Breast Cancer Polygenic Risk Score Validation and Effects of Variable Imputation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Subjects
2.2. Phenotypes
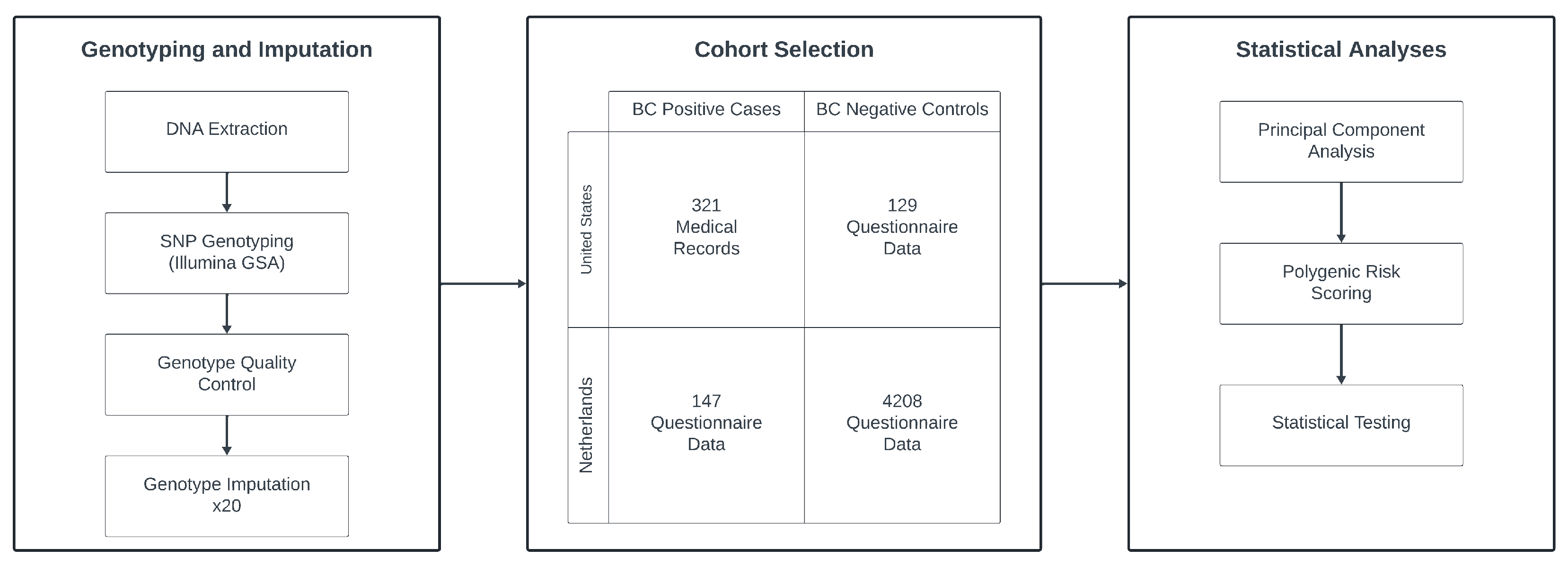
2.3. Genotyping and Imputation
2.4. Statistical Analyses
2.4.1. Genetic Principal Component Analysis
2.4.2. Polygenic Risk Scoring
2.4.3. Statistical Testing
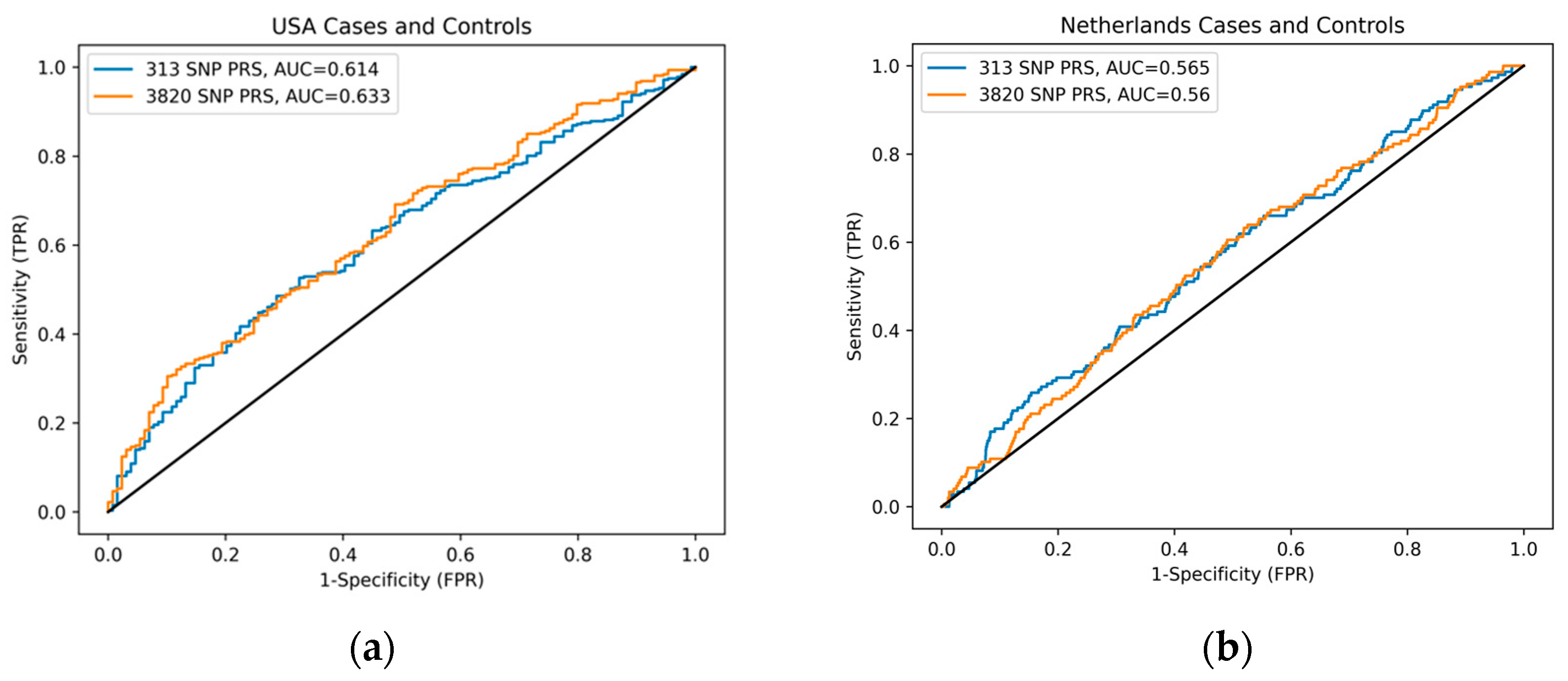
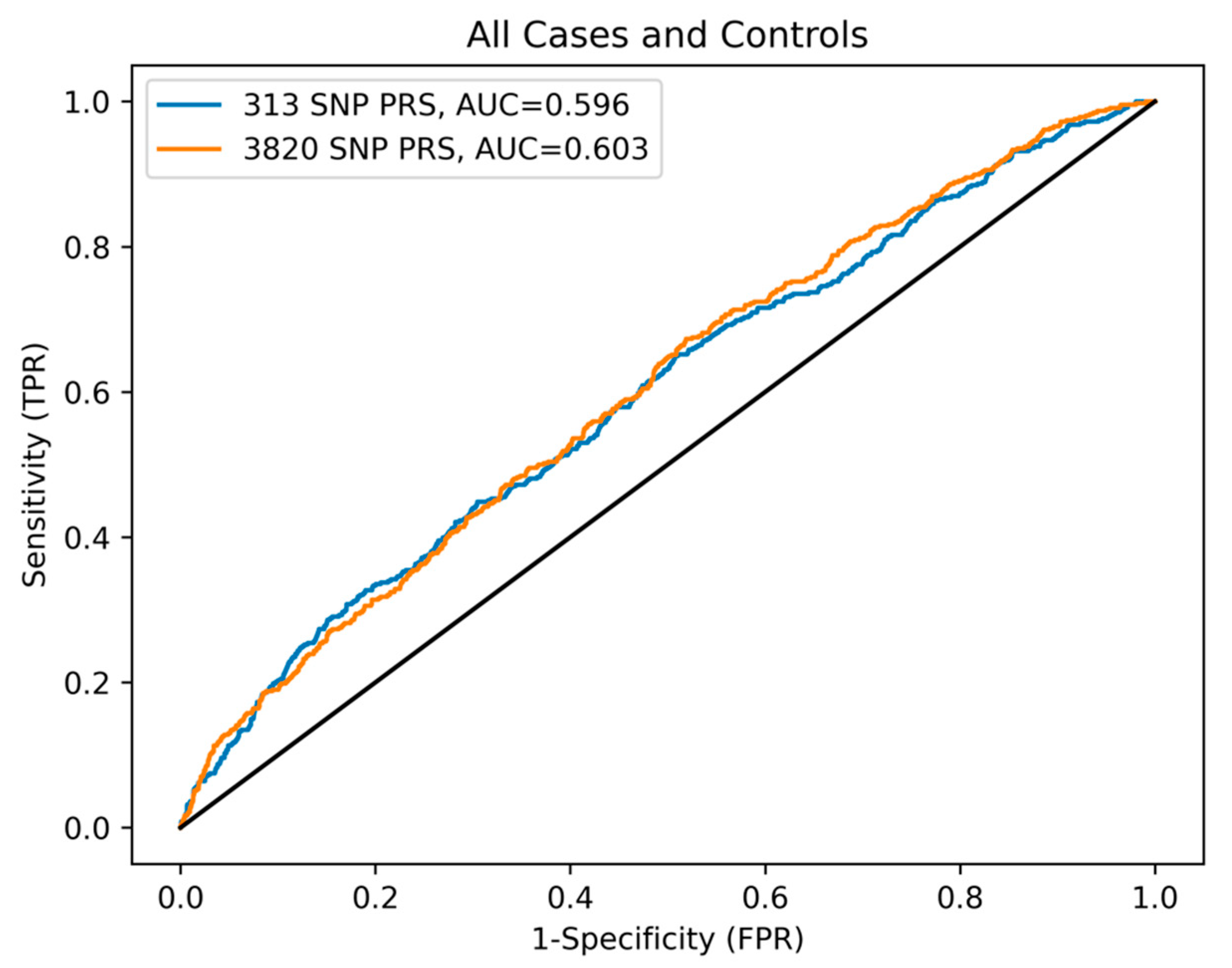
3. Results
3.1. Population Characteristics
3.2. Comparison of PRS Prediction Performance in BC Cases and Controls
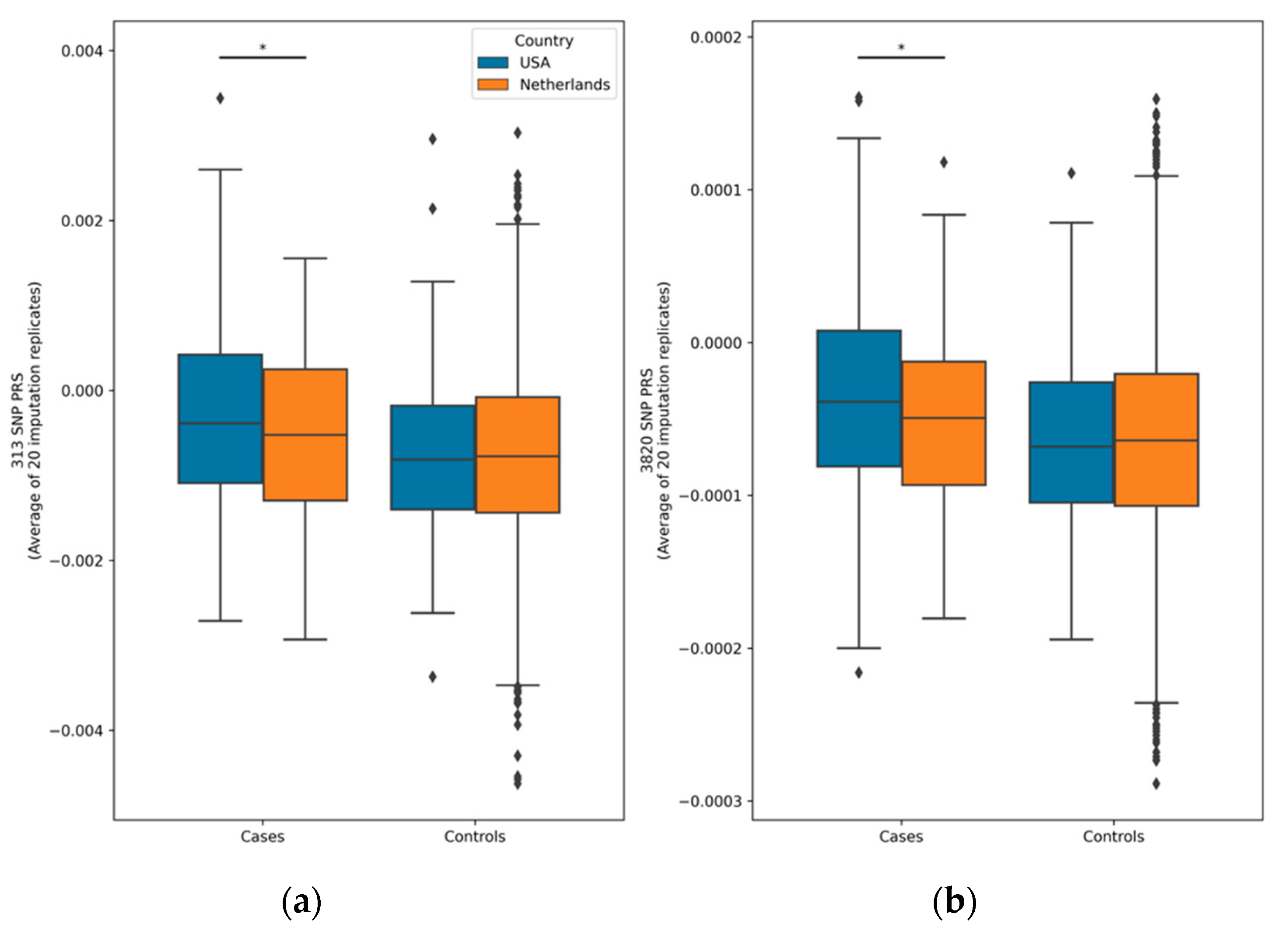
3.3. Assessment of Imputation Replication on PRS Variability
3.4. Relative and Absolute Risk Estimation
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Sample QC | Samples (n) |
|---|---|
| All subjects with genotyped on GSA (US + NL) | 20,723 |
| Callrate < 90% | −224 |
| Multiple DNA measurements same person | −384 |
