Diffuse Gliomas with FGFR3::TACC3 Fusion: Morphological and Molecular Features and Classification Challenges
Abstract
Simple Summary
Abstract
1. Introduction
2. FGFR3::TACC3 Fusion
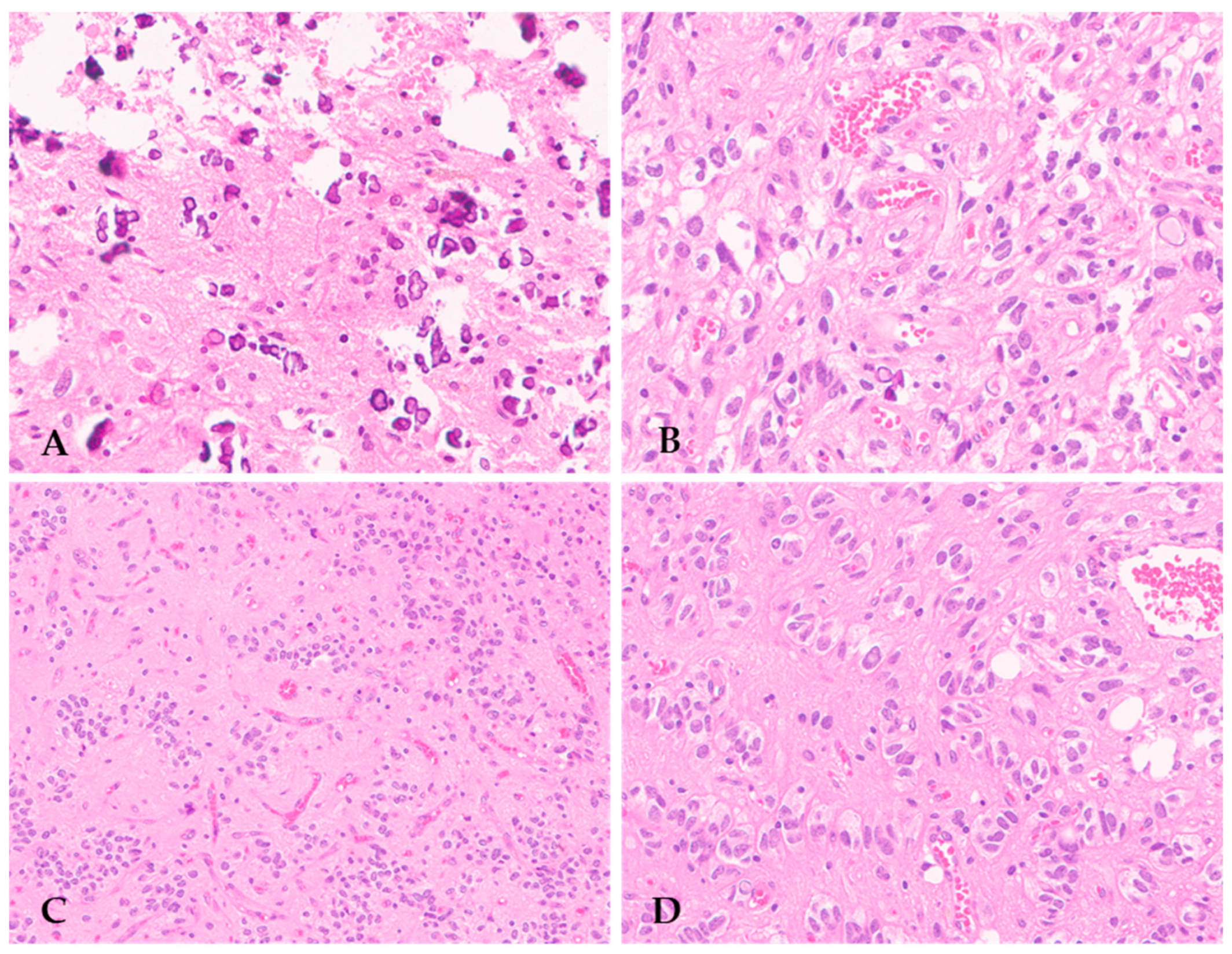
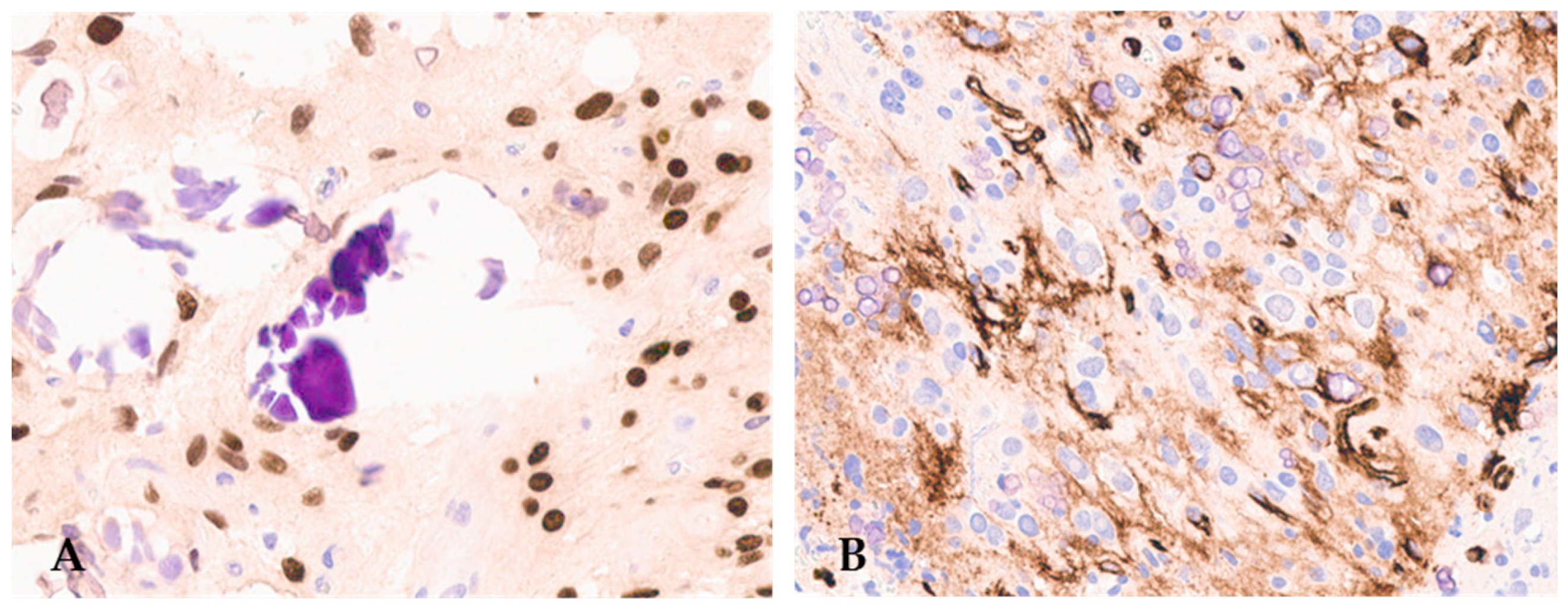
3. Histopathological Features of Diffuse Gliomas with FGFR3::TACC3 Fusion
4. Molecular Features of Diffuse Gliomas with High-Grade Histology and FGFR3::TACC3 Fusion
5. Molecular Features of Diffuse Gliomas with Low-Grade Histology and FGFR3::TACC3 Fusion
6. Histological Differential Diagnosis of F3T3 Gliomas
7. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Ostrom, Q.T.; Price, M.; Neff, C.; Cioffi, G.; Waite, K.A.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2016–2020. Neuro Oncol. 2023, 25, iv1–iv99. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A summary. Neuro Oncol. 2021, 23, 1231–1251. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Giannini, C.; Perry, A.; Reifenberger, G.; Ng, H.K.; Soffietti, R.; Suvà, M.; Sarkar, C.; Wick, W.; Aldape, K.; et al. Glioblastoma, IDH-wildtype. In Central Nervous System Tumours, 5th ed.; Brat, D.J., von Deimling, A., Eds.; IARC Press: Lyon, France, 2021. [Google Scholar]
- Whitfield, B.T.; Huse, J.T. Classification of adult-type diffuse gliomas: Impact of the World Health Organization 2021 update. Brain Pathol. 2022, 32, e13062. [Google Scholar] [CrossRef] [PubMed]
- Harmer, N.J.; Ilag, L.L.; Mulloy, B.; Pellegrini, L.; Robinson, C.V.; Blundell, T.L. Towards a resolution of the stoichiometry of the fibroblast growth factor (FGF)-FGF receptor-heparin complex. J. Mol. Biol. 2004, 339, 821–834. [Google Scholar] [CrossRef] [PubMed]
- Latko, M.; Czyrek, A.; Porębska, N.; Kucińska, M.; Otlewski, J.; Zakrzewska, M.; Opaliński, Ł. Cross-Talk between Fibroblast Growth Factor Receptors and Other Cell Surface Proteins. Cells 2019, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Costa, R.; Carneiro, B.A.; Taxter, T.; Tavora, F.A.; Kalyan, A.; Pai, S.A.; Chae, Y.K.; Giles, F.J. FGFR3-TACC3 fusion in solid tumors: Mini review. Oncotarget 2016, 7, 55924–55938. [Google Scholar] [CrossRef] [PubMed]
- Helsten, T.; Elkin, S.; Arthur, E.; Tomson, B.N.; Carter, J.; Kurzrock, R. The FGFR Landscape in Cancer: Analysis of 4,853 Tumors by Next-Generation Sequencing. Clin. Cancer Res. 2016, 22, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Gu, W.; Yang, J.; Wang, Y.; Xu, J.; Wang, X.; Du, F.; Hu, X.; Guo, H.; Song, C.; Tao, R.; et al. Comprehensive identification of FGFR1-4 alterations in 5 557 Chinese patients with solid tumors by next-generation sequencing. Am. J. Cancer Res. 2021, 11, 3893–3906. [Google Scholar] [PubMed]
