Human GST P1-1 Redesigned for Enhanced Catalytic Activity with the Anticancer Prodrug Telcyta and Improved Thermostability
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Protein Expression and Purification
2.3. Activity Measurements with CDNB
2.4. Telcyta Calibration Curve
2.5. Thin-Layer Chromatography to Assay Catalytic Activity of Gst P1-1 Variants with Telcyta
2.6. Thermostability
2.7. Machine Learning Procedure
2.8. Homology Modeling
3. Results
3.1. Expression and Purification of GST P1-1 Proteins
3.2. Telcyta Quantification
3.3. Catalytic Activity with Telcyta of Wildtype GST P1-1 Homologs
3.4. Site-Directed Point Mutations of Human GST P1-1
3.5. Designed Variants of Human GST P1-1 for Machine-Learning
3.6. Thermostability
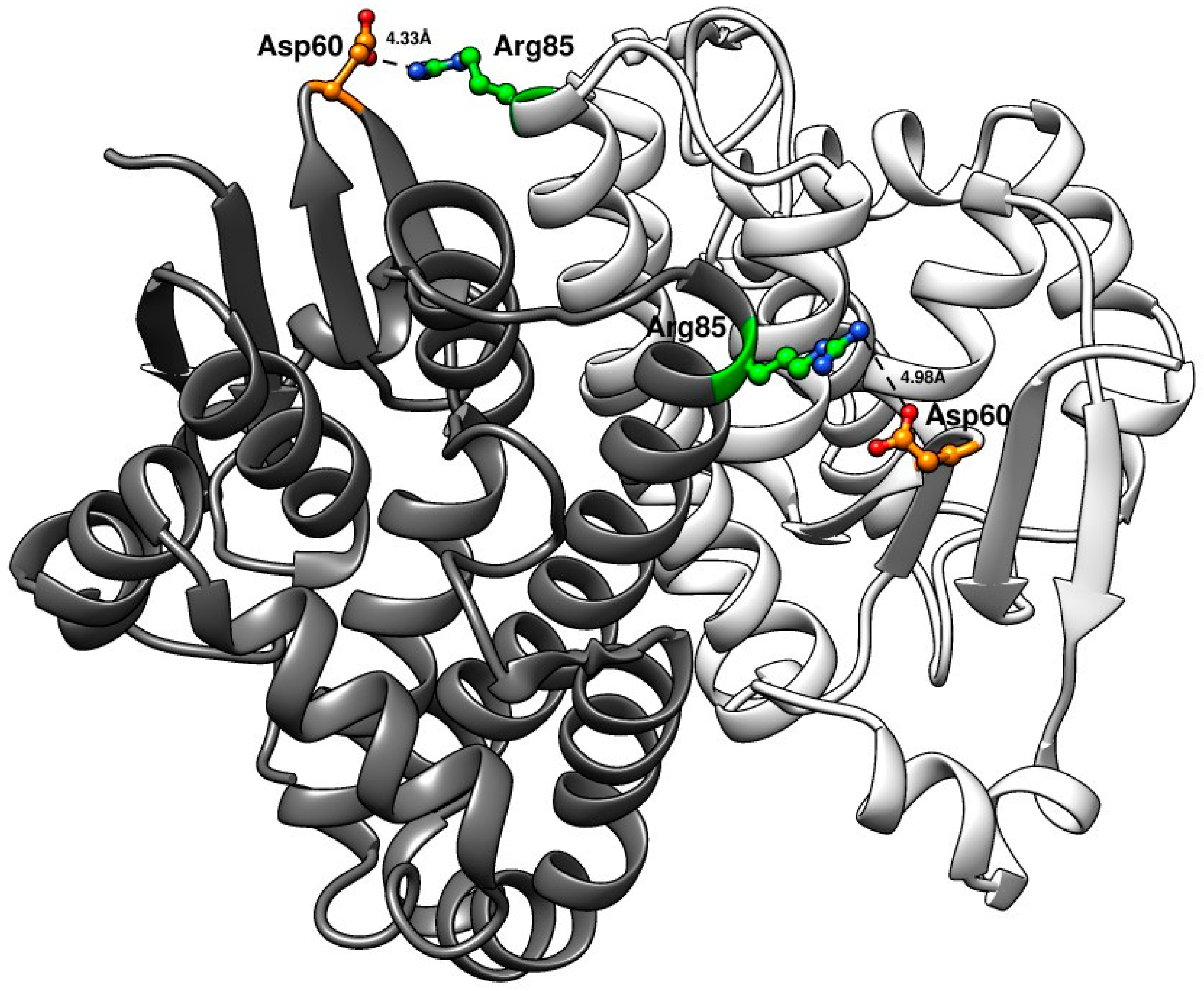
3.7. Homology Modeling
4. Discussion
4.1. Examination of the Enzymatic Activity of Natural GST P1-1 Homologs against Substrate Telcyta
4.2. Engineering of Human GST P1-1 and Catalytic Activity with Telcyta
4.3. Redesign of Gst P1-1 via Machine-Learning Libraries
4.4. Thermostability
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Estimated Activity Relative to Wildtype GST P1-1 | ||
|---|---|---|
| Variants | Telcyta (Score) | CDNB (µmol min−1 mg−1) |
| Human P1-1 | +++ | 106 ± 4 |
| V1 (T35S-Q40L-A46S-Q85R-Y109H) | ++++ | 20.5 ± 1.2 |
| V2 (Q40M-E41Q-A46S-Y109H-V200L) | ++++ | 19.2 ± 0.7 |
| V3 (Q40L-S43P-Q85K-Y109H-V200L) | ++++ | 20.6 ± 0.5 |
| V4 (T35S-E41Q-Q85K-S106T-Y109H) | ++++(+) | 20.7 ± 0.6 |
| V5 (Q40M-S43P-Q85R-Y109H-S185C) | ++++(+) | 17.5 ± 0.5 |
| V6 (Q85R-C102S-S106T-Y109H-V200L) | ++++(+) | 28.2 ± 0.3 |
| V7 (A46S-S106T-Y109H-S185C-V200A) | ++++ | 19.7 ± 0.2 |
| V8 (Q40L-E41Q-Q84P-Y109H-V200A) | ++++ | 21.3 ± 1.3 |
| V9 (T35S-S43P-C102S-Y109H-V200A) | ++++ | 17.9 ± 1.1 |
| V10 (Q40M-Q84P-Q85K-C102S-Y109H) | ++++ | 20.3 ± 0.4 |
| V11 (T35S-Q84P-Y109H-S185C-V200L) | ++++ | 21.9 ± 0.3 |
| V401 (Q85R-Y109H) | ++++ | 20.2 ± 0.7 |
| Estimated Activity Relative to Wildtype GST P1-1 | ||
|---|---|---|
| Variants | Telcyta (Score) | CDNB (µmol min−1 mg−1) |
| Human P1-1 | +++ | 106 ± 4 |
| V201 (T35S-Q40L-E41Q-Q84P-Q85K-S106T-Y109H) | ++++ | 22.4 ± 1.1 |
| V202 (T35S-Q40L-E41Q-Q85K-S106T-Y109H-S185C) | ++++ | 22.2 ± 1.3 |
| V203 (T35S-E41Q-Q84P-Q85K-S106T-Y109H-S185C) | ++++ | 14.0 ± 0.2 |
| V204 (T35S-Q40L-E41Q-Q84P-Q85K-S106T-Y109H-S185C) | ++++ | 14.1 ± 0.5 |
| V205 (E41Q-Q84P-Q85K-S106T-Y109H-S185C) | ++++ | 13.1 ± 1.0 |
| V206 (Q40L-E41Q-Q84P-Q85P-S106T-Y109H-S185C) | ++++ | 14.6 ± 0.4 |
References
- Cole, S.P.; Bhardwaj, G.; Gerlach, J.H.; Mackie, J.E.; Grant, C.E.; Almquist, K.C.; Stewart, A.J.; Kurz, E.U.; Duncan, A.M.V.; Deeley, R.G. Overexpression of a transporter gene in a multidrug-resistant human lung cancer cell line. Science 1992, 258, 1650–1654. [Google Scholar] [CrossRef]
