisman, F.E.; Grünhagen, D.J.; Verhoef, C.; Groot Koerkamp, B. Response to Letter Entitled: Re: Predicting 10-Year Survival after Resection of Colorectal Liver Metastases; an International Study Including Biomarkers and Perioperative Treatment. Eur. J. Cancer 2023, 188, 80. [Google Scholar] [CrossRef]
- Bertsimas, D.; Margonis, G.A.; Sujichantararat, S.; Boerner, T.; Ma, Y.; Wang, J.; Kamphues, C.; Sasaki, K.; Tang, S.; Gagniere, J.; et al. Using Artificial Intelligence to Find the Optimal Margin Width in Hepatectomy for Colorectal Cancer Liver Metastases. JAMA Surg. 2022, 157, e221819. [Google Scholar] [CrossRef]
- Bao, X.; Wang, K.; Liu, M.; Li, B.; Wang, H.; Jin, K.; Yan, X.; Zhang, H.; Bao, Q.; Xu, D.; et al. Characterization of Genomic Alterations in Colorectal Liver Metastasis and Their Prognostic Value. Front. Cell Dev. Biol. 2022, 9, 760618. [Google Scholar] [CrossRef] [PubMed]
- Lam, C.S.N.; Bharwani, A.A.; Chan, E.H.Y.; Chan, V.H.Y.; Au, H.L.H.; Ho, M.K.; Rashed, S.; Kwong, B.M.H.; Fang, W.; Ma, K.W.; et al. A Machine Learning Model for Colorectal Liver Metastasis Post-Hepatectomy Prognostications. Hepatobiliary Surg. Nutr. 2023, 12, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Reijonen, P.; Nordin, A.; Savikko, J.; Poussa, T.; Arola, J.; Isoniemi, H. Histopathological Helsinki Score of Colorectal Liver Metastases Predicts Survival after Liver Resection. Apmis 2023, 131, 249–261. [Google Scholar] [CrossRef]
- Margonis, G.A.; Sasaki, K.; Gholami, S.; Kim, Y.; Andreatos, N.; Rezaee, N.; Deshwar, A.; Buettner, S.; Allen, P.J.; Kingham, T.P.; et al. Genetic and Morphological Evaluation (GAME) Score for Patients with Colorectal Liver Metastases. Br. J. Surg. 2018, 105, 1210–1220. [Google Scholar] [CrossRef]
- Paredes, A.Z.; Hyer, J.M.; Tsilimigras, D.I.; Moro, A.; Bagante, F.; Guglielmi, A.; Ruzzenente, A.; Alexandrescu, S.; Makris, E.A.; Poultsides, G.A.; et al. A Novel Machine-Learning Approach to Predict Recurrence after Resection of Colorectal Liver Metastases. Ann. Surg. Oncol. 2020, 27, 5139–5147. [Google Scholar] [CrossRef] [PubMed]
- Frühling, P.; Urdzik, J.; Strömberg, C.; Isaksson, B. Composite Score: Prognostic Tool to Predict Survival in Patients Undergoing Surgery for Colorectal Liver Metastases. BJS Open 2021, 5, zrab104. [Google Scholar] [CrossRef]
- Taghavi, M.; Trebeschi, S.; Simões, R.; Meek, D.B.; Beckers, R.C.J.; Lambregts, D.M.J.; Verhoef, C.; Houwers, J.B.; van der Heide, U.A.; Beets-Tan, R.G.H.; et al. Machine Learning-Based Analysis of CT Radiomics Model for Prediction of Colorectal Metachronous Liver Metastases. Abdom. Radiol. 2021, 46, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Brudvik, K.W.; Jones, R.P.; Giuliante, F.; Shindoh, J.; Passot, G.; Chung, M.H.; Song, J.; Li, L.; Dagenborg, V.J.; Fretland, Å.A.; et al. RAS Mutation Clinical Risk Score to Predict Survival after Resection of Colorectal Liver Metastases. Ann. Surg. 2019, 269, 120–126. [Google Scholar] [CrossRef]
- Moaven, O.; Tavolara, T.E.; Valenzuela, C.D.; Cheung, T.T.; Corvera, C.U.; Cha, C.H.; Stauffer, J.A.; Niazi, M.K.K.; Gurcan, M.N.; Shen, P. Machine Learning Models for Predicting the Outcomes of Surgical Treatment of Colorectal Liver Metastases. J. Am. Coll. Surg. 2023, 236, 884–893. [Google Scholar] [CrossRef]
- Villard, C.; Abdelrafee, A.; Habib, M.; Ndegwa, N.; Jorns, C.; Sparrelid, E.; Allard, M.A.; Adam, R. Prediction of Survival in Patients with Colorectal Liver Metastases- Development and Validation of a Prognostic Score Model. Eur. J. Surg. Oncol. 2022, 48, 2432–2439. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Chang, W.; Ren, L.; Chen, J.; Tang, W.; Liu, T.; Jian, M.; Liu, Y.; Wei, Y.; Xu, J. Comprehensive Evaluation of Relapse Risk (CERR) Score for Colorectal Liver Metastases: Development and Validation. Oncologist 2020, 25, e1031–e1041. [Google Scholar] [CrossRef]
- Chen, F.L.; Wang, Y.Y.; Liu, W.; Xing, B.C. Prognostic Factors in Colorectal Liver Metastases Patients with Various Tumor Numbers Treated by Liver Resection: A Single-Center, Retrospective Study. World J. Surg. Oncol. 2022, 20, 237. [Google Scholar] [CrossRef] [PubMed]
- Dai, S.; Ye, Y.; Kong, X.; Li, J.; Ding, K. A Predictive Model for Early Recurrence of Colorectal-Cancer Liver Metastases Based on Clinical Parameters. Gastroenterol. Rep. 2021, 9, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Liu, J.M.; Wang, K.; Wang, H.W.; Xing, B.C. Recurrent Colorectal Liver Metastasis Patients Could Benefit from Repeat Hepatic Resection. BMC Surg. 2021, 21, 327. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.Y.; Lin, H.C.; Liu, J.; Wang, D.S.; Yuan, Y.F.; Li, B.K.; Zheng, Y.; Wu, X.J.; Chen, G.; Wang, F.H.; et al. A Novel Prognostic Nomogram for Colorectal Cancer Liver Metastasis Patients with Recurrence after Hepatectomy. Cancer Med. 2021, 10, 1535–1544. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, T.; Xu, Z.; Liu, F.; Cai, S.; Wang, L.; Xu, Y. Risk Scoring System for Recurrence after Simultaneous Resection of Colorectal Cancer Liver Metastasis. Ann. Transl. Med. 2021, 9, 966. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Margonis, G.A.; Moro, A.; Wang, J.; Wagner, D.; Gagnière, J.; Shin, J.K.; D’Silva, M.; Sahara, K.; Miyata, T.; et al. Nontumor Related Risk Score: A New Tool to Improve Prediction of Prognosis after Hepatectomy for Colorectal Liver Metastases. Surgery 2022, 171, 1580–1587. [Google Scholar] [CrossRef] [PubMed]
- Huiskens, J.; Schadde, E.; Lang, H.; Malago, M.; Petrowsky, H.; de Santibañes, E.; Oldhafer, K.; van Gulik, T.M.; Olthof, P.B. Avoiding Postoperative Mortality after ALPPS–Development of a Tumor-Specific Risk Score for Colorectal Liver Metastases. HPB 2019, 21, 898–905. [Google Scholar] [CrossRef]
