A Comprehensive Understanding of Post-Translational Modification of Sox2 via Acetylation and O-GlcNAcylation in Colorectal Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Culture Conditions
2.2. Drugs
2.3. RNAi
2.4. DNA and microRNA Transfection
2.5. Transfection and Lentiviral Transduction
2.6. Reverse-Transcription Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR) for miRNA Quantitation
2.7. Western Blot Analysis
2.8. RNA Extraction and qPCR
2.9. Immunohistochemistry of Patient CRC Tissues
2.10. Immunofluorescence and Confocal Imaging
2.11. Immunoprecipitation
2.12. Cell Fractionation
2.13. Expression Profile Datasets
2.14. Gene Expression Analysis for mRNA Array and microRNA Array
2.15. Immunoprecipitation for Mass Spectrometric Analysis
2.16. In-Gel Digestion
2.17. LC-MS/MS Analysis
2.18. LC-MS/MS Data Analysis for PTMs
2.19. Survival Analysis
2.20. RNA-seq and Bioinformatics Analysis of Patient-Derived Organoids
2.21. Statistical Analysis
3. Results
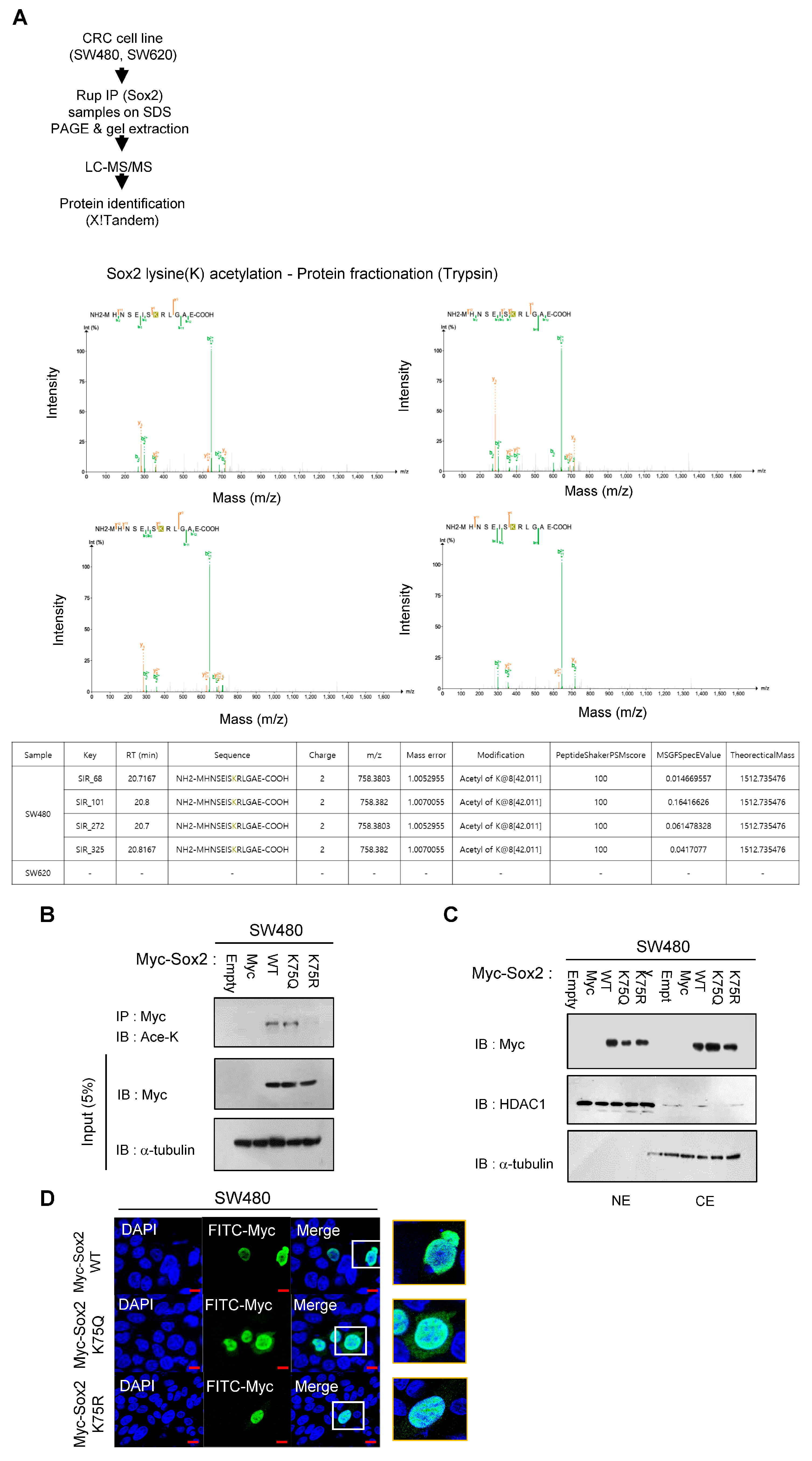
3.1. K75-Lysine Acetylation Induces Proteasomal Degradation of Sox2
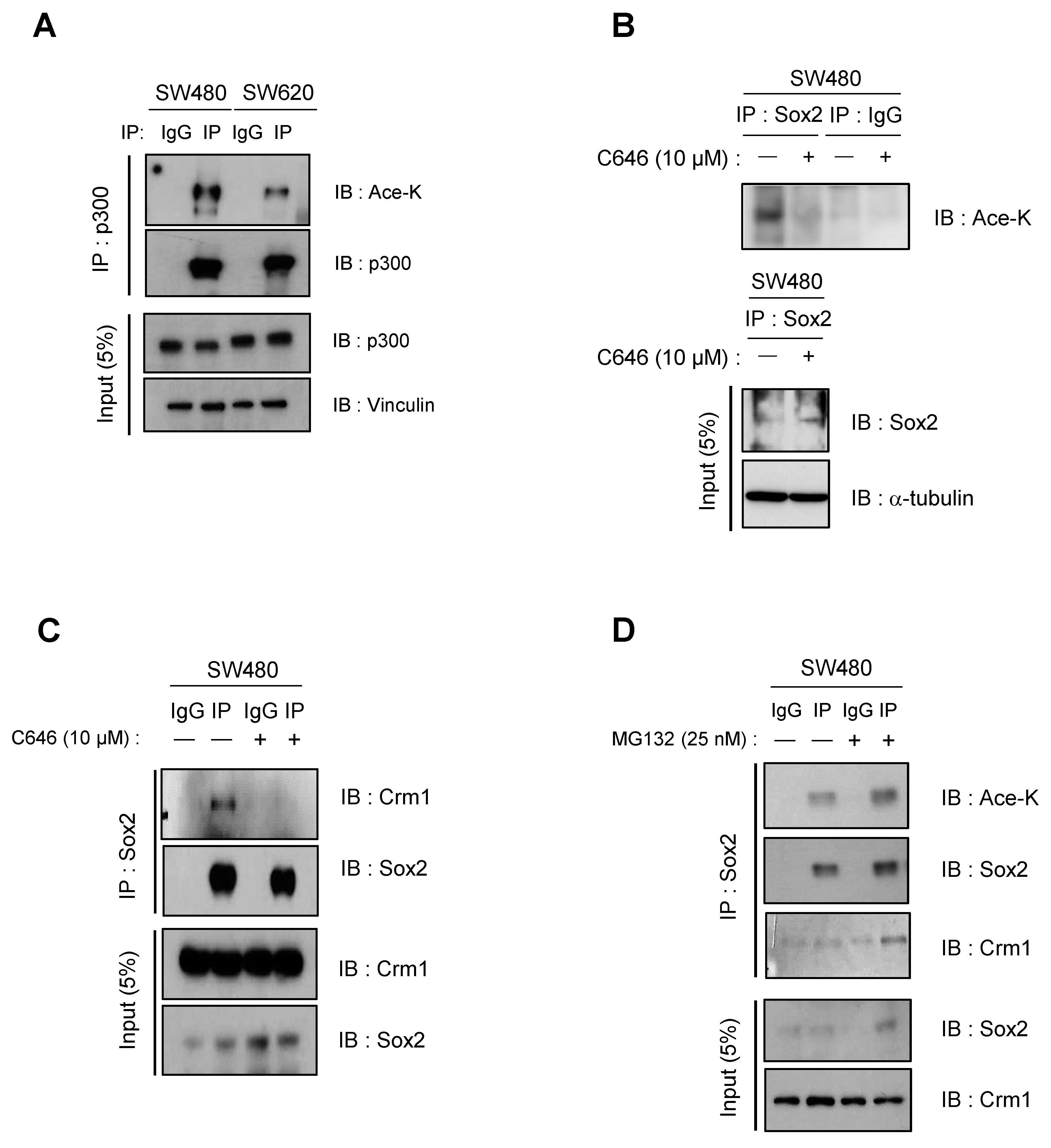
3.2. CBP/p300-Mediated Acetylation Contributes to Nuclear Export and Degradation of Sox2
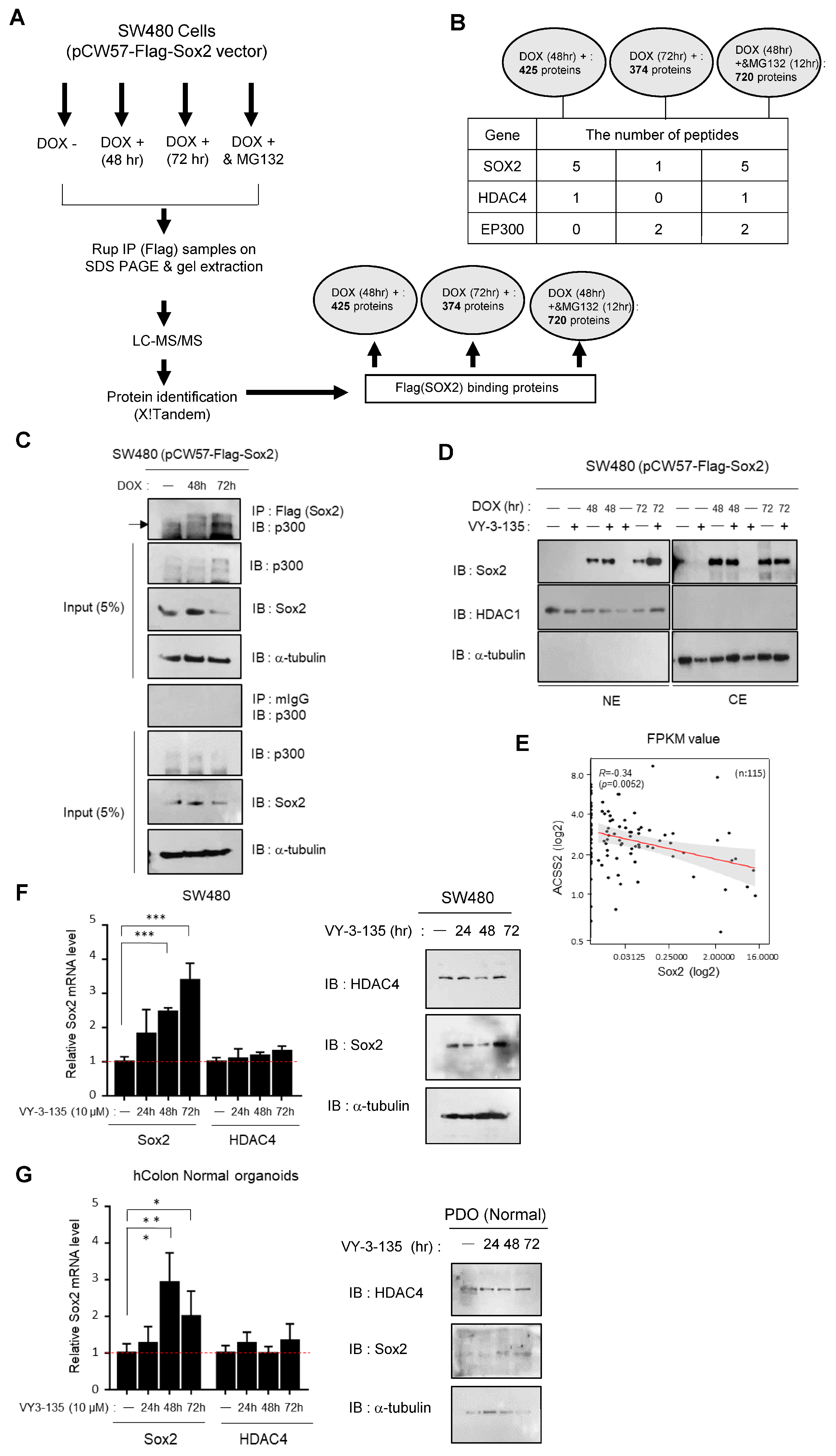
3.3. ACSS2-Mediated Modulation of p300/CBP Activity Regulates Sox2 Expression and Stability
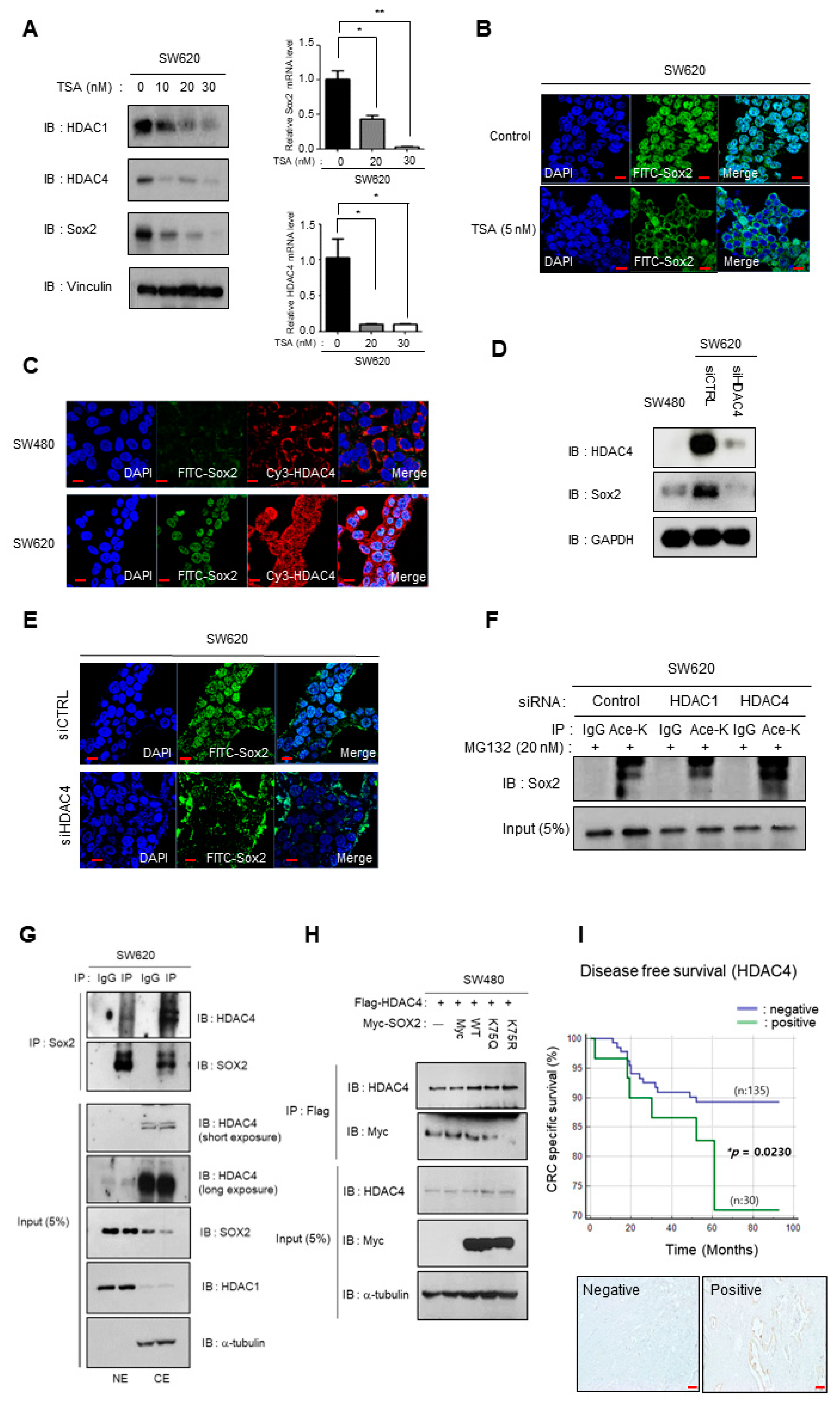
3.4. Depletion of HDAC4 Induces Cytoplasmic Shift and Downregulation of Sox2 Expression
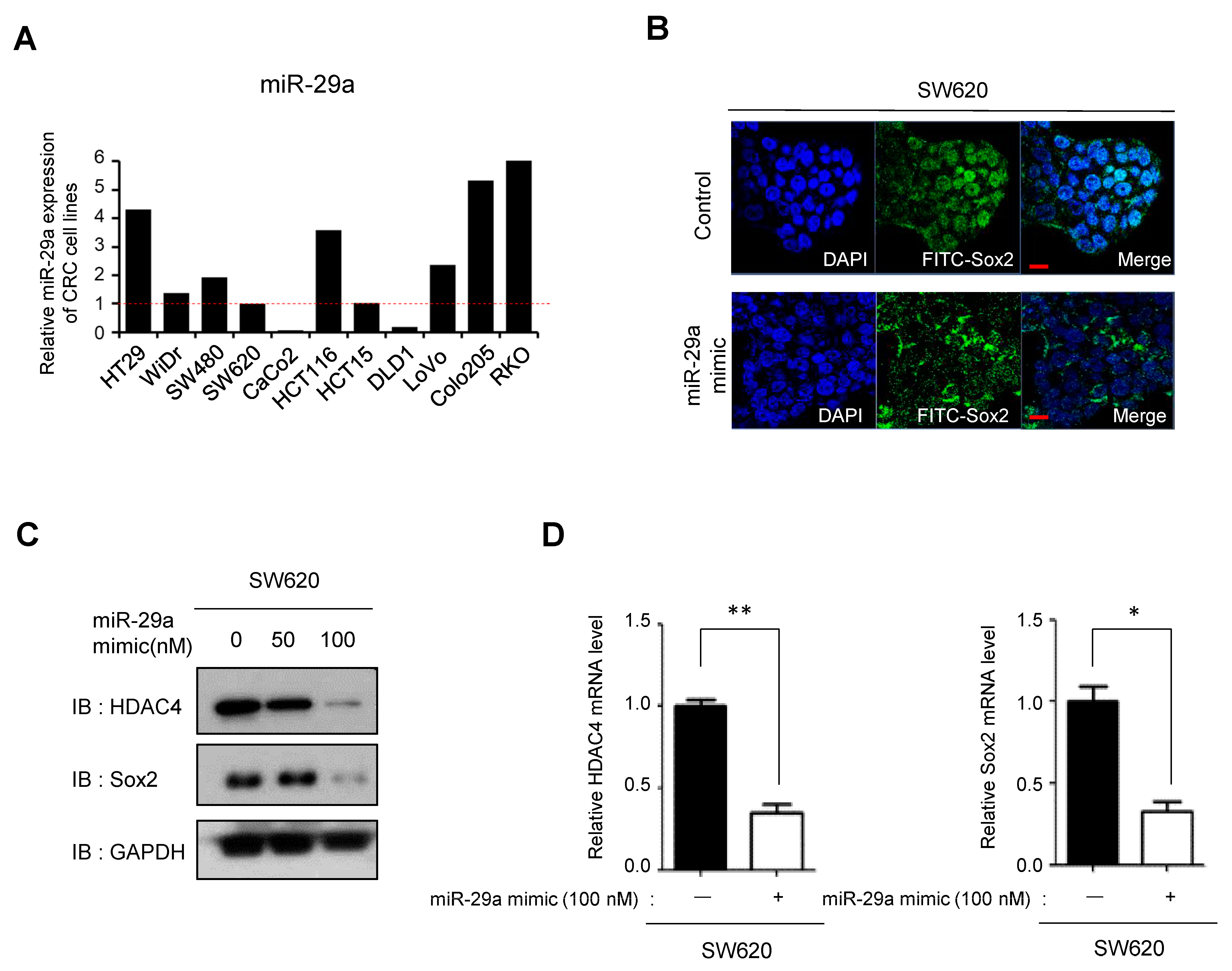
3.5. miR29a-Mediated Suppression of HDAC4 Reduces Sox2 Expression
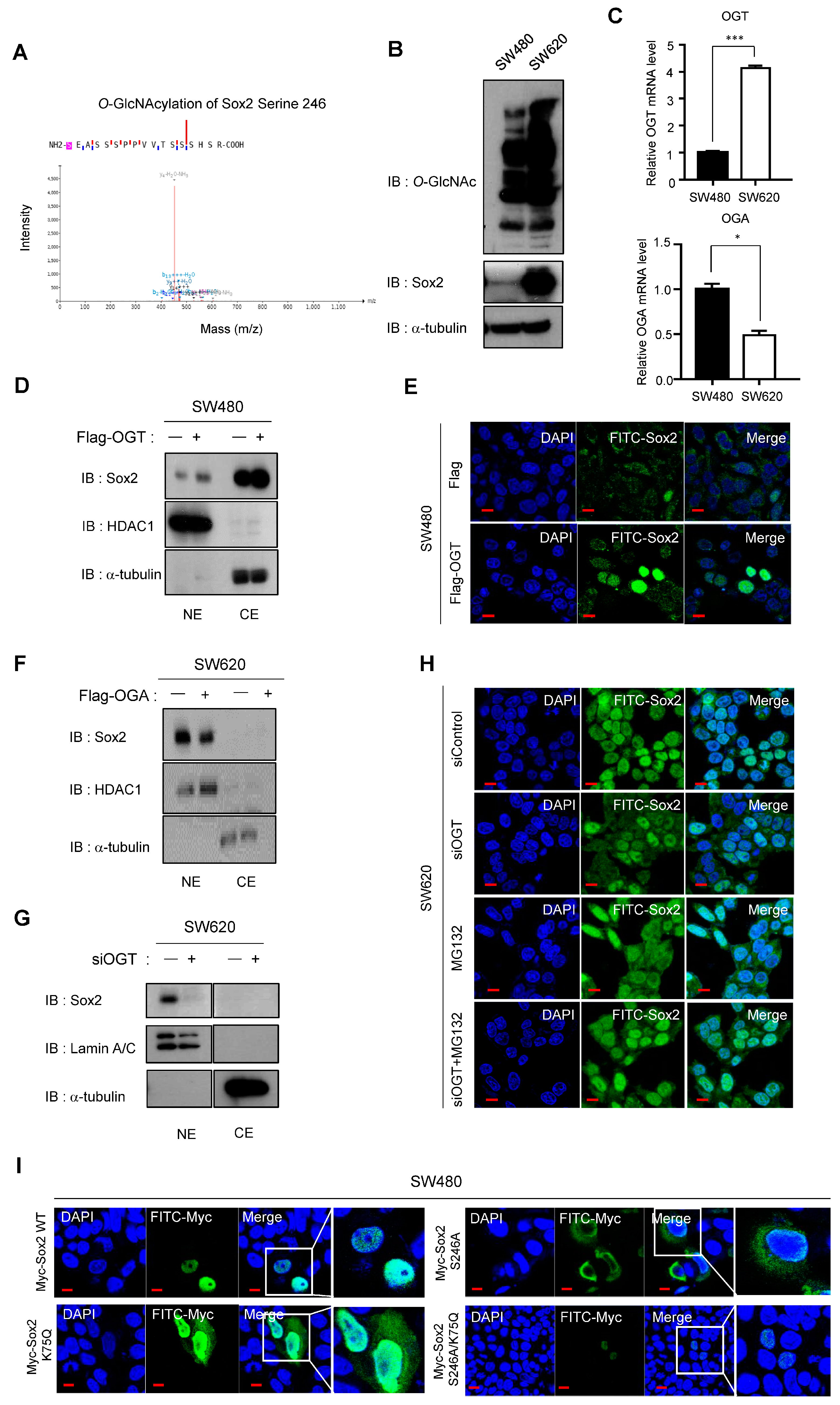
3.6. O-GlcNAcylation and the Related Enzymes OGT and OGA Play Important Roles in Sox2 Regulation
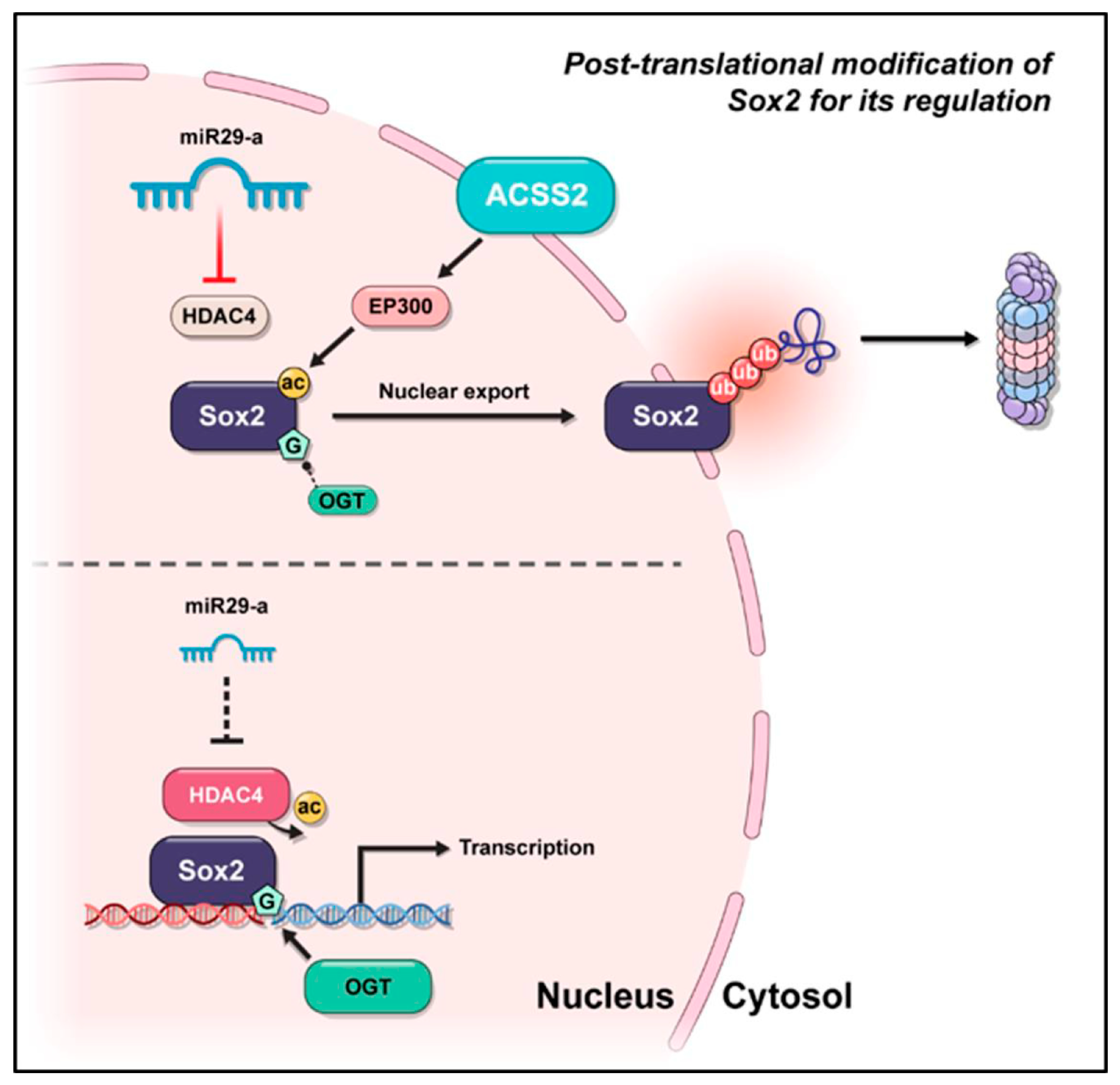
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Džunić, M.; Petkovic, I.; Cvetanovic, A.; Vrbic, S.; Pejcic, I. Current and future targets and therapies in metastatic colorectal cancer. J. BUON. 2019, 24, 1785–1792. [Google Scholar]
