Multiple Myeloma Derived Extracellular Vesicle Uptake by Monocyte Cells Stimulates IL-6 and MMP-9 Secretion and Promotes Cancer Cell Migration and Proliferation
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials
2.1. Cell Culture
2.2. Preparation of Conditioned Media for EV Isolation
2.3. Ultracentrifugation and Iodixonol Density Gradient for EV Isolation
2.4. Western Blot
2.5. Nanoparticle Tracking Analysis
2.6. Transmission Electron Microscopy
2.7. Cell Viability Quantification
2.7.1. Trypan Blue
2.7.2. Flow Cytometry
2.8. EV Quantification
2.9. EV Uptake of THP-1 Cells and Conditioned Media Collection
2.10. Confocal Microscopy of EV Uptake
2.11. ELISA Assay
2.12. Migration Assay
2.13. Proliferation Assay
2.14. Preparation of Samples for Mass Spectrometry
2.15. Mass Spectrometry and Analysis
2.16. Statistical Analysis
3. Results
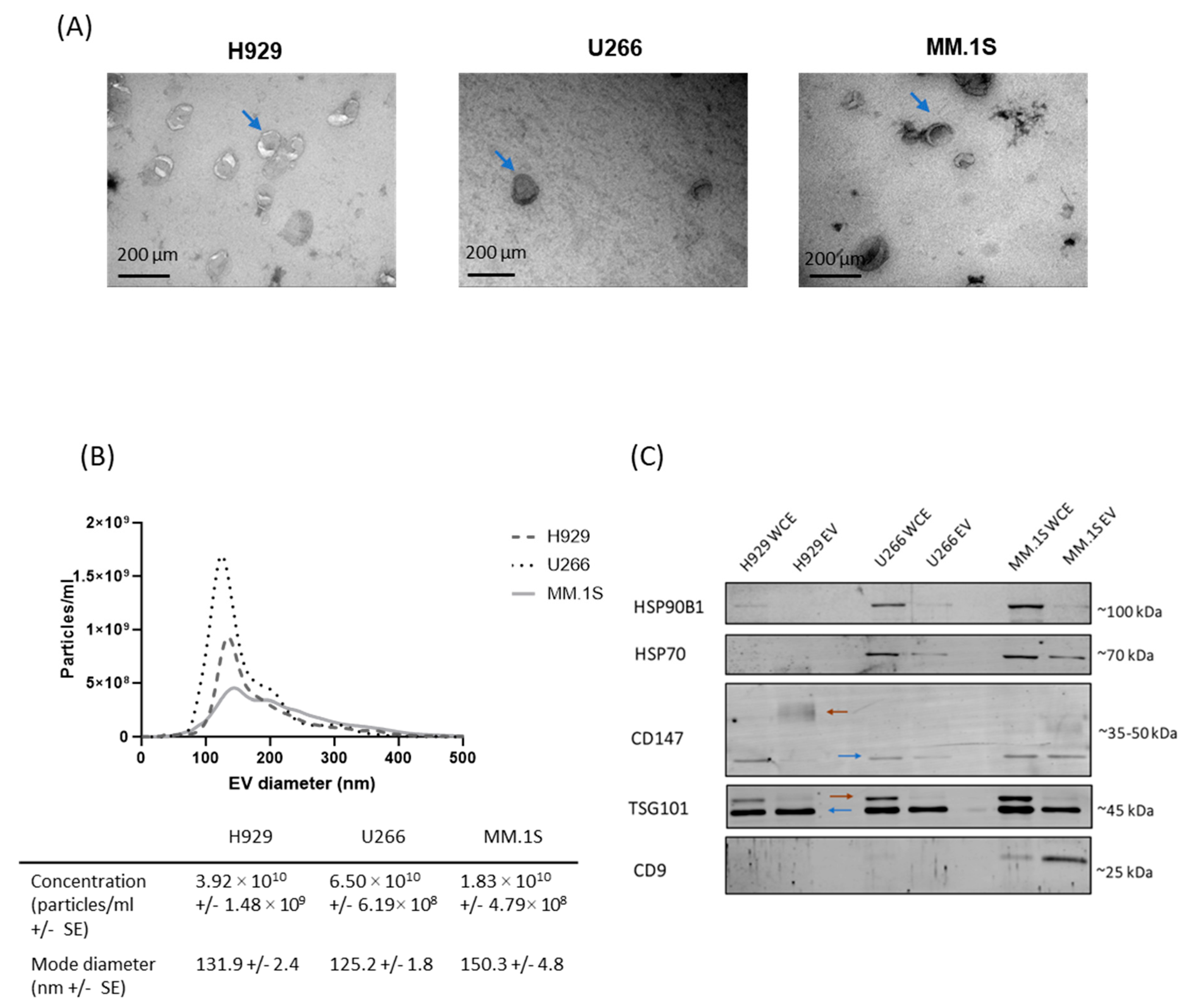
3.1. Confirmation of EV Isolation from MM Cell Lines
3.2. EV Uptake by THP-1 Monocyte Cells Is Dose and Cell Line Dependent
3.3. H929 EV Uptake by THP-1 Cells Is Energy-Dependent
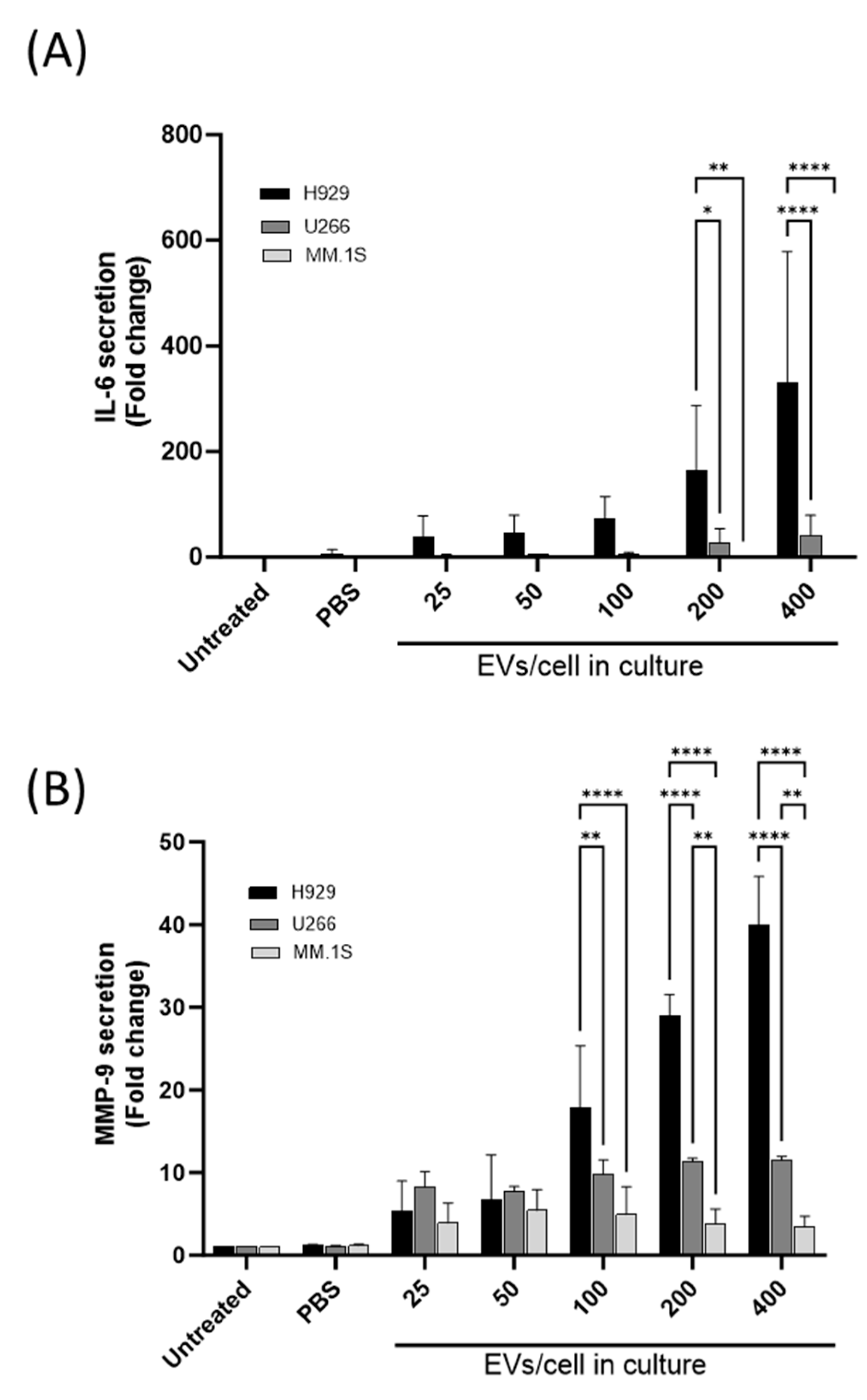
3.4. Differential IL-6 and MMP-9 Secretion from THP-1 Cells following Co-Culture with MM-Derived EVs
3.5. MM-Derived EVs Alter THP-1 Conditioned Media to Promote Migration and Proliferation of MM Cells
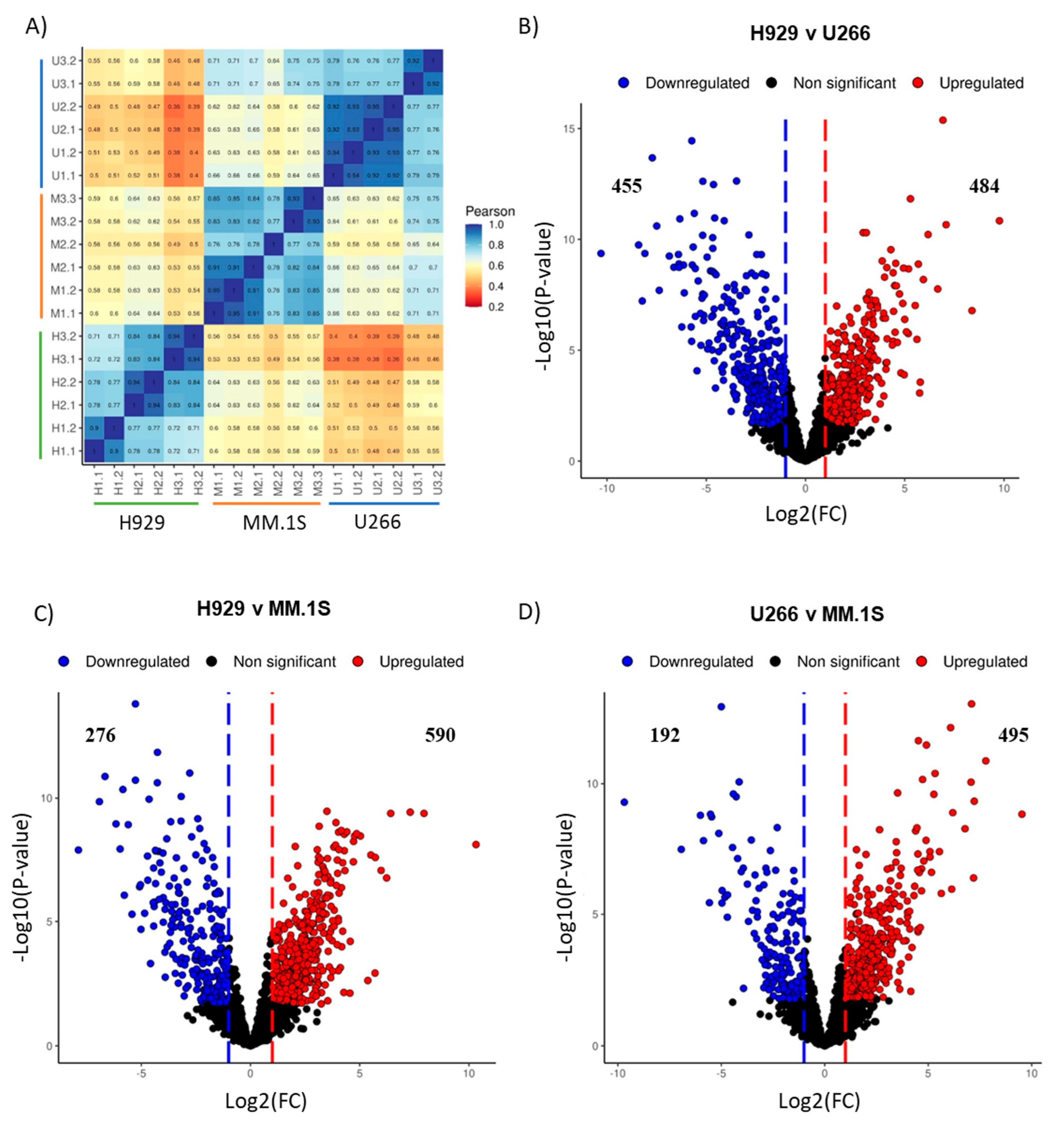
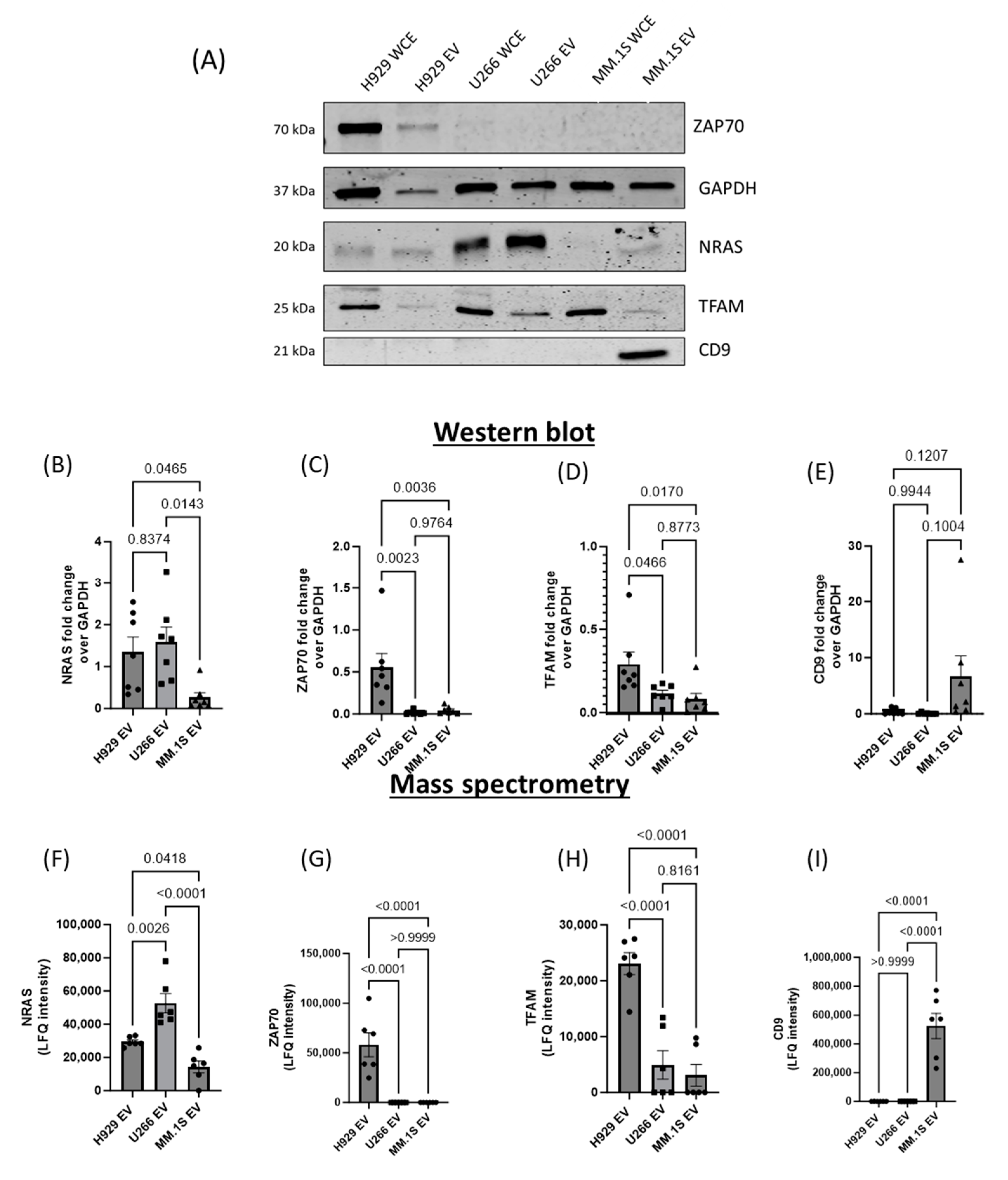
3.6. Mass Spectrometry Analysis of MM EV Content
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Boyle, E.M.; Davies, F.E.; Leleu, X.; Morgan, G.J. Understanding the multiple biological aspects leading to myeloma. Haematologica 2014, 99, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Thorsteinsdottir, S.; Dickman, P.W.; Landgren, O.; Blimark, C.; Hultcrantz, M.; Turesson, I.; Bjorkholm, M.; Kristinsson, S.Y. Dramatically improved survival in multiple myeloma patients in the recent decade: Results from a Swedish population-based study. Haematologica 2018, 103, e412–e415. [Google Scholar] [CrossRef]
- Lemaire, M.; Deleu, S.; De Bruyne, E.; Van Valckenborgh, E.; Menu, E.; Vanderkerken, K. The microenvironment and molecular biology of the multiple myeloma tumor. Adv. Cancer Res. 2011, 110, 19–42. [Google Scholar] [CrossRef]
