Use of Droplet Digital Polymerase Chain Reaction to Identify Biomarkers for Differentiation of Benign and Malignant Renal Masses
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Design
2.2. Plasma Samples
2.3. RNA Isolation
2.4. PCR Analyses
2.4.1. cDNA Synthesis
2.4.2. ddPCR
2.4.3. Conventional Real-Time PCR (RT-PCR)
2.4.4. Selection of Normalizers
2.4.5. Limit of Quantification
2.5. Statistical Analyses
3. Results
3.1. Study Cohort
3.2. Candidate Biomarkers
3.3. PCR
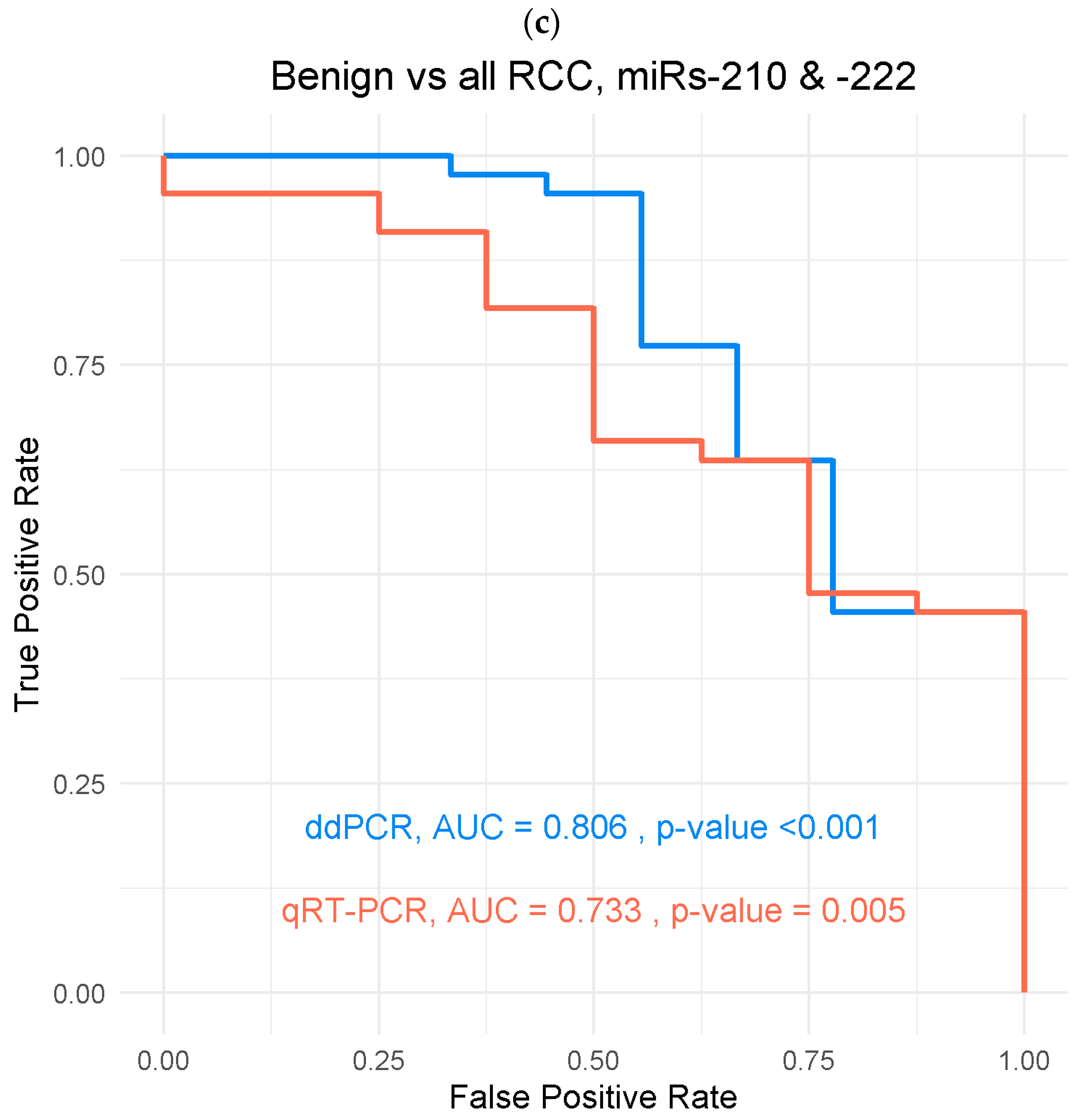
3.3.1. Benign Versus RCC
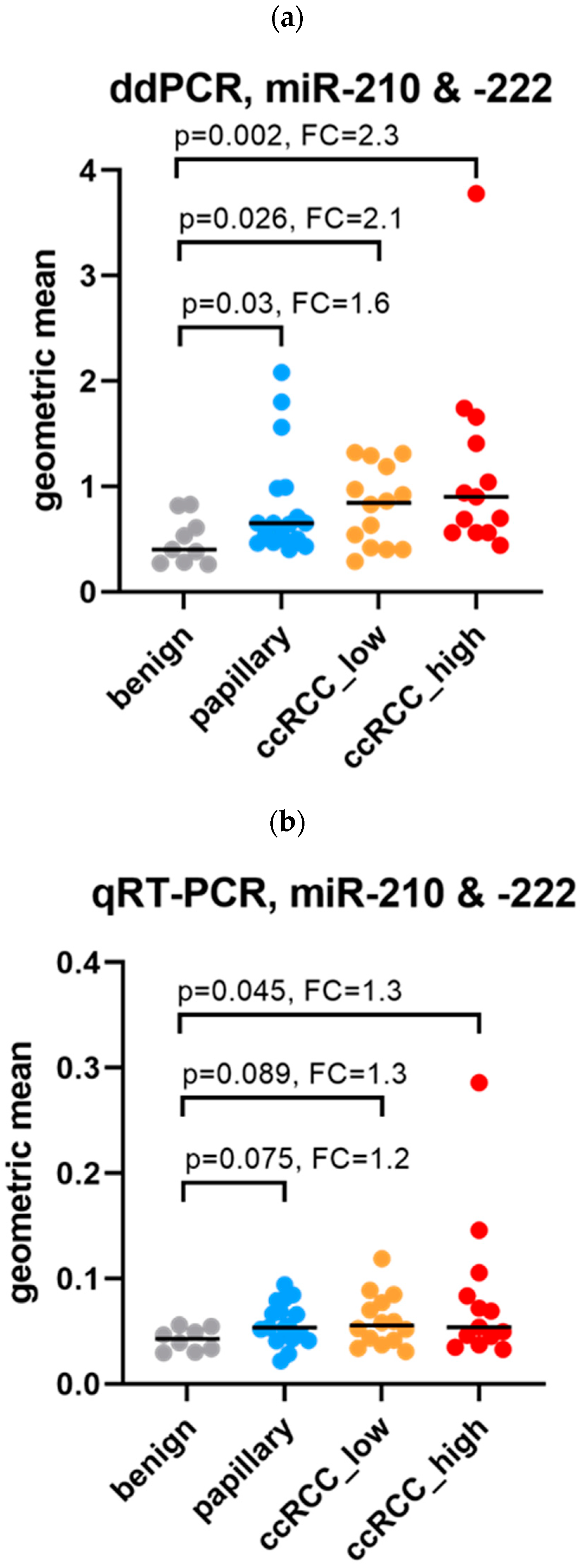
3.3.2. Subgroup Comparisons
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Jonasch, E.; Gao, J.; Rathmell, W.K. Renal Cell Carcinoma. BMJ 2014, 349, g4797. [Google Scholar] [CrossRef] [PubMed]
- Patel, H.D.; Johnson, M.H.; Pierorazio, P.M.; Sozio, S.M.; Sharma, R.; Iyoha, E.; Bass, E.B.; Allaf, M.E. Diagnostic Accuracy and Risks of Biopsy in the Diagnosis of a Renal Mass Suspicious for Localized Renal Cell Carcinoma: Systematic Review of the Literature. J. Urol. 2016, 195, 1340–1347. [Google Scholar] [CrossRef] [PubMed]
- Dieckmann, K.-P.; Radtke, A.; Geczi, L.; Matthies, C.; Anheuser, P.; Eckardt, U.; Sommer, J.; Zengerling, F.; Trenti, E.; Pichler, R.; et al. Serum Levels of MicroRNA-371a-3p (M371 Test) as a New Biomarker of Testicular Germ Cell Tumors: Results of a Prospective Multicentric Study. J. Clin. Oncol. 2019, 37, 1412–1423. [Google Scholar] [CrossRef] [PubMed]
- Silva-Santos, R.M.; Costa-Pinheiro, P.; Luis, A.; Antunes, L.; Lobo, F.; Oliveira, J.; Henrique, R.; Jerónimo, C. MicroRNA Profile: A Promising Ancillary Tool for Accurate Renal Cell Tumour Diagnosis. Br. J. Cancer 2013, 109, 2646–2653. [Google Scholar] [CrossRef]
- Moynihan, M.J.; Sullivan, T.B.; Burks, E.; Schober, J.; Calabrese, M.; Fredrick, A.; Kalantzakos, T.; Warrick, J.; Canes, D.; Raman, J.D.; et al. MicroRNA Profile in Stage I Clear Cell Renal Cell Carcinoma Predicts Progression to Metastatic Disease. Urol. Oncol. 2020, 38, 799.e11. [Google Scholar] [CrossRef]
- Li, M.; Li, L.; Zheng, J.; Li, Z.; Li, S.; Wang, K.; Chen, X. Liquid Biopsy at the Frontier in Renal Cell Carcinoma: Recent Analysis of Techniques and Clinical Application. Mol. Cancer 2023, 22, 37. [Google Scholar] [CrossRef] [PubMed]