- Bai, L.; Yan, X.L.; Lu, Y.X.; Meng, Q.; Rong, Y.M.; Ye, L.F.; Pan, Z.Z.; Xing, B.C.; Wang, D.S. Circulating Lipid- and Inflammation-Based Risk (CLIR) Score: A Promising New Model for Predicting Outcomes in Complete Colorectal Liver Metastases Resection. Ann. Surg. Oncol. 2022, 29, 4308–4323. [Google Scholar] [CrossRef]
- Fang, C.; Huang, Y.; Chen, C.; Nie, D.; Lin, J.; Xiao, Z.; Li, S.; Liu, S.; Luo, R.; Lin, H.; et al. The Prognostic Value of Serum Apolipoprotein A-I Level and Neutrophil-to-Lymphocyte Ratio in Colorectal Cancer Liver Metastasis. J. Oncol. 2022, 2022, 9149788. [Google Scholar] [CrossRef] [PubMed]
- Qin, S.; Hu, H.; Cui, R.; Lin, J.; Liu, Y.; Wang, Y.; Chen, Y.; Liu, G. A Prognostic Nomogram for Intrahepatic Progression-Free Survival in Patients with Colorectal Liver Metastases after Ultrasound-Guided Percutaneous Microwave Ablation. Int. J. Hyperth. 2022, 39, 144–154. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, Y.; Kopetz, S.; Tran Cao, H.S.; Panettieri, E.; De Bellis, M.; Nishioka, Y.; Hwang, H.; Wang, X.; Tzeng, C.W.D.; Chun, Y.S.; et al. Contour Prognostic Model for Predicting Survival after Resection of Colorectal Liver Metastases: Development and Multicentre Validation Study Using Largest Diameter and Number of Metastases with RAS Mutation Status. Br. J. Surg. 2021, 108, 968–975. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Wu, T.; Zhou, J.; Chen, X.; Dong, C.; Guo, Z.; Yang, R.; Liang, R.; Feng, Q.; Hu, R.; et al. Establishment and Verification of Prognostic Model and CeRNA Network Analysis for Colorectal Cancer Liver Metastasis. BMC Med. Genom. 2023, 16, 99. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Mao, R.; Zhao, J.; Bi, X.; Li, Z.; Huang, Z.; Zhang, Y.; Zhou, J.; Zhao, H.; Cai, J. Nomograms Incorporating Preoperative RDW Level for the Prediction of Postoperative Complications and Survival in Colorectal Liver Metastases after Resection. Ann. Palliat. Med. 2021, 10, 4143–4158. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Wu, Y.; Feng, Y.; Lin, Z.; Zhang, N.; Yu, B.; Mao, A.; Zhang, T.; Zhu, W.; Wang, L. A Population-Based Predictive Model Identifying Optimal Candidates for Primary and Metastasis Resection in Patients with Colorectal Cancer with Liver Metastatic. Front. Oncol. 2022, 12, 899659. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Bai, W.; Zhou, J.; Dong, Q.; Zhang, J. Effect of Tumour Size Ratio on Liver Recurrence-Free Survival of Patients Undergoing Hepatic Resection for Colorectal Liver Metastases. BMC Cancer 2022, 22, 103. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, W.; Xu, Y.; Li, Y.H.; Xing, B.C. A Prognostic Scoring System to Predict Survival Outcome of Resectable Colorectal Liver Metastases in This Modern Era. Ann. Surg. Oncol. 2021, 28, 7709–7718. [Google Scholar] [CrossRef] [PubMed]
- Moro, A.; Mehta, R.; Tsilimigras, D.I.; Sahara, K.; Paredes, A.Z.; Bagante, F.; Guglielmi, A.; Alexandrescu, S.; Poultsides, G.A.; Sasaki, K.; et al. Prognostic Factors Differ According to KRAS Mutational Status: A Classification and Regression Tree Model to Define Prognostic Groups after Hepatectomy for Colorectal Liver Metastasis. Surgery 2020, 168, 497–503. [Google Scholar] [CrossRef]
- Chen, Q.; Mao, R.; Zhao, J.; Bi, X.; Li, Z.; Huang, Z.; Zhang, Y.; Zhou, J.; Zhao, H.; Cai, J. Upgraded Nomograms for the Prediction of Complications and Survival in Patients with Colorectal Liver Metastases Treated with Neoadjuvant Chemotherapy Followed by Hepatic Resection. Ann. Transl. Med. 2021, 9, 265. [Google Scholar] [CrossRef]
- Yao, J.; Chen, Q.; Deng, Y.; Zhao, J.; Bi, X.; Li, Z.; Huang, Z.; Zhang, Y.; Zhou, J.; Zhao, H.; et al. Nomograms Predicting Primary Lymph Node Metastases and Prognosis for Synchronous Colorectal Liver Metastasis with Simultaneous Resection of Colorectal Cancer and Liver Metastases. Ann. Palliat. Med. 2021, 10, 4220–4231. [Google Scholar] [CrossRef]
- Kazi, M.; Patkar, S.; Patel, P.; Kunte, A.; Desouza, A.; Saklani, A.; Goel, M. Simultaneous Resection of Synchronous Colorectal Liver Metastasis: Feasibility and Development of a Prediction Model. Ann. Hepato-Biliary-Pancreat. Surg. 2023, 27, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Zheng, N.; Wen, R.; Sui, J.; Zhang, W. Preoperative Nomogram to Predict Survival Following Colorectal Cancer Liver Metastasis Simultaneous Resection. J. Gastrointest. Oncol. 2021, 12, 556–567. [Google Scholar] [CrossRef]
- Imai, K.; Allard, M.A.; Castro Benitez, C.; Vibert, E.; Sa Cunha, A.; Cherqui, D.; Castaing, D.; Bismuth, H.; Baba, H.; Adam, R. Nomogram for Prediction of Prognosis in Patients with Initially Unresectable Colorectal Liver Metastases. Br. J. Surg. 2016, 103, 590–599. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Zhang, Y.; Li, X.; Huang, Z.; Zhao, H.; Cai, J. Nomogram Incorporating Preoperative Testing Markers for the Prediction of Early Recurrence for Colorectal Liver Metastases with Neoadjuvant Chemotherapy Followed by Hepatectomy. J. Cancer 2022, 13, 1758–1767. [Google Scholar] [CrossRef] [PubMed]
- Cheng, X.; Li, Y.; Chen, D.; Xu, X.; Liu, F.; Zhao, F. Nomogram Predicting the Survival of Young-Onset Patients with Colorectal Cancer Liver Metastases. Diagnostics 2022, 12, 1395. [Google Scholar] [CrossRef]
- Kulik, U.; Plohmann-Meyer, M.; Gwiasda, J.; Kolb, J.; Meyer, D.; Kaltenborn, A.; Lehner, F.; Klempnauer, J.; Schrem, H. Proposal of Two Prognostic Models for the Prediction of 10-Year Survival after Liver Resection for Colorectal Metastases. HPB Surg. 2018, 2018, 5618581. [Google Scholar] [CrossRef]
- Bai, L.; Lin, Z.Y.; Lu, Y.X.; Chen, Q.; Zhou, H.; Meng, Q.; Lin, C.P.; Huang, W.L.; Wan, Y.L.; Pan, Z.Z.; et al. The Prognostic Value of Preoperative Serum Lactate Dehydrogenase Levels in Patients Underwent Curative-Intent Hepatectomy for Colorectal Liver Metastases: A Two-Center Cohort Study. Cancer Med. 2021, 10, 8005–8019. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, X.; Lin, H.; Sun, X.; Fong, W.P.; Wu, X.; Pan, Z.; Yuan, Y.; Liang, J.; Wang, D.; et al. A Prehepatectomy Circulating Exosomal Microrna Signature Predicts the Prognosis and Adjuvant Chemotherapeutic Benefits in Colorectal Liver Metastasis. Cancers 2021, 13, 4258. [Google Scholar] [CrossRef]