- Kwiecien, I.; Rutkowska, E.; Raniszewska, A.; Rzepecki, P.; Domagala-Kulawik, J. Modulation of the immune response by heterogeneous monocytes and dendritic cells in lung cancer. World J. Clin. Oncol. 2021, 12, 966–982. [Google Scholar] [CrossRef] [PubMed]
- Qian, B.Z.; Li, J.; Zhang, H.; Kitamura, T.; Zhang, J.; Campion, L.R.; Kaiser, E.A.; Snyder, L.A.; Pollard, J.W. CCL2 recruits inflammatory monocytes to facilitate breast-tumour metastasis. Nature 2011, 475, 222–225. [Google Scholar] [CrossRef]
- Auffray, C.; Sieweke, M.H.; Geissmann, F. Blood monocytes: Development, heterogeneity, and relationship with dendritic cells. Annu. Rev. Immunol. 2009, 27, 669–692. [Google Scholar] [CrossRef]
- Brooks, N.; Stojanovska, L.; Grant, P.; Apostolopoulos, V.; McDonald, C.F.; Pouniotis, D.S. Characterization of blood monocyte phenotype in patients with endometrial cancer. Int. J. Gynecol. Cancer 2012, 22, 1500–1508. [Google Scholar] [CrossRef] [PubMed]
- Mytar, B.; Baj-Krzyworzeka, M.; Majka, M.; Stankiewicz, D.; Zembala, M. Human monocytes both enhance and inhibit the growth of human pancreatic cancer in SCID mice. Anticancer. Res. 2008, 28, 187–192. [Google Scholar]
- Vuk-Pavlovic, S.; Bulur, P.A.; Lin, Y.; Qin, R.; Szumlanski, C.L.; Zhao, X.; Dietz, A.B. Immunosuppressive CD14+HLA-DRlow/- monocytes in prostate cancer. Prostate 2010, 70, 443–455. [Google Scholar] [CrossRef]
- Yona, S.; Kim, K.W.; Wolf, Y.; Mildner, A.; Varol, D.; Breker, M.; Strauss-Ayali, D.; Viukov, S.; Guilliams, M.; Misharin, A.; et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity 2013, 38, 79–91. [Google Scholar] [CrossRef]
- Brocke-Heidrich, K.; Kretzschmar, A.K.; Pfeifer, G.; Henze, C.; Loffler, D.; Koczan, D.; Thiesen, H.J.; Burger, R.; Gramatzki, M.; Horn, F. Interleukin-6-dependent gene expression profiles in multiple myeloma INA-6 cells reveal a Bcl-2 family-independent survival pathway closely associated with Stat3 activation. Blood 2004, 103, 242–251. [Google Scholar] [CrossRef]