| Heterozygosity | −208 |
| Sex problems | −310 |
| IBD problems | −172 |
| 19,425 | |
| Sample filters | |
| Select only female members | 11,580 |
| Remove Non-European samples defined by 1000 Genomes PC projection. | 11,225 |
| Select women with a valid case control status | 6997 |
| 20 × 1000 genomes imputation for 6997 individuals with random seed 1 to 20 | |
| Selected sample for analysis (unrelated people only, keeping one of multiple related) | 4805 * |
| Genotyping QC Steps | SNPs (n) |
|---|---|
| Illumina GSA SNPs | 669,317 |
| Poor quality X Chr clustering | −271 |
| Parametric Y Chr | −6 |
| Duplicate markers | −1246 |
| Absent allele markers (A/C/T/G) | −3503 |
| Total | 664,291 |
| SNP QC | |
| Call rate < 95% | −5531 |
| HWE < 0.001 | −6688 |
| MAF < 0.005 | −130,095 |
| Mendel errors ≥ 5% | −82 |
| 370 duplicate samples more than 5% genotype difference in SNP | −152 |
| Palindromic, >2 alleles, allele frequency difference over 0.10, and removal of MIT + chrY | −2819 |
| Total (pre-imputation and aligned) | 518,924 |
| US Cases | Bin | Subjects (n) | AVG Z-Score | RR (%) | AR (%) | US Controls | Bin | Subjects (n) | AVG Z-Score | RR (%) | AR (%) |
| 313-PRS | ≤25% | 2 | −1.91 | 2.8 | 1.2 | 313-PRS | ≤25% | 1 | −2.56 | 0.5 | 0.9 |
| 25–50% | 131 | −0.57 | 28.4 | 2.1 | 25–50% | 72 | −0.61 | 27.1 | 2.0 | ||
| 50–75% | 165 | 0.90 | 81.6 | 3.7 | 50–75% | 54 | 0.74 | 77 | 3.5 | ||
| >75% | 23 | 2.58 | 99.5 | 7.2 | >75% | 2 | 3.18 | 99.9 | 8.3 | ||
| US Cases | Bin | Subjects (n) | AVG Z-score | RR (%) | AR (%) | US Controls | Bin | Subjects (n) | AVG Z-score | RR (%) | AR (%) |
| 3820-PRS | ≤25% | 2 | −2.27 | 1.2 | 0.9 | 3820-PRS | ≤25% | 6 | −1.92 | 2.7 | 1.0 |
| 25–50% | 106 | −0.60 | 27.4 | 1.9 | 25–50% | 60 | −0.64 | 26.1 | 1.9 | ||