- Xu, D.; Wang, Y.Y.; Yan, X.L.; Li, J.; Wang, K.; Xing, B.C. Development of a Model to Predict Pathologic Response to Chemotherapy in Patients with Colorectal Liver Metastases. J. Gastrointest. Oncol. 2021, 12, 1498–1508. [Google Scholar] [CrossRef]
- Sasaki, K.; Morioka, D.; Conci, S.; Margonis, G.A.; Sawada, Y.; Ruzzenente, A.; Kumamoto, T.; Iacono, C.; Andreatos, N.; Guglielmi, A.; et al. The Tumor Burden Score: A New “Metro-Ticket” Prognostic Tool for Colorectal Liver Metastases Based on Tumor Size and Number of Tumors. Ann. Surg. 2018, 267, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Wada, Y.; Shimada, M.; Morine, Y.; Ikemoto, T.; Saito, Y.; Baba, H.; Mori, M.; Goel, A. A Transcriptomic Signature That Predicts Cancer Recurrence after Hepatectomy in Patients with Colorectal Liver Metastases. Eur. J. Cancer 2022, 163, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.J.; Lim, T.W.; Kang, S.H.; Park, P.J.; Choi, S.B.; Lee, S.I.; Min, B.W.; Kim, W.B. Development and Validation of Novel Scoring System for the Prediction of Disease Recurrence Following Resection of Colorectal Liver Metastasis. Asian J. Surg. 2020, 43, 438–446. [Google Scholar] [CrossRef] [PubMed]
- Dupré, A.; Berhane, S.; Chan, A.W.H.; Rivoire, M.; Chong, C.C.N.; Lai, P.B.S.; Cucchetti, A.; Poston, G.J.; Malik, H.Z.; Johnson, P.J. Multicentre Validation of a Clinical Prognostic Score Integrating the Systemic Inflammatory Response to the Host for Patients Treated with Curative-Intent for Colorectal Liver Metastases: The Liverpool Score. Eur. J. Surg. Oncol. 2019, 45, 999–1004. [Google Scholar] [CrossRef] [PubMed]
- Qi, L.; Liang, J.Y.; Li, Z.W.; Xi, S.Y.; Lai, Y.N.; Gao, F.; Zhang, X.R.; Wang, D.S.; Hu, M.T.; Cao, Y.; et al. Deep Learning-Derived Spatial Organization Features on Histology Images Predicts Prognosis in Colorectal Liver Metastasis Patients after Hepatectomy. iScience 2023, 26, 107702. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Liu, G.J.; Zhang, Z.Y.; Wu, W.; Meng, Y.F.; Wang, S.; Yang, W.; Yan, K. Nomogram Including Chemotherapy Response for Prediction of Intrahepatic Progression-Free Survival in Patients with Colorectal Liver Metastasis through Chemotherapy Followed by Radiofrequency Ablation. Int. J. Hyperth. 2021, 38, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Dasari, B.V.M.; Raptis, D.; Syn, N.; Serrablo, A.; Ramia, J.M.; Laurenzi, A.; Sturesson, C.; Pawlik, T.M.; Siriwardena, A.K.; Lesurtel, M. Development and Validation of a Novel Risk Score to Predict Overall Survival Following Surgical Clearance of Bilobar Colorectal Liver Metastases. BJS Open 2023, 7, zrad085. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Lu, Z.; Yan, J.; Xue, D.; He, X.; Huang, W.; Sun, Q.; Zhao, W.; Li, F. Construction of a Prognostic Signature Associated with Liver Metastases for Prognosis and Immune Response Prediction in Colorectal Cancer. Front. Oncol. 2023, 13, 1234045. [Google Scholar] [CrossRef] [PubMed]
- Amygdalos, I.; Müller-Franzes, G.; Bednarsch, J.; Czigany, Z.; Ulmer, T.F.; Bruners, P.; Kuhl, C.; Neumann, U.P.; Truhn, D.; Lang, S.A. Novel Machine Learning Algorithm Can Identify Patients at Risk of Poor Overall Survival Following Curative Resection for Colorectal Liver Metastases. J. Hepatobiliary Pancreat. Sci. 2023, 30, 602–614. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Li, Q.; Yu, H. Prognosis of Resectable Colorectal Liver Metastases after Surgery Associated with Pathological Features of Primary Tumor. Front. Oncol. 2023, 13, 1181522. [Google Scholar] [CrossRef]
- Wu, Q.; Wang, W.J.; Huang, Y.Q.; Fang, S.Y.; Guan, Y.J. Nomograms for Estimating Survival in Patients with Liver-Only Colorectal Metastases: A Retrospective Study. Int. J. Surg. 2018, 60, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Chen, Q.; Li, C.; Chen, J.; Cai, J.; Li, Y.; Zhao, H. Nomogram Predicting Early Recurrence Defined by the Minimum P Value Approach for Colorectal Liver Metastasis Patients Receiving Colorectal Cancer Resection with Simultaneous Liver Metastasis Resection: Development and Validation. J. Gastrointest. Oncol. 2023, 14, 1279–1292. [Google Scholar] [CrossRef]
- Berardi, G.; Chou, J.; Gonen, M.; Balachandran, V.P.; Drebin, J.; Jarnagin, W.R.; Kingham, T.P.; Soares, K.C.; Wei, A.; D’Angelica, M. A Model to Predict Treatment Failure in Patients Undergoing Upfront Surgery for Resectable Colorectal Liver Metastases. Ann. Surg. Oncol. 2023, 30, 2820–2827. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, K.; Han, Y.; Liang, J.Y.; Li, Y.H.; Xing, B.C. Nomogram Predicted Disease Free Survival for Colorectal Liver Metastasis Patients with Preoperative Chemotherapy Followed by Hepatic Resection. Eur. J. Surg. Oncol. 2019, 45, 2070–2077. [Google Scholar] [CrossRef] [PubMed]
- Welsh, F.K.S.; Tekkis, P.P.; O’Rourke, T.; John, T.G.; Rees, M. Quantification of Risk of a Positive (R1) Resection Margin Following Hepatic Resection for Metastatic Colorectal Cancer: An Aid to Clinical Decision-Making. Surg. Oncol. 2008, 17, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Famularo, S.; Milana, F.; Cimino, M.; Franchi, E.; Giuffrida, M.; Costa, G.; Procopio, F.; Donadon, M.; Torzilli, G. Upfront Surgery versus Neoadjuvant Perioperative Chemotherapy for Resectable Colorectal Liver Metastases: A Machine-Learning Decision Tree to Identify the Best Potential Candidates under a Parenchyma-Sparing Policy. Cancers 2023, 15, 613. [Google Scholar] [CrossRef] [PubMed]
- He, M.; Jia, Z.; Hu, L.; Wu, H. Development and Validation of a Nomogram to Predict Which Patients with Colorectal Cancer Liver Metastases Would Benefit from Primary Tumor Resection. Int. J. Color. Dis. 2023, 38, 144. [Google Scholar] [CrossRef]