- Lou, N.; Ruan, A.-M.; Qiu, B.; Bao, L.; Xu, Y.-C.; Zhao, Y.; Sun, R.-L.; Zhang, S.-T.; Xu, G.-H.; Ruan, H.-L.; et al. miR-144-3p as a Novel Plasma Diagnostic Biomarker for Clear Cell Renal Cell Carcinoma. Urol. Oncol. 2017, 35, 36.e7. [Google Scholar] [CrossRef]
- Marchioni, M.; Rivas, J.G.; Autran, A.; Socarras, M.; Albisinni, S.; Ferro, M.; Schips, L.; Scarpa, R.M.; Papalia, R.; Esperto, F. Biomarkers for Renal Cell Carcinoma Recurrence: State of the Art. Curr. Urol. Rep. 2021, 22, 31. [Google Scholar] [CrossRef]
- Hindson, C.M.; Chevillet, J.R.; Briggs, H.A.; Gallichotte, E.N.; Ruf, I.K.; Hindson, B.J.; Vessella, R.L.; Tewari, M. Absolute Quantification by Droplet Digital PCR versus Analog Real-Time PCR. Nat. Methods 2013, 10, 1003–1005. [Google Scholar] [CrossRef]
- Kirschner, M.B.; Kao, S.C.; Edelman, J.J.; Armstrong, N.J.; Vallely, M.P.; van Zandwijk, N.; Reid, G. Haemolysis during Sample Preparation Alters microRNA Content of Plasma. PLoS ONE 2011, 6, e24145. [Google Scholar] [CrossRef]
- Rogers, C.J.; Kyubwa, E.M.; Lukaszewicz, A.I.; Starbird, M.A.; Nguyen, M.; Copeland, B.T.; Yamada-Hanff, J.; Menon, N. Observation of Unique Circulating miRNA Signatures in Non-Human Primates Exposed to Total-Body vs. Whole Thorax Lung Irradiation. Radiat. Res. 2021, 196, 547–559. [Google Scholar] [CrossRef]
- Laeremans, T.; D’haese, S.; Aernout, J.; Barbé, K.; Pannus, P.; Rutsaert, S.; Vancutsem, E.; Vanham, G.; Necsoi, C.; Spiegelaere, W.D.; et al. Qualitative Plasma Viral Load Determination as a Tool for Screening of Viral Reservoir Size in PWH. AIDS 2022, 36, 1761–1768. [Google Scholar] [CrossRef]
- Cirillo, P.D.R.; Margiotti, K.; Fabiani, M.; Barros-Filho, M.C.; Sparacino, D.; Cima, A.; Longo, S.A.; Cupellaro, M.; Mesoraca, A.; Giorlandino, C. Multi-Analytical Test Based on Serum miRNAs and Proteins Quantification for Ovarian Cancer Early Detection. PLoS ONE 2021, 16, e0255804. [Google Scholar] [CrossRef]
- Floren, C.; Wiedemann, I.; Brenig, B.; Schütz, E.; Beck, J. Species Identification and Quantification in Meat and Meat Products Using Droplet Digital PCR (ddPCR). Food Chem. 2015, 173, 1054–1058. [Google Scholar] [CrossRef] [PubMed]