| 50–75% | 173 | 0.70 | 75.5 | 3.5 | 50–75% | 60 | 0.60 | 72.6 | 3.3 | ||
| >75% | 40 | 2.22 | 98.6 | 7.0 | >75% | 3 | 2.34 | 99 | 7.4 | ||
| NL Cases | Bin | Subjects (n) | AVG Z-score | RR (%) | AR (%) | NL Controls | Bin | Subjects (n) | AVG Z-score | RR (%) | AR (%) |
| 313-PRS | ≤25% | 3 | −2.03 | 2.1 | 1.7 | 313-PRS | ≤25% | 142 | −2.28 | 2.51 | 1.6 |
| 25–50% | 68 | −0.52 | 30.2 | 2.4 | 25–50% | 2220 | −0.61 | 27.1 | 2.4 | ||
| 50–75% | 73 | 0.93 | 82.4 | 3.4 | 50–75% | 1776 | 0.82 | 79.4 | 3.3 | ||
| >75% | 3 | 2.25 | 98.8 | 4.5 | >75% | 70 | 2.51 | 99.4 | 4.8 | ||
| NL Cases | Bin | Subjects (n) | AVG Z-score | RR (%) | AR (%) | NL Controls | Bin | Subjects (n) | AVG Z-score | RR (%) | AR (%) |
| 3820-PRS | ≤25% | 2 | −1.82 | 3.4 | 1.9 | 3820-PRS | ≤25% | 169 | −2.19 | 1.4 | 1.7 |
| 25–50% | 56 | −0.69 | 24.5 | 2.4 | 25–50% | 1939 | −0.68 | 24.8 | 2.4 | ||
| 50–75% | 80 | 0.67 | 74.9 | 3.2 | 50–75% | 1961 | 0.69 | 75.5 | 3.2 | ||
| >75% | 9 | 2.17 | 98.5 | 4.3 | >75% | 139 | 2.24 | 98.7 | 4.4 |
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA A Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- National Center for Health Statistics (US). Table 5, Age-adjusted death rates for selected causes of death, by sex, race, and Hispanic origin: United States, selected years 1950–2018. In Health, United States, 2019 [Internet]; National Center for Health Statistics (US): Hyattsville, MD, USA, 2021. [Google Scholar]
- Siu, A.L. Screening for Breast Cancer: U.S. Preventive Services Task Force Recommendation Statement. Ann. Intern. Med. 2016, 164, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Key Statistics for Breast Cancer. Available online: https://www.cancer.org/cancer/types/breast-cancer/about/how-common-is-breast-cancer.html (accessed on 10 November 2023).