- Kattan, M.W.; Gönen, M.; Jarnagin, W.R.; DeMatteo, R.; D’Angelica, M.; Weiser, M.; Blumgart, L.H.; Fong, Y. A Nomogram for Predicting Disease-Specific Survival after Hepatic Resection for Metastatic Colorectal Cancer. Ann. Surg. 2008, 247, 282–287. [Google Scholar] [CrossRef]
- Wensink, G.E.; Bolhuis, K.; Elferink, M.A.G.; Fijneman, R.J.A.; Kranenburg, O.; Borel Rinkes, I.H.M.; Koopman, M.; Swijnenburg, R.J.; Vink, G.R.; Hagendoorn, J.; et al. Predicting Early Extrahepatic Recurrence after Local Treatment of Colorectal Liver Metastases. Br. J. Surg. 2023, 110, 362–371. [Google Scholar] [CrossRef]
- Fendler, W.P.; Ilhan, H.; Paprottka, P.M.; Jakobs, T.F.; Heinemann, V.; Bartenstein, P.; Khalaf, F.; Ezziddin, S.; Hacker, M.; Haug, A.R. Nomogram Including Pretherapeutic Parameters for Prediction of Survival after SIRT of Hepatic Metastases from Colorectal Cancer. Eur. Radiol. 2015, 25, 2693–2700. [Google Scholar] [CrossRef]
- Marfà, S.; Marti, J.; Reyes, A.; Casals, G.; Fernández-Varo, G.; Carvajal, S.; García-Valdecasas, J.C.; Fuster, J.; Jiménez, W. Metastatic Tissue Proteomic Profiling Predicts 5-Year Outcomes in Patients with Colorectal Liver Metastases. Transl. Oncol. 2016, 9, 445–452. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.; Zhou, S.C.; Zhu, Z.X.; Chen, J.H.; Liang, J.W. Survival Nomograms for Simultaneous Resection of Primary and Hepatic Lesions without Neoadjuvant Chemotherapy in Patients with Resectable Colorectal Liver Metastasis. Cancer Innov. 2023, 2, 240–252. [Google Scholar] [CrossRef] [PubMed]
- Endo, Y.; Alaimo, L.; Moazzam, Z.; Woldesenbet, S.; Lima, H.A.; Yang, J.; Munir, M.M.; Shaikh, C.F.; Azap, L.; Katayama, E.; et al. Optimal Policy Tree to Assist in Adjuvant Therapy Decision-Making after Resection of Colorectal Liver Metastases. Surgery 2024, 175, 645–653. [Google Scholar] [CrossRef] [PubMed]
- Rees, M.; Tekkis, P.P.; Welsh, F.K.S.; O’Rourke, T.; John, T.G. Evaluation of Long-Term Survival after Hepatic Resection for Metastatic Colorectal Cancer: A Multifactorial Model of 929 Patients. Ann. Surg. 2008, 247, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Zakaria, S.; Donohue, J.H.; Que, F.G.; Farnell, M.B.; Schleck, C.D.; Ilstrup, D.M.; Nagorney, D.M. Hepatic Resection for Colorectal Metastases: Value for Risk Scoring Systems? Ann. Surg. 2007, 246, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Tan, M.C.B.; Castaldo, E.T.; Gao, F.; Chari, R.S.; Linehan, D.C.; Wright, J.K.; Hawkins, W.G.; Siegel, B.A.; Delbeke, D.; Pinson, C.W.; et al. A Prognostic System Applicable to Patients with Resectable Liver Metastasis from Colorectal Carcinoma Staged by Positron Emission Tomography with [18F]Fluoro-2-Deoxy-D-Glucose: Role of Primary Tumor Variables. J. Am. Coll. Surg. 2008, 206, 857–868. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.R.S.; Chagpar, R.B.; Callender, G.G.; Brown, R.E.; Gilbert, J.E.; Martin, R.C.G.; McMasters, K.M.; Scoggins, C.R. Recurrence Following Hepatectomy for Metastatic Colorectal Cancer: Development of a Model That Predicts Patterns of Recurrence and Survival. Ann. Surg. Oncol. 2012, 19, 139–144. [Google Scholar] [CrossRef] [PubMed]
- Takeda, Y.; Mise, Y.; Matsumura, M.; Hasegawa, K.; Yoshimoto, J.; Imamura, H.; Noro, T.; Yamamoto, J.; Ishizuka, N.; Inoue, Y.; et al. Accuracy of Modern Clinical Risk Score Including RAS Status Changes Based on Whether Patients Received Perioperative Chemotherapy for Colorectal Liver Metastases. World J. Surg. 2021, 45, 2176–2184. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Liu, W.; Yan, X.L.; Li, J.; Xing, B.C. Long-Term Postoperative Survival Prediction in Patients with Colorectal Liver Metastasis. Oncotarget 2017, 8, 79927–79934. [Google Scholar] [CrossRef]
- Spelt, L.; Nilsson, J.; Andersson, R.; Andersson, B. Artificial Neural Networks-A Method for Prediction of Survival Following Liver Resection for Colorectal Cancer Metastases. Eur. J. Surg. Oncol. 2013, 39, 648–654. [Google Scholar] [CrossRef]
- Dindo, D.; Demartines, N.; Clavien, P.-A. Classification of Surgical Complications. Ann. Surg. 2004, 240, 205–213. [Google Scholar] [CrossRef] [PubMed]
- Taylor, A.; Primrose, J.; Langeberg, W.; Kelsh, M.; Mowat, F.; Alexander, D.; Choti, M.; Poston, G.; Kanas, G. Survival after Liver Resection in Metastatic Colorectal Cancer: Review and Meta-Analysis of Prognostic Factors. Clin. Epidemiol. 2012, 4, 283–301. [Google Scholar] [CrossRef] [PubMed]
- Ai, X.; Tao, M.; Wang, H.; Li, J.; Sun, T.; Xiu, D. Analysis of Survival Factors after Hepatic Resection for Colorectal Cancer Liver Metastases: Does the R1 Margin Matter? Front. Surg. 2023, 9, 1020240. [Google Scholar] [CrossRef] [PubMed]
- Dueland, S.; Smedman, T.M.; Syversveen, T.; Grut, H.; Hagness, M.; Line, P.-D. Long-Term Survival, Prognostic Factors, and Selection of Patients with Colorectal Cancer for Liver Transplant. JAMA Surg. 2023, 158, e232932. [Google Scholar] [CrossRef] [PubMed]
- Brudvik, K.W.; Kopetz, S.E.; Li, L.; Conrad, C.; Aloia, T.A.; Vauthey, J.-N. Meta-Analysis of KRAS Mutations and Survival after Resection of Colorectal Liver Metastases. Br. J. Surg. 2015, 102, 1175–1183. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.-Q.; Wang, X.-Y.; Wu, W.-X.; Chen, Y.-X.; Wang, J.; Zhang, X.; Qian, Y.; Du, S.-S.; Sun, J.; Zeng, Z.-C. Molecular Mechanisms Investigation for Liver Metastasis of Colorectal Cancer by Combined Bioinformatic Gene Expression Profile Analysis. Cancer Treat. Res. Commun. 2023, 35, 100694. [Google Scholar] [CrossRef] [PubMed]
- Moons, K.G.M.; Wolff, R.F.; Riley, R.D.; Whiting, P.F.; Westwood, M.; Collins, G.S.; Reitsma, J.B.; Kleijnen, J.; Mallett, S. PROBAST: A Tool to Assess Risk of Bias and Applicability of Prediction Model Studies: Explanation and Elaboration. Ann. Intern. Med. 2019, 170, W1. [Google Scholar] [CrossRef] [PubMed]