- Nappi, A.; Nasti, G.; Romano, C.; Berretta, M.; Ottaiano, A. Metastatic Colorectal Cancer: Prognostic and Predictive Factors. Curr. Med. Chem. 2020, 27, 2779–2791. [Google Scholar] [CrossRef]
- Kim, S.; Lee, J.H.; Park, E.J.; Lee, H.S.; Baik, S.H.; Jeon, T.J.; Lee, K.Y.; Ryu, Y.H.; Kang, J. Prediction of Microsatellite Instability in Colorectal Cancer Using a Machine Learning Model Based on PET/CT Radiomics. Yonsei Med. J. 2023, 64, 320–326. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, Y.; Uchikawa, M.; Kondoh, H. Pairing SOX off: With partners in the regulation of embryonic development. Trends Genet. 2000, 16, 182–187. [Google Scholar] [CrossRef]
- Han, X.; Fang, X.; Lou, X.; Hua, D.; Ding, W.; Foltz, G.; Hood, L.; Yuan, Y.; Lin, B. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PLoS ONE 2012, 7, e41335. [Google Scholar] [CrossRef] [PubMed]
- Novak, D.; Hüser, L.; Elton, J.J.; Umansky, V.; Altevogt, P.; Utikal, J. SOX2 in development and cancer biology. In Seminars in Cancer Biology; Academic Press: Cambridge, MA, USA, 2020; pp. 74–82. [Google Scholar]
- Lundberg, I.V.; Löfgren Burström, A.; Edin, S.; Eklöf, V.; Öberg, Å.; Stenling, R.; Palmqvist, R.; Wikberg, M.L.; Hoffmann, A.-C. SOX2 expression is regulated by BRAF and contributes to poor patient prognosis in colorectal cancer. PLoS ONE 2014, 9, e101957. [Google Scholar] [CrossRef] [PubMed]
- Kamachi, Y.; Kondoh, H. Sox proteins: Regulators of cell fate specification and differentiation. Development 2013, 140, 4129–4144. [Google Scholar] [CrossRef] [PubMed]
- Baltus, G.A.; Kowalski, M.P.; Zhai, H.; Tutter, A.V.; Quinn, D.; Wall, D.; Kadam, S. Acetylation of sox2 induces its nuclear export in embryonic stem cells. Stem Cells 2009, 27, 2175–2184. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, A.; Olivier-Van Stichelen, S.; Baldini, S.F.; Dehennaut, V.; Toillon, R.-A.; Le Bourhis, X.; El Yazidi-Belkoura, I.; Lefebvre, T. Silencing the nucleocytoplasmic O-GlcNAc transferase reduces proliferation, adhesion, and migration of cancer and fetal human colon cell lines. Front. Endocrinol. 2016, 7, 46. [Google Scholar] [CrossRef]