- Tucureanu, M.M.; Rebleanu, D.; Constantinescu, C.A.; Deleanu, M.; Voicu, G.; Butoi, E.; Calin, M.; Manduteanu, I. Lipopolysaccharide-induced inflammation in monocytes/macrophages is blocked by liposomal delivery of G(i)-protein inhibitor. Int. J. Nanomed. 2018, 13, 63–76. [Google Scholar] [CrossRef]
- Lokhorst, H.M.; Lamme, T.; de Smet, M.; Klein, S.; de Weger, R.A.; van Oers, R.; Bloem, A.C. Primary tumor cells of myeloma patients induce interleukin-6 secretion in long-term bone marrow cultures. Blood 1994, 84, 2269–2277. [Google Scholar] [CrossRef]
- Van Valckenborgh, E.; Croucher, P.I.; De Raeve, H.; Carron, C.; De Leenheer, E.; Blacher, S.; Devy, L.; Noel, A.; De Bruyne, E.; Asosingh, K.; et al. Multifunctional role of matrix metalloproteinases in multiple myeloma: A study in the 5T2MM mouse model. Am. J. Pathol. 2004, 165, 869–878. [Google Scholar] [CrossRef] [PubMed]
- Tkach, M.; Thery, C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell 2016, 164, 1226–1232. [Google Scholar] [CrossRef] [PubMed]
- Battistelli, M.; Falcieri, E. Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication. Biology 2020, 9, 21. [Google Scholar] [CrossRef] [PubMed]
- Nishida-Aoki, N.; Tominaga, N.; Takeshita, F.; Sonoda, H.; Yoshioka, Y.; Ochiya, T. Disruption of Circulating Extracellular Vesicles as a Novel Therapeutic Strategy against Cancer Metastasis. Mol. Ther. 2017, 25, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Lopes, R.; Caetano, J.; Barahona, F.; Pestana, C.; Ferreira, B.V.; Lourenco, D.; Queiros, A.C.; Bilreiro, C.; Shemesh, N.; Beck, H.C.; et al. Multiple Myeloma-Derived Extracellular Vesicles Modulate the Bone Marrow Immune Microenvironment. Front. Immunol. 2022, 13, 909880. [Google Scholar] [CrossRef]
- Peinado, H.; Aleckovic, M.; Lavotshkin, S.; Matei, I.; Costa-Silva, B.; Moreno-Bueno, G.; Hergueta-Redondo, M.; Williams, C.; Garcia-Santos, G.; Ghajar, C.; et al. Melanoma exosomes educate bone marrow progenitor cells toward a pro-metastatic phenotype through MET. Nat. Med. 2012, 18, 883–891. [Google Scholar] [CrossRef]
- Colombo, M.; Giannandrea, D.; Lesma, E.; Basile, A.; Chiaramonte, R. Extracellular Vesicles Enhance Multiple Myeloma Metastatic Dissemination. Int. J. Mol. Sci. 2019, 20, 3236. [Google Scholar] [CrossRef]