- Steyerberg, E.W.; Vickers, A.J.; Cook, N.R.; Gerds, T.; Gonen, M.; Obuchowski, N.; Pencina, M.J.; Kattan, M.W. Assessing the Performance of Prediction Models: A Framework for Traditional and Novel Measures. Epidemiology 2010, 21, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Andaur Navarro, C.L.; Damen, J.A.A.; van Smeden, M.; Takada, T.; Nijman, S.W.J.; Dhiman, P.; Ma, J.; Collins, G.S.; Bajpai, R.; Riley, R.D.; et al. Systematic Review Identifies the Design and Methodological Conduct of Studies on Machine Learning-Based Prediction Models. J. Clin. Epidemiol. 2023, 154, 8–22. [Google Scholar] [CrossRef]
- He, Y.; Ong, Y.; Li, X.; Din, F.V.; Brown, E.; Timofeeva, M.; Wang, Z.; Farrington, S.M.; Campbell, H.; Dunlop, M.G.; et al. Performance of Prediction Models on Survival Outcomes of Colorectal Cancer with Surgical Resection: A Systematic Review and Meta-Analysis. Surg. Oncol. 2019, 29, 196–202. [Google Scholar] [CrossRef]
- Steyerberg, E.W. Clinical Prediction Models; Statistics for Biology and Health; Springer International Publishing: Cham, Switzerland, 2019; ISBN 978-3-030-16398-3. [Google Scholar]
| First Author (Year) | Data Collection | Model Type | Univariate Screening of Predictors | Outcome(s) | Patients (n) | Internal/External Validation | Missing Data | Risk Groups |
|---|---|---|---|---|---|---|---|---|
| Buisman (2022) [20] | retrospective | Cox regression | no | OS | 4112 | Cross-validation | Multiple imputation | yes (4 groups) |
| Bertsimas (2022) [21] | retrospective | RF, OPT | no | OS and intrahepatic recurrence | 761 | IV: Split sample/external validation cohort | Complete case analysis | no |
| Bao (2021) [22] | retrospective | NGS, Cox and LASSO regression | yes | OS | 144 | External validation (gene signature only) | No information | yes (2 groups) |
| Lam (2023) [23] | retrospective | Cox and LASSO regression | yes | OS and RFS | 572 | Split sample | Multiple imputation | no |
| Reijonen (2023) [24] | retrospective | Cox regression | yes | OS and DFS | 816 | Not performed | The final sum of their risk score points was estimated using the mean of the evaluable predictors | yes (3 groups) |
| Margonis (2018) [25] | retrospective | Cox regression | yes | OS | 502 (development), 747 (validation) | External validation | No information | yes (3 groups) |
| Paredes (2020) [26] | retrospective | Mixed-effects logistic regression | no | Recurrence | 703 (development), 703 (validation) | Split sample, bootstrapping | Multiple imputation | yes (3 groups) |
| Fruhling (2021) [27] | retrospective | Cox regression | yes | OS | 1212 | Cross-validation | Multiple imputation | yes (3 groups) |
| Taghavi (2021) [28] | retrospective | RF | no | Development of metachronous metastases | 70 (development), 21 (validation) | Split sample, cross-validation | Single imputation | no |
| Brudvik (2019) [29] | retrospective | Cox regression | no | OS, RFS | 564 (development), 608 (validation) | External validation | Complete case analysis | no |
| Moaven (2023) [30] | retrospective | GBT and LRB in a leave-one-out cross-validation | no | OS, recurrence | 1004 | Cross-validation, bootstrapping | Variables with more than 20% missing data were eliminated from the model | yes (3 groups) |
| Villard (2022) [31] | retrospective | Cox regression | no | OS | 1013 (development), 391 (validation) | External validation | Multiple imputation | yes (4 groups) |
| Chen (2020) [32] | retrospective | Cox regression | no | RFS | 787 (cohort 1), 162 (cohort 2) | IV: Bootstrapping/temporal validation | Complete case analysis | yes (3 groups) |
| Chen (2022) [33] | retrospective | Cox regression | yes | OS | 1095 | Not performed | Multiple imputation | no |
| Dai (2021) [34] | retrospective | Logistic regression | yes | Early recurrence within 6 months | 150 (development), 52 (validation) | Split sample | Complete case analysis | no |
| Liu (2021) [35] | retrospective | Cox regression | yes | OS after recurrence | 867 | Bootstrapping | No information | yes (2 groups) |
| Liang (2021) [36] | retrospective | Cox regression | yes | Post-recurrence survival | 251 (development), 125 (validation) | Split sample, bootstrapping | Complete case analysis | yes (3 groups) |
| Wu (2021) [37] | retrospective | Cox regression | yes | Recurrence, PFS | 229 (development), 128 (validation) | Temporal validation | Complete case analysis | yes (3 groups) |
| Sasaki (2022) [38] | prospective | Cox regression | yes | OS | 1205 (development), 1307 + 1058 (validation) | External validation | No information | yes (3 groups) |
| Huiskens (2019) [39] | retrospective | Logistic regression | yes | 90-day mortality (after stage 2) | 486 | Not performed | Complete case analysis | yes (3 groups) |
| Bai (2022) [40] | retrospective | Cox regression | yes | OS and RFS | 341 (development), 325 (validation) | External validation | Complete case analysis | yes (3 groups) |
| Fang (2022) [41] | retrospective | Cox regression | yes | OS | 237 | Not performed | Complete case analysis | yes (3 groups) |
| Qin (2022) [42] | prospective | Cox regression | yes | ihPFS | 314 | Not performed | No information | yes (3 groups) |
| Kawaguchi (2021) [43] | prospective | Cox regression | yes | OS | 810 (development), 673 (validation) | External validation | Complete case analysis | no |
| Zhang (2023) [44] | retrospective | Cox and LASSO regression | yes | OS | 415 (development), 207 (validation) | IV: Split sample/External validation cohort | No information | yes (2 groups) |
| Chen (2021) [45] | retrospective | Logistic and Cox regression | yes | Postoperative complications, PFS, OS | 380 | Not performed | Complete case analysis | yes (3 groups) |
| Jin (2022) [46] | retrospective | Cox regression | yes | CSS | 881 (development), 169 (validation) | IV: Split sample/External validation cohort | Complete case analysis | yes (2 groups) |
