Clinical and Therapeutic Intervention of Hypereosinophilia in the Era of Molecular Diagnosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Epidemiology
3. Etiology
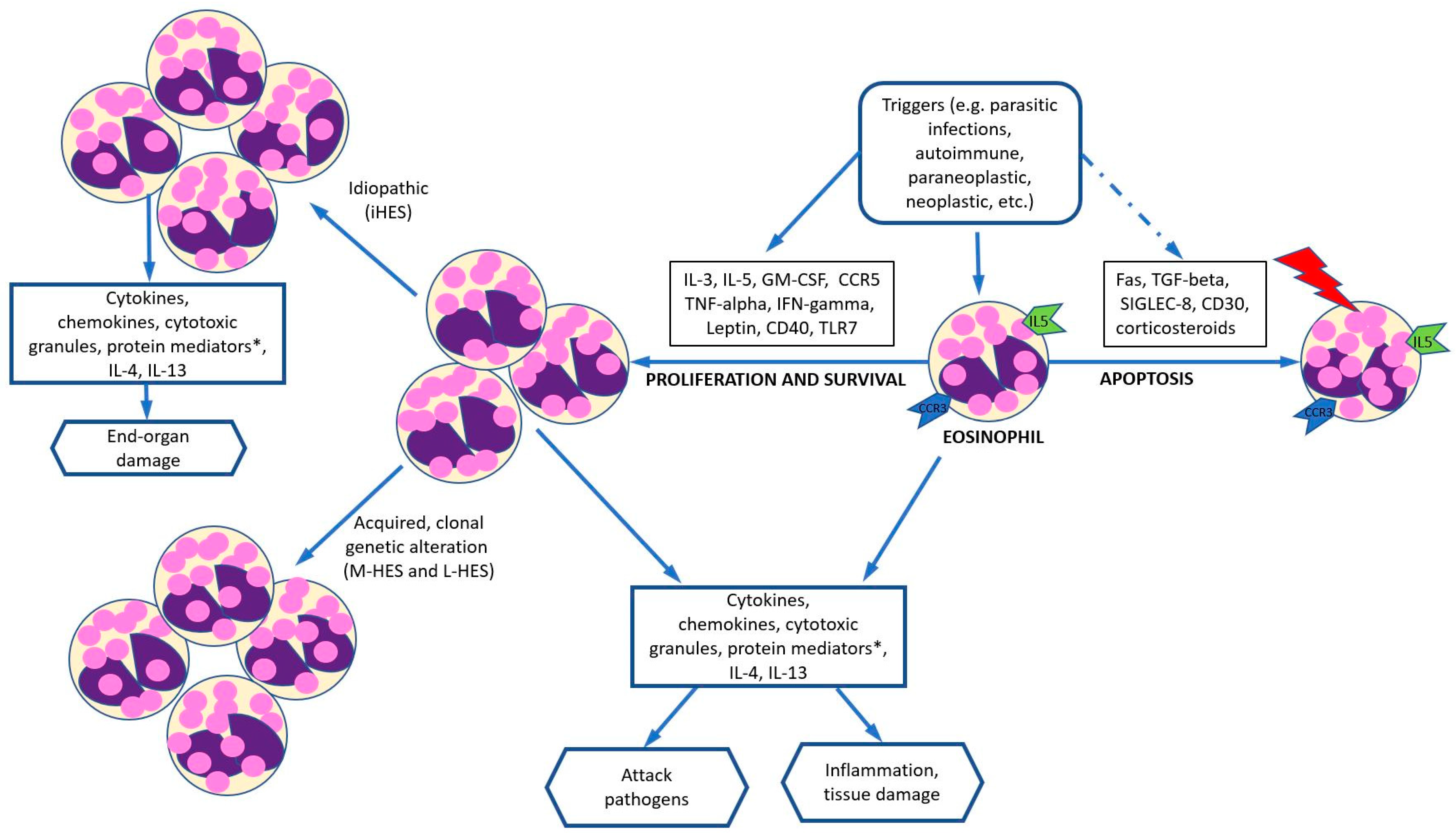
4. Pathogenesis
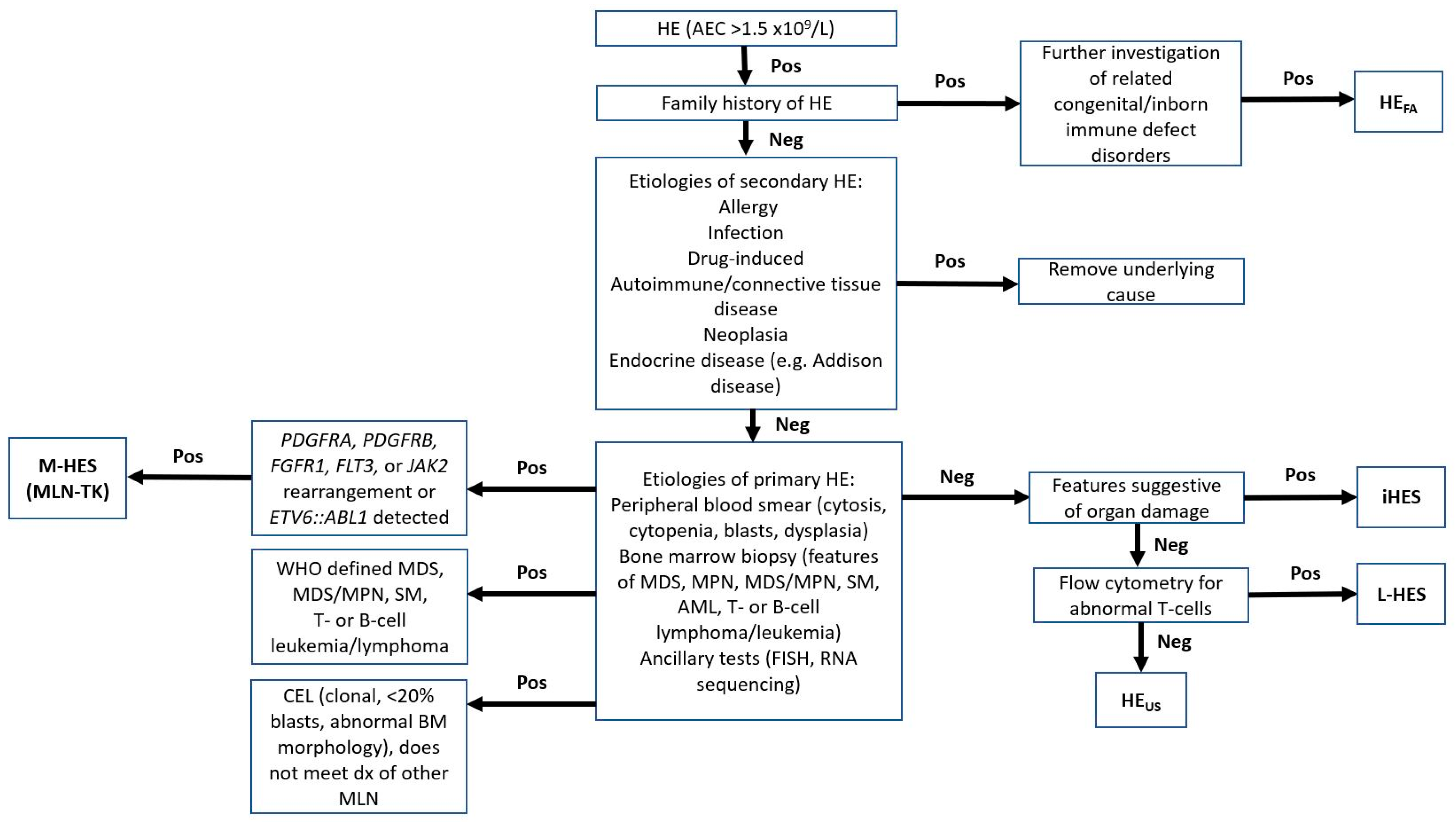
5. Diagnosis
5.1. Clinical
5.2. Laboratory and Pathology
5.3. Potential Genetic Determinants and Biomarkers
6. Differential Diagnosis
6.1. Myeloproliferative Variants of Hypereosinophilia (M-HES)
| Partners | Concurrent Mutations | Typical Clinical Association | Reference (PMID) | |
|---|---|---|---|---|
| PDGFRA * | FIP1L1, KIF5B, CDK5RAP2, STRN, ETV6, BCR, TNKS2 | N/A | MPN or MDS/MPN typically in chronic phase and less frequently in blast phase of myeloid or lymphoid lineage | [34,35,36,37,38,39,40,41] |
| PDGFRB | WDR48, CAPR1N1, TPM3, PDE4DIP, SPTBN, PRKG2, GOLGA4, TNIP1, HIP1, HECW1, KANK1, CCDC6, SART3, GIT2, ERC1, BIN2, NIN, CCDC88C, TP53BP1, NDE1, RABEP1, SPECC1, MYO18A, COL1A1, DTD1 | N/A | Commonly CMML with eosinophilia and less commonly MDS/MPN with neutrophilia (formerly aCML), and CEL (or MPN with eosinophilia) | [42,43,44] |
| FGFR1 | ZMYM2, FGFR1OP, TRIM24, MYO18A, HERVK, FGFR1OP2, RANBP2, LRRFIP1, CUX1, CPSF6, BCR, TPR, CEP43, CNTRL | Concurrent mutations involving RUNX1; associated with increased proliferation of the clone and poor outcome | Variable phenotype including precursor B-cell, T-cell, myeloid or MPAL or MPN or MDS/MPN with associated eosinophilia, rarely B-ALL | [45,46,47,48,49,50,51,52] |
| JAK2 | PCM1, ETV6, BCR | ASXL1, BCOR, ETV6, RUNX1, SRSF2, TET2, and TP53 | MPN, ALL, AML | [53] |
| FLT3 | BCR, ZMYM2, TRIP11, SPTBN1, GOLGB1, CCDC88C, ZBTB44, MYO18A | ASXL1, SETBP1, U2AF1, STAT5B, TP53, SRSF2, TET2, RUNX1, and PTPN11 | Extramedullary involvement with T-ALL, MyeS, or rarely with mixed-phenotype features including B-cell, T-cell, or myeloid lineage disease. | [31,54,55] |
| ETV6::ABL1 | ETV6::ABL1 | N/A | MDS/MPN with neutrophilia, CEL, or other MDS/MPN. | [56,57,58,59] |
| Other | ETV6::FGFR2; ETV6::LYN; ETV6::NTRK3; RANBP2::ALK; BCR::RET and FGFR1OP::RET | N/A | MDS/MPN, often with notable eosinophilia, ±monocytosis, T-cell differentiation is more common such as T-ALL or PTCL, mast cell proliferations and/or bone marrow fibrosis | [60,61,62,63,64,65] |
6.2. T-Lymphocytic Variants of HE (L-HES)
6.3. Idiopathic HES (iHES)
6.4. Hypereosinophilia of Undetermined Significance (HEUS)
6.5. Familial HE/HES
6.6. Specific or Defined Syndromes Associated with HE
6.7. Organ-Restricted HE Conditions
6.8. Secondary/Reactive HE
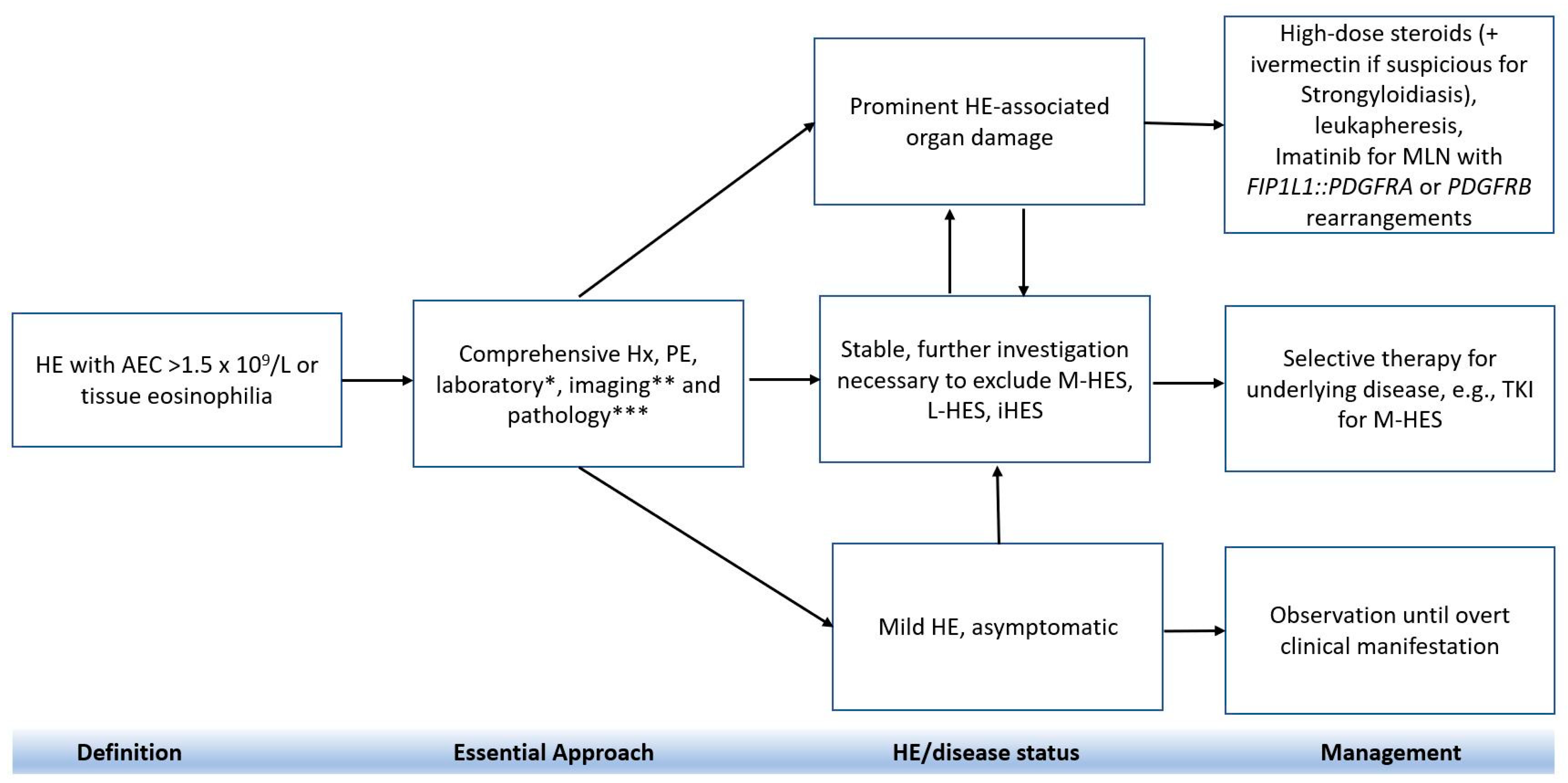
7. Treatment
7.1. Conventional Therapy
7.2. Second-line Therapy
7.3. Disease-Specific Treatment
7.3.1. iHES
7.3.2. Malignancy Associated HE
7.3.3. Localized HE
7.4. Novel Approaches and Clinical Trials
7.5. Allogeneic Hematopoietic Stem Cell Transplant
| Pathway | Name | Nature/Function | Reference |
|---|---|---|---|
| IL-5 inhibitor | Mepolizumab and Reslizumab | Humanized monoclonal anti-IL-5 antibody | [105,106,117] |
| Benralizumab | IL-5 receptor antagonist, focusing on eosinophils, basophils, and their precursors through antibody-dependent cell-mediated cytotoxicity | [117] | |
| SIGLEC-8 inhibitor | Lirentelimab | Afucosylated antibody against Siglec-8 leading to decrease eosinophils and basophils and inactivate mast cells | [117] |
| IL-4 inhibitor | Dupilumab | IL-4 antibody, reduces IL-4 and IL-13 signaling, and blocks exotaxin-mediated tissue migration of eosinophils | [118,119] |
| TKI | Imatinib | BCR-ABL1 inhibitor | [35,71] |
| Ruxolitinib | JAK2 inhibitor | [111] | |
| STAT5 inhibitor | Stafiba | Actively inhibiting the SH2 domains of STAT5a and STAT5b | [120] |
| IgE inhibitor | Omalizumab | Monoclonal anti-immunoglobulin E | [66] |
8. Conclusions and Future Direction
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Marichal, T.; Mesnil, C.; Bureau, F. Homeostatic Eosinophils: Characteristics and Functions. Front. Med. 2017, 4, 101. [Google Scholar] [CrossRef]
- Wen, T.; Rothenberg, M.E. The Regulatory Function of Eosinophils. Microbiol. Spectr. 2016, 4, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Shomali, W.; Gotlib, J. World Health Organization-defined eosinophilic disorders: 2022 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2022, 97, 129–148. [Google Scholar] [CrossRef] [PubMed]
- Klion, A.D. Approach to the patient with suspected hypereosinophilic syndrome. Hematology 2022, 2022, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Liacouras, C.A.; Furuta, G.T.; Hirano, I.; Atkins, D.; Attwood, S.E.; Bonis, P.A.; Burks, A.W.; Chehade, M.; Collins, M.H.; Dellon, E.S.; et al. Eosinophilic esophagitis: Updated consensus recommendations for children and adults. J. Allergy Clin. Immunol. 2011, 128, 3–20.e26; quiz 21–22. [Google Scholar] [CrossRef] [PubMed]
- Valent, P.; Klion, A.D.; Roufosse, F.; Simon, D.; Metzgeroth, G.; Leiferman, K.M.; Schwaab, J.; Butterfield, J.H.; Sperr, W.R.; Sotlar, K.; et al. Proposed refined diagnostic criteria and classification of eosinophil disorders and related syndromes. Allergy 2023, 78, 47–59. [Google Scholar] [CrossRef] [PubMed]
- Sunadome, H.; Sato, S.; Matsumoto, H.; Murase, K.; Kawaguchi, T.; Tabara, Y.; Chin, K.; Matsuda, F.; Hirai, T. Similar distribution of peripheral blood eosinophil counts in European and East Asian populations from investigations of large-scale general population studies: The Nagahama Study. Eur. Respir. J. 2021, 57, 2004101. [Google Scholar] [CrossRef] [PubMed]