- Livshits, M.A.; Khomyakova, E.; Evtushenko, E.G.; Lazarev, V.N.; Kulemin, N.A.; Semina, S.E.; Generozov, E.V.; Govorun, V.M. Isolation of exosomes by differential centrifugation: Theoretical analysis of a commonly used protocol. Sci. Rep. 2015, 5, 17319. [Google Scholar] [CrossRef]
- Breuza, L.; Poux, S.; Estreicher, A.; Famiglietti, M.L.; Magrane, M.; Tognolli, M.; Bridge, A.; Baratin, D.; Redaschi, N.; UniProt, C. The UniProtKB guide to the human proteome. Database 2016, 2016, bav120. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell Proteom. 2014, 13, 2513–2526. [Google Scholar] [CrossRef]
- Tyanova, S.; Cox, J. Perseus: A Bioinformatics Platform for Integrative Analysis of Proteomics Data in Cancer Research. Methods Mol. Biol. 2018, 1711, 133–148. [Google Scholar] [CrossRef]
- Thery, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef]
- Jiao, J.; Sun, K.; Walker, W.P.; Bagher, P.; Cota, C.D.; Gunn, T.M. Abnormal regulation of TSG101 in mice with spongiform neurodegeneration. Biochim. Biophys. Acta 2009, 1792, 1027–1035. [Google Scholar] [CrossRef] [PubMed]
- Costa-Silva, B.; Aiello, N.M.; Ocean, A.J.; Singh, S.; Zhang, H.; Thakur, B.K.; Becker, A.; Hoshino, A.; Mark, M.T.; Molina, H.; et al. Pancreatic cancer exosomes initiate pre-metastatic niche formation in the liver. Nat. Cell Biol. 2015, 17, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Tesic Mark, M.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Huang, A.C.; Zhang, W.; Zhang, G.; Wu, M.; Xu, W.; Yu, Z.; Yang, J.; Wang, B.; Sun, H.; et al. Exosomal PD-L1 contributes to immunosuppression and is associated with anti-PD-1 response. Nature 2018, 560, 382–386. [Google Scholar] [CrossRef] [PubMed]
- Poggio, M.; Hu, T.; Pai, C.C.; Chu, B.; Belair, C.D.; Chang, A.; Montabana, E.; Lang, U.E.; Fu, Q.; Fong, L.; et al. Suppression of Exosomal PD-L1 Induces Systemic Anti-tumor Immunity and Memory. Cell 2019, 177, 414–427.e13. [Google Scholar] [CrossRef]
- Marar, C.; Starich, B.; Wirtz, D. Extracellular vesicles in immunomodulation and tumor progression. Nat. Immunol. 2021, 22, 560–570. [Google Scholar] [CrossRef]
