Tumour Size and Overall Survival in a Cohort of Patients with Unifocal Glioblastoma: A Uni- and Multivariable Prognostic Modelling and Resampling Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethical Approval
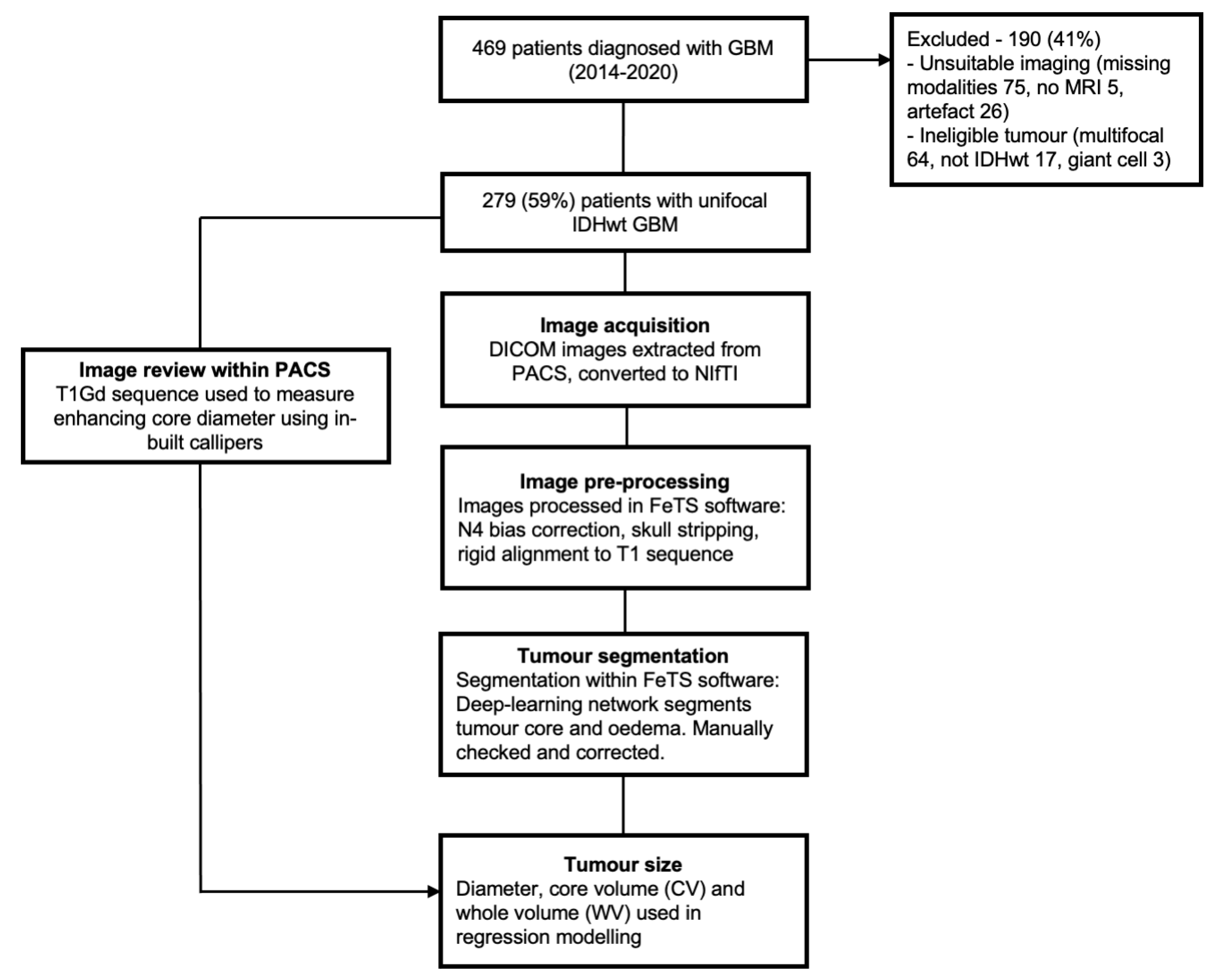
2.2. Patient Selection and Clinical Predictor Definitions
2.3. Data Preparation
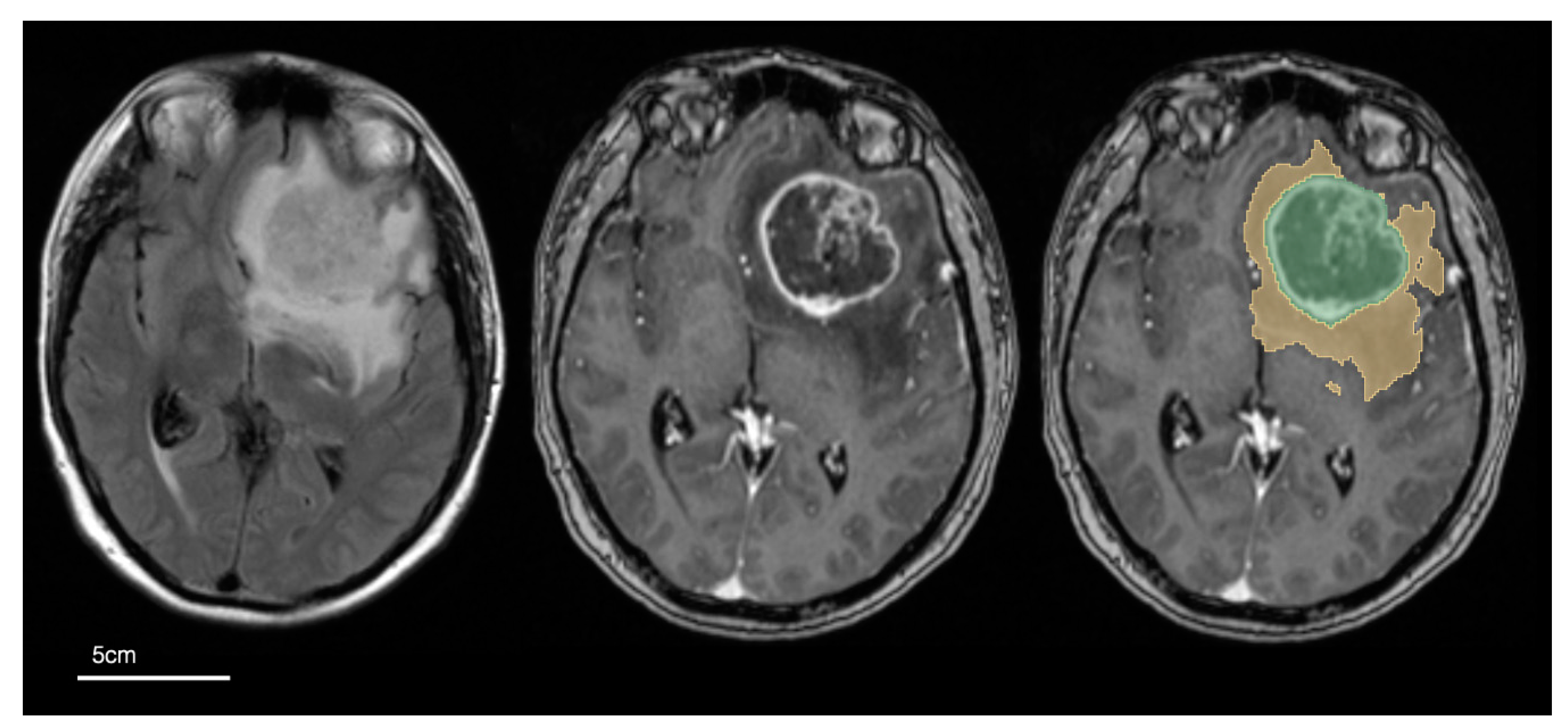
2.4. Image Pre-Processing and Tumour Segmentation
2.5. Statistical Analysis
3. Results
3.1. Demographics of the Study Population
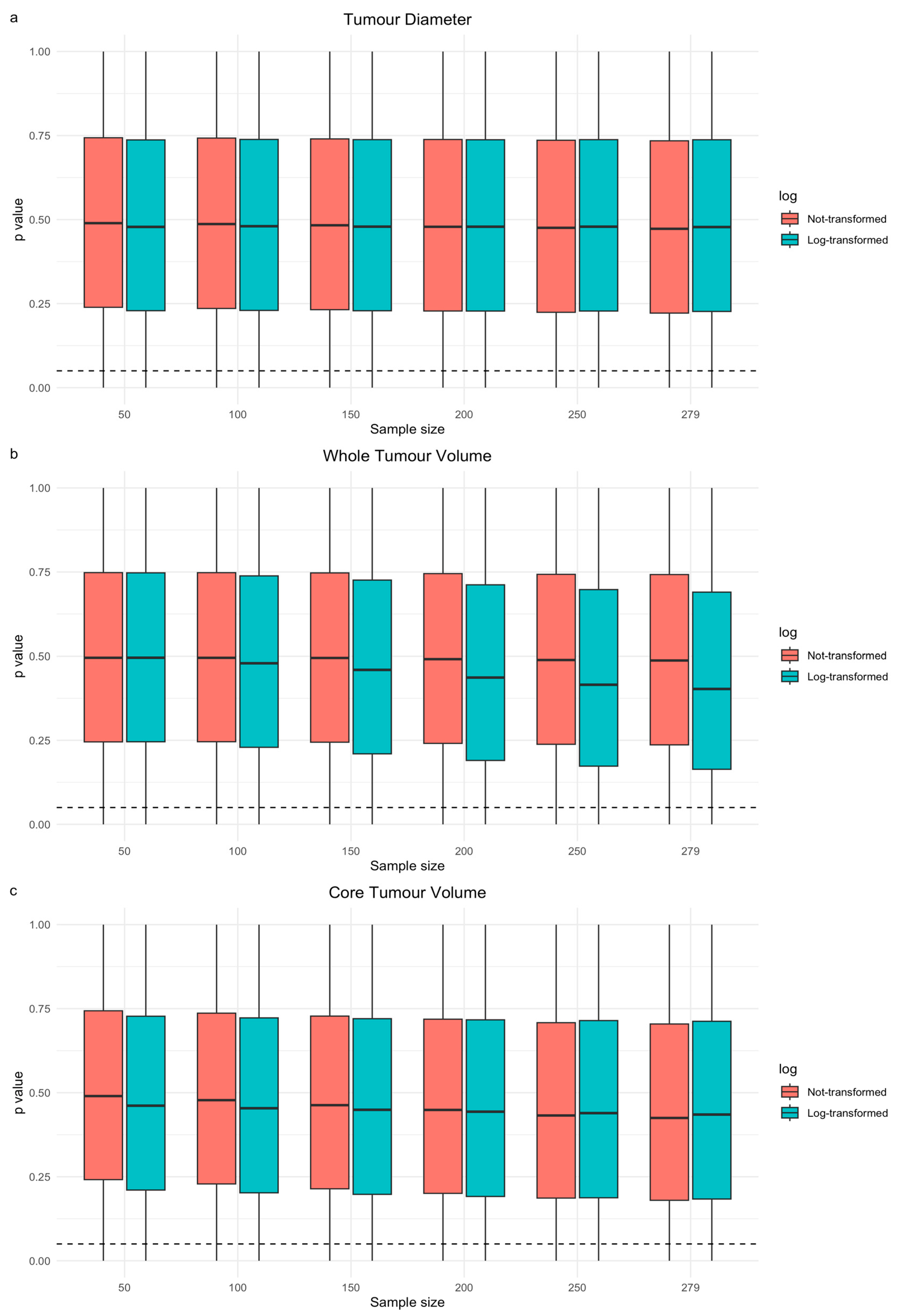
3.2. Segmentations and Univariable Cox Models of Tumour Size
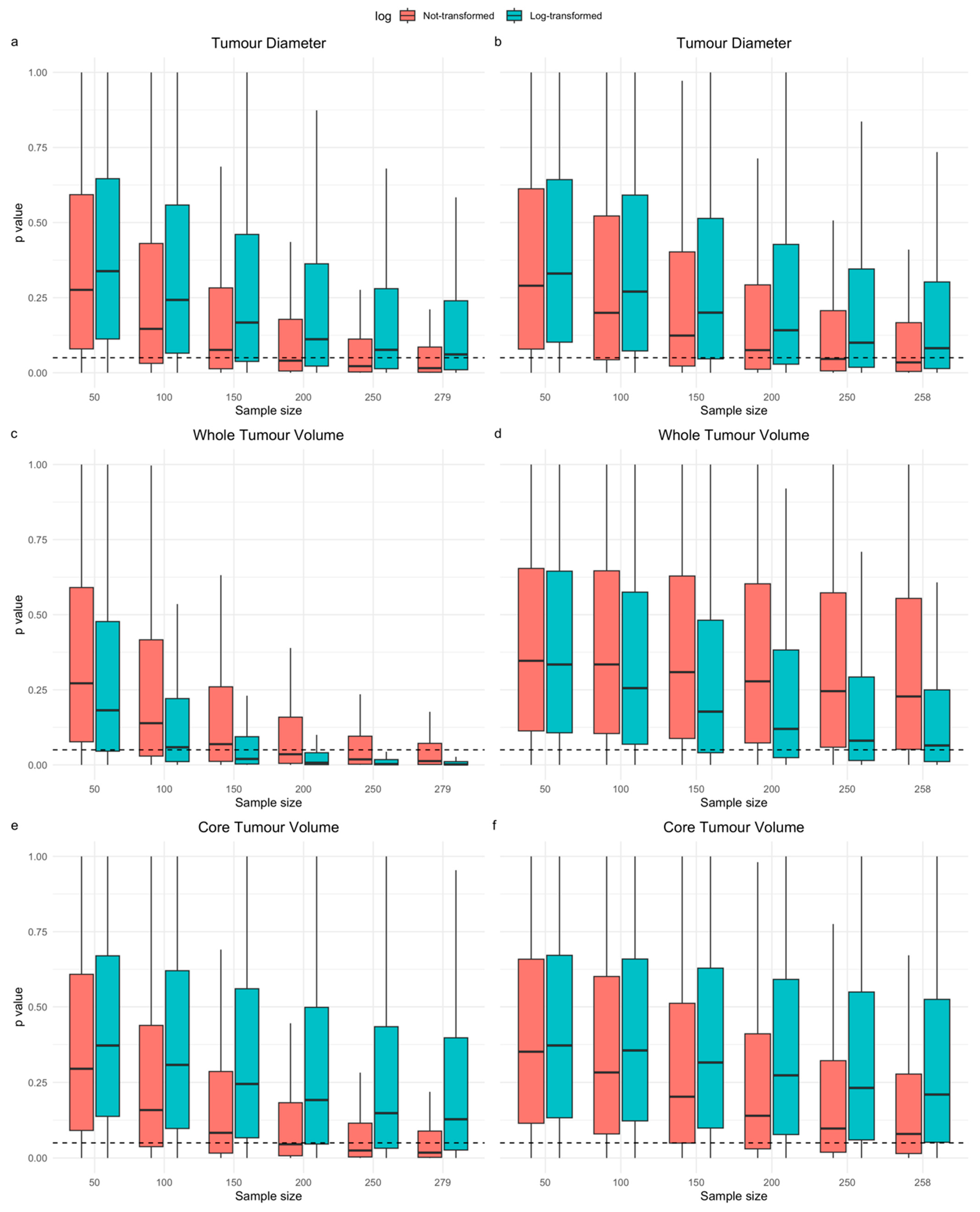
3.3. Resampling Study
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.B.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus Concomitant and Adjuvant Temozolomide for Glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef] [PubMed]
- Stupp, R.; Hegi, M.E.; Mason, W.P.; van den Bent, M.J.; Taphoorn, M.J.; Janzer, R.C.; Ludwin, S.K.; Allgeier, A.; Fisher, B.; Belanger, K.; et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009, 10, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Mowforth, O.D.; Brannigan, J.; El Khoury, M.; Sarathi, C.I.P.; Bestwick, H.; Bhatti, F.; Mair, R. Personalised therapeutic approaches to glioblastoma: A systematic review. Front. Med. 2023, 10, 1166104. [Google Scholar] [CrossRef] [PubMed]