- Crane, M.M.; Chang, C.M.; Kobayashi, M.G.; Weller, P.F. Incidence of myeloproliferative hypereosinophilic syndrome in the United States and an estimate of all hypereosinophilic syndrome incidence. J. Allergy Clin. Immunol. 2010, 126, 179–181. [Google Scholar] [CrossRef] [PubMed]
- Ruan, G.J.; Smith, C.J.; Day, C.; Harmsen, W.S.; Zblewski, D.L.; Alkhateeb, H.; Begna, K.; Al-Kali, A.; Litzow, M.R.; Hogan, W.; et al. A population-based study of chronic eosinophilic leukemia-not otherwise specified in the United States. Am. J. Hematol. 2020, 95, E257–E260. [Google Scholar] [CrossRef]
- Williams, K.W.; Ware, J.; Abiodun, A.; Holland-Thomas, N.C.; Khoury, P.; Klion, A.D. Hypereosinophilia in Children and Adults: A Retrospective Comparison. J. Allergy Clin. Immunol. Pract. 2016, 4, 941–947.e941. [Google Scholar] [CrossRef]
- Kaur, P.; Khan, W.A. Myeloid/Lymphoid Neoplasms with Eosinophilia and Platelet Derived Growth Factor Receptor Alpha (PDGFRA) Rearrangement. Brisb. (AU) Exon Publ. 2022, 8, 129–149. [Google Scholar] [CrossRef]
- Hu, Z.; Wang, W.; Thakral, B.; Chen, Z.; Estrov, Z.; Bueso-Ramos, C.E.; Verstovsek, S.; Medeiros, L.J.; Wang, S.A. Lymphocytic variant of hypereosinophilic syndrome: A report of seven cases from a single institution. Cytom. B Clin. Cytom. 2021, 100, 352–360. [Google Scholar] [CrossRef] [PubMed]
- Klion, A. Hypereosinophilic syndrome: Approach to treatment in the era of precision medicine. Hematology 2018, 2018, 326–331. [Google Scholar] [CrossRef] [PubMed]
- Ackerman, S.J.; Bochner, B.S. Mechanisms of eosinophilia in the pathogenesis of hypereosinophilic disorders. Immunol. Allergy Clin. N. Am. 2007, 27, 357–375. [Google Scholar] [CrossRef] [PubMed]
- Akuthota, P.; Weller, P.F. Spectrum of Eosinophilic End-Organ Manifestations. Immunol. Allergy Clin. N. Am. 2015, 35, 403–411. [Google Scholar] [CrossRef] [PubMed]
- Grueso-Navarro, E.; Navarro, P.; Laserna-Mendieta, E.J.; Lucendo, A.J.; Arias-González, L. Blood-Based Biomarkers for Eosinophilic Esophagitis and Concomitant Atopic Diseases: A Look into the Potential of Extracellular Vesicles. Int. J. Mol. Sci. 2023, 24, 3669. [Google Scholar] [CrossRef] [PubMed]
- Bélanger, É.; Madore, A.-M.; Boucher-Lafleur, A.-M.; Simon, M.-M.; Kwan, T.; Pastinen, T.; Laprise, C. Eosinophil microRNAs Play a Regulatory Role in Allergic Diseases Included in the Atopic March. Int. J. Mol. Sci. 2020, 21, 9011. [Google Scholar] [CrossRef] [PubMed]
- Potaczek, D.P.; Kabesch, M. Current concepts of IgE regulation and impact of genetic determinants. Clin. Exp. Allergy 2012, 42, 852–871. [Google Scholar] [CrossRef] [PubMed]
- Moffatt, M.F.; Gut, I.G.; Demenais, F.; Strachan, D.P.; Bouzigon, E.; Heath, S.; von Mutius, E.; Farrall, M.; Lathrop, M.; Cookson, W.O.C.M. A Large-Scale, Consortium-Based Genomewide Association Study of Asthma. N. Engl. J. Med. 2010, 363, 1211–1221. [Google Scholar] [CrossRef]
- Kabesch, M. Novel Asthma-Associated Genes From Genome-Wide Association Studies: What Is Their Significance? Chest 2010, 137, 909–915. [Google Scholar] [CrossRef] [PubMed]
- Weidinger, S.; Baurecht, H.; Naumann, A.; Novak, N. Genome-wide association studies on IgE regulation: Are genetics of IgE also genetics of atopic disease? Curr. Opin. Allergy Clin. Immunol. 2010, 10, 408–417. [Google Scholar] [CrossRef] [PubMed]
- Andiappan, A.K.; Wang, D.Y.; Anantharaman, R.; Parate, P.N.; Suri, B.K.; Low, H.Q.; Li, Y.; Zhao, W.; Castagnoli, P.; Liu, J.; et al. Genome-Wide Association Study for Atopy and Allergic Rhinitis in a Singapore Chinese Population. PLoS ONE 2011, 6, e19719. [Google Scholar] [CrossRef] [PubMed]
- Ramasamy, A.; Curjuric, I.; Coin, L.J.; Kumar, A.; McArdle, W.L.; Imboden, M.; Leynaert, B.; Kogevinas, M.; Schmid-Grendelmeier, P.; Pekkanen, J.; et al. A genome-wide meta-analysis of genetic variants associated with allergic rhinitis and grass sensitization and their interaction with birth order. J. Allergy Clin. Immunol. 2011, 128, 996–1005. [Google Scholar] [CrossRef] [PubMed]
- Esparza-Gordillo, J.; Weidinger, S.; Fölster-Holst, R.; Bauerfeind, A.; Ruschendorf, F.; Patone, G.; Rohde, K.; Marenholz, I.; Schulz, F.; Kerscher, T.; et al. A common variant on chromosome 11q13 is associated with atopic dermatitis. Nat. Genet. 2009, 41, 596–601. [Google Scholar] [CrossRef] [PubMed]
- Klion, A.D. Recent advances in the diagnosis and treatment of hypereosinophilic syndromes. Hematol. Am. Soc. Hematol. Educ. Program. 2005, 2005, 209–214. [Google Scholar] [CrossRef] [PubMed]
- Leru, P.M. Eosinophilic disorders: Evaluation of current classification and diagnostic criteria, proposal of a practical diagnostic algorithm. Clin. Transl. Allergy 2019, 9, 36. [Google Scholar] [CrossRef] [PubMed]
- Cheah, C.Y.; Burbury, K.; Apperley, J.F.; Huguet, F.; Pitini, V.; Gardembas, M.; Ross, D.M.; Forrest, D.; Genet, P.; Rousselot, P.; et al. Patients with myeloid malignancies bearing PDGFRB fusion genes achieve durable long-term remissions with imatinib. Blood 2014, 123, 3574–3577. [Google Scholar] [CrossRef] [PubMed]
- Saultz, J.N.; Kaffenberger, B.H.; Taylor, M.; Heerema, N.A.; Klisovic, R. Novel Chromosome 5 Inversion Associated With PDGFRB Rearrangement in Hypereosinophilic Syndrome. JAMA Dermatol. 2016, 152, 1391–1393. [Google Scholar] [CrossRef] [PubMed]
- Bain, B.J.; Ahmad, S. Should myeloid and lymphoid neoplasms with PCM1-JAK2 and other rearrangements of JAK2 be recognized as specific entities? Br. J. Haematol. 2014, 166, 809–817. [Google Scholar] [CrossRef] [PubMed]
- Tang, G.; Sydney Sir Philip, J.K.; Weinberg, O.; Tam, W.; Sadigh, S.; Lake, J.I.; Margolskee, E.M.; Rogers, H.J.; Miranda, R.N.; Bueso-Ramos, C.C.; et al. Hematopoietic neoplasms with 9p24/JAK2 rearrangement: A multicenter study. Mod. Pathol. 2019, 32, 490–498. [Google Scholar] [CrossRef]