- Jiang, M.; Xu, B.; Li, X.; Shang, Y.; Chu, Y.; Wang, W.; Chen, D.; Wu, N.; Hu, S.; Zhang, S. O-GlcNAcylation promotes colorectal cancer metastasis via the miR-101-O-GlcNAc/EZH2 regulatory feedback circuit. Oncogene 2019, 38, 301–316. [Google Scholar] [CrossRef]
- Myers, S.A.; Peddada, S.; Chatterjee, N.; Friedrich, T.; Tomoda, K.; Krings, G.; Thomas, S.; Maynard, J.; Broeker, M.; Thomson, M. SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. Elife 2016, 5, e10647. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.S.; Gupta, V.K.; Dauer, P.; Kesh, K.; Hadad, R.; Giri, B.; Chandra, A.; Dudeja, V.; Slawson, C.; Banerjee, S. O-GlcNAc modification of Sox2 regulates self-renewal in pancreatic cancer by promoting its stability. Theranostics 2019, 9, 3410. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.-J.; Suk, J.-E.; Patrick, C.; Bae, E.-J.; Cho, J.-H.; Rho, S.; Hwang, D.; Masliah, E.; Lee, S.-J. Direct transfer of α-synuclein from neuron to astroglia causes inflammatory responses in synucleinopathies. J. Biol. Chem. 2010, 285, 9262–9272. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef]
- Craig, R.; Beavis, R.C. TANDEM: Matching proteins with tandem mass spectra. Bioinformatics 2004, 20, 1466–1467. [Google Scholar] [CrossRef]
- Tabb, D.L.; Fernando, C.G.; Chambers, M.C. MyriMatch: Highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J. Proteome Res. 2007, 6, 654–661. [Google Scholar] [CrossRef]
- Kim, S.; Pevzner, P.A. MS-GF+ makes progress towards a universal database search tool for proteomics. Nat. Commun. 2014, 5, 5277. [Google Scholar] [CrossRef]
- Vaudel, M.; Barsnes, H.; Berven, F.S.; Sickmann, A.; Martens, L. SearchGUI: An open-source graphical user interface for simultaneous OMSSA and X! Tandem searches. Proteomics 2011, 11, 996–999. [Google Scholar] [CrossRef] [PubMed]