- Gergely, F.; Kidd, D.; Jeffers, K.; Wakefield, J.G.; Raff, J.W. D-TACC: A novel centrosomal protein required for normal spindle function in the early Drosophila embryo. EMBO J. 2000, 19, 241–252. [Google Scholar] [CrossRef]
- Mata, D.A.; Benhamida, J.K.; Lin, A.L.; Vanderbilt, C.M.; Yang, S.R.; Villafania, L.B.; Ferguson, D.C.; Jonsson, P.; Miller, A.M.; Tabar, V.; et al. Genetic and epigenetic landscape of IDH-wildtype glioblastomas with FGFR3-TACC3 fusions. Acta Neuropathol. Commun. 2020, 8, 186. [Google Scholar] [CrossRef]
- Métais, A.; Tauziède-Espariat, A.; Garcia, J.; Appay, R.; Uro-Coste, E.; Meyronet, D.; Maurage, C.A.; Vandenbos, F.; Rigau, V.; Chiforeanu, D.C.; et al. Clinico-pathological and epigenetic heterogeneity of diffuse gliomas with FGFR3::TACC3 fusion. Acta Neuropathol. Commun. 2023, 11, 14. [Google Scholar] [CrossRef] [PubMed]
- Picca, A.; Sansone, G.; Santonocito, O.S.; Mazzanti, C.M.; Sanson, M.; Di Stefano, A.L. Diffuse Gliomas with FGFR3-TACC3 Fusions: Oncogenic Mechanisms, Hallmarks, and Therapeutic Perspectives. Cancers 2023, 15, 5555. [Google Scholar] [CrossRef] [PubMed]
- Singh, D.; Chan, J.M.; Zoppoli, P.; Niola, F.; Sullivan, R.; Castano, A.; Liu, E.M.; Reichel, J.; Porrati, P.; Pellegatta, S.; et al. Transforming fusions of FGFR and TACC genes in human glioblastoma. Science 2012, 337, 1231–1235. [Google Scholar] [CrossRef] [PubMed]
- Parker, B.C.; Engels, M.; Annala, M.; Zhang, W. Emergence of FGFR family gene fusions as therapeutic targets in a wide spectrum of solid tumours. J. Pathol. 2014, 232, 4–15. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.L.; Picca, A.; Saragoussi, E.; Bielle, F.; Ducray, F.; Villa, C.; Eoli, M.; Paterra, R.; Bellu, L.; Mathon, B.; et al. Clinical, molecular, and radiomic profile of gliomas with FGFR3-TACC3 fusions. Neuro Oncol. 2020, 22, 1614–1624. [Google Scholar] [CrossRef]
- Lasorella, A.; Sanson, M.; Iavarone, A. FGFR-TACC gene fusions in human glioma. Neuro Oncol. 2017, 19, 475–483. [Google Scholar] [CrossRef] [PubMed]
- Frattini, V.; Pagnotta, S.M.; Tala; Fan, J.J.; Russo, M.V.; Lee, S.B.; Garofano, L.; Zhang, J.; Shi, P.; Lewis, G.; et al. A metabolic function of FGFR3-TACC3 gene fusions in cancer. Nature 2018, 553, 222–227. [Google Scholar] [CrossRef]
- Kurobe, M.; Kojima, T.; Nishimura, K.; Kandori, S.; Kawahara, T.; Yoshino, T.; Ueno, S.; Iizumi, Y.; Mitsuzuka, K.; Arai, Y.; et al. Development of RNA-FISH Assay for Detection of Oncogenic FGFR3-TACC3 Fusion Genes in FFPE Samples. PLoS ONE 2016, 11, e0165109. [Google Scholar] [CrossRef]