- Tewarie, I.A.; Senders, J.T.; Kremer, S.; Devi, S.; Gormley, W.B.; Arnaout, O.; Smith, T.R.; Broekman, M.L.D. Survival prediction of glioblastoma patients—Are we there yet? A systematic review of prognostic modeling for glioblastoma and its clinical potential. Neurosurg. Rev. 2021, 44, 2047–2057. [Google Scholar] [CrossRef] [PubMed]
- Currie, S.; Fatania, K.; Matthew, R.; Wurdak, H.; Chakrabarty, A.; Murray, L.; Short, S. A Comprehensive Clinical Review of Adult-Type Diffuse Glioma Incorporating the 2021 World Health Organization Classification. Neurographics 2022, 12, 43–70. [Google Scholar] [CrossRef]
- Henker, C.; Kriesen, T.; Glass, Ä.; Schneider, B.; Piek, J. Volumetric quantification of glioblastoma: Experiences with different measurement techniques and impact on survival. J. Neuro-Oncol. 2017, 135, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Menze, B.H.; Jakab, A.; Bauer, S.; Kalpathy-Cramer, J.; Farahani, K.; Kirby, J.; Burren, Y.; Porz, N.; Slotboom, J.; Wiest, R.; et al. The Multimodal Brain Tumor Image Segmentation Benchmark (BRATS). IEEE Trans. Med. Imaging 2015, 34, 1993–2024. [Google Scholar] [CrossRef] [PubMed]
- Rudie, J.D.; Rauschecker, A.M.; Bryan, R.N.; Davatzikos, C.; Mohan, S. Emerging Applications of Artificial Intelligence in Neuro-Oncology. Radiology 2019, 290, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Forghani, R. Precision Digital Oncology: Emerging Role of Radiomics-based Biomarkers and Artificial Intelligence for Advanced Imaging and Characterization of Brain Tumors. Radiol. Imaging Cancer 2020, 2, e190047. [Google Scholar] [CrossRef]