- Tang, G.; Tam, W.; Short, N.J.; Bose, P.; Wu, D.; Hurwitz, S.N.; Bagg, A.; Rogers, H.J.; Hsi, E.D.; Quesada, A.E.; et al. Myeloid/lymphoid neoplasms with FLT3 rearrangement. Mod. Pathol. 2021, 34, 1673–1685. [Google Scholar] [CrossRef]
- Shao, H.; Wang, W.; Song, J.; Tang, G.; Zhang, X.; Tang, Z.; Srivastava, J.; Shah, B.; Medeiros, L.J.; Zhang, L. Myeloid/lymphoid neoplasms with eosinophilia and FLT3 rearrangement. Leuk. Res. 2020, 99, 106460. [Google Scholar] [CrossRef] [PubMed]
- Reiter, A.; Schwaab, J.; DeAngelo, D.J.; Gotlib, J.; Deininger, M.W.; Pettit, K.M.; Alvarez-Twose, I.; Vannucchi, A.M.; Panse, J.; Platzbecker, U.; et al. Efficacy and safety of avapritinib in previously treated patients with advanced systemic mastocytosis. Blood Adv. 2022, 6, 5750–5762. [Google Scholar] [CrossRef] [PubMed]
- Score, J.; Curtis, C.; Waghorn, K.; Stalder, M.; Jotterand, M.; Grand, F.H.; Cross, N.C. Identification of a novel imatinib responsive KIF5B-PDGFRA fusion gene following screening for PDGFRA overexpression in patients with hypereosinophilia. Leukemia 2006, 20, 827–832. [Google Scholar] [CrossRef] [PubMed]
- Walz, C.; Curtis, C.; Schnittger, S.; Schultheis, B.; Metzgeroth, G.; Schoch, C.; Lengfelder, E.; Erben, P.; Müller, M.C.; Haferlach, T.; et al. Transient response to imatinib in a chronic eosinophilic leukemia associated with ins(9;4)(q33;q12q25) and a CDK5RAP2-PDGFRA fusion gene. Genes. Chromosomes Cancer 2006, 45, 950–956. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.E.; Grand, F.H.; Musto, P.; Clark, A.; Murphy, J.; Perla, G.; Minervini, M.M.; Stewart, J.; Reiter, A.; Cross, N.C. Two novel imatinib-responsive PDGFRA fusion genes in chronic eosinophilic leukaemia. Br. J. Haematol. 2007, 138, 77–81. [Google Scholar] [CrossRef] [PubMed]
- Baxter, E.J.; Hochhaus, A.; Bolufer, P.; Reiter, A.; Fernandez, J.M.; Senent, L.; Cervera, J.; Moscardo, F.; Sanz, M.A.; Cross, N.C. The t(4;22)(q12;q11) in atypical chronic myeloid leukaemia fuses BCR to PDGFRA. Hum. Mol. Genet. 2002, 11, 1391–1397. [Google Scholar] [CrossRef] [PubMed]
- Safley, A.M.; Sebastian, S.; Collins, T.S.; Tirado, C.A.; Stenzel, T.T.; Gong, J.Z.; Goodman, B.K. Molecular and cytogenetic characterization of a novel translocation t(4;22) involving the breakpoint cluster region and platelet-derived growth factor receptor-alpha genes in a patient with atypical chronic myeloid leukemia. Genes Chromosomes Cancer 2004, 40, 44–50. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Ali, S.M.; Ohgami, R.S.; Campregher, P.V.; Frampton, G.M.; Yelensky, R.; Elvin, J.A.; Palma, N.A.; Erlich, R.; Vergilio, J.A.; et al. Comprehensive genomic profiling identifies a novel TNKS2-PDGFRA fusion that defines a myeloid neoplasm with eosinophilia that responded dramatically to imatinib therapy. Blood Cancer J. 2015, 5, e278. [Google Scholar] [CrossRef] [PubMed]
- Metzgeroth, G.; Walz, C.; Score, J.; Siebert, R.; Schnittger, S.; Haferlach, C.; Popp, H.; Haferlach, T.; Erben, P.; Mix, J.; et al. Recurrent finding of the FIP1L1-PDGFRA fusion gene in eosinophilia-associated acute myeloid leukemia and lymphoblastic T-cell lymphoma. Leukemia 2007, 21, 1183–1188. [Google Scholar] [CrossRef] [PubMed]
- Schwaab, J.; Jawhar, M.; Naumann, N.; Schmitt-Graeff, A.; Fabarius, A.; Horny, H.P.; Cross, N.C.; Hofmann, W.K.; Reiter, A.; Metzgeroth, G. Diagnostic challenges in the work up of hypereosinophilia: Pitfalls in bone marrow core biopsy interpretation. Ann. Hematol. 2016, 95, 557–562. [Google Scholar] [CrossRef] [PubMed]
- Curtis, C.E.; Grand, F.H.; Waghorn, K.; Sahoo, T.P.; George, J.; Cross, N.C. A novel ETV6-PDGFRB fusion transcript missed by standard screening in a patient with an imatinib responsive chronic myeloproliferative disease. Leukemia 2007, 21, 1839–1841. [Google Scholar] [CrossRef] [PubMed]
- Steer, E.J.; Cross, N.C. Myeloproliferative disorders with translocations of chromosome 5q31-35: Role of the platelet-derived growth factor receptor Beta. Acta Haematol. 2002, 107, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Bain, B.J.; Fletcher, S.H. Chronic eosinophilic leukemias and the myeloproliferative variant of the hypereosinophilic syndrome. Immunol. Allergy Clin. N. Am. 2007, 27, 377–388. [Google Scholar] [CrossRef] [PubMed]
- Roumiantsev, S.; Krause, D.S.; Neumann, C.A.; Dimitri, C.A.; Asiedu, F.; Cross, N.C.; Van Etten, R.A. Distinct stem cell myeloproliferative/T lymphoma syndromes induced by ZNF198-FGFR1 and BCR-FGFR1 fusion genes from 8p11 translocations. Cancer Cell 2004, 5, 287–298. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhai, Y.P.; Tang, Y.M.; Wang, L.P.; Wan, P.J. Identification of a novel partner gene, TPR, fused to FGFR1 in 8p11 myeloproliferative syndrome. Genes Chromosomes Cancer 2012, 51, 890–897. [Google Scholar] [CrossRef] [PubMed]
- Popovici, C.; Zhang, B.; Grégoire, M.J.; Jonveaux, P.; Lafage-Pochitaloff, M.; Birnbaum, D.; Pébusque, M.J. The t(6;8)(q27;p11) translocation in a stem cell myeloproliferative disorder fuses a novel gene, FOP, to fibroblast growth factor receptor 1. Blood 1999, 93, 1381–1389. [Google Scholar] [CrossRef] [PubMed]
- Vizmanos, J.L.; Hernández, R.; Vidal, M.J.; Larráyoz, M.J.; Odero, M.D.; Marín, J.; Ardanaz, M.T.; Calasanz, M.J.; Cross, N.C. Clinical variability of patients with the t(6;8)(q27;p12) and FGFR1OP-FGFR1 fusion: Two further cases. Hematol. J. 2004, 5, 534–537. [Google Scholar] [CrossRef] [PubMed]
- Strati, P.; Tang, G.; Duose, D.Y.; Mallampati, S.; Luthra, R.; Patel, K.P.; Hussaini, M.; Mirza, A.S.; Komrokji, R.S.; Oh, S.; et al. Myeloid/lymphoid neoplasms with FGFR1 rearrangement. Leuk. Lymphoma 2018, 59, 1672–1676. [Google Scholar] [CrossRef]