- Akdogan, B.; Gudeloglu, A.; Inci, K.; Gunay, L.M.; Koni, A.; Ozen, H. Prevalence and Predictors of Benign Lesions in Renal Masses Smaller than 7 Cm Presumed to Be Renal Cell Carcinoma. Clin. Genitourin. Cancer 2012, 10, 121–125. [Google Scholar] [CrossRef]
- Gorin, M.A.; Rowe, S.P.; Baras, A.S.; Solnes, L.B.; Ball, M.W.; Pierorazio, P.M.; Pavlovich, C.P.; Epstein, J.I.; Javadi, M.S.; Allaf, M.E. Prospective Evaluation of (99m)Tc-Sestamibi SPECT/CT for the Diagnosis of Renal Oncocytomas and Hybrid Oncocytic/Chromophobe Tumors. Eur. Urol. 2016, 69, 413–416. [Google Scholar] [CrossRef] [PubMed]
- Moch, H.; Amin, M.B.; Berney, D.M.; Compérat, E.M.; Gill, A.J.; Hartmann, A.; Menon, S.; Raspollini, M.R.; Rubin, M.A.; Srigley, J.R.; et al. The 2022 World Health Organization Classification of Tumours of the Urinary System and Male Genital Organs-Part A: Renal, Penile, and Testicular Tumours. Eur. Urol. 2022, 82, 458–468. [Google Scholar] [CrossRef] [PubMed]
- Leveridge, M.J.; Mattar, K.; Kachura, J.; Jewett, M.A.S. Assessing Outcomes in Probe Ablative Therapies for Small Renal Masses. J. Endourol. 2010, 24, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Akram, F.; Atique, N.; Haq, I.U.; Ahmed, Z.; Jabbar, Z.; Nawaz, A.; Aqeel, A.; Akram, R. MicroRNA, a Promising Biomarker for Breast and Ovarian Cancer: A Review. Curr. Protein Pept. Sci. 2021, 22, 599–619. [Google Scholar] [CrossRef]
- Kabzinski, J.; Maczynska, M.; Majsterek, I. MicroRNA as a Novel Biomarker in the Diagnosis of Head and Neck Cancer. Biomolecules 2021, 11, 844. [Google Scholar] [CrossRef]
- Wulfken, L.M.; Moritz, R.; Ohlmann, C.; Holdenrieder, S.; Jung, V.; Becker, F.; Herrmann, E.; Walgenbach-Brünagel, G.; von Ruecker, A.; Müller, S.C.; et al. MicroRNAs in Renal Cell Carcinoma: Diagnostic Implications of Serum miR-1233 Levels. PLoS ONE 2011, 6, e25787. [Google Scholar] [CrossRef] [PubMed]
- Fedorko, M.; Pacik, D.; Wasserbauer, R.; Juracek, J.; Varga, G.; Ghazal, M.; Nussir, M.I. MicroRNAs in the Pathogenesis of Renal Cell Carcinoma and Their Diagnostic and Prognostic Utility as Cancer Biomarkers. Int. J. Biol. Markers 2016, 31, e26–e37. [Google Scholar] [CrossRef]