- Peeken, J.C.; Hesse, J.; Haller, B.; Kessel, K.A.; Nüsslin, F.; Combs, S.E. Semantic imaging features predict disease progression and survival in glioblastoma multiforme patients. Strahlenther. Onkol. 2018, 194, 580–590. [Google Scholar] [CrossRef]
- Lacroix, M.; Abi-Said, D.; Fourney, D.R.; Gokaslan, Z.L.; Shi, W.; DeMonte, F.; Lang, F.F.; McCutcheon, I.E.; Hassenbusch, S.J.; Holland, E.; et al. A multivariate analysis of 416 patients with glioblastoma multiforme: Prognosis, extent of resection, and survival. J. Neurosurg. 2001, 95, 190–198. [Google Scholar] [CrossRef] [PubMed]
- Compter, I.; Verduin, M.; Shi, Z.; Woodruff, H.C.; Smeenk, R.J.; Rozema, T.; Leijenaar, R.T.H.; Monshouwer, R.; Eekers, D.B.P.; Hoeben, A.; et al. Deciphering the glioblastoma phenotype by computed tomography radiomics. Radiother. Oncol. J. Eur. Soc. Ther. Radiol. Oncol. 2021, 160, 132–139. [Google Scholar] [CrossRef] [PubMed]
- Senders, J.T.; Staples, P.; Mehrtash, A.; Cote, D.J.; Taphoorn, M.J.B.; Reardon, D.A.; Gormley, W.B.; Smith, T.R.; Broekman, M.L.; Arnaout, O. An Online Calculator for the Prediction of Survival in Glioblastoma Patients Using Classical Statistics and Machine Learning. Neurosurgery 2020, 86, E184–E192. [Google Scholar] [CrossRef] [PubMed]