- Baer, C.; Muehlbacher, V.; Kern, W.; Haferlach, C.; Haferlach, T. Molecular genetic characterization of myeloid/lymphoid neoplasms associated with eosinophilia and rearrangement of PDGFRA, PDGFRB, FGFR1 or PCM1-JAK2. Haematologica 2018, 103, e348–e350. [Google Scholar] [CrossRef] [PubMed]
- Vega, F.; Medeiros, L.J.; Bueso-Ramos, C.E.; Arboleda, P.; Miranda, R.N. Hematolymphoid neoplasms associated with rearrangements of PDGFRA, PDGFRB, and FGFR1. Am. J. Clin. Pathol. 2015, 144, 377–392. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.A.; Hasserjian, R.P.; Tam, W.; Tsai, A.G.; Geyer, J.T.; George, T.I.; Foucar, K.; Rogers, H.J.; Hsi, E.D.; Rea, B.A.; et al. Bone marrow morphology is a strong discriminator between chronic eosinophilic leukemia, not otherwise specified and reactive idiopathic hypereosinophilic syndrome. Haematologica 2017, 102, 1352–1360. [Google Scholar] [CrossRef] [PubMed]
- Alaggio, R.; Amador, C.; Anagnostopoulos, I.; Attygalle, A.D.; Araujo, I.B.O.; Berti, E.; Bhagat, G.; Borges, A.M.; Boyer, D.; Calaminici, M.; et al. The 5th edition of the World Health Organization Classification of Haematolymphoid Tumours: Lymphoid Neoplasms. Leukemia 2022, 36, 1720–1748. [Google Scholar] [CrossRef] [PubMed]
- Chao, A.K.; Meyer, J.A.; Lee, A.G.; Hecht, A.; Tarver, T.; Van Ziffle, J.; Koegel, A.K.; Golden, C.; Braun, B.S.; Sweet-Cordero, E.A.; et al. Fusion driven JMML: A novel CCDC88C-FLT3 fusion responsive to sorafenib identified by RNA sequencing. Leukemia 2020, 34, 662–666. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Paliga, A.; Hobbs, E.; Moore, S.; Olson, S.; Long, N.; Dao, K.T.; Tyner, J.W. Two myeloid leukemia cases with rare FLT3 fusions. Cold Spring Harb. Mol. Case Stud. 2018, 4, a003079. [Google Scholar] [CrossRef] [PubMed]
- Andreasson, P.; Johansson, B.; Carlsson, M.; Jarlsfelt, I.; Fioretos, T.; Mitelman, F.; Höglund, M. BCR/ABL-negative chronic myeloid leukemia with ETV6/ABL fusion. Genes. Chromosomes Cancer 1997, 20, 299–304. [Google Scholar] [CrossRef]
- Keung, Y.K.; Beaty, M.; Steward, W.; Jackle, B.; Pettnati, M. Chronic myelocytic leukemia with eosinophilia, t(9;12)(q34;p13), and ETV6-ABL gene rearrangement: Case report and review of the literature. Cancer Genet. Cytogenet. 2002, 138, 139–142. [Google Scholar] [CrossRef] [PubMed]
- Nand, R.; Bryke, C.; Kroft, S.H.; Divgi, A.; Bredeson, C.; Atallah, E. Myeloproliferative disorder with eosinophilia and ETV6-ABL gene rearrangement: Efficacy of second-generation tyrosine kinase inhibitors. Leuk. Res. 2009, 33, 1144–1146. [Google Scholar] [CrossRef] [PubMed]
- Yao, J.; Xu, L.; Aypar, U.; Meyerson, H.J.; Londono, D.; Gao, Q.; Baik, J.; Dietz, J.; Benayed, R.; Sigler, A.; et al. Myeloid/lymphoid neoplasms with eosinophilia/basophilia and ETV6-ABL1 fusion: Cell-of-origin and response to tyrosine kinase inhibition. Haematologica 2021, 106, 614–618. [Google Scholar] [CrossRef] [PubMed]
- Carll, T.; Patel, A.; Derman, B.; Hyjek, E.; Lager, A.; Wanjari, P.; Segal, J.; Odenike, O.; Fidai, S.; Arber, D. Diagnosis and treatment of mixed phenotype (T-myeloid/lymphoid) acute leukemia with novel ETV6-FGFR2 rearrangement. Blood Adv. 2020, 4, 4924–4928. [Google Scholar] [CrossRef]
- Telford, N.; Alexander, S.; McGinn, O.J.; Williams, M.; Wood, K.M.; Bloor, A.; Saha, V. Myeloproliferative neoplasm with eosinophilia and T-lymphoblastic lymphoma with ETV6-LYN gene fusion. Blood Cancer J. 2016, 6, e412. [Google Scholar] [CrossRef] [PubMed]
- Röttgers, S.; Gombert, M.; Teigler-Schlegel, A.; Busch, K.; Gamerdinger, U.; Slany, R.; Harbott, J.; Borkhardt, A. ALK fusion genes in children with atypical myeloproliferative leukemia. Leukemia 2010, 24, 1197–1200. [Google Scholar] [CrossRef] [PubMed]
- Maesako, Y.; Izumi, K.; Okamori, S.; Takeoka, K.; Kishimori, C.; Okumura, A.; Honjo, G.; Akasaka, T.; Ohno, H. inv(2)(p23q13)/RAN-binding protein 2 (RANBP2)-ALK fusion gene in myeloid leukemia that developed in an elderly woman. Int. J. Hematol. 2014, 99, 202–207. [Google Scholar] [CrossRef] [PubMed]
- Ballerini, P.; Struski, S.; Cresson, C.; Prade, N.; Toujani, S.; Deswarte, C.; Dobbelstein, S.; Petit, A.; Lapillonne, H.; Gautier, E.F.; et al. RET fusion genes are associated with chronic myelomonocytic leukemia and enhance monocytic differentiation. Leukemia 2012, 26, 2384–2389. [Google Scholar] [CrossRef] [PubMed]
- Forghieri, F.; Morselli, M.; Potenza, L.; Maccaferri, M.; Pedrazzi, L.; Paolini, A.; Bonacorsi, G.; Artusi, T.; Giacobbi, F.; Corradini, G.; et al. Chronic eosinophilic leukaemia with ETV6-NTRK3 fusion transcript in an elderly patient affected with pancreatic carcinoma. Eur. J. Haematol. 2011, 86, 352–355. [Google Scholar] [CrossRef] [PubMed]
- Cross, N.C.P.; Hoade, Y.; Tapper, W.J.; Carreno-Tarragona, G.; Fanelli, T.; Jawhar, M.; Naumann, N.; Pieniak, I.; Lübke, J.; Ali, S.; et al. Recurrent activating STAT5B N642H mutation in myeloid neoplasms with eosinophilia. Leukemia 2019, 33, 415–425. [Google Scholar] [CrossRef] [PubMed]
- Roufosse, F.; Cogan, E.; Goldman, M. Recent advances in pathogenesis and management of hypereosinophilic syndromes. Allergy 2004, 59, 673–689. [Google Scholar] [CrossRef] [PubMed]
- Simon, H.U.; Plötz, S.G.; Dummer, R.; Blaser, K. Abnormal clones of T cells producing interleukin-5 in idiopathic eosinophilia. N. Engl. J. Med. 1999, 341, 1112–1120. [Google Scholar] [CrossRef] [PubMed]