- Bielle, F.; Di Stefano, A.L.; Meyronet, D.; Picca, A.; Villa, C.; Bernier, M.; Schmitt, Y.; Giry, M.; Rousseau, A.; Figarella-Branger, D.; et al. Diffuse gliomas with FGFR3-TACC3 fusion have characteristic histopathological and molecular features. Brain Pathol. 2018, 28, 674–683. [Google Scholar] [CrossRef]
- Parker, B.C.; Annala, M.J.; Cogdell, D.E.; Granberg, K.J.; Sun, Y.; Ji, P.; Li, X.; Gumin, J.; Zheng, H.; Hu, L.; et al. The tumorigenic FGFR3-TACC3 gene fusion escapes miR-99a regulation in glioblastoma. J. Clin. Investig. 2013, 123, 855–865. [Google Scholar] [CrossRef]
- Priesterbach-Ackley, L.P.; van Kuik, J.; Tops, B.B.J.; Lasorella, A.; Iavarone, A.; van Hecke, W.; Robe, P.A.; Wesseling, P.; de Leng, W.W.J. RT-PCR assay to detect FGFR3::TACC3 fusions in formalin-fixed, paraffin-embedded glioblastoma samples. Neurooncol. Pract. 2024, 11, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Schittenhelm, J.; Ziegler, L.; Sperveslage, J.; Mittelbronn, M.; Capper, D.; Burghardt, I.; Poso, A.; Biskup, S.; Skardelly, M.; Tabatabai, G. FGFR3 overexpression is a useful detection tool for FGFR3 fusions and sequence variations in glioma. Neurooncol. Pract. 2021, 8, 209–221. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, A.L.; Fucci, A.; Frattini, V.; Labussiere, M.; Mokhtari, K.; Zoppoli, P.; Marie, Y.; Bruno, A.; Boisselier, B.; Giry, M.; et al. Detection, Characterization, and Inhibition of FGFR-TACC Fusions in IDH Wild-type Glioma. Clin. Cancer Res. 2015, 21, 3307–3317. [Google Scholar] [CrossRef]
- Broggi, G.; Piombino, E.; Altieri, R.; Romano, C.; Certo, F.; Barbagallo, G.M.V.; Vigneri, P.; Condorelli, D.; Colarossi, L.; Colarossi, C.; et al. Glioblastoma, IDH-Wild Type With FGFR3-TACC3 Fusion: When Morphology May Reliably Predict the Molecular Profile of a Tumor. A Case Report and Literature Review. Front. Neurol. 2022, 13, 823015. [Google Scholar] [CrossRef] [PubMed]
- Gilani, A.; Davies, K.D.; Kleinschmidt-DeMasters, B.K. Can adult IDH-wildtype glioblastomas with FGFR3:TACC3 fusions be reliably predicted by histological features? Clin. Neuropathol. 2021, 40, 165–167. [Google Scholar] [CrossRef] [PubMed]