- Zhao, A.; Li, G.; Péoc’h, M.; Genin, C.; Gigante, M. Serum miR-210 as a Novel Biomarker for Molecular Diagnosis of Clear Cell Renal Cell Carcinoma. Exp. Mol. Pathol. 2013, 94, 115–120. [Google Scholar] [CrossRef] [PubMed]
- Iwamoto, H.; Kanda, Y.; Sejima, T.; Osaki, M.; Okada, F.; Takenaka, A. Serum miR-210 as a Potential Biomarker of Early Clear Cell Renal Cell Carcinoma. Int. J. Oncol. 2014, 44, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.Y.; Zhang, H.; Du, S.M.; Li, J.; Wen, X.H. Expression of microRNA-210 in Tissue and Serum of Renal Carcinoma Patients and Its Effect on Renal Carcinoma Cell Proliferation, Apoptosis, and Invasion. Genet. Mol. Res. 2016, 15, 15017746. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Ni, M.; Su, Y.; Wang, H.; Zhu, S.; Zhao, A.; Li, G. MicroRNAs in Serum Exosomes as Potential Biomarkers in Clear-Cell Renal Cell Carcinoma. Eur. Urol. Focus 2018, 4, 412–419. [Google Scholar] [CrossRef]
- Teixeira, A.L.; Ferreira, M.; Silva, J.; Gomes, M.; Dias, F.; Santos, J.I.; Maurício, J.; Lobo, F.; Medeiros, R. Higher Circulating Expression Levels of miR-221 Associated with Poor Overall Survival in Renal Cell Carcinoma Patients. Tumour Biol. 2014, 35, 4057–4066. [Google Scholar] [CrossRef]
- Teixeira, A.L.; Dias, F.; Ferreira, M.; Gomes, M.; Santos, J.I.; Lobo, F.; Maurício, J.; Machado, J.C.; Medeiros, R. Combined Influence of EGF+61G>A and TGFB+869T>C Functional Polymorphisms in Renal Cell Carcinoma Progression and Overall Survival: The Link to Plasma Circulating MiR-7 and MiR-221/222 Expression. PLoS ONE 2014, 10, e0103258. [Google Scholar] [CrossRef]
- Youssef, Y.M.; White, N.M.A.; Grigull, J.; Krizova, A.; Samy, C.; Mejia-Guerrero, S.; Evans, A.; Yousef, G.M. Accurate Molecular Classification of Kidney Cancer Subtypes Using microRNA Signature. Eur. Urol. 2011, 59, 721–730. [Google Scholar] [CrossRef]
- Xiao, C.-T.; Lai, W.-J.; Zhu, W.-A.; Wang, H. MicroRNA Derived from Circulating Exosomes as Noninvasive Biomarkers for Diagnosing Renal Cell Carcinoma. Onco Targets Ther. 2020, 13, 10765–10774. [Google Scholar] [CrossRef]
