FXR Agonism with Bile Acid Mimetic Reduces Pre-Clinical Triple-Negative Breast Cancer Burden
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Cell Lines and Media
2.3. Proliferation Assay
2.4. Cell Viability Assay
2.5. Migration Assay
2.6. Western Immunoblot
2.7. Xenograft Model
2.8. In Silico Analysis
2.9. RNA-Seq and Pathway Analysis of MDA-MB-231 Tumors
2.10. Gene Set Enrichment Analysis (GSEA) of MDA-MB-231 Tumor RNA
2.11. Statistical Analysis
3. Results
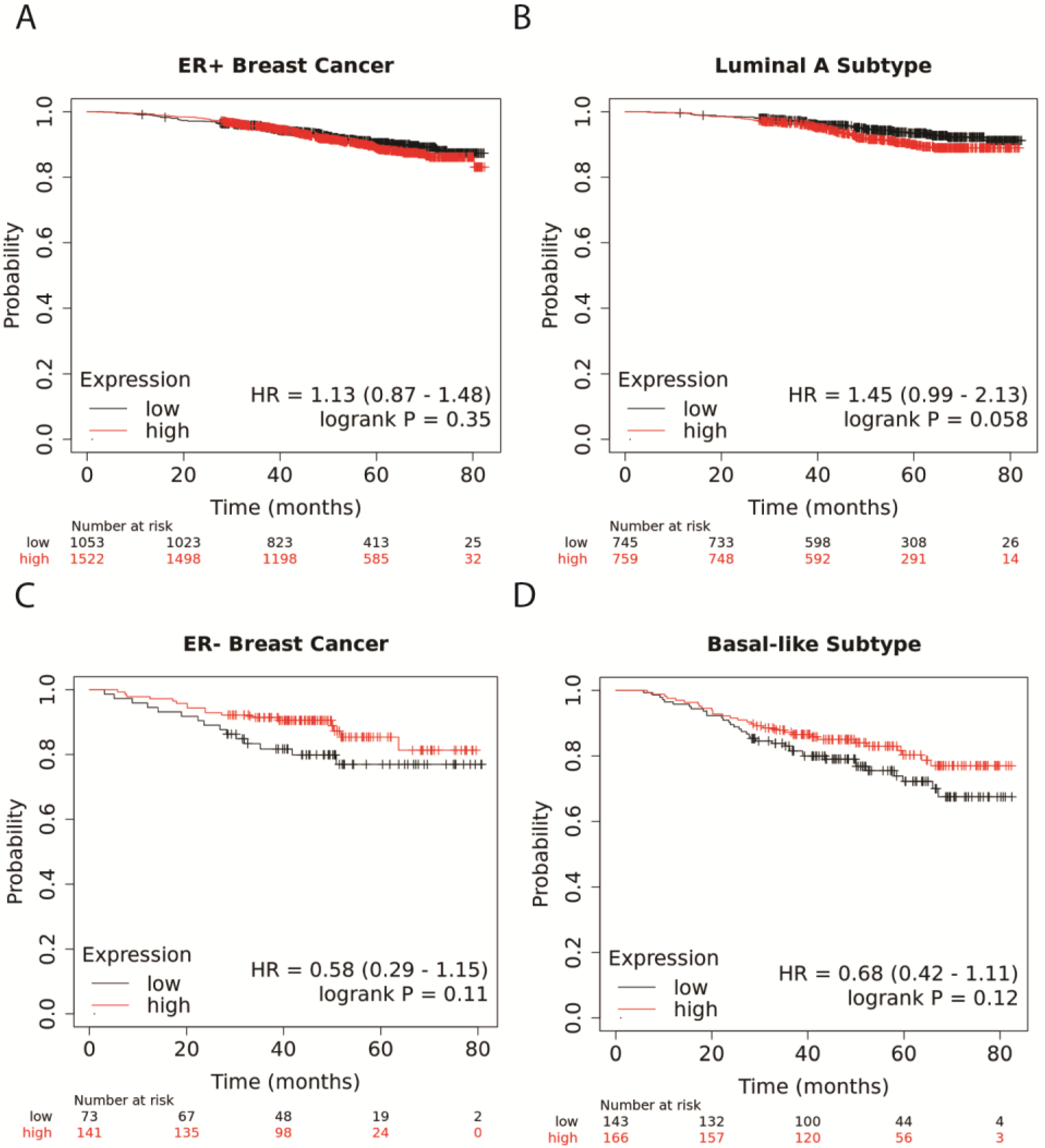
3.1. Higher NR1H4 Expression Is Associated with Greater Survival in Patients with ER− and Basal-like BC Subtypes
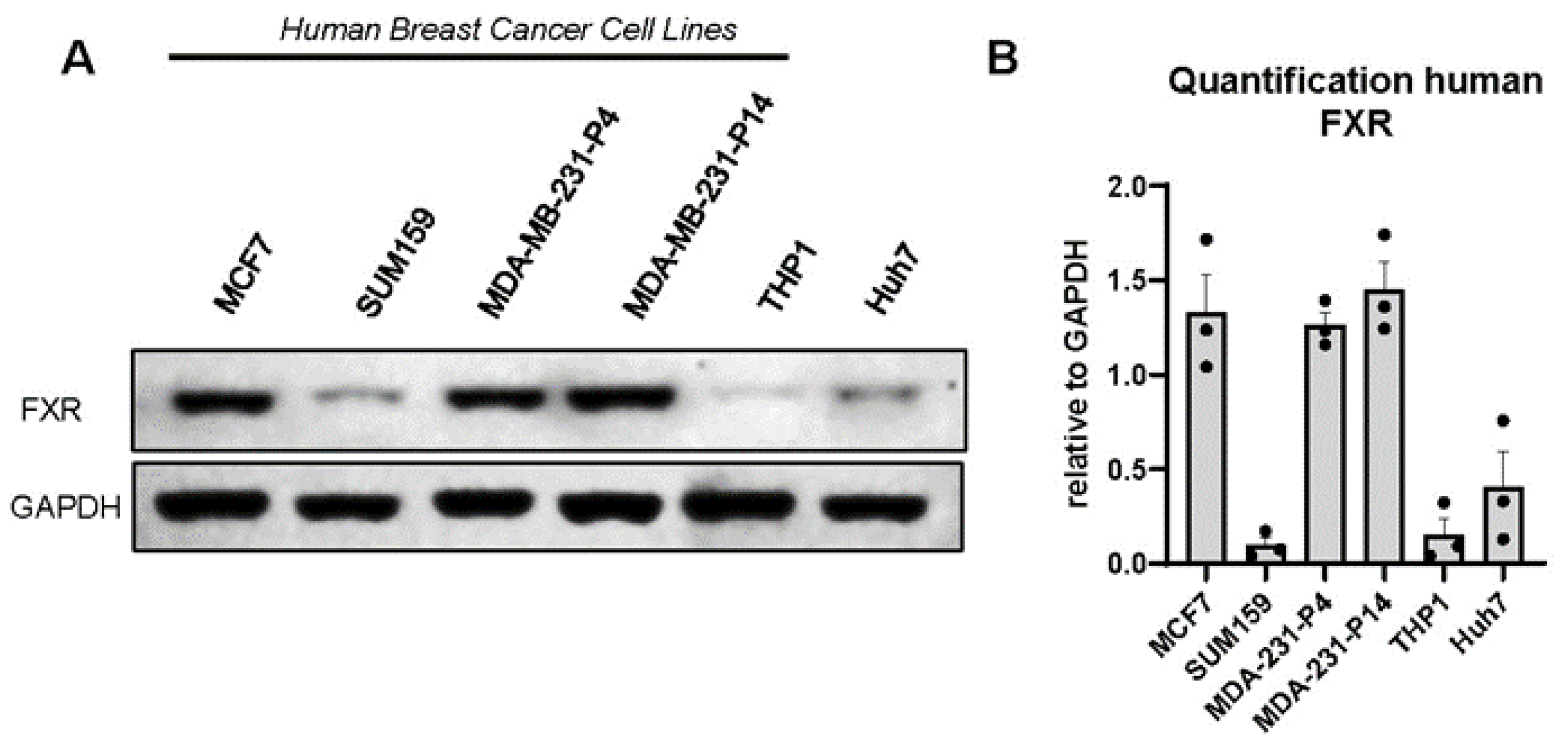
3.2. FXR Is Expressed in Human and Murine Triple-Negative Breast Cancer Cell Lines
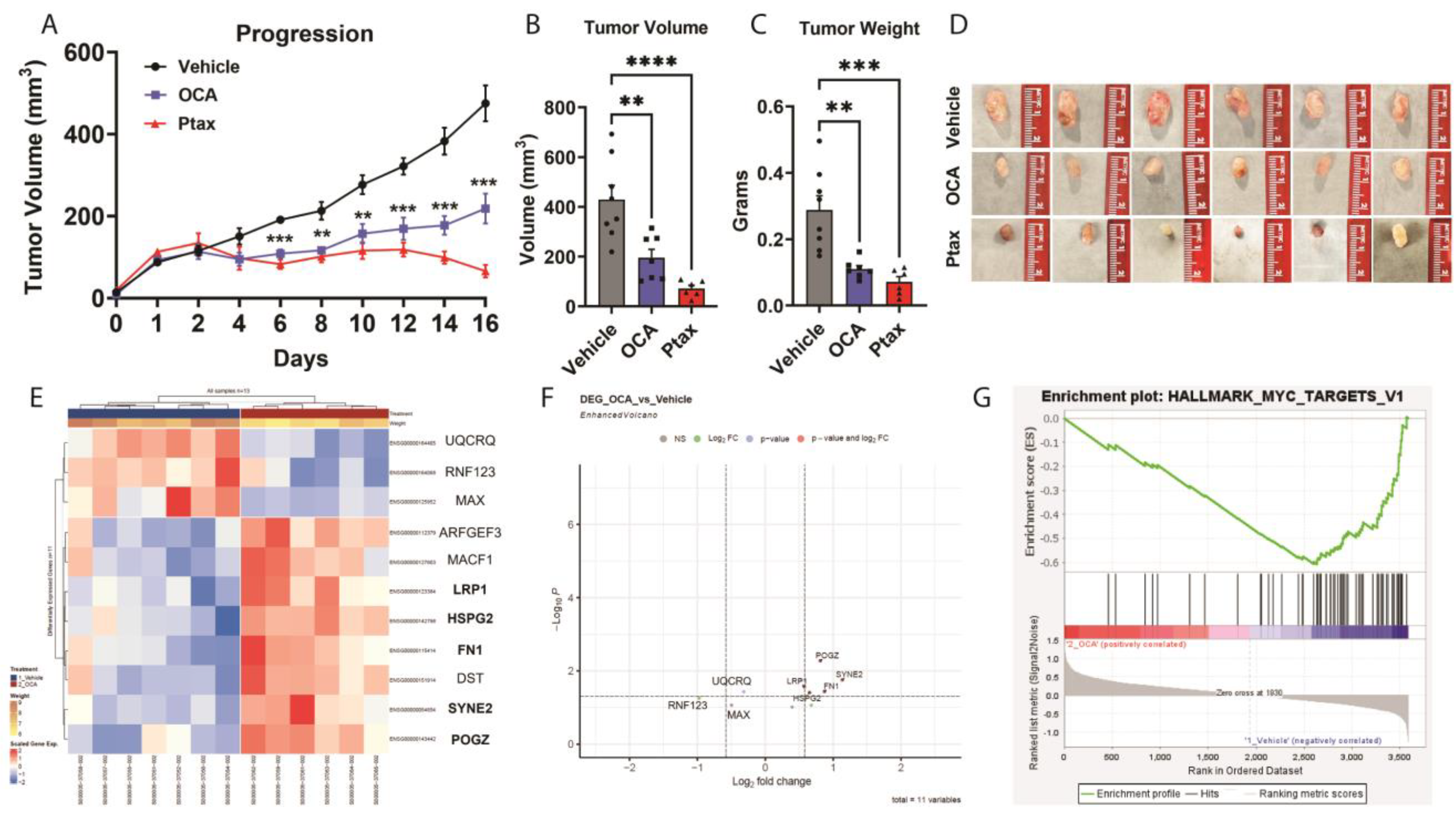
3.3. OCA Treatment Reduced TNBC Tumor Growth in Xenograft Model
3.4. Transcriptomic and GSEA Analysis of OCA-Treated TNBC Tumors Reveal Downregulation of Hallmark MYC Target Gene Set
3.5. Agonism of FXR Reduces Triple-Negative Breast Cancer Cell Proliferation, Viability, and Migration
3.6. TGR5 Bile Acid Receptor Agonism Failed to Reduce Triple-Negative Breast Cancer Cell Proliferation, Viability, and Migration
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA A Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Sipe, L.M.; Chaib, M.; Pingili, A.K.; Pierre, J.F.; Makowski, L. Microbiome, bile acids, and obesity: How microbially modified metabolites shape anti-tumor immunity. Immunol. Rev. 2020, 295, 220–239. [Google Scholar] [CrossRef]
- Bohm, M.S.; Sipe, L.M.; Pye, M.E.; Davis, M.J.; Pierre, J.F.; Makowski, L. The role of obesity and bariatric surgery-induced weight loss in breast cancer. Cancer Metastasis Rev. 2022, 41, 673–695. [Google Scholar] [CrossRef] [PubMed]
- Mikó, E.; Vida, A.; Kovács, T.; Ujlaki, G.; Trencsényi, G.; Márton, J.; Sári, Z.; Kovács, P.; Boratkó, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. et Biophys. Acta (BBA) Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef] [PubMed]