- Sankatsing, V.D.V.; van Ravesteyn, N.T.; Heijnsdijk, E.A.M.; Looman, C.W.N.; van Luijt, P.A.; Fracheboud, J.; den Heeten, G.J.; Broeders, M.J.M.; de Koning, H.J. The effect of population-based mammography screening in Dutch municipalities on breast cancer mortality: 20 years of follow-up. Int. J. Cancer 2017, 141, 671–677. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Gangnon, R.E.; Sprague, B.L.; Stout, N.K.; Alagoz, O.; Weedon-Fekjær, H.; Holford, T.R.; Trentham-Dietz, A. The contribution of mammography screening to breast cancer incidence trends in the United States: An updated age-period-cohort model. Cancer Epidemiol. Biomark. Prev. 2015, 24, 905–912. [Google Scholar] [CrossRef] [PubMed]
- Economopoulou, P.; Dimitriadis, G.; Psyrri, A. Beyond BRCA: New hereditary breast cancer susceptibility genes. Cancer Treat. Rev. 2015, 41, 1–8. [Google Scholar] [CrossRef]
- Ford, D.; Easton, D.F.; Peto, J. Estimates of the gene frequency of BRCA1 and its contribution to breast and ovarian cancer incidence. Am. J. Hum. Genet. 1995, 57, 1457–1462. [Google Scholar] [PubMed]
- CHEK2 Breast Cancer Case-Control Consortium. CHEK2*1100delC and susceptibility to breast cancer: A collaborative analysis involving 10,860 breast cancer cases and 9,065 controls from 10 studies. Am. J. Hum. Genet. 2004, 74, 1175–1182. [Google Scholar] [CrossRef]
- Thompson, D.; Duedal, S.; Kirner, J.; McGuffog, L.; Last, J.; Reiman, A.; Byrd, P.; Taylor, M.; Easton, D.F. Cancer risks and mortality in heterozygous ATM mutation carriers. J. Natl. Cancer Inst. 2005, 97, 813–822. [Google Scholar] [CrossRef]
- Mack, T.M.; Hamilton, A.S.; Press, M.F.; Diep, A.; Rappaport, E.B. Heritable breast cancer in twins. Br. J. Cancer 2002, 87, 294–300. [Google Scholar] [CrossRef][Green Version]
- Möller, S.; Mucci, L.A.; Harris, J.R.; Scheike, T.; Holst, K.; Halekoh, U.; Adami, H.O.; Czene, K.; Christensen, K.; Holm, N.V.; et al. The Heritability of Breast Cancer among Women in the Nordic Twin Study of Cancer. Cancer Epidemiol. Biomark. Prev. 2016, 25, 145–150. [Google Scholar] [CrossRef] [PubMed]
- Lichtenstein, P.; Holm, N.V.; Verkasalo, P.K.; Iliadou, A.; Kaprio, J.; Koskenvuo, M.; Pukkala, E.; Skytthe, A.; Hemminki, K. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N. Engl. J. Med. 2000, 343, 78–85. [Google Scholar] [CrossRef] [PubMed]
- Slunecka, J.L.; van der Zee, M.D.; Beck, J.J.; Johnson, B.N.; Finnicum, C.T.; Pool, R.; Hottenga, J.J.; de Geus, E.J.C.; Ehli, E.A. Implementation and implications for polygenic risk scores in healthcare. Hum. Genom. 2021, 15, 46. [Google Scholar] [CrossRef]
- Khera, A.V.; Chaffin, M.; Aragam, K.G.; Haas, M.E.; Roselli, C.; Choi, S.H.; Natarajan, P.; Lander, E.S.; Lubitz, S.A.; Ellinor, P.T.; et al. Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat. Genet. 2018, 50, 1219–1224. [Google Scholar] [CrossRef] [PubMed]
- Mavaddat, N.; Michailidou, K.; Dennis, J.; Lush, M.; Fachal, L.; Lee, A.; Tyrer, J.P.; Chen, T.H.; Wang, Q.; Bolla, M.K.; et al. Polygenic Risk Scores for Prediction of Breast Cancer and Breast Cancer Subtypes. Am. J. Hum. Genet. 2019, 104, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.F.; Dias, R.; Evans, D.; Salfati, E.L.; Liu, S.; Wineinger, N.E.; Torkamani, A. Genotype imputation and variability in polygenic risk score estimation. Genome Med. 2020, 12, 100. [Google Scholar] [CrossRef] [PubMed]