- Wang, X.-G.; Zhu, Y.-W.; Wang, T.; Chen, B.; Xing, J.-C.; Xiao, W. MiR-483-5p Downregulation Contributed to Cell Proliferation, Metastasis, and Inflammation of Clear Cell Renal Cell Carcinoma. Kaohsiung J. Med. Sci. 2021, 37, 192–199. [Google Scholar] [CrossRef]
- Xiao, W.; Wang, C.; Chen, K.; Wang, T.; Xing, J.; Zhang, X.; Wang, X. MiR-765 Functions as a Tumour Suppressor and Eliminates Lipids in Clear Cell Renal Cell Carcinoma by Downregulating PLP2. EBioMedicine 2020, 51, 102622. [Google Scholar] [CrossRef]
- Zhang, W.-B.; Pan, Z.-Q.; Yang, Q.-S.; Zheng, X.-M. Tumor Suppressive miR-509-5p Contributes to Cell Migration, Proliferation and Antiapoptosis in Renal Cell Carcinoma. Ir. J. Med. Sci. 2013, 182, 621–627. [Google Scholar] [CrossRef]
- Dias, F.; Teixeira, A.L.; Nogueira, I.; Morais, M.; Maia, J.; Bodo, C.; Ferreira, M.; Silva, A.; Vilhena, M.; Lobo, J.; et al. Extracellular Vesicles Enriched in Hsa-miR-301a-3p and Hsa-miR-1293 Dynamics in Clear Cell Renal Cell Carcinoma Patients: Potential Biomarkers of Metastatic Disease. Cancers 2020, 12, 1450. [Google Scholar] [CrossRef]
- Chanudet, E.; Wozniak, M.B.; Bouaoun, L.; Byrnes, G.; Mukeriya, A.; Zaridze, D.; Brennan, P.; Muller, D.C.; Scelo, G. Large-Scale Genome-Wide Screening of Circulating microRNAs in Clear Cell Renal Cell Carcinoma Reveals Specific Signatures in Late-Stage Disease. Int. J. Cancer 2017, 141, 1730–1740. [Google Scholar] [CrossRef]
- Zhao, J.; Lei, T.; Xu, C.; Li, H.; Ma, W.; Yang, Y.; Fan, S.; Liu, Y. MicroRNA-187, down-Regulated in Clear Cell Renal Cell Carcinoma and Associated with Lower Survival, Inhibits Cell Growth and Migration Though Targeting B7-H3. Biochem. Biophys. Res. Commun. 2013, 438, 439–444. [Google Scholar] [CrossRef] [PubMed]
- Yadav, S.; Khandelwal, M.; Seth, A.; Saini, A.K.; Dogra, P.N.; Sharma, A. Serum microRNA Expression Profiling: Potential Diagnostic Implications of a Panel of Serum microRNAs for Clear Cell Renal Cell Cancer. Urology 2017, 104, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Huang, G.; Li, X.; Chen, Z.; Wang, J.; Zhang, C.; Chen, X.; Peng, X.; Liu, K.; Zhao, L.; Lai, Y.; et al. A Three-microRNA Panel in Serum: Serving as a Potential Diagnostic Biomarker for Renal Cell Carcinoma. Pathol. Oncol. Res. 2020, 26, 2425–2434. [Google Scholar] [CrossRef] [PubMed]