- Režen, T.; Rozman, D.; Kovács, T.; Kovács, P.; Sipos, A.; Bai, P.; Mikó, E. The role of bile acids in carcinogenesis. Cell. Mol. Life Sci. 2022, 79, 1–39. [Google Scholar] [CrossRef] [PubMed]
- Chiang, J.Y.; Ferrell, J.M. Discovery of farnesoid X receptor and its role in bile acid metabolism. Mol. Cell. Endocrinol. 2022, 548, 111618. [Google Scholar] [CrossRef] [PubMed]
- Schumacher, J.D.; Guo, G.L. Pharmacologic Modulation of Bile Acid-FXR-FGF15/FGF19 Pathway for the Treatment of Nonalcoholic Steatohepatitis. Handb Exp Pharmacol 2019, 256, 325–357. [Google Scholar] [PubMed]
- Huang, X.; Fan, M.; Huang, W. Pleiotropic roles of FXR in liver and colorectal cancers. Mol. Cell. Endocrinol. 2022, 543, 111543. [Google Scholar] [CrossRef]
- Kao, C.-C.; Lai, C.-R.; Lin, Y.-H.; Chen, T.-M.; Tsai, Y.-L.; Tsai, W.-C.; Ong, T.-Y.; Wang, H.-H.; Wu, S.-T.; Chen, Y. GW4064 inhibits migration and invasion through cathepsin B and MMP2 downregulation in human bladder cancer. Chem. Interactions 2024, 110869. [Google Scholar] [CrossRef]
- Golonka, R.M.; Yeoh, B.S.; Saha, P.; Tian, Y.; Chiang, J.Y.; Patterson, A.D.; Gewirtz, A.T.; Joe, B.; Vijay-Kumar, M. Sex Dimorphic Effects of Bile Acid Metabolism in Liver Cancer in Mice. Cell. Mol. Gastroenterol. Hepatol. 2024, 17, 719–735. [Google Scholar] [CrossRef]
- Stofan, M.; Guo, G.L. Bile Acids and FXR: Novel Targets for Liver Diseases. Front. Med. 2020, 7, 544. [Google Scholar] [CrossRef] [PubMed]
- Pingili, A.K.; Chaib, M.; Sipe, L.M.; Miller, E.J.; Teng, B.; Sharma, R.; Yarbro, J.R.; Asemota, S.; Al Abdallah, Q.; Mims, T.S.; et al. Immune checkpoint blockade reprograms systemic immune landscape and tumor microenvironment in obesity-associated breast cancer. Cell Rep. 2021, 35, 109285. [Google Scholar] [CrossRef] [PubMed]
- Chaib, M.; Sipe, L.M.; Yarbro, J.R.; Bohm, M.S.; Counts, B.R.; Tanveer, U.; Pingili, A.K.; Daria, D.; Marion, T.N.; Carson, J.A.; et al. PKC agonism restricts innate immune suppression, promotes antigen cross-presentation and synergizes with agonistic CD40 antibody therapy to activate CD8+ T cells in breast cancer. Cancer Lett. 2022, 531, 98–108. [Google Scholar] [CrossRef] [PubMed]