- Bonsergent, E.; Grisard, E.; Buchrieser, J.; Schwartz, O.; Thery, C.; Lavieu, G. Quantitative characterization of extracellular vesicle uptake and content delivery within mammalian cells. Nat. Commun. 2021, 12, 1864. [Google Scholar] [CrossRef]
- Verdera, H.C.; Gitz-Francois, J.J.; Schiffelers, R.M.; Vader, P. Cellular uptake of extracellular vesicles is mediated by clathrin-independent endocytosis and macropinocytosis. J. Control. Release 2017, 266, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Tu, C.; Zhang, J.; Wang, J. Inhibition of multiple myeloma-derived exosomes uptake suppresses the functional response in bone marrow stromal cell. Int. J. Oncol. 2019, 54, 1061–1070. [Google Scholar] [CrossRef]
- Khan, I.; Steeg, P.S. Endocytosis: A pivotal pathway for regulating metastasis. Br. J. Cancer 2021, 124, 66–75. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Morgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef] [PubMed]
- Christianson, H.C.; Svensson, K.J.; van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed]
- Tosato, G.; Jones, K.D. Interleukin-1 induces interleukin-6 production in peripheral blood monocytes. Blood 1990, 75, 1305–1310. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Hiura, K.; Wilde, J.; Shioyasono, A.; Moriyama, K.; Hashimoto, T.; Kido, S.; Oshima, T.; Shibata, H.; Ozaki, S.; et al. Osteoclasts enhance myeloma cell growth and survival via cell-cell contact: A vicious cycle between bone destruction and myeloma expansion. Blood 2004, 104, 2484–2491. [Google Scholar] [CrossRef] [PubMed]
- Grigorieva, I.; Thomas, X.; Epstein, J. The bone marrow stromal environment is a major factor in myeloma cell resistance to dexamethasone. Exp. Hematol. 1998, 26, 597–603. [Google Scholar]
- Damiano, J.S.; Cress, A.E.; Hazlehurst, L.A.; Shtil, A.A.; Dalton, W.S. Cell adhesion mediated drug resistance (CAM-DR): Role of integrins and resistance to apoptosis in human myeloma cell lines. Blood 1999, 93, 1658–1667. [Google Scholar] [CrossRef]
- Hideshima, T.; Chauhan, D.; Podar, K.; Schlossman, R.L.; Richardson, P.; Anderson, K.C. Novel therapies targeting the myeloma cell and its bone marrow microenvironment. Semin. Oncol. 2001, 28, 607–612. [Google Scholar] [CrossRef]