- Vaudel, M.; Burkhart, J.M.; Zahedi, R.P.; Oveland, E.; Berven, F.S.; Sickmann, A.; Martens, L.; Barsnes, H. PeptideShaker enables reanalysis of MS-derived proteomics data sets. Nat. Biotechnol. 2015, 33, 22–24. [Google Scholar] [CrossRef] [PubMed]
- Poojan, S.; Bae, S.-H.; Min, J.-W.; Lee, E.Y.; Song, Y.; Kim, H.Y.; Sim, H.W.; Kang, E.-K.; Kim, Y.-H.; Lee, H.-O. Cancer cells undergoing epigenetic transition show short-term resistance and are transformed into cells with medium-term resistance by drug treatment. Exp. Mol. Med. 2020, 52, 1102–1115. [Google Scholar] [CrossRef]
- Neumann, J.; Bahr, F.; Horst, D.; Kriegl, L.; Engel, J.; Mejías-Luque, R.; Gerhard, M.; Kirchner, T.; Jung, A. SOX2 expression correlates with lymph-node metastases and distant spread in right-sided colon cancer. BMC Cancer 2011, 11, 518. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.R.; Coyaud, E.; Laurent, E.M.; St-Germain, J.; Van de Laar, E.; Tsao, M.-S.; Raught, B.; Moghal, N. Identification of the SOX2 interactome by BioID reveals EP300 as a mediator of SOX2-dependent squamous differentiation and lung squamous cell carcinoma growth. Mol. Cell. Proteom. 2017, 16, 1864–1888. [Google Scholar] [CrossRef] [PubMed]
- Thompson, P.R.; Wang, D.; Wang, L.; Fulco, M.; Pediconi, N.; Zhang, D.; An, W.; Ge, Q.; Roeder, R.G.; Wong, J. Regulation of the p300 HAT domain via a novel activation loop. Nat. Struct. Mol. Biol. 2004, 11, 308–315. [Google Scholar] [CrossRef] [PubMed]
- Mariño, G.; Pietrocola, F.; Eisenberg, T.; Kong, Y.; Malik, S.A.; Andryushkova, A.; Schroeder, S.; Pendl, T.; Harger, A.; Niso-Santano, M. Regulation of autophagy by cytosolic acetyl-coenzyme A. Mol. Cell 2014, 53, 710–725. [Google Scholar] [CrossRef] [PubMed]