- Sipe, L.M.; Chaib, M.; Korba, E.B.; Jo, H.; Lovely, M.C.; Counts, B.R.; Tanveer, U.; Holt, J.R.; Clements, J.C.; A John, N.; et al. Response to immune checkpoint blockade improved in pre-clinical model of breast cancer after bariatric surgery. eLife 2022, 11, e79143. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.E.; Ahmed, U.; Saad, S.M.; Anwar, A.; Khan, K.M.; Tan, E.W.; Tan, K.O. New synthetic phenylquinazoline derivatives induce apoptosis by targeting the pro-survival members of the BCL-2 family. Bioorganic Med. Chem. Lett. 2022, 67, 128731. [Google Scholar]
- Chaib, M.; Holt, J.R.; Fisher, E.L.; Sipe, L.M.; Bohm, M.S.; Joseph, S.C.; Simmons, B.W.; Simon, S.E.; Yarbro, J.R.; Tanveer, U.; et al. Protein kinase C delta regulates mononuclear phagocytes and hinders response to immunotherapy in cancer. Sci. Adv. 2023, 9, eadd3231. [Google Scholar] [CrossRef] [PubMed]
- Györffy, B.; Lanczky, A.; Eklund, A.C.; Denkert, C.; Budczies, J.; Li, Q.; Szallasi, Z. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res. Treat. 2009, 123, 725–731. [Google Scholar] [CrossRef] [PubMed]
- Győrffy, B. Survival analysis across the entire transcriptome identifies biomarkers with the highest prognostic power in breast cancer. Comput. Struct. Biotechnol. J. 2021, 19, 4101–4109. [Google Scholar] [CrossRef] [PubMed]
- Clough, E.; Barrett, T.; E Wilhite, S.; Ledoux, P.; Evangelista, C.; Kim, I.F.; Tomashevsky, M.; A Marshall, K.; Phillippy, K.H.; Sherman, P.M.; et al. NCBI GEO: Archive for gene expression and epigenomics data sets: 23-year update. Nucleic Acids Res. 2023, 52, D138–D144. [Google Scholar] [CrossRef]
- Chang, A.; Jeske, L.; Ulbrich, S.; Hofmann, J.; Koblitz, J.; Schomburg, I.; Neumann-Schaal, M.; Jahn, D.; Schomburg, D. BRENDA, the ELIXIR core data resource in 2021: New developments and updates. Nucleic Acids Res. 2021, 49, D498–D508. [Google Scholar] [CrossRef]