- Sanai, N.; Alvarez-Buylla, A.; Berger, M.S. Neural Stem Cells and the Origin of Gliomas. N. Engl. J. Med. 2005, 353, 811–822. [Google Scholar] [CrossRef]
- Laug, D.; Glasgow, S.M.; Deneen, B. A glial blueprint for gliomagenesis. Nat. Rev. Neurosci. 2018, 19, 393–403. [Google Scholar] [CrossRef] [PubMed]
- Soltani, M.; Bonakdar, A.; Shakourifar, N.; Babaie, R.; Raahemifar, K. Efficacy of Location-Based Features for Survival Prediction of Patients With Glioblastoma Depending on Resection Status. Front. Oncol. 2021, 11, 661123. [Google Scholar] [CrossRef] [PubMed]
- Park, M.; Lee, S.K.; Chang, J.H.; Kang, S.G.; Kim, E.H.; Kim, S.H.; Song, M.K.; Ma, B.G.; Ahn, S.S. Elderly patients with newly diagnosed glioblastoma: Can preoperative imaging descriptors improve the predictive power of a survival model? J. Neuro-Oncol. 2017, 134, 423–431. [Google Scholar] [CrossRef]
- Li, Y.M.; Suki, D.; Hess, K.; Sawaya, R. The influence of maximum safe resection of glioblastoma on survival in 1229 patients: Can we do better than gross-total resection? J. Neurosurg. 2016, 124, 977–988. [Google Scholar] [CrossRef] [PubMed]
- Molinaro, A.M.; Hervey-Jumper, S.; Morshed, R.A.; Young, J.; Han, S.J.; Chunduru, P.; Zhang, Y.; Phillips, J.J.; Shai, A.; Lafontaine, M.; et al. Association of Maximal Extent of Resection of Contrast-Enhanced and Non-Contrast-Enhanced Tumor with Survival Within Molecular Subgroups of Patients with Newly Diagnosed Glioblastoma. JAMA Oncol. 2020, 6, 495–503. [Google Scholar] [CrossRef]