- Lefèvre, G.; Copin, M.C.; Staumont-Sallé, D.; Avenel-Audran, M.; Aubert, H.; Taieb, A.; Salles, G.; Maisonneuve, H.; Ghomari, K.; Ackerman, F.; et al. The lymphoid variant of hypereosinophilic syndrome: Study of 21 patients with CD3-CD4+ aberrant T-cell phenotype. Medicine 2014, 93, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Shi, M.; Rech, K.L.; Otteson, G.E.; Horna, P.; Olteanu, H.; Pardanani, A.; Chen, D.; Jevremovic, D. Prevalence and spectrum of T-cell lymphoproliferative disorders in patients with Hypereosinophilia: A reference laboratory experience. Ann. Diagn. Pathol. 2020, 44, 151412. [Google Scholar] [CrossRef] [PubMed]
- Requena, G.; van den Bosch, J.; Akuthota, P.; Kovalszki, A.; Steinfeld, J.; Kwon, N.; Van Dyke, M.K. Clinical Profile and Treatment in Hypereosinophilic Syndrome Variants: A Pragmatic Review. J. Allergy Clin. Immunol. Pract. 2022, 10, 2125–2134. [Google Scholar] [CrossRef] [PubMed]
- D’Souza, A.; Pardanani, A.; Tefferi, A. Lymphoma-associated versus lymphocytic-variant hypereosinophilia. Leuk. Lymphoma 2012, 53, 2103–2104. [Google Scholar] [CrossRef] [PubMed]
- Novembre, E.; Mori, F.; Arcangeli, F.; Cianferoni, A.; Bernardini, R.; Pucci, N.; Annunziato, F.; Parronchi, P.; De Martino, M.; Vierucci, A. High intracytoplasmatic levels of Il-4 and Il-5 in a patient with Gleichs syndrome: Case report. Int. J. Immunopathol. Pharmacol. 2006, 19, 935–938. [Google Scholar] [CrossRef] [PubMed]
- Cogan, E.; Schandene, L.; Crusiaux, A.; Cochaux, P.; Velu, T.; Goldman, M. Clonal Proliferation of Type 2 Helper T Cells in a Man with the Hypereosinophilic Syndrome. N. Engl. J. Med. 1994, 330, 535–538. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.A.; Xi, L.; Cauff, B.; DeZure, A.; Freeman, A.F.; Hambleton, S.; Kleiner, G.; Leahy, T.R.; O’Sullivan, M.; Makiya, M.; et al. Somatic STAT5b gain-of-function mutations in early onset nonclonal eosinophilia, urticaria, dermatitis, and diarrhea. Blood 2017, 129, 650–653. [Google Scholar] [CrossRef] [PubMed]
- Walker, S.; Wang, C.; Walradt, T.; Hong, B.S.; Tanner, J.R.; Levinsohn, J.L.; Goh, G.; Subtil, A.; Lessin, S.R.; Heymann, W.R.; et al. Identification of a gain-of-function STAT3 mutation (p.Y640F) in lymphocytic variant hypereosinophilic syndrome. Blood 2016, 127, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Umrau, K.; Naganuma, K.; Gao, Q.; Dogan, A.; Kizaki, M.; Roshal, M.; Liu, Y.; Yabe, M. Activating STAT5B mutations can cause both primary hypereosinophilia and lymphocyte-variant hypereosinophilia. Leuk. Lymphoma 2023, 64, 238–241. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, C. What we have learned about lymphocytic variant hypereosinophilic syndrome: A systematic literature review. Clin. Immunol. 2022, 237, 108982. [Google Scholar] [CrossRef] [PubMed]
- Gotlib, J. World Health Organization-defined eosinophilic disorders: 2017 update on diagnosis, risk stratification, and management. Am. J. Hematol. 2017, 92, 1243–1259. [Google Scholar] [CrossRef]
- Radin, M.; Bertero, L.; Roccatello, D.; Sciascia, S. Severe Multi-Organ Failure and Hypereosinophilia: When to Call It “Idiopathic”? J. Investig. Med. High. Impact Case Rep. 2018, 6, 2324709618758347. [Google Scholar] [CrossRef] [PubMed]
- Rosenfeld, K.; Stodell, M.A. Eosinophilic fasciitis in a father and son. Ann. Rheum. Dis. 1994, 53, 281. [Google Scholar] [CrossRef] [PubMed]
- Thomson, G.T.; MacDougall, B.; Watson, P.H.; Chalmers, I.M. Eosinophilic fasciitis in a pair of siblings. Arthritis Rheum. 1989, 32, 96–99. [Google Scholar] [CrossRef] [PubMed]
- Collins, M.H.; Blanchard, C.; Abonia, J.P.; Kirby, C.; Akers, R.; Wang, N.; Putnam, P.E.; Jameson, S.C.; Assa’ad, A.H.; Konikoff, M.R.; et al. Clinical, pathologic, and molecular characterization of familial eosinophilic esophagitis compared with sporadic cases. Clin. Gastroenterol. Hepatol. 2008, 6, 621–629. [Google Scholar] [CrossRef] [PubMed]
- Rioux, J.D.; Stone, V.A.; Daly, M.J.; Cargill, M.; Green, T.; Nguyen, H.; Nutman, T.; Zimmerman, P.A.; Tucker, M.A.; Hudson, T.; et al. Familial eosinophilia maps to the cytokine gene cluster on human chromosomal region 5q31-q33. Am. J. Hum. Genet. 1998, 63, 1086–1094. [Google Scholar] [CrossRef]
- Klion, A.D.; Law, M.A.; Riemenschneider, W.; McMaster, M.L.; Brown, M.R.; Horne, M.; Karp, B.; Robinson, M.; Sachdev, V.; Tucker, E.; et al. Familial eosinophilia: A benign disorder? Blood 2004, 103, 4050–4055. [Google Scholar] [CrossRef] [PubMed]
- Williams, K.W.; Milner, J.D.; Freeman, A.F. Eosinophilia Associated with Disorders of Immune Deficiency or Immune Dysregulation. Immunol. Allergy Clin. N. Am. 2015, 35, 523–544. [Google Scholar] [CrossRef] [PubMed]
- Khoury, P.; Herold, J.; Alpaugh, A.; Dinerman, E.; Holland-Thomas, N.; Stoddard, J.; Gurprasad, S.; Maric, I.; Simakova, O.; Schwartz, L.B.; et al. Episodic angioedema with eosinophilia (Gleich syndrome) is a multilineage cell cycling disorder. Haematologica 2015, 100, 300–307. [Google Scholar] [CrossRef]
- Nguyen, Y.; Guillevin, L. Eosinophilic Granulomatosis with Polyangiitis (Churg-Strauss). Semin. Respir. Crit. Care Med. 2018, 39, 471–481. [Google Scholar] [CrossRef] [PubMed]
- MacCarrick, G.; Black, J.H.; Bowdin, S.; El-Hamamsy, I.; Frischmeyer-Guerrerio, P.A.; Guerrerio, A.L.; Sponseller, P.D.; Loeys, B.; Dietz, H.C. Loeys–Dietz syndrome: A primer for diagnosis and management. Genet. Med. 2014, 16, 576–587. [Google Scholar] [CrossRef] [PubMed]
- Gouda, P.; Kay, R.; Habib, M.; Aziz, A.; Aziza, E.; Welsh, R. Clinical features and complications of Loeys-Dietz syndrome: A systematic review. Int. J. Cardiol. 2022, 362, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Montgomery, N.D.; Dunphy, C.H.; Mooberry, M.; Laramore, A.; Foster, M.C.; Park, S.I.; Fedoriw, Y.D. Diagnostic complexities of eosinophilia. Arch. Pathol. Lab. Med. 2013, 137, 259–269. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaki, M.; Shimamoto, Y.; Tokioka, T.; Suga, K.; Sueoka, E.; Ono, K.; Sano, M.; Yamaguchi, M. T-cell type non-Hodgkin’s lymphoma associated with eosinophilia. Rinsho Ketsueki 1991, 32, 874–878. [Google Scholar] [PubMed]