| Zhai (2022) [47] | retrospective | Cox regression | yes | Liver RFS | 147 | Not performed | Complete case analysis | yes (3 groups) |
| Liu (2021) [48] | retrospective | Cox regression | yes | PFS | 532 (development), 237 (validation) | External validation | No information | yes (2 groups) |
| Moro (2020) [49] | retrospective | CART analysis | no | OS | 1123 | Bootstrapping | Multiple imputation | yes (4 groups) |
| Chen (2021) [50] | retrospective | Logistic and Cox regression | yes | Complications, PFS, OS | 169 | Not performed | Complete case analysis | yes (3 groups) |
| Yao (2021) [51] | retrospective | Logistic and Cox regression | yes | Presence of LN metastases, PFS | 241 | Not performed | Complete case analysis | no |
| Kazi (2023) [52] | retrospective | Logistic and Cox regression | yes | Serious complications | 92 | Bootstrapping | No information | yes (4 groups) |
| Meng (2021) [53] | retrospective | Cox regression | yes | OS | 174 (development), 60 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Imai (2016) [54] | prospective | Cox regression | yes | OS | 439 | Not performed | No information | yes (4 groups) |
| Chen (2022) [55] | retrospective | Logistic regression | yes | Early recurrence (<11 months) | 144 (development), 40 (validation) | Another cohort from the same hospital | Complete case analysis | no |
| Cheng (2022) [56] | retrospective | Cox regression | yes | CSS | 1314 (development), 560 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Kulik (2018) [57] | retrospective | Logistic regression | yes | OS | 965 | Not performed | Complete case analysis | no |
| Bai (2021) [58] | retrospective | Cox regression | yes | OS | 490 | Not performed | Complete case analysis | yes (7 and 6 groups) |
| Wang (2021) [59] | retrospective | Cox and LASSO regression | no | OS | 113 (development), 114 (validation), 168 (external validation) | IV: Split sample/external validation cohort | Complete case analysis | yes (2 groups) |
| Xu (2021) [60] | retrospective | Logistic regression | yes | Major pathologic response to chemotherapy | 241 (development), 241 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Sasaki (2018) [61] | retrospective | A priori selection of predictors and interactions | no | OS | 604 (development) | External validation | No information | yes (3 groups) |
| Wada (2022) [62] | retrospective | Cox and LASSO regression | no | Recurrence | 169 (development), 151 (validation) | External validation | No information | yes (2 groups) |
| Kim (2020) [63] | retrospective | Cox regression | yes | Recurrence | 197 (development), 98 (validation) | Split sample | No information | yes (2 groups) |
| Dupre (2019) [64] | prospective | Cox regression | yes | OS | 364 (development), 219 (validation) | External validation | No information | yes (2 groups) |
| Qi (2023) [65] | retrospective | Automated tissue classification and quantification of CRLM SOFs derived from histology images with deep learning and Cox regression | yes | OS | 433 (development), 403 (validation) | External validation | Complete case analysis | yes (SOF scoring system 2 groups, SOF-CRS 3 groups) |
| Wu (2021) [66] | retrospective | Cox regression | yes | PFS | 158 | Not performed | Complete case analysis | yes (3 groups) |
| Dasari (2023) [67] | retrospective | Cox and LASSO regression | yes | OS | 927 (development), 309 (validation) | Split sample | Complete case analysis | yes (5 groups) |
| Liu (2023) [68] | retrospective | Cox and LASSO regression | yes | OS | 295 (development), 295 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Amygdalos (2023) [69] | retrospective | GBT with the Top6 selected predictors | no | OS | 389 (development), 98 (validation) | Split sample | Complete case analysis | yes (2 groups) |
| Chen (2023) [70] | retrospective | Cox regression | yes | OS | 85 | Not performed | Complete case analysis | yes (3 groups) |
| Wu (2018) [71] | retrospective | Cox regression | yes | OS and CSS | 4825 (development), 4790 (validation) | Split sample | Complete case analysis | no |
| Deng (2023) [72] | retrospective | Logistic regression | yes | Early recurrence (<13 months) | 323 (development), 71 (validation) | External validation | Complete case analysis | no |
| Berardi (2023) [73] | prospective | Logistic regression | yes | Treatment failure (recurrence or death within 12 months) | 535 (development), 248 (validation) | Split sample | No information | yes (2 groups) |
| Liu (2019) [74] | retrospective | Cox regression | yes | DFS | 447 (development), 117 (validation) | External validation | No information | yes (3 groups) |
| Welsh (2008) [75] | prospective | Logistic regression | yes | R1 resection margin | 911 | Bootstrapping | Single (median) imputation | no |
| Famularo (2023) [76] | prospective | Survival RF to estimate the best possible treatment, then CART was used to develop a decision tree | no | OS | 448 | Cross-validation | Multiple imputation | yes (7 groups) |
| He (2023) [77] | retrospective | Logistic regression | yes | Benefit from upfront surgery (survival > 15 months) | 572 (development), 242 (validation) | Split sample | Complete case analysis | no |
| Kattan (2008) [78] | retrospective | Cox regression | yes | DSS | 1477 | Bootstrapping | No information | no |
| Wensink (2023) [79] | retrospective | Cox regression | no | Early extrahepatic recurrence (at 6 and 12 months) | 1077 | Bootstrapping and internal–external cross-validation | Multiple imputation | yes (4 groups) |
| Fendler (2015) [80] | retrospective | Cox regression | yes | OS | 100 (development), 25 (validation) | IV: Bootstrapping/external validation cohort | No information | no |
| Marfa (2016) [81] | prospective | CART analysis | no | OS | 57 (development), 28 (validation) | Split sample | No information | yes (2 groups) |