- Wangaryattawanich, P.; Hatami, M.; Wang, J.; Thomas, G.; Flanders, A.; Kirby, J.; Wintermark, M.; Huang, E.S.; Bakhtiari, A.S.; Luedi, M.M.; et al. Multicenter imaging outcomes study of The Cancer Genome Atlas glioblastoma patient cohort: Imaging predictors of overall and progression-free survival. Dev. Oncol. 2015, 17, 1525–1537. [Google Scholar] [CrossRef]
- Harrell, F.E. Regression Modeling Strategies: With Applications to Linear Models, Logistic and Ordinal Regression, and Survival Analysis, 2nd ed.; Springer Series in Statistics; Springer: Cham, Switzerland, 2015; ISBN 9783319194257. [Google Scholar]
- Riley, R.D.; van der Windt, D.; Croft, P.; Moons, K.G.M. Prognosis Research in Healthcare: Concepts, Methods, and Impact; Riley, R.D., van der Windt, D.A., Croft, P., Moons, K.G.M., Eds.; Oxford University Press: Oxford, UK, 2019; ISBN 9780198796619. [Google Scholar]
- Currie, S.; Fatania, K.; Frood, R.; Whitehead, R.; Start, J.; Lee, M.-T.; McDonald, B.; Rankeillor, K.; Roberts, P.; Chakrabarty, A.; et al. Imaging Spectrum of the Developing Glioblastoma: A Cross-Sectional Observation Study. Curr. Oncol. 2023, 30, 6682–6698. [Google Scholar] [CrossRef]
- Van Rossum, G.; Drake, F.L. Python 3 Reference Manual; CreateSpace: Scotts Valley, CA, USA, 2009; ISBN 1441412697. [Google Scholar]
- Icometrix, Dicom2nifti. Available online: https://icometrix.github.io/dicom2nifti/ (accessed on 31 January 2022).