- Bulusu, V.; Tumanov, S.; Michalopoulou, E.; van den Broek, N.J.; MacKay, G.; Nixon, C.; Dhayade, S.; Schug, Z.T.; Voorde, J.V.; Blyth, K. Acetate recapturing by nuclear acetyl-CoA synthetase 2 prevents loss of histone acetylation during oxygen and serum limitation. Cell Rep. 2017, 18, 647–658. [Google Scholar] [CrossRef] [PubMed]
- Cox, J.L.; Mallanna, S.K.; Luo, X.; Rizzino, A. Sox2 uses multiple domains to associate with proteins present in Sox2-protein complexes. PLoS ONE 2010, 5, e15486. [Google Scholar] [CrossRef] [PubMed]
- Fang, X.; Yu, W.; Li, L.; Shao, J.; Zhao, N.; Chen, Q.; Ye, Z.; Lin, S.-C.; Zheng, S.; Lin, B. ChIP-seq and functional analysis of the SOX2 gene in colorectal cancers. Omics A J. Integr. Biol. 2010, 14, 369–384. [Google Scholar] [CrossRef]
- Saigusa, S.; Tanaka, K.; Toiyama, Y.; Yokoe, T.; Okugawa, Y.; Ioue, Y.; Miki, C.; Kusunoki, M. Correlation of CD133, OCT4, and SOX2 in rectal cancer and their association with distant recurrence after chemoradiotherapy. Ann. Surg. Oncol. 2009, 16, 3488–3498. [Google Scholar] [CrossRef]
- Takeda, K.; Mizushima, T.; Yokoyama, Y.; Hirose, H.; Wu, X.; Qian, Y.; Ikehata, K.; Miyoshi, N.; Takahashi, H.; Haraguchi, N. Sox2 is associated with cancer stem-like properties in colorectal cancer. Sci. Rep. 2018, 8, 17639. [Google Scholar] [CrossRef]
- Wang, H.; Li, C.; Jian, Z.; Ou, Y.; Ou, J. TGF-β1 reduces miR-29a expression to promote tumorigenicity and metastasis of cholangiocarcinoma by targeting HDAC4. PLoS ONE 2015, 10, e0136703. [Google Scholar] [CrossRef]
- Itkonen, H.M.; Gorad, S.S.; Duveau, D.Y.; Martin, S.E.; Barkovskaya, A.; Bathen, T.F.; Moestue, S.A.; Mills, I.G. Inhibition of O-GlcNAc transferase activity reprograms prostate cancer cell metabolism. Oncotarget 2016, 7, 12464. [Google Scholar] [CrossRef]
- Burén, S.; Gomes, A.L.; Teijeiro, A.; Fawal, M.-A.; Yilmaz, M.; Tummala, K.S.; Perez, M.; Rodriguez-Justo, M.; Campos-Olivas, R.; Megías, D. Regulation of OGT by URI in response to glucose confers c-MYC-dependent survival mechanisms. Cancer Cell 2016, 30, 290–307. [Google Scholar] [CrossRef]