- Drysdale, R.; E Cook, C.; Petryszak, R.; Baillie-Gerritsen, V.; Barlow, M.; Gasteiger, E.; Gruhl, F.; Haas, J.; Lanfear, J.; Lopez, R.; et al. The ELIXIR Core Data Resources: Fundamental infrastructure for the life sciences. Bioinformatics 2020, 36, 2636–2642. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Aksoy, B.A.; Dogrusoz, U.; Dresdner, G.; Gross, B.E.; Sumer, S.O.; Sun, Y.; Jacobsen, A.; Sinha, R.; Larsson, E.; et al. Integrative Analysis of Complex Cancer Genomics and Clinical Profiles Using the cBioPortal. Sci. Signal. 2013, 6, pl1. [Google Scholar] [CrossRef] [PubMed]
- Cerami, E.; Gao, J.; Dogrusoz, U.; Gross, B.E.; Sumer, S.O.; Aksoy, B.A.; Jacobsen, A.; Byrne, C.J.; Heuer, M.L.; Larsson, E.; et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012, 2, 401–404. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Kundra, R.; Mastrogiacomo, B.; Tran, T.N.; Sikina, L.; Mazor, T.; Li, X.; Ochoa, A.; Zhao, G.; Lai, B.; et al. Analysis and Visualization of Longitudinal Genomic and Clinical Data from the AACR Project GENIE Biopharma Collaborative in cBioPortal. Cancer Res 2023, 83, 3861–3867. [Google Scholar] [CrossRef] [PubMed]
- Lánczky, A.; Győrffy, B. Web-Based Survival Analysis Tool Tailored for Medical Research (KMplot): Development and Implementation. J. Med Internet Res. 2021, 23, e27633. [Google Scholar] [CrossRef] [PubMed]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Bonfield, J.K.; Liddle, J.; Marshall, J.; Ohan, V.; Pollard, M.O.; Whitwham, A.; Keane, T.; McCarthy, S.A.; Davies, R.M.; et al. Twelve years of SAMtools and BCFtools. GigaScience 2021, 10, giab008. [Google Scholar] [CrossRef] [PubMed]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [PubMed]
- R Foundation. R Foundation for Statistical Computing; R Foundation: Vienna, Austria, 2014. [Google Scholar]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2016, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Gu, Z.; Eils, R.; Schlesner, M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 2016, 32, 2847–2849. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Liberzon, A.; Birger, C.; Thorvaldsdóttir, H.; Ghandi, M.; Mesirov, J.P.; Tamayo, P. The Molecular Signatures Database Hallmark Gene Set Collection. Cell Syst. 2015, 1, 417–425. [Google Scholar] [CrossRef] [PubMed]