- Tusong, H.; Maolakuerban, N.; Guan, J.; Rexiati, M.; Wang, W.-G.; Azhati, B.; Nuerrula, Y.; Wang, Y.-J. Functional Analysis of Serum microRNAs miR-21 and miR-106a in Renal Cell Carcinoma. Cancer Biomark. 2017, 18, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-K.; Luo, L.; Du, Z.-J.; Zhang, G.-M.; Sun, L.-J. MiR429 Expression Level in Renal Cell Cancer and Its Correlation with the Prognosis of Patients. J. BUON 2017, 22, 1428–1433. [Google Scholar] [PubMed]
- Zhang, Q.; Di, W.; Dong, Y.; Lu, G.; Yu, J.; Li, J.; Li, P. High Serum miR-183 Level Is Associated with Poor Responsiveness of Renal Cancer to Natural Killer Cells. Tumour Biol. 2015, 36, 9245–9249. [Google Scholar] [CrossRef] [PubMed]
- Heinemann, F.G.; Tolkach, Y.; Deng, M.; Schmidt, D.; Perner, S.; Kristiansen, G.; Müller, S.C.; Ellinger, J. Serum miR-122-5p and miR-206 Expression: Non-Invasive Prognostic Biomarkers for Renal Cell Carcinoma. Clin. Epigenetics 2018, 10, 11. [Google Scholar] [CrossRef]
- Song, W.; Chen, Y.; Zhu, G.; Xie, H.; Yang, Z.; Li, L. Exosome-Mediated miR-9-5p Promotes Proliferation and Migration of Renal Cancer Cells Both in Vitro and in Vivo by Targeting SOCS4. Biochem. Biophys. Res. Commun. 2020, 529, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Deng, X.; Zhang, J. Identification of Dysregulated Serum miR-508-3p and miR-885-5p as Potential Diagnostic Biomarkers of Clear Cell Renal Carcinoma. Mol. Med. Rep. 2019, 20, 5075–5083. [Google Scholar] [CrossRef]
- Fedorko, M.; Stanik, M.; Iliev, R.; Redova-Lojova, M.; Machackova, T.; Svoboda, M.; Pacik, D.; Dolezel, J.; Slaby, O. Combination of MiR-378 and MiR-210 Serum Levels Enables Sensitive Detection of Renal Cell Carcinoma. Int. J. Mol. Sci. 2015, 16, 23382–23389. [Google Scholar] [CrossRef]
- Hauser, S.; Wulfken, L.M.; Holdenrieder, S.; Moritz, R.; Ohlmann, C.-H.; Jung, V.; Becker, F.; Herrmann, E.; Walgenbach-Brünagel, G.; von Ruecker, A.; et al. Analysis of Serum microRNAs (miR-26a-2*, miR-191, miR-337-3p and miR-378) as Potential Biomarkers in Renal Cell Carcinoma. Cancer Epidemiol. 2012, 36, 391–394. [Google Scholar] [CrossRef]