- Verstraeten, A.S.; De Weerdt, A.; van Den Eynden, G.; Van Marck, E.; Snoeckx, A.; Jorens, P.G. Excessive eosinophilia as paraneoplastic syndrome in a patient with non-small-cell lung carcinoma: A case report and review of the literature. Acta Clin. Belg. 2011, 66, 293–297. [Google Scholar] [CrossRef] [PubMed]
- Pandit, R.; Scholnik, A.; Wulfekuhler, L.; Dimitrov, N. Non-small-cell lung cancer associated with excessive eosinophilia and secretion of interleukin-5 as a paraneoplastic syndrome. Am. J. Hematol. 2007, 82, 234–237. [Google Scholar] [CrossRef] [PubMed]
- McNair, P.J.; Marshall, R.N.; Matheson, J.A. Important features associated with acute anterior cruciate ligament injury. N. Z. Med. J. 1990, 103, 537–539. [Google Scholar] [PubMed]
- Kuang, F.L.; De Melo, M.S.; Makiya, M.; Kumar, S.; Brown, T.; Wetzler, L.; Ware, J.M.; Khoury, P.; Collins, M.H.; Quezado, M.; et al. Benralizumab Completely Depletes Gastrointestinal Tissue Eosinophils and Improves Symptoms in Eosinophilic Gastrointestinal Disease. J. Allergy Clin. Immunol. Pract. 2022, 10, 1598–1605.e1592. [Google Scholar] [CrossRef] [PubMed]
- O’Connell, E.M.; Nutman, T.B. Eosinophilia in Infectious Diseases. Immunol. Allergy Clin. N. Am. 2015, 35, 493–522. [Google Scholar] [CrossRef] [PubMed]
- Prin, L.; Lefebvre, P.; Gruart, V.; Capron, M.; Storme, L.; Formstecher, P.; Loiseau, S.; Capron, A. Heterogeneity of human eosinophil glucocorticoid receptor expression in hypereosinophilic patients: Absence of detectable receptor correlates with resistance to corticotherapy. Clin. Exp. Immunol. 1989, 78, 383–389. [Google Scholar] [PubMed]
- Stokes, K.; Yoon, P.; Makiya, M.; Gebreegziabher, M.; Holland-Thomas, N.; Ware, J.; Wetzler, L.; Khoury, P.; Klion, A.D. Mechanisms of glucocorticoid resistance in hypereosinophilic syndromes. Clin. Exp. Allergy 2019, 49, 1598–1604. [Google Scholar] [CrossRef] [PubMed]
- Tzankov, A.; Reichard, K.K.; Hasserjian, R.P.; Arber, D.A.; Orazi, A.; Wang, S.A. Updates on eosinophilic disorders. Virchows Arch. 2023, 482, 85–97. [Google Scholar] [CrossRef]
- Khoury, P.; Desmond, R.; Pabon, A.; Holland-Thomas, N.; Ware, J.M.; Arthur, D.C.; Kurlander, R.; Fay, M.P.; Maric, I.; Klion, A.D. Clinical features predict responsiveness to imatinib in platelet-derived growth factor receptor-alpha-negative hypereosinophilic syndrome. Allergy 2016, 71, 803–810. [Google Scholar] [CrossRef] [PubMed]
- Cohen, P.; Pagnoux, C.; Mahr, A.; Arène, J.-P.; Mouthon, L.; Le Guern, V.; André, M.-H.; Gayraud, M.; Jayne, D.; Blöckmans, D.; et al. Churg-Strauss syndrome with poor-prognosis factors: A prospective multicenter trial comparing glucocorticoids and six or twelve cyclophosphamide pulses in forty-eight patients. Arthritis Care Res. 2007, 57, 686–693. [Google Scholar] [CrossRef] [PubMed]
- Parrillo, J.E.; Fauci, A.S.; Wolff, S.M. Therapy of the hypereosinophilic syndrome. Ann. Intern. Med. 1978, 89, 167–172. [Google Scholar] [CrossRef] [PubMed]
- Maritati, F.; Alberici, F.; Oliva, E.; Urban, M.L.; Palmisano, A.; Santarsia, F.; Andrulli, S.; Pavone, L.; Pesci, A.; Grasselli, C.; et al. Methotrexate versus cyclophosphamide for remission maintenance in ANCA-associated vasculitis: A randomised trial. PLoS ONE 2017, 12, e0185880. [Google Scholar] [CrossRef] [PubMed]
- Wechsler, M.E.; Akuthota, P.; Jayne, D.; Khoury, P.; Klion, A.; Langford, C.A.; Merkel, P.A.; Moosig, F.; Specks, U.; Cid, M.C.; et al. Mepolizumab or Placebo for Eosinophilic Granulomatosis with Polyangiitis. N. Engl. J. Med. 2017, 376, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Kuang, F.L.; Fay, M.P.; Ware, J.; Wetzler, L.; Holland-Thomas, N.; Brown, T.; Ortega, H.; Steinfeld, J.; Khoury, P.; Klion, A.D. Long-Term Clinical Outcomes of High-Dose Mepolizumab Treatment for Hypereosinophilic Syndrome. J. Allergy Clin. Immunol. Pract. 2018, 6, 1518–1527.e1515. [Google Scholar] [CrossRef] [PubMed]
- Dworetzky, S.I.; Hebrank, G.T.; Archibald, D.G.; Reynolds, I.J.; Farwell, W.; Bozik, M.E. The targeted eosinophil-lowering effects of dexpramipexole in clinical studies. Blood Cells Mol. Dis. 2017, 63, 62–65. [Google Scholar] [CrossRef] [PubMed]
- Panch, S.R.; Bozik, M.E.; Brown, T.; Makiya, M.; Prussin, C.; Archibald, D.G.; Hebrank, G.T.; Sullivan, M.; Sun, X.; Wetzler, L.; et al. Dexpramipexole as an oral steroid-sparing agent in hypereosinophilic syndromes. Blood 2018, 132, 501–509. [Google Scholar] [CrossRef] [PubMed]
- Dellon, E.S.; Peterson, K.A.; Murray, J.A.; Falk, G.W.; Gonsalves, N.; Chehade, M.; Genta, R.M.; Leung, J.; Khoury, P.; Klion, A.D.; et al. Anti-Siglec-8 Antibody for Eosinophilic Gastritis and Duodenitis. N. Engl. J. Med. 2020, 383, 1624–1634. [Google Scholar] [CrossRef] [PubMed]
- Hirano, I.; Dellon, E.S.; Hamilton, J.D.; Collins, M.H.; Peterson, K.; Chehade, M.; Schoepfer, A.M.; Safroneeva, E.; Rothenberg, M.E.; Falk, G.W.; et al. Efficacy of Dupilumab in a Phase 2 Randomized Trial of Adults With Active Eosinophilic Esophagitis. Gastroenterology 2020, 158, 111–122.e110. [Google Scholar] [CrossRef] [PubMed]
- Faguer, S.; Groh, M.; Vergez, F.; Hunault-Berger, M.; Duployez, N.; Renaudineau, Y.; Paul, C.; Lefevre, G.; Kahn, J.E. JAK inhibition for CD3(-) CD4(+) lymphocytic-variant hypereosinophilic syndrome. Clin. Immunol. 2023, 251, 109275. [Google Scholar] [CrossRef]