- Jain, P.B.; Guerreiro, P.S.; Canato, S.; Janody, F. The spectraplakin Dystonin antagonizes YAP activity and suppresses tumourigenesis. Sci. Rep. 2019, 9, 1–17. [Google Scholar]
- Siouda, M.; Dujardin, A.D.; Dekeyzer, B.; Schaeffer, L.; Mulligan, P. Chromodomain on Y-like 2 (CDYL2) implicated in mitosis and genome stability regulation via interaction with CHAMP1 and POGZ. Cell. Mol. Life Sci. 2023, 80, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Guddeti, R.K.; Pacharla, H.; Yellapu, N.K.; Karyala, P.; Pakala, S.B. MORC2 and MAX contributes to the expression of glycolytic enzymes, breast cancer cell proliferation and migration. Med Oncol. 2023, 40, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Feigelson, H.S.; Caan, B.; Weinmann, S.; Leonard, A.C.; Powers, J.D.; Yenumula, P.R.; Arterburn, D.E.; Koebnick, C.; Altaye, M.; Schauer, D.P. Bariatric Surgery is Associated with Reduced Risk of Breast Cancer in Both Premenopausal and Postmenopausal Women. Ann. Surg. 2019, 272, 1053–1059. [Google Scholar] [CrossRef]
- Wang, W.; Cheng, Z.; Wang, Y.; Dai, Y.; Zhang, X.; Hu, S. Role of Bile Acids in Bariatric Surgery. Front. Physiol. 2019, 10, 374. [Google Scholar] [CrossRef]
- Giaginis, C.; Karandrea, D.; Alexandrou, P.; Giannopoulou, I.; Tsourouflis, G.; Troungos, C.; Danas, E.; Keramopoulos, A.; Patsouris, E.; Nakopoulou, L.; et al. High Farnesoid X Receptor (FXR) expression is a strong and independent prognosticator in invasive breast carcinoma. Neoplasma 2017, 64, 633–639. [Google Scholar] [CrossRef] [PubMed]
- Barone, I.; Vircillo, V.; Giordano, C.; Gelsomino, L.; Győrffy, B.; Tarallo, R.; Rinaldi, A.; Bruno, G.; Caruso, A.; Romeo, F.; et al. Activation of Farnesoid X Receptor impairs the tumor-promoting function of breast cancer-associated fibroblasts. Cancer Lett. 2018, 437, 89–99. [Google Scholar] [CrossRef] [PubMed]