- Redova, M.; Poprach, A.; Nekvindova, J.; Iliev, R.; Radova, L.; Lakomy, R.; Svoboda, M.; Vyzula, R.; Slaby, O. Circulating miR-378 and miR-451 in Serum Are Potential Biomarkers for Renal Cell Carcinoma. J. Transl. Med. 2012, 10, 55. [Google Scholar] [CrossRef]
- Yang, Q.; Liu, J.; Liang, Y.; Wang, C.; Han, J.; Zhu, L.; Yuan, S.; Sun, Q.; Zhang, H. Correlation between serum level of miRNA-106a expression with clinicopathological characteristics and prognosis of patients with renal cell carcinoma. Zhonghua Yi Xue Yi Chuan Xue Za Zhi 2021, 38, 652–655. [Google Scholar] [CrossRef]
- Huang, G.; Li, H.; Wang, J.; Peng, X.; Liu, K.; Zhao, L.; Zhang, C.; Chen, X.; Lai, Y.; Ni, L. Combination of Tumor Suppressor miR-20b-5p, miR-30a-5p, and miR-196a-5p as a Serum Diagnostic Panel for Renal Cell Carcinoma. Pathol. Res. Pract. 2020, 216, 153152. [Google Scholar] [CrossRef]
- Wang, C.; Hu, J.; Lu, M.; Gu, H.; Zhou, X.; Chen, X.; Zen, K.; Zhang, C.-Y.; Zhang, T.; Ge, J.; et al. A Panel of Five Serum miRNAs as a Potential Diagnostic Tool for Early-Stage Renal Cell Carcinoma. Sci. Rep. 2015, 5, 7610. [Google Scholar] [CrossRef]
- Muramatsu-Maekawa, Y.; Kawakami, K.; Fujita, Y.; Takai, M.; Kato, D.; Nakane, K.; Kato, T.; Tsuchiya, T.; Koie, T.; Miura, Y.; et al. Profiling of Serum Extracellular Vesicles Reveals miRNA-4525 as a Potential Biomarker for Advanced Renal Cell Carcinoma. Cancer Genom. Proteom. 2021, 18, 253–259. [Google Scholar] [CrossRef]
- Zhai, Q.; Zhou, L.; Zhao, C.; Wan, J.; Yu, Z.; Guo, X.; Qin, J.; Chen, J.; Lu, R. Identification of miR-508-3p and miR-509-3p That Are Associated with Cell Invasion and Migration and Involved in the Apoptosis of Renal Cell Carcinoma. Biochem. Biophys. Res. Commun. 2012, 419, 621–626. [Google Scholar] [CrossRef] [PubMed]
- D’Antona, P.; Cattoni, M.; Dominioni, L.; Poli, A.; Moretti, F.; Cinquetti, R.; Gini, E.; Daffrè, E.; Noonan, D.M.; Imperatori, A.; et al. Serum miR-223: A Validated Biomarker for Detection of Early-Stage Non-Small Cell Lung Cancer. Cancer Epidemiol. Biomark. Prev. 2019, 28, 1926–1933. [Google Scholar] [CrossRef] [PubMed]
- Cirillo, P.D.R.; Margiotti, K.; Mesoraca, A.; Giorlandino, C. Quantification of Circulating microRNAs by Droplet Digital PCR for Cancer Detection. BMC Res. Notes 2020, 13, 351. [Google Scholar] [CrossRef] [PubMed]