| Jiang (2023) [82] | retrospective | Cox regression | yes | OSS and CSS | 225 (development), 180 (validation) | External validation | Complete case analysis | no |
| Endo (2023) [83] | retrospective | OPT analysis | no | OS and RFS | 679(development), 679 (validation) | Split sample | Multiple imputation | yes (multiple nodes) |
| Rees (2008) [84] | prospective | Cox regression | yes | CSS | 929 | Bootstrapping | Single (median) imputation | yes (5 groups) |
| Zakaria (2007) [85] | retrospective | Cox regression | yes | DFS, recurrence | 662 | Not performed | Complete case analysis | yes (3 groups) |
| Tan (2008) [86] | retrospective | Cox regression | yes | OS | 296 | Not performed | Multiple imputation | yes (3 groups) |
| Hill (2012) [87] | retrospective | Cox regression | yes | Survival following resection for recurrence | 280 | Bootstrapping | No information | yes (3 groups) |
| Takeda (2021) [88] | retrospective | Cox regression | yes | OS | 341 (development), 309 (validation) | External validation | Complete case analysis | yes (4 groups) |
| Wang (2017) [89] | retrospective | Cox regression | yes | OS | 300 | Not performed | No information | yes (4 groups) |
| Spelt (2013) [90] | retrospective | ANN and Cox regression | yes | OS | 241 | Cross-validation | Multiple imputation | no |
| First Author (Year) | Discrimination (AUC) | Calibration Measures | Calibration: Performance | DCA |
|---|---|---|---|---|
| Buisman (2022) [20] | 0.73 | Calibration curve | Good calibration (MSKCC model)/slight underprediction (Erasmus MC model) | NR |
| Bertsimas (2022) [21] | KRAS-variant: 0.76 (both training and testing)/external validation: 0.78/wild-type, training: 0.79/wild-type, testing: 0.57 | NR | NR | NR |
| Bao (2021) [22] | Mean time-dependent: 0.75 | NR | NR | NR |
| Lam (2023) [23] | 0.65 (both for OS and RFS) | NR | NR | NR |
| Reijonen (2023) [24] | 0.62 (OS) | NR | NR | NR |
| Margonis (2018) [25] | 0.625 | NR | NR | NR |
| Paredes (2020) [26] | Model without KRAS: 0.649–0.662 (validation cohort)/model with KRAS: 0.642–0.667 (validation cohort) | Calibration curve | No KRAS: good calibration/KRAS: fair | NR |
| Fruhling (2021) [27] | 1-, 3-, 5-year OS: 0.71, 0.67, 0.67/internal validation: 0.62 | Calibration curve | Excellent calibration in development cohort | NR |
| Taghavi (2021) [28] | Training: 0.64/validation: 0.71 | NR | NR | NR |
| Brudvik (2019) [29] | Development, 5 -y OS: 0.69/development: 5 y RFS: 0.66 | NR | NR | NR |
| Moaven (2023) [30] | GBT, OS: 0.77/GBT, recurrence: 0.63/LRB, OS: 0.64/LRB, recurrence: 0.57 | NR | NR | NR |
| Villard (2022) [31] | Development: 0.74/validation: 0.69/simplified model, development: 0.74, validation: 0.66 | Calibration curve, CITL, slope, HL test | CITL: 0.36, slope: 0.89 (validation), good overall fit | NR |
| Chen (2020) [32] | Development: 0.69 at 24 months and 0.65 at 33 months/internal validation: 0.63/cohort 2: 0.81 at 15 months | Calibration curve | Good calibration | NR |
| Chen (2022) [33] | 1-, 3-, 5-year OS: 0.828, 0.740, 0.700 in the solitary LM group; 0.747, 0.714, 0.753 in the 2–4 LM group; 0.728, 0.741, 0.792 in the ≥ 5 LM group | Calibration curve | Fair calibration only in the 2–4 LM group | NR |
| Dai (2021) [34] | Training: 0.866/validation: 0.792 | Calibration curve | Poor calibration in the validation cohort | Clinical utility with lift curves |
| Liu (2021) [35] | 0.707 | Calibration curve | Fair | NR |
| Liang (2021) [36] | Training: 0.742/validation: 0.773 | Calibration curve | Fair in both training and validation cohorts | NR |
| Wu (2021) [37] | 0.71 (both neoadjuvant and non-neoadjuvant groups) | NR | NR | NR |
| Sasaki (2022) [38] | Development: 0.61 (model as a continuous variable), 0.60 (model as a categorical variable)/Asian external validation cohort: 0.62 (model as a continuous variable), 0.60 (model as a categorical variable)/European external validation cohort: 0.57 (model as a continuous variable), 0.57 (model as a categorical variable) | NR | NR | NR |
| Huiskens (2019) [39] | Stage 1 model: 0.70/Stage 2 model: 0.72 | H-L test | Stage 1 model: chi-square: 3.5, p = 0.63/Stage 2 model: chi-square: 7.8, p = 0.18 | NR |
| Bai (2022) [40] | 5-year OS, development: 0.721/5-year OS, validation: 0.665/2-year RFS, development: 0.728/2-year RFS, validation: 0.640 | NR | NR | NR |
| Fang (2022) [41] | 0.715 | NR | NR | NR |
| Qin (2022) [42] | 1-, 2-, 3-year ihPFS: 0.695, 0.764, 0.782 | Calibration curve | Fair calibration | yes |
| Kawaguchi (2021) [43] | RAS mutant, development: 0.629/RAS mutant, validation: 0.644/wild type, development: 0.625/wild type, validation: 0.624 | Calibration curve | Fair calibration (development and validation cohort) | NR |
| Zhang (2023) [44] | Risk score: 1, 3, 5 years, training: 0.624, 0.630, 0.662/testing: 0.610, 0.646, 0.688/validation: 0.612, 0.622, 0.652/full model: 0.783, corrected: 0.772 | Calibration curve | Fair calibration | yes |
| Chen (2021) [45] | Complications: 0.658/PFS: 0.676/OS: 0.700 | Calibration curve, HL test | Complications: fair, HL test: chi-square 3.99, p = 0.91/PFS: fair/OS: good | yes (for complications) |
| Jin (2022) [46] | Training: 0.826/validation: 0.820/external validation: 0.763 | Calibration curve | Poor calibration (internal validation), fair (external validation) | yes |
| Zhai (2022) [47] | 0.659 | NR | NR | NR |
| Liu (2021) [48] | Development: 0.696/validation: 0.682 | Calibration curve | Development: fair/validation: poor | NR |
| Moro (2020) [49] | AIC: wtKRAS: 1356, mtKRAS: 1356 | Brier scores after bootstrapping | Brier: 0.1741 (wtKRAS), 0.1793 (mtKRAS) | NR |
| Chen (2021) [50] | Complications: 0.750/PFS: 0.663/OS: 0.684 | Calibration curves and HL test | Complications: fair/PFS: fair/OS: fair | yes |
| Yao (2021) [51] | Presence of LN metastases: 0.655/PFS: 0.656 | Calibration curves and HL test | Presence of LN metastases: fair/PFS: fair | NR |