- Pati, S.; Baid, U.; Edwards, B.; Sheller, M.; Wang, S.-H.; Reina, G.A.; Foley, P.; Gruzdev, A.; Karkada, D.; Davatzikos, C.; et al. Federated Learning Enables Big Data for Rare Cancer Boundary Detection. Nat. Commun. 2022, 13, 7346. [Google Scholar] [CrossRef] [PubMed]
- Davatzikos, C.; Rathore, S.; Bakas, S.; Pati, S.; Bergman, M.; Kalarot, R.; Sridharan, P.; Gastounioti, A.; Jahani, N.; Cohen, E.; et al. Cancer imaging phenomics toolkit: Quantitative imaging analytics for precision diagnostics and predictive modeling of clinical outcome. J. Med. Imaging 2018, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Rohlfing, T.; Zahr, N.M.; Sullivan, E.V.; Pfefferbaum, A. The SRI24 multichannel atlas of normal adult human brain structure. Hum. Brain Mapp. 2009, 31, 798–819. [Google Scholar] [CrossRef]
- Yushkevich, P.A.; Pluta, J.; Wang, H.; Wisse, L.E.M.; Das, S.; Wolk, D. IC-P-174: Fast Automatic Segmentation of Hippocampal Subfields and Medial Temporal Lobe Subregions In 3 Tesla and 7 Tesla T2-Weighted MRI. Alzheimer’s Dement. 2016, 12, 126–127. [Google Scholar] [CrossRef]
- Thakur, S.; Doshi, J.; Pati, S.; Rathore, S.; Sako, C.; Bilello, M.; Ha, S.M.; Shukla, G.; Flanders, A.; Kotrotsou, A.; et al. Brain extraction on MRI scans in presence of diffuse glioma: Multi-institutional performance evaluation of deep learning methods and robust modality-agnostic training. Neuroimage 2020, 220, 117081. [Google Scholar] [CrossRef]
- Isensee, F.; Jaeger, P.F.; Kohl, S.A.A.; Petersen, J.; Maier-Hein, K.H. nnU-Net: A self-configuring method for deep learning-based biomedical image segmentation. Nat. Methods 2021, 18, 203–211. [Google Scholar] [CrossRef]
- Zou, K.H.; Warfield, S.K.; Bharatha, A.; Tempany, C.M.C.; Kaus, M.R.; Haker, S.J.; Wells, W.M.; Jolesz, F.A.; Kikinis, R. Statistical Validation of Image Segmentation Quality Based on a Spatial Overlap Index. Acad. Radiol. 2004, 11, 178–189. [Google Scholar] [CrossRef]
- Therneau, T.M. A Package for Survival Analysis in R 2023. Available online: https://cran.r-project.org/package=survival (accessed on 31 January 2023).
- Perperoglou, A.; Sauerbrei, W.; Abrahamowicz, M.; Schmid, M. A review of spline function procedures in R. BMC Med. Res. Methodol. 2019, 19, 46. [Google Scholar] [CrossRef] [PubMed]
- Verduin, M.; Primakov, S.; Compter, I.; Woodruff, H.C.; van Kuijk, S.M.J.; Ramaekers, B.L.T.; Dorsthorst, M.T.; Revenich, E.G.M.; Laan, M.T.; Pegge, S.A.H.; et al. Prognostic and predictive value of integrated qualitative and quantitative magnetic resonance imaging analysis in glioblastoma. Cancers 2021, 13, 722. [Google Scholar] [CrossRef]
- Henker, C.; Hiepel, M.C.; Kriesen, T.; Scherer, M.; Glass, Ä.; Herold-Mende, C.; Bendszus, M.; Langner, S.; Weber, M.A.; Schneider, B.; et al. Volumetric assessment of glioblastoma and its predictive value for survival. Acta Neurochir. 2019, 161, 1723–1732. [Google Scholar] [CrossRef] [PubMed]
- Iliadis, G.; Kotoula, V.; Chatzisotiriou, A.; Televantou, D.; Eleftheraki, A.G.; Lambaki, S.; Misailidou, D.; Selviaridis, P.; Fountzilas, G. Volumetric and MGMT parameters in glioblastoma patients: Survival analysis. BMC Cancer 2012, 12, 3. [Google Scholar] [CrossRef] [PubMed]