- Bagshawe, K.D.; Springer, C.J.; Searle, F.; Antoniw, P.; Sharma, S.K.; Melton, R.G.; Sherwood, R.F. A cytotoxic agent can be generated selectively at cancer sites. Br. J. Cancer 1988, 58, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Josephy, P.D.; Mannervik, B. Molecular Toxicology, 2nd ed.; Oxford University Press: New York, NY, USA, 2006; pp. 333–364. [Google Scholar]
- Tew, K.D. Glutathione-associated enzymes in anticancer drug resistance. Cancer Res. 2016, 7, 7–9. [Google Scholar] [CrossRef]
- Townsend, D.M.; Tew, K.D. The role of glutathione-S-transferases in anti-cancer drug resistance. Oncogene 2003, 22, 7369–7375. [Google Scholar] [CrossRef] [PubMed]
- Findlay, V.J.; Townsend, D.M.; Saavedra, J.E.; Buzard, G.S.; Citro, M.L.; Keefer, L.K.; Ji, X.; Tew, K.D. Tumor cell responses to a novel glutathione-S-transferases-activated nitric oxide-releasing prodrug. Mol. Pharmacol. 2004, 65, 1070–1079. [Google Scholar] [CrossRef] [PubMed]
- Lyttle, M.H.; Satyam, A.; Hocker, M.D.; Bauer, K.E.; Caldwell, C.G.; Hui, H.C.; Morgan, A.S.; Mergia, A.; Kauvar, L.M. Glutathione-S-transferase activates novel alkylating agents. J. Med. Chem. 1994, 37, 1501–1507. [Google Scholar] [CrossRef] [PubMed]
- Oakley, A.J.; Lo Bello, M.; Battistoni, A.; Ricci, G.; Rossjohn, J.; Villar, H.O.; Parker, M.W. The structures of human glutathione transferase P1-1 in complex with glutathione and various inhibitors at high resolution. J. Mol. Biol. 1997, 274, 84–100. [Google Scholar] [CrossRef] [PubMed]
- Kavanagh, J.J.; Gershenson, D.M.; Choi, H.; Lewis, L.; Patel, K.; Brown, G.L.; Garcia, A.; Spriggs, D.R. Multi-institutional phase 2 study of TLK286 (TELCYTA, a glutathione s-transferase P1-1 activated glutathione analog prodrug) in patients with platinum and paclitaxel refractory or resistant ovarian cancer. Int. J. Gynecol. Cancer 2005, 15, 593–600. [Google Scholar] [CrossRef]
- Vergote, I.; Finkler, N.; del Campo, J.; Lohr, A.; Hunter, J.; Matei, D.; Kavanagh, J.; Vermorken, J.B.; Meng, L.; Jones, M.; et al. Phase 3 randomised study of canfosfamide (Telcyta®, TLK286) versus pegylated liposomal doxorubicin or topotecan as third-line therapy in patients with platinum-refractory or -resistant ovarian cancer. Eur. J. Cancer 2009, 45, 2324–2332. [Google Scholar] [CrossRef] [PubMed]
- Tew, K.D. TLK-286: A novel glutathione S-transferase-activated prodrug. Expert Opin. Investig. Drugs 2005, 14, 1047–1054. [Google Scholar] [CrossRef]
- Dachs, G.U.; Hunt, M.A.; Syddall, S.; Singleton, D.C.; Patterson, A.V. Bystander or no bystander for gene directed enzyme prodrug therapy. Molecules 2009, 14, 4517–4545. [Google Scholar] [CrossRef]