- Sherman, S.; Shats, O.; Fleissner, E.; Bascom, G.; Yiee, K.; Copur, M.; Crow, K.; Rooney, J.; Mateen, Z.; Ketcham, M.A.; et al. Multicenter breast cancer collaborative registry. Cancer Inform. 2011, 10, 217–226. [Google Scholar] [CrossRef]
- Kittelsrud, J.; Ehli, E.A.; Petersen, V.; Jung, T.; Beck, J.J.; Willemsen, G.; Boomsma, D.; Davies, G. Avera Twin Register: Growing through Online Consenting and Survey Collection. Twin Res. Human. Genet. 2019, 22, 686–690. [Google Scholar] [CrossRef] [PubMed]
- Ligthart, L.; van Beijsterveldt, C.E.M.; Kevenaar, S.T.; de Zeeuw, E.; van Bergen, E.; Bruins, S.; Pool, R.; Helmer, Q.; van Dongen, J.; Hottenga, J.J.; et al. The Netherlands Twin Register: Longitudinal Research Based on Twin and Twin-Family Designs. Twin Res. Hum. Genet. 2019, 22, 623–636. [Google Scholar] [CrossRef]
- Beck, J.J.; Hottenga, J.J.; Mbarek, H.; Finnicum, C.T.; Ehli, E.A.; Hur, Y.M.; Martin, N.G.; de Geus, E.J.C.; Boomsma, D.I.; Davies, G.E. Genetic Similarity Assessment of Twin-Family Populations by Custom-Designed Genotyping Array. Twin Res. Hum. Genet. 2019, 22, 210–219. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef] [PubMed]
- Manichaikul, A.; Mychaleckyj, J.C.; Rich, S.S.; Daly, K.; Sale, M.; Chen, W.M. Robust relationship inference in genome-wide association studies. Bioinformatics 2010, 26, 2867–2873. [Google Scholar] [CrossRef] [PubMed]
- Consortium, G.P.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Browning, B.L.; Zhou, Y.; Browning, S.R. A One-Penny Imputed Genome from Next-Generation Reference Panels. Am. J. Hum. Genet. 2018, 103, 338–348. [Google Scholar] [CrossRef] [PubMed]
- Price, A.L.; Weale, M.E.; Patterson, N.; Myers, S.R.; Need, A.C.; Shianna, K.V.; Ge, D.; Rotter, J.I.; Torres, E.; Taylor, K.D. Long-range LD can confound genome scans in admixed populations. Am. J. Hum. Genet. 2008, 83, 132–135. [Google Scholar] [CrossRef] [PubMed]
- Pain, O.; Gillett, A.C.; Austin, J.C.; Folkersen, L.; Lewis, C.M. A tool for translating polygenic scores onto the absolute scale using summary statistics. Eur. J. Hum. Genet. 2022, 30, 339–348. [Google Scholar] [CrossRef] [PubMed]
- SEER*Explorer: An Interactive Website for SEER Cancer Statistics [Internet]. Surveillance Research Program, National Cancer Institute. 10 November 2023. Available online: https://seer.cancer.gov/statistics-network/explorer/ (accessed on 12 November 2023).
- Schwartz, P.H.; Meslin, E.M. The ethics of information: Absolute risk reduction and patient understanding of screening. J. Gen. Intern. Med. 2008, 23, 867–870. [Google Scholar] [CrossRef] [PubMed]
- van den Broek, J.J.; Schechter, C.B.; van Ravesteyn, N.T.; Janssens, A.; Wolfson, M.C.; Trentham-Dietz, A.; Simard, J.; Easton, D.F.; Mandelblatt, J.S.; Kraft, P.; et al. Personalizing Breast Cancer Screening Based on Polygenic Risk and Family History. J. Natl. Cancer Inst. 2021, 113, 434–442. [Google Scholar] [CrossRef]
- Jiang, X.; Finucane, H.K.; Schumacher, F.R.; Schmit, S.L.; Tyrer, J.P.; Han, Y.; Michailidou, K.; Lesseur, C.; Kuchenbaecker, K.B.; Dennis, J.; et al. Shared heritability and functional enrichment across six solid cancers. Nat. Commun. 2019, 10, 431. [Google Scholar] [CrossRef]