- Martelli, F.M.; Marastoni, E.; Barresi, V. A diffuse glioma with oligodendroglial-like cells and extensive calcifications. Brain Pathol. 2024, 34, e13187. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Lopes Abath Neto, O.; Bale, T.A.; Benhamida, J.; Mata, D.; Turakulov, R.; Abdullaev, Z.; Marker, D.; Ketchum, C.; Chung, H.J.; et al. DNA methylation analysis of glioblastomas harboring FGFR3-TACC3 fusions identifies a methylation subclass with better patient survival. Acta Neuropathol. 2022, 144, 155–157. [Google Scholar] [CrossRef] [PubMed]
- Capper, D.; Stichel, D.; Sahm, F.; Jones, D.T.W.; Schrimpf, D.; Sill, M.; Schmid, S.; Hovestadt, V.; Reuss, D.E.; Koelsche, C.; et al. Practical implementation of DNA methylation and copy-number-based CNS tumor diagnostics: The Heidelberg experience. Acta Neuropathol. 2018, 136, 181–210. [Google Scholar] [CrossRef] [PubMed]
- Bale, T.A.; Sait, S.F.; Benhamida, J.; Ptashkin, R.; Haque, S.; Villafania, L.; Sill, M.; Sadowska, J.; Akhtar, R.B.; Liechty, B.; et al. Malignant transformation of a polymorphous low grade neuroepithelial tumor of the young (PLNTY). Acta Neuropathol. 2021, 141, 123–125. [Google Scholar] [CrossRef]
- Chen, Y.; Tian, T.; Guo, X.; Zhang, F.; Fan, M.; Jin, H.; Liu, D. Polymorphous low-grade neuroepithelial tumor of the young: Case report and review focus on the radiological features and genetic alterations. BMC Neurol. 2020, 20, 123. [Google Scholar] [CrossRef]
- Granberg, K.J.; Annala, M.; Lehtinen, B.; Kesseli, J.; Haapasalo, J.; Ruusuvuori, P.; Yli-Harja, O.; Visakorpi, T.; Haapasalo, H.; Nykter, M.; et al. Strong FGFR3 staining is a marker for FGFR3 fusions in diffuse gliomas. Neuro Oncol. 2017, 19, 1206–1216. [Google Scholar] [CrossRef] [PubMed]
- Huse, J.T.; Snuderl, M.; Jones, D.T.; Brathwaite, C.D.; Altman, N.; Lavi, E.; Saffery, R.; Sexton-Oates, A.; Blumcke, I.; Capper, D.; et al. Polymorphous low-grade neuroepithelial tumor of the young (PLNTY): An epileptogenic neoplasm with oligodendroglioma-like components, aberrant CD34 expression, and genetic alterations involving the MAP kinase pathway. Acta Neuropathol. 2017, 133, 417–429. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.; Severson, E.; Gay, L.; Vergilio, J.A.; Elvin, J.; Suh, J.; Daniel, S.; Covert, M.; Frampton, G.M.; Hsu, S.; et al. Comprehensive Genomic Profiling of 282 Pediatric Low- and High-Grade Gliomas Reveals Genomic Drivers, Tumor Mutational Burden, and Hypermutation Signatures. Oncologist 2017, 22, 1478–1490. [Google Scholar] [CrossRef] [PubMed]
- Lazo De La Vega, L.; Comeau, H.; Sallan, S.; Al-Ibraheemi, A.; Gupta, H.; Li, Y.Y.; Tsai, H.K.; Kang, W.; Ward, A.; Church, A.J.; et al. Rare FGFR Oncogenic Alterations in Sequenced Pediatric Solid and Brain Tumors Suggest FGFR Is a Relevant Molecular Target in Childhood Cancer. JCO Precis. Oncol. 2022, 6, e2200390. [Google Scholar] [CrossRef] [PubMed]