- Palpan Flores, A.; Vivancos Sanchez, C.; Roda, J.M.; Cerdán, S.; Barrios, A.J.; Utrilla, C.; Royo, A.; Gandía González, M.L. Assessment of Pre-operative Measurements of Tumor Size by MRI Methods as Survival Predictors in Wild Type IDH Glioblastoma. Front. Oncol. 2020, 10, 1662. [Google Scholar] [CrossRef] [PubMed]
- Fuster-Garcia, E.; Juan-Albarracín, J.; García-Ferrando, G.A.; Martí-Bonmatí, L.; Aparici-Robles, F.; García-Gómez, J.M. Improving the estimation of prognosis for glioblastoma patients by MR based hemodynamic tissue signatures. NMR Biomed. 2018, 31, e4006. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Jiang, H.; Chen, X.; Bai, J.; Cui, Y.; Ren, X.; Chen, X.; Wang, J.; Zeng, W.; Lin, S. Identifying the survival subtypes of glioblastoma by quantitative volumetric analysis of MRI. J. Neuro-Oncol. 2014, 119, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.; Ahn, K.J.; Nam, Y.; Jang, J.; Shin, N.Y.; Choi, H.S.; Jung, S.L.; Kim, B.S. Analysis of heterogeneity of peritumoral T2 hyperintensity in patients with pretreatment glioblastoma: Prognostic value of MRI-based radiomics. Eur. J. Radiol. 2019, 120, 108642. [Google Scholar] [CrossRef] [PubMed]
- Kickingereder, P.; Neuberger, U.; Bonekamp, D.; Piechotta, P.L.; Götz, M.; Wick, A.; Sill, M.; Kratz, A.; Shinohara, R.T.; Jones, D.T.W.; et al. Radiomic subtyping improves disease stratification beyond key molecular, clinical, and standard imaging characteristics in patients with glioblastoma. Dev. Oncol. 2018, 20, 848–857. [Google Scholar] [CrossRef]
- Lutterbach, J.; Sauerbrei, W.; Guttenberger, R. Multivariate analysis of prognostic factors in patients with glioblastoma. Strahlenther. Onkol. 2003, 179, 8–15. [Google Scholar] [CrossRef]
- Steyerberg, E.W. Clinical Prediction Models: A Practical Approach to Development, Validation, and Updating; Statistics for Biology and Health; Springer: New York, NY, USA, 2009; ISBN 9780387772448. [Google Scholar]
| Demographic | Value |
|---|---|
| Age, years–median (IQR) | 62 (55–68) |
| Gender–no. female (%) | 108 (39%) |
| Surgical treatment–no. (%) | |
| Biopsy | 71 (25%) |
| 100% resected a | 57 (20%) |
| ≥90% resected a | 86 (31%) |
| <90% resected a | 65 (23%) |
| Adjuvant oncology treatment–no. (%) | |
| No Stupp | 150 (54%) |
| Full Stupp b | 58 (21%) |
| Partial Stupp c | 71 (25%) |
| MGMT methylation–no. (% of known) d | 103 (40%) |
| Overall survival, months–median (95% CI) | 12 (11–14) |
| Maximum tumour diameter, cm–median (IQR) | 4.4 (3.3–5.4) |
| Core volume, cm3–median (IQR) | 28.1 (12.6–50.3) |
| Whole volume, cm3–median (IQR) | 103 (45.6–160) |
| Whole Volume (WV) | Core Volume (CV) | Tumour Diameter | ||||
|---|---|---|---|---|---|---|
| WV | log(WV) | CV | log(CV) | Diameter | log(Diameter) | |
| C (95% CI) | 0.5 (0.46–0.54) | 0.5 (0.46–0.54) | 0.5 (0.46–0.54) | 0.5 (0.46–0.54) | 0.5 (0.46–0.54) | 0.5 (0.46–0.54) |
| HR (95% CI) | 1 (1–1) | 1.1 (0.81–1.6) | 1 (1–1) | 0.95 (0.71–1.3) | 1 (0.93–1.1) | 0.94 (0.43–2) |
| p value | 0.784 | 0.475 | 0.539 | 0.704 | 0.745 | 0.875 |
| Tumour Diameter | Whole Volume (WV) | Core Volume (CV) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Diameter | log(Diameter) | WV | log(WV) | CV | log(CV) | |||||||
| Variable | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p | HR (95% CI) | p |
| Age | 1.01 (0.92–1.10) | 0.91 | 0.86 (0.39–1.88) | 0.70 | 1.00 (1.00–1.00) | 0.90 | 1.09 (0.78–1.5) | 0.61 | 1.00 (1.00–1.01) | 0.56 | 0.93 (0.7–1.23) | 0.6 |