- Girisa, S.; Henamayee, S.; Parama, D.; Rana, V.; Dutta, U.; Kunnumakkara, A.B. Targeting Farnesoid X receptor (FXR) for developing novel therapeutics against cancer. Mol. Biomed. 2021, 2, 1–23. [Google Scholar]
- Singh, N.; Yadav, M.; Singh, A.K.; Kumar, H.; Dwivedi, S.K.D.; Mishra, J.S.; Gurjar, A.; Manhas, A.; Chandra, S.; Yadav, P.N.; et al. Synthetic FXR Agonist GW4064 Is a Modulator of Multiple G Protein–Coupled Receptors. Mol. Endocrinol. 2014, 28, 659–673. [Google Scholar] [CrossRef]
- Swales, K.E.; Korbonits, M.; Carpenter, R.; Walsh, D.T.; Warner, T.D.; Bishop-Bailey, D. The Farnesoid X Receptor Is Expressed in Breast Cancer and Regulates Apoptosis and Aromatase Expression. Cancer Res 2006, 66, 10120–10126. [Google Scholar] [CrossRef] [PubMed]
- Baker, P.; Wilton, J.; Jones, C.; Stenzel, D.; Watson, N.; Smith, G. Bile acids influence the growth, oestrogen receptor and oestrogen-regulated proteins of MCF-7 human breast cancer cells. Br. J. Cancer 1992, 65, 566–572. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Barone, I.; Vircillo, V.; Panza, S.; Malivindi, R.; Gelsomino, L.; Pellegrino, M.; Rago, V.; Mauro, L.; Lanzino, M.; et al. Activated FXR Inhibits Leptin Signaling and Counteracts Tumor-promoting Activities of Cancer-Associated Fibroblasts in Breast Malignancy. Sci. Rep. 2016, 6, 21782. [Google Scholar] [CrossRef] [PubMed]
- Giordano, C.; Catalano, S.; Panza, S.; Vizza, D.; Barone, I.; Bonofiglio, D.; Gelsomino, L.; Rizza, P.; Fuqua, S.A.W.; Andò, S. Farnesoid X receptor inhibits tamoxifen-resistant MCF-7 breast cancer cell growth through downregulation of HER2 expression. Oncogene 2011, 30, 4129–4140. [Google Scholar] [CrossRef]
- Sepich-Poore, G.D.; Zitvogel, L.; Straussman, R.; Hasty, J.; Wargo, J.A.; Knight, R. The microbiome and human cancer. Science 2021, 371, 1331. [Google Scholar] [CrossRef]
- Gihawi, A.; Ge, Y.; Lu, J.; Puiu, D.; Xu, A.; Cooper, C.S.; Brewer, D.S.; Pertea, M.; Salzberg, S.L. Major data analysis errors invalidate cancer microbiome findings. mBio 2023, 14, e0160723. [Google Scholar] [CrossRef]