- Qaddoumi, I.; Orisme, W.; Wen, J.; Santiago, T.; Gupta, K.; Dalton, J.D.; Tang, B.; Haupfear, K.; Punchihewa, C.; Easton, J.; et al. Genetic alterations in uncommon low-grade neuroepithelial tumors: BRAF, FGFR1, and MYB mutations occur at high frequency and align with morphology. Acta Neuropathol. 2016, 131, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Riva, G.; Cima, L.; Villanova, M.; Ghimenton, C.; Sina, S.; Riccioni, L.; Munari, G.; Fassan, M.; Giangaspero, F.; Eccher, A. Low-grade neuroepithelial tumor: Unusual presentation in an adult without history of seizures. Neuropathology 2018, 38, 557–560. [Google Scholar] [CrossRef] [PubMed]
- McDonald, M.F.; Athukuri, P.; Anand, A.; Gopakumar, S.; Jalali, A.; Patel, A.J.; Rao, G.; Goodman, J.C.; Lu, H.C.; Mandel, J.J. Varied histomorphology and clinical outcomes of FGFR3-TACC3 fusion gliomas. Neurosurg. Focus 2022, 53, E16. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, S.D.; Zhou, S.; Huse, J.T.; de Groot, J.F.; Xiu, J.; Subramaniam, D.S.; Mehta, S.; Gatalica, Z.; Swensen, J.; Sanai, N.; et al. Targetable Gene Fusions Associate With the IDH Wild-Type Astrocytic Lineage in Adult Gliomas. J. Neuropathol. Exp. Neurol. 2018, 77, 437–442. [Google Scholar] [CrossRef]
- Solomon, D.A.; Varlet, P.; Blumcke, I.; Capper, D.; Gupta, H. Ganglioglioma. In WHO Classification of Tumours, Central Nervous System Tumours, 5th ed.; von Deimling, A., Figarella-Branger, D., Eds.; IARC Press: Lyon, France, 2021. [Google Scholar]
- Rosenblum, M.K.; Ellison, D.W.; Huse, J.T.; Blumcke, I. Polymorphous low-grade neuroepithelial tumour of the young. In WHO Classification of Tumours, Central Nervous System Tumours, 5th ed.; Reifenberger, G., Perry, A., Eds.; IARC Press: Lyon, France, 2021. [Google Scholar]
- Golub, D.; Lynch, D.G.; Pan, P.C.; Liechty, B.; Slocum, C.; Bale, T.; Pisapia, D.J.; Juthani, R. Polymorphous low-grade neuroepithelial tumor of the young with FGFR3-TACC3 fusion mimicking high-grade glioma: Case report and series of high-grade correlates. Front. Oncol. 2023, 13, 1307591. [Google Scholar] [CrossRef]

| Case [Refs.] | Age | Sex | +7/−10 | pTERT | EGFR Ampl | MC | t-SNE Cluster | Resection | Adjuvant Therapy | FU | Length (Mo) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 [30] | 15 | F | no | NA | no | no match | NA | gross total | no | alive | 34 |
| 2 [20] | 74 | F | yes | mut | no | NA | NA | biopsy | NA | NA | NA |