- Hathi, D.; Chanswangphuwana, C.; Cho, N.; Fontana, F.; Maji, D.; Ritchey, J.; O’Neal, J.; Ghai, A.; Duncan, K.; Akers, W.J.; et al. Ablation of VLA4 in multiple myeloma cells redirects tumor spread and prolongs survival. Sci. Rep. 2022, 12, 30. [Google Scholar] [CrossRef]
- Abe, M.; Hiura, K.; Ozaki, S.; Kido, S.; Matsumoto, T. Vicious cycle between myeloma cell binding to bone marrow stromal cells via VLA-4-VCAM-1 adhesion and macrophage inflammatory protein-1alpha and MIP-1beta production. J. Bone Miner. Metab. 2009, 27, 16–23. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, H.; He, J.; Lin, P.; Tong, Q.; Yang, J. Myeloma cells shift osteoblastogenesis to adipogenesis by inhibiting the ubiquitin ligase MURF1 in mesenchymal stem cells. Sci. Signal 2020, 13, eaay8203. [Google Scholar] [CrossRef] [PubMed]
- Joshi, S.; Singh, A.R.; Zulcic, M.; Bao, L.; Messer, K.; Ideker, T.; Dutkowski, J.; Durden, D.L. Rac2 controls tumor growth, metastasis and M1-M2 macrophage differentiation in vivo. PLoS ONE 2014, 9, e95893. [Google Scholar] [CrossRef] [PubMed]
- Ohi, M.D. Structural and functional analyses of the spliceosome requires a multi-disciplinary approach. Methods 2017, 125, 1. [Google Scholar] [CrossRef] [PubMed]
- Climente-Gonzalez, H.; Porta-Pardo, E.; Godzik, A.; Eyras, E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep. 2017, 20, 2215–2226. [Google Scholar] [CrossRef] [PubMed]
- Bae, J.; Leo, C.P.; Hsu, S.Y.; Hsueh, A.J. MCL-1S, a splicing variant of the antiapoptotic BCL-2 family member MCL-1, encodes a proapoptotic protein possessing only the BH3 domain. J. Biol. Chem. 2000, 275, 25255–25261. [Google Scholar] [CrossRef] [PubMed]
- Pavlyukov, M.S.; Yu, H.; Bastola, S.; Minata, M.; Shender, V.O.; Lee, Y.; Zhang, S.; Wang, J.; Komarova, S.; Wang, J.; et al. Apoptotic Cell-Derived Extracellular Vesicles Promote Malignancy of Glioblastoma Via Intercellular Transfer of Splicing Factors. Cancer Cell 2018, 34, 119–135.e10. [Google Scholar] [CrossRef] [PubMed]
- O’Connor, B.P.; Danhorn, T.; De Arras, L.; Flatley, B.R.; Marcus, R.A.; Farias-Hesson, E.; Leach, S.M.; Alper, S. Regulation of toll-like receptor signaling by the SF3a mRNA splicing complex. PLoS Genet. 2015, 11, e1004932. [Google Scholar] [CrossRef]