- Winter, G. Harnessing evolution to make medicines (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2019, 58, 14438–14445. [Google Scholar] [CrossRef]
- Carter, P.J.; Rajpal, A. Designing antibodies as therapeutics. Cell 2022, 185, 2789–2805. [Google Scholar] [CrossRef]
- Jin, S.; Sun, Y.; Liang, X.; Gu, X.; Ning, J.; Xu, Y.; Chen, S.; Pan, L. Emerging new therapeutic antibody derivatives for cancer treatment. Signal. Transduct. Target. Ther. 2022, 7, 39. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Modén, O.; Mannervik, B. Structure-based redesign of GST A2-2 for enhanced catalytic efficiency with azathioprine. Chem. Biol. 2012, 19, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Keefe, A.D.; Szostak, J.W. Functional proteins from a random-sequence library. Nature 2001, 410, 715–718. [Google Scholar] [CrossRef] [PubMed]
- Govindarajan, S.; Mannervik, B.; Silverman, A.J.; Wright, K.; Regitsky, D.; Hegazy, U.; Purcell, J.T.; Welch, M.; Minshull, J.; Gustafsson, C. Mapping of amino acid substitutions conferring herbicide resistance in wheat glutathione transferase. ACS Synth. Biol. 2015, 4, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Johansson, A.-S.; Stenberg, G.; Widersten, M.; Mannervik, B. Structure-activity relationship and thermal stability of human glutathione transferase P1-1 governed by the H-site residue 105. J. Mol. Biol. 1998, 278, 687–698. [Google Scholar] [CrossRef]
- Ismail, A.; Lewis, E.; Sjödin, B.; Mannervik, B. Characterization of dog glutathione transferase P1-1, an enzyme relevant to veterinary medicine. Int. J. Mol. Sci. 2021, 22, 4079. [Google Scholar] [CrossRef]
- Welch, M.; Govindarajan, S.; Ness, J.E.; Villalobos, A.; Gurney, A.; Minshull, J.; Gustafsson, C. Design parameters to control synthetic gene expression in Escherichia coli. PLoS ONE 2009, 4, e7002. [Google Scholar] [CrossRef]
- Kolm, R.H.; Stenberg, G.; Widersten, M.; Mannervik, B. High-level bacterial expression of human glutathione transferase P1-1 encoded by semisynthetic DNA. Protein Expr. Purif. 1995, 6, 265–271. [Google Scholar] [CrossRef]
- Habig, W.H.; Pabst, M.J.; Jakoby, W.B. Glutathione S-transferases. The first enzymatic step in mercapturic acid formation. J. Biol. Chem. 1974, 249, 7130–7139. [Google Scholar] [CrossRef] [PubMed]
- Gustafsson, C.; Govindarajan, S.; Minshull, J.S. Systems and Methods for Biopolymer Engineering. US Patent 8,005,620, 23 August 2011. [Google Scholar]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Systems and Methods for Biopolymer Engineering. US Patent 8,635,029, 21 January 2014. [Google Scholar]