| 3 [20] | 59 | F | yes | mut | no | NA | NA | gross total | NA | NA | NA |
| 4 [20] | 72 | F | no | wt | no | NA | NA | gross total | NA | NA | NA |
| 5 [31] | 14 | F | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 6 [32] | NA | M | no | wt | no | NA | NA | NA | NA | NA | NA |
| 7 [32] | NA | F | no | wt | no | NA | NA | NA | NA | NA | NA |
| 8 [33] | 17 | F | no | wt | no | NA | NA | gross total | NA | alive | 89 |
| 9 [34] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 10 [34] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 11 [34] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 12 [35] | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 13 [27] | 53 | F | yes | mut | no | NA | NA | partial resection | yes | alive | 25 |
| 14 [38] | 66 | F | no | mut | no | NA | NA | biopsy | yes | dead | 12 |
| 15 [38] | 28 | F | no | wt | no | NA | NA | gross total | no | alive | 14 |
| 16 [12] | 65 | NA | yes | mut | no | GBM IDH-wt (v 11b4); | GB | NA | NA | NA | NA |
| no match (v 12.5) | |||||||||||
| 17 [12] | 56 | NA | no | mut | no | no match | GB | NA | NA | NA | NA |
| (v 11b4; v 12.5) | |||||||||||
| 18 [12] | 77 | NA | yes | mut | no | no match | GB | NA | NA | NA | NA |
| (v 11b4; v 12.5) | |||||||||||
| 19 [12] | 45 | NA | no | mut | no | no match | GB | NA | yes | dead | 11 |
| (v 11b4; v 12.5) | |||||||||||
| 20 [12] | 24 | NA | yes | mut | no | no match | GB | NA | NA | NA | NA |
| (v11b4; v 12.5) | |||||||||||
| 21 [12] | 4 | NA | no | wt | no | no match | GG | NA | NA | NA | NA |
| (v 11b4; v 12.5) | |||||||||||
| 22 [12] | 12 | NA | no | wt | no | GG (v 11b4); | GG | NA | NA | NA | NA |
| GG (v 12.5) | |||||||||||
| 23 [12] | 72 | NA | no | mut | no | no match (v 11b4); | GG | NA | yes | dead | 66 |
| GG (v 12.5) | |||||||||||
| 24 [12] | 10 | NA | no | wt | no | no match (v 11b4); | GG | NA | NA | NA | NA |
| GG (v 12.5) | |||||||||||
| 25 [12] | 38 | NA | yes | mut | no | no match (v 11b4); | GG | NA | NA | NA | NA |
| GG (v 12.5) | |||||||||||
| 26 [12] | 13 | NA | no | wt | no | no match | DNET | NA | NA | NA | NA |
| (v11b4; v 12.5) | |||||||||||
| 27 [12] | 29 | NA | no | wt | no | no match | DNET | NA | yes | alive | 37 |
| (v 11b4; v 12.5) | |||||||||||
| 28 [12] | 6 | NA | no | wt | no | LGG, DNET (v 11b4) DNET (v 12.5) | DNET | NA | NA | NA | NA |
| 29 [12] | 1 | NA | no | wt | no | no match | DNET | NA | NA | NA | NA |
| 30 | 68 | F | yes | mut | no | GBM (v 11b4) | NA | gross total | yes | alive | 14 |
| no match (v 12.5) | |||||||||||
| 31 [36] | 6 | M | NA | NA | NA | NA | NA | NA | NA | NA | NA |