- Cai, P.; Liu, S.; Duan, L.; Huo, L.; Wu, D.; Chen, S.; Yang, R.; Yang, X. Sustained Response to Ruxolitinib of Eosinophilia-Associated Myeloproliferative Neoplasm with Translocation t(8;9)(p21;p24). Acta Haematol. 2023, 146, 397–400. [Google Scholar] [CrossRef] [PubMed]
- Kawashima, I.; Kirito, K. Chronic neutrophilic leukemia/chronic eosinophilic leukemia. Rinsho Ketsueki 2023, 64, 1326–1334. [Google Scholar] [CrossRef] [PubMed]
- Cooper, M.A.; Akard, L.P.; Thompson, J.M.; Dugan, M.J.; Jansen, J. Hypereosinophilic syndrome: Long-term remission following allogeneic stem cell transplant in spite of transient eosinophilia post-transplant. Am. J. Hematol. 2005, 78, 33–36. [Google Scholar] [CrossRef]
- Juvonen, E.; Volin, L.; Koponen, A.; Ruutu, T. Allogeneic blood stem cell transplantation following non-myeloablative conditioning for hypereosinophilic syndrome. Bone Marrow Transplant. 2002, 29, 457–458. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Archimbaud, E.; Guyotat, D.; Guillaume, C.; Godard, J.; Fiere, D. Hypereosinophilic syndrome with multiple organ dysfunction treated by allogeneic bone marrow transplantation. Am. J. Hematol. 1988, 27, 302–303. [Google Scholar] [CrossRef] [PubMed]
- Pitlick, M.M.; Li, J.T.; Pongdee, T. Current and emerging biologic therapies targeting eosinophilic disorders. World Allergy Organ. J. 2022, 15, 100676. [Google Scholar] [CrossRef]
- Bernstein, J.S.; Wechsler, M.E. Eosinophilic respiratory disorders and the impact of biologics. Curr. Opin. Pulm. Med. 2023, 29, 202–208. [Google Scholar] [CrossRef]
- Du, X.; Chen, Y.; Chang, J.; Sun, X.; Zhang, Y.; Zhang, M.; Maurer, M.; Li, Y.; Zhao, Z.; Tong, X. Dupilumab as a novel steroid-sparing treatment for hypereosinophilic syndrome. JAAD Case Rep. 2022, 29, 106–109. [Google Scholar] [CrossRef] [PubMed]
- Eckhardt, K.S.; Münzel, T.; Gräb, J.; Berg, T. Stafiba: A STAT5-Selective Small-Molecule Inhibitor. Chembiochem 2023, 24, e202200553. [Google Scholar] [CrossRef] [PubMed]
| Primary/Clonal HE | Secondary/Reactive HE | ||
|---|---|---|---|
| Category | Example | Category | Example |
| Myeloproliferative variants of HE (M-HES) | AML ALL MPN (CEL) MLN-TK (PDGFRA, PDGFRB, FGFR1, JAK2, FLT3 rearrangement, and ETV6::ABL1 fusion and other variants) | Fungal, parasite, protozoal, viral, or mycobacterial infection | Systemic fungal infection (Coccidioides), helminth infection (tissue invasion phase), ectoparasites, Sarcocystis and Cystoisospora, HIV, COVID-19, tuberculosis (rare) |
| T-lymphocytic variant (L-HES) | L-HES | Atopic/Allergy | Atopic dermatitis, chronic rhinosinusitis, asthma, drug hypersensitivity |
| Idiopathic HE | Autoimmune and immune dysregulation | Connective tissue disorders, UC, Crohn’s disease, IgG4-related disease, sarcoidosis | |
| Category | Example | Neoplasm | Leukemia, lymphoma (cHL, T-cell), solid tumor |
| Idiopathic HES (iHES) | HE with organ damage | Therapy/medication | Radiation, interleukin or GM-CSF therapy |
| HE of unknown significance (HEUS) | HE without organ damage and unknown etiology | Other | Addison’s disease, cholesterol emboli |
| Organ-restricted HE | Eosinophilic esophagitis, Eosinophilic gastrointestinal disorders, Eosinophilic dermatitis, Chronic eosinophilic pneumonia | Family HE/Inborn errors in immunity (IEIs) * | Omenn syndrome, Wiskott-Aldrich syndrome, Netherton syndrome, and Hyper IgE syndrome (DOCK8 deficiency), Loeys-Dietz syndrome |
| Symptoms and Signs | Functional Tests | Tissue Biopsy | |
|---|---|---|---|
| Cardiac (myocarditis) | Dyspnea, arrythmia, ischemic attack | Serum troponin ECG, EchoCG, MRI | N/A |
| Lung | Dyspnea, hypoxemia, eosinophilic pleural effusions | CXR, chest CT, pulmonary function testing | Bronchoalveolar lavage, Lung bx, Pleural fluid cytology |
| GI tract (eosinophilic esophagitis, gastritis, enteritis, colitis) | Esophagus: reflux, dysphagia Bowel: abdominal pain, GI bleed, ischemia | Serum LFT, amylase, lipase | Endoscopic tissue bx |
| Cutaneous | Urticaria, angioedema, rash, erythematous papules, or nodules | N/A | Skin bx |
| Renal | Chronic UTI-like symptoms | Serum creatinine level, Urine eosinophils | Kidney bx |
| Neurologic | Peripheral neuropathy, TIA, stroke | Head MRI, Head CT, Nerve conduction studies | Nerve biopsy (rarely performed) |
| Clinical | Laboratory/ Pathology | Molecular/ Genetic Study | |
|---|---|---|---|
| Secondary (reactive) | |||
| Allergy | Including atopic or non-atopic diseases: ABPA, asthma, allergic rhinitis, ECRS, NARES, food allergies, atopic dermatitis, drug allergies (e.g., DRESS), eosinophilic otitis media, eosinophilic laryngitis | Elevated IgE level | N/A |
| Infection | Infected microorganism related signs and symptoms Parasitic (Toxocara, Toxoplasma, Strongyloides, Ascariasis, Trichinella, Echinococcus, scabies, microfilariae), Fungal (Coccidioides), Viral (HIV, HCV) | Positive culture of microorganisms, elevated viral load or antibody titers, identification of parasites | PCR or NGS positive for specific microorganisms |
| Autoimmune | Connective tissue disorders, sarcoidosis, IBD, bullous pemphigoid, systemic vasculitis, granulomatosis with polyangiitis, eosinophilic granulomatosis with polyangiitis (Churg–Strauss syndrome) | Depending on disease type, presence of rheumatoid factor, ANA, anti-dsDNA, etc. | N/A |
| Immunodeficiency | Hyper IgE syndrome (Job syndrome), Omenn syndrome | Markedly elevated IgE level | STAT3 mutations (Job syndrome); RAG1 and RAG2 mutations (Omenn syndrome) |
| Organ specific HE | Esophagitis (dyspepsia, dysphasia, reflux), gastroenteritis, cystitis, pneumonia (cough), dermatologic conditions (rash, pruritis) | Tissue infiltration by eosinophils, infectious or neoplastic etiologies have been excluded | N/A |