- Jiang, M.; Qiu, Z.; Zhang, S.; Fan, X.; Cai, X.; Xu, B.; Li, X.; Zhou, J.; Zhang, X.; Chu, Y. Elevated O-GlcNAcylation promotes gastric cancer cells proliferation by modulating cell cycle related proteins and ERK 1/2 signaling. Oncotarget 2016, 7, 61390. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.-B.; Han, X.; Li, M.-D.; Singh, J.P.; Qian, K.; Azarhoush, S.; Zhao, L.; Bennett, A.M.; Samuel, V.T.; Wu, J. O-GlcNAc transferase/host cell factor C1 complex regulates gluconeogenesis by modulating PGC-1α stability. Cell Metab. 2012, 16, 226–237. [Google Scholar] [CrossRef]
- Mu, W.-L.; Wang, Y.-J.; Xu, P.; Hao, D.-L.; Liu, X.-Z.; Wang, T.-T.; Chen, F.; Chen, H.-Z.; Lv, X.; Liu, D.-P. Sox2 deacetylation by Sirt1 is involved in mouse somatic reprogramming. Stem Cells 2015, 33, 2135–2147. [Google Scholar] [CrossRef] [PubMed]
- Yoon, D.S.; Choi, Y.; Jang, Y.; Lee, M.; Choi, W.J.; Kim, S.-H.; Lee, J.W. SIRT1 directly regulates SOX2 to maintain self-renewal and multipotency in bone marrow-derived mesenchymal stem cells. Stem Cells 2014, 32, 3219–3231. [Google Scholar] [CrossRef]
- Hur, H.; Kim, Y.B.; Ham, I.H.; Lee, D. Loss of ACSS2 expression predicts poor prognosis in patients with gastric cancer. J. Surg. Oncol. 2015, 112, 585–591. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.M.; Kim, J.H.; Oh, H.J.; Park, H.E.; Lee, T.H.; Cho, N.-Y.; Kang, G.H. Downregulation of acetyl-CoA synthetase 2 is a metabolic hallmark of tumor progression and aggressiveness in colorectal carcinoma. Mod. Pathol. 2017, 30, 267–277. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Collins, L.B.; Wali, A.; Bigler, R.; Sun, W.; Bultman, S.J. The Warburg effect dictates the mechanism of butyrate-mediated histone acetylation and cell proliferation. Mol. Cell 2012, 48, 612–626. [Google Scholar] [CrossRef]