- Ferracin, M.; Lupini, L.; Salamon, I.; Saccenti, E.; Zanzi, M.V.; Rocchi, A.; Da Ros, L.; Zagatti, B.; Musa, G.; Bassi, C.; et al. Absolute Quantification of Cell-Free microRNAs in Cancer Patients. Oncotarget 2015, 6, 14545–14555. [Google Scholar] [CrossRef] [PubMed]



| Clinicopathologic Findings | Clear Cell n = 27 | Papillary n = 17 | Benign n = 9 | p Value |
|---|---|---|---|---|
| Stage/type | ||||
| pT1-pT2 | 14 | 15 | - | |
| pT3-pT4 | 13 | 2 | - | |
| AML | - | - | 7 | |
| other | - | - | 2 | |
| Sex | 0.743 | |||
| Female | 11 (41) | 5 (29) | 4 (44) | |
| Male | 16 (59) | 12 (71) | 5 (56) | |
| Age at diagnosis | 60 (53–68) | 66 (57–73) | 51 (42–59) | 0.013 |
| BMI at surgery | 29.2 (25.2–34.3) | 27.5 (24.2–32.3) | 29.8 (24.7–32.3) | 0.773 |
| Tumor size, cm | 6.3 (3.4–9.0) | 3.1 (2.6–4.9) | 2.6 (1.7–3.2) | 0.009 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hayden, J.P.; Wiggins, A.; Sullivan, T.; Kalantzakos, T.; Hooper, K.; Moinzadeh, A.; Rieger-Christ, K. Use of Droplet Digital Polymerase Chain Reaction to Identify Biomarkers for Differentiation of Benign and Malignant Renal Masses. Cancers 2024, 16, 787. https://doi.org/10.3390/cancers16040787
Hayden JP, Wiggins A, Sullivan T, Kalantzakos T, Hooper K, Moinzadeh A, Rieger-Christ K. Use of Droplet Digital Polymerase Chain Reaction to Identify Biomarkers for Differentiation of Benign and Malignant Renal Masses. Cancers. 2024; 16(4):787. https://doi.org/10.3390/cancers16040787
Chicago/Turabian StyleHayden, Joshua P., Adam Wiggins, Travis Sullivan, Thomas Kalantzakos, Kailey Hooper, Alireza Moinzadeh, and Kimberly Rieger-Christ. 2024. "Use of Droplet Digital Polymerase Chain Reaction to Identify Biomarkers for Differentiation of Benign and Malignant Renal Masses" Cancers 16, no. 4: 787. https://doi.org/10.3390/cancers16040787
APA StyleHayden, J. P., Wiggins, A., Sullivan, T., Kalantzakos, T., Hooper, K., Moinzadeh, A., & Rieger-Christ, K. (2024). Use of Droplet Digital Polymerase Chain Reaction to Identify Biomarkers for Differentiation of Benign and Malignant Renal Masses. Cancers, 16(4), 787. https://doi.org/10.3390/cancers16040787