| Therapy/ Medication | Radiation, IL-2, IL-3, IL-5, or GM-CSF | N/A | N/A |
| Endocrine disorders | Addison’s disease | Decreased aldosterone, increased ACTH | N/A |
| Rare diseases | Gleich syndrome (episodic angioedema, eosinophilia, polyclonal IgM) Eosinophilia-myalgia syndrome | Eosinophilia, polyclonal IgM for Gleich syndrome | N/A |
| Other | GvHD, cholesterol embolization, radiation exposure | GvHD specific findings | Post-engraftment analysis for GvHD |
| Primary (clonal) | |||
| MLN-TK | Variable, male predominance, hepatosplenomegaly, anemia; Good response to imatinib or other TKI, Variable steroid response | Concurrent or subsequent myeloid and lymphoid neoplasms; increased serum B12, thrombocytopenia, dysplastic eosinophils ± myelofibrosis, leukoerythroblastosis | FISH, RT-PCR or RNA sequencing for PDGFRA (CHIC deletion), PDGFRB, FGFR1, JAK2, or FLT3 fusions, and ETV6::ABL1 fusion |
| CEL | Asymptomatic or symptomatic (B-symptoms), systemic involvement | Eosinophilia > 1.5 × 109/L on at least two occasions over an interval of 4+ weeks, clonality identified, abnormal BM morphology, <20% blasts. Excludes: CHIP, MPN, MDS/MPN, MDS, MLN-TK, SM and AML with inv(16). Tissue eosinophilic infiltrate can be seen | Clonal abnormalities, e.g., mutations involving ASXl1, DNMT3A, EZH2, TET2, SRSF2, SETBP1, and CBL (VAF ≥ 10%, more than one mutation preferred) |
| KIT-mutated SM | Depends on affected organ or tissue, asymptomatic to pruritic (skin), diarrhea (GI), organomegaly | Increased serum tryptase | PCR or NGS for c-KIT mutation |
| Lymphoid variant of HE (L-HES) | Male = Female, may manifest with skin lesions, GI symptoms, or obstructive lung disease; potential progression to T-cell lymphoma, rare cardiac involvement, responds to steroids with good outcome | Abnormal T-cell population (often sCD3−/CD4+), increased IL-4 and IL-5 levels, increased serum IgE, increased TARC (thymus activation regulated chemokine) | Clonal TCR gene rearrangement detected by PCR |
| Paraneoplastic HE | |||
| AML | Cytopenia related signs and symptoms (e.g., pallor, infection, bleeding) | Circulating blasts ± HE; bone marrow with increased blasts (can be < 20%), immature eosinophilic precursors | inv(16), t(16;16)/CBFB::MYH11 |
| B-ALL | Cytopenia related signs and symptoms (e.g., pallor, infection, bleeding), ±lymphadenopathy or splenomegaly | PB and BM loaded with B-lymphoblasts and increased eosinophils | t(5;14)(q31.1;q32.3) /IGH::IL3 |
| MDS | Asymptomatic, weakness, fatigue | Cytopenia, thrombocytosis seen in MDS with del(5q); bone marrow dysplasia ≥ 10% of each lineage, ±increased blasts | Del(5q)/-5, del(7q)/-7, +8, del(20q)/-20, del(17p)/-17, KMT2A/MLL rearrangement SF3B1 or TP53 mutation * |
| MDS/MPN | ±Splenomegaly | Leukocytosis, commonly monocytosis or thrombocytosis, BM with mixed myelodysplastic and myeloproliferative features | Cytogenetic alterations related to MDS and/or MPN, molecular changes related to SF3B1, JAK2, CALR, MPL can be identified |
| SM, MPN other than CEL | Pruritis for SM, splenomegaly | Leukocytosis for CML; elevated serum tryptase level for SM | BCR::ABL1 for CML, c-KIT D816V or other variants for SM, JAK2, CALR, or MPL mutation |
| cHL | B-symptoms, lymphadenopathy | Tissue biopsy with Reed-Sternberg or Hodgkin cells positive for CD30, CD15, dim PAX-5 and MUM1 and negative for CD20, and CD45 | B-cell gene rearrangement by PCR |
| LCH | Lytic bone lesions, skin lesions | Tissue biopsy with Langerhans cell proliferation and eosinophilic infiltrate, positive for CD1a, S100, Langerin, +/− BRAF | 40% with BRAF V600E |
| T-cell neoplasms | AILT, PTCL | Sheets of abnormal proliferation of neoplastic T-cells, along with reactive histiocytes, or plasma cells and EBV+ B-cells (AILT) | TET2, ROA mutation |
| Non-hematologic malignancies | Adenocarcinoma of the lung, gastrointestinal tract, pancreas, thyroid, genital and skin tumors | Solid tumor confirmed by tissue biopsy, elevated cancer markers (e.g., CEA, CA19.9, TSH) | Genomic study for gene alteration specific to the tumor |
| Idiopathic | |||
| iHES | Variable, mild to intensive pruritus, angioedema, accompany with organ damage related signs or symptoms | HE or tissue eosinophilic infiltrate, does not fulfill the dx criteria of reactive or neoplastic HE + eosinophilic organ damage | No clonal abnormality |
| HEUS | Nonspecific or asymptomatic; no evidence of eosinophilic organ damage-associated signs and symptoms | HE or tissue eosinophilic infiltrate, not fulfilling the dx criteria of reactive or neoplastic HE | None |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nguyen, L.; Saha, A.; Kuykendall, A.; Zhang, L. Clinical and Therapeutic Intervention of Hypereosinophilia in the Era of Molecular Diagnosis. Cancers 2024, 16, 1383. https://doi.org/10.3390/cancers16071383
Nguyen L, Saha A, Kuykendall A, Zhang L. Clinical and Therapeutic Intervention of Hypereosinophilia in the Era of Molecular Diagnosis. Cancers. 2024; 16(7):1383. https://doi.org/10.3390/cancers16071383
Chicago/Turabian StyleNguyen, Lynh, Aditi Saha, Andrew Kuykendall, and Ling Zhang. 2024. "Clinical and Therapeutic Intervention of Hypereosinophilia in the Era of Molecular Diagnosis" Cancers 16, no. 7: 1383. https://doi.org/10.3390/cancers16071383
APA StyleNguyen, L., Saha, A., Kuykendall, A., & Zhang, L. (2024). Clinical and Therapeutic Intervention of Hypereosinophilia in the Era of Molecular Diagnosis. Cancers, 16(7), 1383. https://doi.org/10.3390/cancers16071383