- Ling, R.; Chen, G.; Tang, X.; Liu, N.; Zhou, Y.; Chen, D. Acetyl-CoA synthetase 2 (ACSS2): A review with a focus on metabolism and tumor development. Discov. Oncol. 2022, 13, 58. [Google Scholar] [CrossRef]
- Fischle, W.; Kiermer, V.; Dequiedt, F.; Verdin, E. The emerging role of class II histone deacetylases. Biochem. Cell Biol. 2001, 79, 337–348. [Google Scholar] [CrossRef]
- Wilson, A.J.; Byun, D.-S.; Nasser, S.; Murray, L.B.; Ayyanar, K.; Arango, D.; Figueroa, M.; Melnick, A.; Kao, G.D.; Augenlicht, L.H. HDAC4 promotes growth of colon cancer cells via repression of p21. Mol. Biol. Cell 2008, 19, 4062–4075. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.-S.; Yang, X.-Z.; Wen, Y.-F.; Mai, S.-J.; Wang, M.-H.; Zhang, M.-Y.; Zheng, X.S.; Wang, H.-Y. Overexpressed HDAC4 is associated with poor survival and promotes tumor progression in esophageal carcinoma. Aging 2016, 8, 1236. [Google Scholar] [CrossRef]
- Pan, F.; Mao, H.; Bu, F.; Tong, X.; Li, J.; Zhang, S.; Liu, X.; Wang, L.; Wu, L.; Chen, R. Sp1-mediated transcriptional activation of miR-205 promotes radioresistance in esophageal squamous cell carcinoma. Oncotarget 2017, 8, 5735. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, X. MiR-103a-3p Contributes to the Progression of Colorectal Cancer by Regulating GREM2 Expression. Yonsei Med. J. 2022, 63, 520–529. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Wang, K.; Wei, Y.; Yao, Q.; Zhang, Q.; Qu, H.; Zhu, G. lncRNA-MIAT regulates cell biological behaviors in gastric cancer through a mechanism involving the miR-29a-3p/HDAC4 axis. Oncol. Rep. 2017, 38, 3465–3472. [Google Scholar] [CrossRef]
- Slawson, C.; Copeland, R.; Hart, G.W. O-GlcNAc signaling: A metabolic link between diabetes and cancer? Trends Biochem. Sci. 2010, 35, 547–555. [Google Scholar] [CrossRef] [PubMed]
- Van Hoof, D.; Muñoz, J.; Braam, S.R.; Pinkse, M.W.; Linding, R.; Heck, A.J.; Mummery, C.L.; Krijgsveld, J. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell 2009, 5, 214–226. [Google Scholar] [CrossRef]