| Kazi (2023) [52] | 0.692 | Calibration table | Good calibration (small group numbers) | NR |
| Meng (2021) [53] | 1 yr OS, training: 0.788/3 yr OS, validation: 0.702/3 yr OS, training: 0.752/3 yr OS, validation: 0.848 | Calibration curve | 1 yr OS: fair, 3 yr OS: good (small numbers) | NR |
| Imai (2016) [54] | 0.66 | Calibration curve | 3 and 5 yr OS: fair | NR |
| Chen (2022) [55] | Development: 0.754/validation: 0.882 | Calibration curve, HL test | HL: chi-square: 1.36, p = 0.998, calibration curve: good calibration in development and validation cohorts | yes |
| Cheng (2022) [56] | Training: 0.709/validation: 0.735 | Calibration curve | CSS: fair in training and validation/OS: fair in training and validation | NR |
| Kulik (2018) [57] | Preoperative: 0.716/preop- and perioperative: 0.761 | NR | NR | NR |
| Bai (2021) [58] | LDH-CRS: 0.674/mCRS: 0.681 | NR | NR | NR |
| Wang (2021) [59] | 1st score, 1, 3, 5 yr OS, training: 0.84, 0.73, 0.70/1, 3, 5 yr OS, int. validation: 0.75, 0.70, 0.70/1, 3, 5 yr OS, ext. validation: 0.77, 0.78, 0.72/2nd score, 3 yr OS, training: 0.76/5 yr OS, training: 0.75/3 yr OS, validation: 0.74/5 yr OS, validation: 0.66 | Calibration curve | Merged score: fair | NR |
| Xu (2021) [60] | Training: 0.746/validation: 0.764 | Calibration curve, slope, intercept | Validation: fair, calibration slope 1.09, intercept: −0.006 | NR |
| Sasaki (2018) [61] | 0.669 | NR | NR | NR |
| Wada (2022) [62] | Training: 0.83/validation: 0.81/mixed model: 0.85 | NR | NR | NR |
| Kim (2020) [63] | Training: 0.824/validation: 0.898 | H-L test | p = 0.831 | NR |
| Dupre (2019) [64] | Preoperative: 0.619/postoperative: 0.637 | NR | NR | NR |
| Qi (2023) [65] | SOF, 5 yr: 0.63/SOF, 8 yr: 0.74/combined, 5 yr: 0.69/combined, 8 yr: 0.79 | Calibration curve | Fair calibration | NR |
| Wu (2021) [66] | 0.705 | Calibration curve | Fair calibration | NR |
| Dasari (2023) [67] | Development, 1, 2, 3, 5 yr: 0.756, 0.745, 0.706, 0.698/validation, 1, 2, 3, 5 yr: 0.679, 0.659, 0.678, 0.732 | NR | NR | NR |
| Liu (2023) [68] | DEG risk score, development, 5 yr: 0.74/validation, 5 yr: 0.64/mixed model: 0.69 | Calibration curve | Good calibration | yes |
| Amygdalos (2023) [69] | 0.70 | NR | NR | NR |
| Chen (2023) [70] | 0.732 | Calibration curve | Fair | NR |
| Wu (2018) [71] | OS, 1 and 3 yr: 0.621,0.661/CSS, 1 and 3 yr: 0.621,0.660 | Calibration curve | Fair in training and validation, both for OS and CSS | NR |
| Deng (2023) [72] | Training: 0.720/validation: 0.740 | Calibration curve, HL test | Training: fair calibration, chi-square 4.97, p = 0.7612/validation: poor calibration, chi: 3.89, p = 0.8671 | yes (utility in a narrow range of thresholds) |
| Berardi (2023) [73] | Training: 0.68/validation: 0.60 | Calibration curve | Fair | NR |
| Liu (2019) [74] | Development: 0.675/validation: 0.77 | Calibration curve | Development: 1 yr poor, 3 yr good/validation: 1 yr poor, 3 yr poor, 5 yr poor | NR |
| Welsh (2008) [75] | 0.781 | Calibration plot, HL test | Validation: chi-square = 6.03, p = 0.196 | NR |
| Famularo (2023) [76] | RF model: 0.66 | NR | NR | NR |
| He (2023) [77] | Training: 0.801/validation: 0.739 | Calibration curve, slope, intercept | Development: good calibration/validation: fair calibration, slope: 1.0, intercept 0.0 | yes |
| Kattan (2008) [78] | Optimism-corrected: 0.612 | Calibration curve | Fair | NR |
| Wensink (2023) [79] | Optimism-corrected, 6 m: 0.643, 12 m: 0.641 | Calibration curve, slope | Fair at 6 and 12 months, optimism-corrected slope: 0.86 | yes |
| Fendler (2015) [80] | Training 0.81/validation: 0.83 | NR | NR | NR |
| Marfa (2016) [81] | Training: 0.903 | NR | NR | NR |
| Jiang (2023) [82] | CSS, training, 1 and 3 yr: 0.77, 0.70/validation, 1 and 3 yr: 0.72, 0.68/OS, training, 1 and 3 yr 0.78, 0.70/validation, 1 and 3 yr: 0.74, 0.70 | Calibration curve | Training: fair, validation poor | yes (superior to AJCC stage) |
| Endo (2023) [83] | OS-OPT, training: 0.68/testing: 0.69/RFS-OPT, training: 0.68/testing: 0.69 | NR | NR | NR |
| Rees (2008) [84] | Preoperative: 0.781/postoperative: 0.805 | H-L test | Preoperative: chi-square: 8.125; p = 0.087/postoperative: chi-square: 7.453, p = 0.114 | NR |
| Zakaria (2007) [85] | DSS: 0.61/recurrence: 0.58 | NR | NR | NR |
| Tan (2008) [86] | 0.59 | NR | NR | NR |
| Hill (2012) [87] | Apparent: 0.69/optimism-corrected: 0.67 | NR | NR | NR |
| Takeda (2021) [88] | Development: 0.65 | NR | NR | NR |
| Wang (2017) [89] | 0.642 | NR | NR | NR |
| Spelt (2013) [90] | ANN: 0.72/Cox model: 0.66 | NR | NR | NR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kokkinakis, S.; Ziogas, I.A.; Llaque Salazar, J.D.; Moris, D.P.; Tsoulfas, G. Clinical Prediction Models for Prognosis of Colorectal Liver Metastases: A Comprehensive Review of Regression-Based and Machine Learning Models. Cancers 2024, 16, 1645. https://doi.org/10.3390/cancers16091645
Kokkinakis S, Ziogas IA, Llaque Salazar JD, Moris DP, Tsoulfas G. Clinical Prediction Models for Prognosis of Colorectal Liver Metastases: A Comprehensive Review of Regression-Based and Machine Learning Models. Cancers. 2024; 16(9):1645. https://doi.org/10.3390/cancers16091645
Chicago/Turabian StyleKokkinakis, Stamatios, Ioannis A. Ziogas, Jose D. Llaque Salazar, Dimitrios P. Moris, and Georgios Tsoulfas. 2024. "Clinical Prediction Models for Prognosis of Colorectal Liver Metastases: A Comprehensive Review of Regression-Based and Machine Learning Models" Cancers 16, no. 9: 1645. https://doi.org/10.3390/cancers16091645
APA StyleKokkinakis, S., Ziogas, I. A., Llaque Salazar, J. D., Moris, D. P., & Tsoulfas, G. (2024). Clinical Prediction Models for Prognosis of Colorectal Liver Metastases: A Comprehensive Review of Regression-Based and Machine Learning Models. Cancers, 16(9), 1645. https://doi.org/10.3390/cancers16091645