- Pollyea, D.A.; Harris, C.; Rabe, J.L.; Hedin, B.R.; De Arras, L.; Katz, S.; Wheeler, E.; Bejar, R.; Walter, M.J.; Jordan, C.T.; et al. Myelodysplastic syndrome-associated spliceosome gene mutations enhance innate immune signaling. Haematologica 2019, 104, e388–e392. [Google Scholar] [CrossRef]
- Lv, Z.; Wang, Z.; Luo, L.; Chen, Y.; Han, G.; Wang, R.; Xiao, H.; Li, X.; Hou, C.; Feng, J.; et al. Spliceosome protein Eftud2 promotes colitis-associated tumorigenesis by modulating inflammatory response of macrophage. Mucosal Immunol. 2019, 12, 1164–1173. [Google Scholar] [CrossRef]
- De Arras, L.; Alper, S. Limiting of the innate immune response by SF3A-dependent control of MyD88 alternative mRNA splicing. PLoS Genet. 2013, 9, e1003855. [Google Scholar] [CrossRef]
- De Arras, L.; Laws, R.; Leach, S.M.; Pontis, K.; Freedman, J.H.; Schwartz, D.A.; Alper, S. Comparative genomics RNAi screen identifies Eftud2 as a novel regulator of innate immunity. Genetics 2014, 197, 485–496. [Google Scholar] [CrossRef] [PubMed]
- Khalife, J.; Ghose, J.; Martella, M.; Viola, D.; Rocci, A.; Troadec, E.; Terrazas, C.; Satoskar, A.R.; Gunes, E.G.; Dona, A.; et al. MiR-16 regulates crosstalk in NF-κB tolerogenic inflammatory signaling between myeloma cells and bone marrow macrophages. JCI Insight 2019, 4, e129348. [Google Scholar] [CrossRef] [PubMed]
- Deutsch, E.W.; Bandeira, N.; Perez-Riverol, Y.; Sharma, V.; Carver, J.J.; Mendoza, L.; Kundu, D.J.; Wang, S.; Bandla, C.; Kamatchinathan, S.; et al. The ProteomeXchange consortium at 10 years: 2023 update. Nucleic Acids Res. 2023, 51, D1539–D1548. [Google Scholar] [CrossRef] [PubMed]





| H929/U266 | U266/MM1s | H929/MM1s | ||||
|---|---|---|---|---|---|---|
| log2 (FC) | FDR | log2 (FC) | FDR | log2 (FC) | FDR | |
| MYL4 | 3.89384 | 8.03 × 10−7 | 1.31732 | 0.002187 | 5.211161 | 6.02× 10−10 |
| H1F0 | 3.390922 | 3.47× 10−5 | 1.402214 | 0.022585 | 4.793137 | 9.78× 10−7 |
| VAT1 | 2.964245 | 0.000402 | 4.253141 | 6.16× 10−05 | 7.217386 | 8.10× 10−7 |
| ITGB1 | 2.866007 | 0.000122 | 2.102115 | 0.002692 | 4.968122 | 1.99× 10−6 |