| USA | Netherlands | Total (USA and Netherlands) | ||||
|---|---|---|---|---|---|---|
| Controls | Cases | Controls | Cases | Controls | Cases | |
| Sample Size | 129 | 321 | 4208 | 147 | 4337 | 468 |
| Average Age | 42.60 (15.21) | 57.19 (12.14) | 45.34 (16.84) | 56.86 (9.76) | 45.25 (16.80) | 57.08 (11.43) |
| 313-SNP PRS Avg. % | 48.51 (12.00) | 53.66 (13.56) | 48.06 (12.63) | 50.95 (12.11) | 48.07 (12.61) | 52.81 (13.17) |
| 3820-SNP PRS Avg. % | 49.64 (12.94) | 56.61 (14.46) | 49.81 (14.16) | 52.94 (13.47) | 49.81 (14.32) | 55.45 (14.32) |
| US Allele Frequency | NL Allele Frequency | Summary Statistics | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| rsID | Chr | BP | A1 | A2 | Cases | Controls | Cases | Controls | R2 * | Effect Allele | Effect Size |
| rs13291323 | 9 | 6185360 | C | T | 0.032 | 0.058 | 0.087 | 0.061 | 0.920 | C | 0.0046 |
| rs7113140 | 11 | 123053078 | T | C | 0.488 | 0.434 | 0.371 | 0.428 | 1.000 | T | 0.0019 |
| rs12296461 | 12 | 116347863 | A | G | 0.542 | 0.552 | 0.655 | 0.553 | 0.970 | A | 0.0048 |
| rs12907670 | 15 | 63742901 | G | A | 0.832 | 0.857 | 0.913 | 0.869 | 0.980 | G | 0.0102 |
| rs4984247 | 15 | 63758647 | C | T | 0.850 | 0.892 | 0.926 | 0.884 | 0.970 | C | 0.0044 |
| rs34853502 | 16 | 53865368 | A | AG | 0.319 | 0.279 | 0.216 | 0.258 | 0.990 | A | 0.0145 |
| rs13049602 | 21 | 33501003 | C | T | 0.826 | 0.810 | 0.731 | 0.780 | 0.961 | C | 0.0082 |
| US | NL | ||||
|---|---|---|---|---|---|
| SNP Panel | Cases | Controls | Cases | Controls | |
| 313-SNP PRS | Relative Risk distribution of PRS | 54.40% | 39.40% | 59.10% | 49.60% |
| Absolute Risk distribution of PRS | 2.60% | 2.20% | 2.80% | 2.60% | |
| 3820-SNP PRS | Relative Risk distribution of PRS | 55.20% | 36.70% | 58.30% | 49.60% |
| Absolute Risk distribution of PRS | 2.60% | 2.10% | 2.80% | 2.60% | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beck, J.J.; Slunecka, J.L.; Johnson, B.N.; Van Asselt, A.J.; Finnicum, C.T.; Ageton, C.; Krie, A.; Nickles, H.; Cowan, K.; Maxwell, J.; et al. Breast Cancer Polygenic Risk Score Validation and Effects of Variable Imputation. Cancers 2024, 16, 1578. https://doi.org/10.3390/cancers16081578
Beck JJ, Slunecka JL, Johnson BN, Van Asselt AJ, Finnicum CT, Ageton C, Krie A, Nickles H, Cowan K, Maxwell J, et al. Breast Cancer Polygenic Risk Score Validation and Effects of Variable Imputation. Cancers. 2024; 16(8):1578. https://doi.org/10.3390/cancers16081578
Chicago/Turabian StyleBeck, Jeffrey J., John L. Slunecka, Brandon N. Johnson, Austin J. Van Asselt, Casey T. Finnicum, Cheryl Ageton, Amy Krie, Heidi Nickles, Kenneth Cowan, Jessica Maxwell, and et al. 2024. "Breast Cancer Polygenic Risk Score Validation and Effects of Variable Imputation" Cancers 16, no. 8: 1578. https://doi.org/10.3390/cancers16081578
APA StyleBeck, J. J., Slunecka, J. L., Johnson, B. N., Van Asselt, A. J., Finnicum, C. T., Ageton, C., Krie, A., Nickles, H., Cowan, K., Maxwell, J., Boomsma, D. I., de Geus, E., Ehli, E. A., & Hottenga, J.-J. (2024). Breast Cancer Polygenic Risk Score Validation and Effects of Variable Imputation. Cancers, 16(8), 1578. https://doi.org/10.3390/cancers16081578