| Gender | 1.00 (0.92–1.10) | 0.93 | 0.87 (0.40–1.89) | 0.73 | 1.00 (1.00–1.00) | 0.99 | 1.09 (0.78–1.5) | 0.61 | 1.00 (1.00–1.01) | 0.71 | 0.91 (0.69–1.22) | 0.54 |
| Type of surgery | 1.14 (1.02–1.26) | 0.016 | 2.4 (0.96–5.98) | 0.063 | 1.00 (1.00–1.00) | 0.013 | 1.90 (1.28–2.82) | 0.001 | 1.01 (1.00–1.01) | 0.018 | 1.29 (0.93–1.79) | 0.13 |
| Adjuvant oncology treatment | 1.00 (0.91–1.09) | 0.93 | 0.82 (0.37–1.79) | 0.61 | 1.00 (1.00–1.00) | 0.99 | 1.05 (0.75–1.47) | 0.76 | 1.00 (1.00–1.01) | 0.67 | 0.92 (0.69–1.23) | 0.58 |
| MGMT methylation | 1.02 (0.93–1.12) | 0.70 | 0.96 (0.43–2.18) | 0.93 | 1.00 (1.00–1.00) | 0.98 | 1.10 (0.78–1.5) | 0.60 | 1.00 (1.00–1.01) | 0.71 | 0.94 (0.70–1.26) | 0.68 |
| Age + Gender + Surgery + Oncology + MGMTa | 1.12 (1.01–1.25) | 0.032 | 2.3 (0.91–6.01) | 0.076 | 1.00 (1.00–1.00) | 0.24 | 1.45 (0.98–2.14) | 0.06 | 1.00 (1.00–1.01) | 0.072 | 1.24 (0.89–1.7) | 0.20 |
| Adjusted for Operation Type | Adjusted for Age + Gender + Surgery + Oncology + MGMT | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Sample Size | Tumour Diameter | Whole Volume (WV) | Core Volume (CV) | Sample Size | Tumour Diameter | Whole Volume (WV) | Core Volume (CV) | ||||||
| Diameter | log(Diameter) | WV | log(WV) | CV | log(CV) | Diameter | log(Diameter) | WV | log(WV) | CV | log(CV) | ||
| 50 | 19.01 | 14.78 | 19.45 | 26.30 | 17.15 | 11.93 | 50 | 19.53 | 16.39 | 14.95 | 15.54 | 14.87 | 12.93 |
| 100 | 31.24 | 21.54 | 32.15 | 47.22 | 28.95 | 16.28 | 100 | 26.74 | 20.39 | 16.13 | 20.97 | 19.15 | 13.78 |
| 150 | 42.94 | 28.75 | 44.56 | 64.84 | 40.92 | 21.16 | 150 | 35.14 | 26.01 | 18.29 | 27.80 | 25.15 | 16.32 |
| 200 | 53.50 | 36.03 | 55.58 | 77.80 | 51.63 | 26.03 | 200 | 43.47 | 32.24 | 20.47 | 34.84 | 31.77 | 19.38 |
| 250 | 62.47 | 42.82 | 65.10 | 86.42 | 61.30 | 30.92 | 250 | 51.34 | 38.30 | 23.06 | 41.89 | 38.19 | 22.67 |
| 279 | 67.16 | 46.66 | 69.87 | 89.94 | 66.05 | 33.61 | 258 a | 55.67 | 41.79 | 24.57 | 45.72 | 41.89 | 24.50 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fatania, K.; Frood, R.; Mistry, H.; Short, S.C.; O’Connor, J.; Scarsbrook, A.F.; Currie, S. Tumour Size and Overall Survival in a Cohort of Patients with Unifocal Glioblastoma: A Uni- and Multivariable Prognostic Modelling and Resampling Study. Cancers 2024, 16, 1301. https://doi.org/10.3390/cancers16071301
Fatania K, Frood R, Mistry H, Short SC, O’Connor J, Scarsbrook AF, Currie S. Tumour Size and Overall Survival in a Cohort of Patients with Unifocal Glioblastoma: A Uni- and Multivariable Prognostic Modelling and Resampling Study. Cancers. 2024; 16(7):1301. https://doi.org/10.3390/cancers16071301
Chicago/Turabian StyleFatania, Kavi, Russell Frood, Hitesh Mistry, Susan C. Short, James O’Connor, Andrew F. Scarsbrook, and Stuart Currie. 2024. "Tumour Size and Overall Survival in a Cohort of Patients with Unifocal Glioblastoma: A Uni- and Multivariable Prognostic Modelling and Resampling Study" Cancers 16, no. 7: 1301. https://doi.org/10.3390/cancers16071301
APA StyleFatania, K., Frood, R., Mistry, H., Short, S. C., O’Connor, J., Scarsbrook, A. F., & Currie, S. (2024). Tumour Size and Overall Survival in a Cohort of Patients with Unifocal Glioblastoma: A Uni- and Multivariable Prognostic Modelling and Resampling Study. Cancers, 16(7), 1301. https://doi.org/10.3390/cancers16071301