- Javitt, N.; Budai, K.; Raju, U.; Levitz, M.; Miller, D.; Cahan, A. Breast-gut connection: Origin of chenodeoxycholic acid in breast cyst fluid. Lancet 1994, 343, 633–635. [Google Scholar] [CrossRef] [PubMed]
- Raju, U.; Levitz, M.; Javitt, N.B. Bile Acids in Human Breast Cyst Fluid: The Identification of Lithocholic Acid*. J. Clin. Endocrinol. Metab. 1990, 70, 1030–1034. [Google Scholar] [CrossRef] [PubMed]
- Shively, C.A.; Register, T.C.; Appt, S.E.; Clarkson, T.B.; Uberseder, B.; Clear, K.Y.; Wilson, A.S.; Chiba, A.; Tooze, J.A.; Cook, K.L. Consumption of Mediterranean versus Western Diet Leads to Distinct Mammary Gland Microbiome Populations. Cell Rep. 2018, 25, 47–56.e3. [Google Scholar] [CrossRef] [PubMed]
- Aminian, A.; Wilson, R.; Al-Kurd, A.; Tu, C.; Milinovich, A.; Kroh, M.; Rosenthal, R.J.; Brethauer, S.A.; Schauer, P.R.; Kattan, M.W.; et al. Association of Bariatric Surgery with Cancer Risk and Mortality in Adults With Obesity. JAMA 2022, 327, 2423–2433. [Google Scholar] [CrossRef] [PubMed]
- Flegal, K.M.; Kruszon-Moran, D.; Carroll, M.D.; Fryar, C.D.; Ogden, C.L. Trends in Obesity Among Adults in the United States, 2005 to 2014. JAMA 2016, 315, 2284–2291. [Google Scholar] [CrossRef] [PubMed]
- Kanny, D.; Liu, Y.; Brewer, R.D.; Garvin, W.S.; Balluz, L. Vital Signs: Binge Drinking Prevalence, Frequency, and Intensity among Adults--United States, 2010. MMWR Morb. Mortal Wkly. Rep. 2012, 61, 14–19. [Google Scholar]
- Kohler, B.A.; Sherman, R.L.; Howlader, N.; Jemal, A.; Ryerson, A.B.; Henry, K.A.; Boscoe, F.P.; Cronin, K.A.; Lake, A.; Noone, A.-M.; et al. Annual Report to the Nation on the Status of Cancer, 1975–2011, Featuring Incidence of Breast Cancer Subtypes by Race/Ethnicity, Poverty, and State. J. Natl. Cancer Inst. 2015, 107, djv048. [Google Scholar] [CrossRef]