- Fang, L.; Zhang, L.; Wei, W.; Jin, X.; Wang, P.; Tong, Y.; Li, J.; Du, J.X.; Wong, J. A methylation-phosphorylation switch determines Sox2 stability and function in ESC maintenance or differentiation. Mol. Cell 2014, 55, 537–551. [Google Scholar] [CrossRef]
- Qi, D.; Wang, Q.; Yu, M.; Lan, R.; Li, S.; Lu, F. Mitotic phosphorylation of SOX2 mediated by Aurora kinase A is critical for the stem-cell like cell maintenance in PA-1 cells. Cell Cycle 2016, 15, 2009–2018. [Google Scholar] [CrossRef]
- Alonso, M.M.; Diez-Valle, R.; Manterola, L.; Rubio, A.; Liu, D.; Cortes-Santiago, N.; Urquiza, L.; Jauregi, P.; De Munain, A.L.; Sampron, N. Genetic and epigenetic modifications of Sox2 contribute to the invasive phenotype of malignant gliomas. PLoS ONE 2011, 6, e26740. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, Y.; Kim, D.K.; Park, J.; Park, S.J.; Park, J.J.; Cheon, J.H.; Kim, T.I. A Comprehensive Understanding of Post-Translational Modification of Sox2 via Acetylation and O-GlcNAcylation in Colorectal Cancer. Cancers 2024, 16, 1035. https://doi.org/10.3390/cancers16051035
Seo Y, Kim DK, Park J, Park SJ, Park JJ, Cheon JH, Kim TI. A Comprehensive Understanding of Post-Translational Modification of Sox2 via Acetylation and O-GlcNAcylation in Colorectal Cancer. Cancers. 2024; 16(5):1035. https://doi.org/10.3390/cancers16051035
Chicago/Turabian StyleSeo, Yoojeong, Dong Keon Kim, Jihye Park, Soo Jung Park, Jae Jun Park, Jae Hee Cheon, and Tae Il Kim. 2024. "A Comprehensive Understanding of Post-Translational Modification of Sox2 via Acetylation and O-GlcNAcylation in Colorectal Cancer" Cancers 16, no. 5: 1035. https://doi.org/10.3390/cancers16051035
APA StyleSeo, Y., Kim, D. K., Park, J., Park, S. J., Park, J. J., Cheon, J. H., & Kim, T. I. (2024). A Comprehensive Understanding of Post-Translational Modification of Sox2 via Acetylation and O-GlcNAcylation in Colorectal Cancer. Cancers, 16(5), 1035. https://doi.org/10.3390/cancers16051035