| 32 [37] | 57 | M | NA | NA | NA | NA | NA | NA | NA | alive | 12 |
| 33 [23] | 16 | F | NA | NA | no | NA | NA | NA | NA | NA | NA |
| 34 [23] | 54 | M | NA | NA | no | NA | NA | NA | NA | NA | NA |
| 35 [28] | 71 | F | yes | mut | no | no match (v 11b4) | NA | NA | yes | alive | 69 |
| 36 [28] | 74 | M | yes | wt | no | GBM (v 11b4) | NA | NA | yes | dead | 20 |
| no match (v 12.5) | |||||||||||
| 37 [28] | 68 | F | yes | mut | yes | no match (v 11b4) | NA | NA | yes | dead | 23 |
| 38 [28] | 58 | F | yes | mut | no | no match (v 11b4) | NA | NA | yes | dead | 39 |
| 39 [28] | 70 | M | yes | mut | no | no match (v 11b4) | NA | NA | yes | dead | 10 |
| 40 [28] | 63 | M | yes | wt | no | no match (v 11b4) | NA | NA | yes | alive | 70 |
| 41 [28] | 59 | M | no | mut | no | GBM (v 11b4) | NA | NA | yes | alive | 34 |
| 42 [28] | 72 | M | yes | mut | no | GBM (v 11b4) | NA | NA | yes | alive | 65 |
| 43 [28] | 64 | M | yes | mut | no | GBM (v 11b4) | NA | NA | NA | NA | NA |
| 44 [28] | 70 | F | no | mut | no | GBM (v 11b4) | NA | NA | NA | dead | 1 |
| 45 [28] | 47 | F | yes | mut | yes | GBM (v 11b4) | NA | NA | NA | dead | 1 |
| 46 [28] | 73 | M | yes | wt | no | GBM (v 11b4) | NA | NA | NA | dead | 14 |
| 47 [28] | NA | F | no | mut | no | no match (v 11b4) | NA | NA | NA | dead | NA |
| 48 [28] | 60 | F | yes | mut | no | no match (v 11b4) | NA | NA | NA | NA | NA |
| 49 [28] | 62 | F | yes | mut | no | GBM (v 11b4) | NA | NA | NA | dead | 37 |
| 50 [28] | 58 | M | yes | mut | no | no match (v 11b4) | NA | NA | NA | dead | 14 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marastoni, E.; Mulone, D.; Barresi, V. Diffuse Gliomas with FGFR3::TACC3 Fusion: Morphological and Molecular Features and Classification Challenges. Cancers 2024, 16, 1644. https://doi.org/10.3390/cancers16091644
Marastoni E, Mulone D, Barresi V. Diffuse Gliomas with FGFR3::TACC3 Fusion: Morphological and Molecular Features and Classification Challenges. Cancers. 2024; 16(9):1644. https://doi.org/10.3390/cancers16091644
Chicago/Turabian StyleMarastoni, Elena, Davide Mulone, and Valeria Barresi. 2024. "Diffuse Gliomas with FGFR3::TACC3 Fusion: Morphological and Molecular Features and Classification Challenges" Cancers 16, no. 9: 1644. https://doi.org/10.3390/cancers16091644
APA StyleMarastoni, E., Mulone, D., & Barresi, V. (2024). Diffuse Gliomas with FGFR3::TACC3 Fusion: Morphological and Molecular Features and Classification Challenges. Cancers, 16(9), 1644. https://doi.org/10.3390/cancers16091644