- Kuchenbaecker, K.B.; EMBRACE Study; Neuhausen, S.L.; Robson, M.; Barrowdale, D.; McGuffog, L.; Mulligan, A.M.; Andrulis, I.L.; Spurdle, A.B.; Schmidt, M.K.; et al. Associations of common breast cancer susceptibility alleles with risk of breast cancer subtypes in BRCA1 and BRCA2 mutation carriers. Breast Cancer Res. 2014, 16, 1–27. [Google Scholar] [CrossRef] [PubMed]
- Kovács, S.A.; Fekete, J.T.; Győrffy, B. Predictive biomarkers of immunotherapy response with pharmacological applications in solid tumors. Acta Pharmacol. Sin. 2023, 44, 1879–1889. [Google Scholar] [CrossRef]
- Obidiro, O.; Battogtokh, G.; Akala, E.O. Triple Negative Breast Cancer Treatment Options and Limitations: Future Outlook. Pharmaceutics 2023, 15, 1796. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Joseph, S.C.; Eugin Simon, S.; Bohm, M.S.; Kim, M.; Pye, M.E.; Simmons, B.W.; Graves, D.G.; Thomas-Gooch, S.M.; Tanveer, U.A.; Holt, J.R.; et al. FXR Agonism with Bile Acid Mimetic Reduces Pre-Clinical Triple-Negative Breast Cancer Burden. Cancers 2024, 16, 1368. https://doi.org/10.3390/cancers16071368
Joseph SC, Eugin Simon S, Bohm MS, Kim M, Pye ME, Simmons BW, Graves DG, Thomas-Gooch SM, Tanveer UA, Holt JR, et al. FXR Agonism with Bile Acid Mimetic Reduces Pre-Clinical Triple-Negative Breast Cancer Burden. Cancers. 2024; 16(7):1368. https://doi.org/10.3390/cancers16071368
Chicago/Turabian StyleJoseph, Sydney C., Samson Eugin Simon, Margaret S. Bohm, Minjeong Kim, Madeline E. Pye, Boston W. Simmons, Dillon G. Graves, Stacey M. Thomas-Gooch, Ubaid A. Tanveer, Jeremiah R. Holt, and et al. 2024. "FXR Agonism with Bile Acid Mimetic Reduces Pre-Clinical Triple-Negative Breast Cancer Burden" Cancers 16, no. 7: 1368. https://doi.org/10.3390/cancers16071368
APA StyleJoseph, S. C., Eugin Simon, S., Bohm, M. S., Kim, M., Pye, M. E., Simmons, B. W., Graves, D. G., Thomas-Gooch, S. M., Tanveer, U. A., Holt, J. R., Ponnusamy, S., Sipe, L. M., Hayes, D. N., Cook, K. L., Narayanan, R., Pierre, J. F., & Makowski, L. (2024). FXR Agonism with Bile Acid Mimetic Reduces Pre-Clinical Triple-Negative Breast Cancer Burden. Cancers, 16(7), 1368. https://doi.org/10.3390/cancers16071368