| FUS | 2.791342 | 0.000183 | 1.567522 | 0.004318 | 4.358864 | 8.27× 10−7 |
| TF | 2.231458 | 0.043132 | 2.657278 | 0.01826 | 4.888736 | 1.73× 10−7 |
| ITGA4 | 2.120799 | 0.000634 | 4.002313 | 0.002133 | 6.123113 | 5.79× 10−5 |
| MANF | 1.975773 | 0.01541 | 2.34326 | 0.005678 | 4.319033 | 1.79× 10−6 |
| SRPK2;SRPK3 | 1.822525 | 0.040219 | 2.117563 | 0.001355 | 3.940088 | 0.000108 |
| KIF4A | 1.800351 | 5.92× 10−05 | 1.826726 | 0.005663 | 3.627077 | 2.95× 10−5 |
| MAPRE2 | 1.676595 | 0.004901 | 1.742989 | 0.006958 | 3.419584 | 1.98× 10−5 |
| NDRG3 | 1.543764 | 0.011512 | 1.886626 | 0.050681 | 3.43039 | 0.000854 |
| HIST1H1D | 1.417215 | 0.000121 | 2.381263 | 0.029233 | 3.798478 | 0.002065 |
| EXOC4 | 1.184284 | 0.01465 | 1.865372 | 0.010685 | 3.049656 | 7.39× 10−5 |
| RAB8B | 1.104097 | 0.005696 | 1.514335 | 0.02753 | 2.618432 | 0.00118 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sheridan, R.; Brennan, K.; Bazou, D.; O’Gorman, P.; Matallanas, D.; Mc Gee, M.M. Multiple Myeloma Derived Extracellular Vesicle Uptake by Monocyte Cells Stimulates IL-6 and MMP-9 Secretion and Promotes Cancer Cell Migration and Proliferation. Cancers 2024, 16, 1011. https://doi.org/10.3390/cancers16051011
Sheridan R, Brennan K, Bazou D, O’Gorman P, Matallanas D, Mc Gee MM. Multiple Myeloma Derived Extracellular Vesicle Uptake by Monocyte Cells Stimulates IL-6 and MMP-9 Secretion and Promotes Cancer Cell Migration and Proliferation. Cancers. 2024; 16(5):1011. https://doi.org/10.3390/cancers16051011
Chicago/Turabian StyleSheridan, Rebecca, Kieran Brennan, Despina Bazou, Peter O’Gorman, David Matallanas, and Margaret M. Mc Gee. 2024. "Multiple Myeloma Derived Extracellular Vesicle Uptake by Monocyte Cells Stimulates IL-6 and MMP-9 Secretion and Promotes Cancer Cell Migration and Proliferation" Cancers 16, no. 5: 1011. https://doi.org/10.3390/cancers16051011
APA StyleSheridan, R., Brennan, K., Bazou, D., O’Gorman, P., Matallanas, D., & Mc Gee, M. M. (2024). Multiple Myeloma Derived Extracellular Vesicle Uptake by Monocyte Cells Stimulates IL-6 and MMP-9 Secretion and Promotes Cancer Cell Migration and Proliferation. Cancers, 16(5), 1011. https://doi.org/10.3390/cancers16051011