- Liao, J.; Warmuth, M.K.; Govindarajan, S.; Ness, J.E.; Wang, R.P.; Gustafsson, C.; Minshull, J. Engineering proteinase K using machine learning and synthetic genes. BMC Biotechnol. 2007, 7, 16. [Google Scholar] [CrossRef]
- Dayhoff, M.O.; Schwartz, R.M.; Orcutt, B.C. A model of evolutionary change in proteins. In Atlas of Protein Sequence and Structure; Dayhoff, M.O., Ed.; Supplement 3; National Biomedical Research Foundation: Washington, DC, USA, 1978; Volume 5, pp. 345–352. [Google Scholar]
- Casari, G.; Sander, C.; Valencia, A. A method to predict functional residues in proteins. Nat. Struct. Mol. Biol. 1995, 2, 171–178. [Google Scholar] [CrossRef]
- Fukami-Kobayashi, K.; Schreiber, D.R.; Benner, S.A. Detecting compensatory covariation signals in protein evolution using reconstructed ancestral sequences. J. Mol. Biol. 2002, 319, 729–743. [Google Scholar] [CrossRef]
- Sali, A.; Blundell, T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993, 234, 779–815. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—A visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed]
- Dourado, F.A.R.D.; Fernandes, A.P.; Ramos, J.M.; Mannervik, B. Mechanism of glutathione transferase P1-1-catalyzed activation of the prodrug canfosfamide (TLK286, Telcyta). Biochemistry 2013, 52, 8069–8078. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Tokuriki, N.; Stricher, F.; Schymkowitz, J.; Serrano, L.; Tawfik, D.S. The stability effects of protein mutations appear to be universally distributed. J. Mol. Biol. 2007, 369, 1318–1332. [Google Scholar] [CrossRef]
- Kupreienko, O.; Pouliou, F.; Konstandinidis, K.; Axarli, I.; Douni, E.; Papageorgiou, A.C.; Labrou, N.E. Inhibition analysis and high-resolution crystal structure of Mus musculus glutathione transferase P1-1. Biomolecules 2023, 13, 613. [Google Scholar] [CrossRef]
- Reinemer, P.; Dirr, H.W.; Ladenstein, R.; Huber, R.; Lo Bello, M.; Federici, G.; Parker, M.W. Three-dimensional structure of class pi glutathione S-transferase from human placenta in complex with S-hexylglutathione at 2.8 A resolution. J. Mol. Biol. 1992, 227, 214–226. [Google Scholar] [CrossRef]
- Ji, X.; Tordova, M.; O’Donnell, R.; Parsons, J.F.; Hayden, J.B.; Gilliland, G.L.; Zimniak, P. Structure and function of the xenobiotic substrate-binding site and location of a potential non-substrate-binding site in a class π glutathione S-transferase. Biochemistry 1997, 36, 9690–9702. [Google Scholar] [CrossRef]
- Bammler, T.K.; Driessen, H.; Finnstrom, N.; Wolf, C.R. Amino acid differences at position 10, 11, and 104 explain the profound catalytic differences between two murine pi-class glutathione S-transferases. Biochemistry 1995, 34, 9000–9008. [Google Scholar] [CrossRef]
- Cai, D.; Chew, J.; Wray, J.; Kumar, V.; Danko, S.; Sambucetti, L.; Gomez, R.; Keck, J.G. GST P1-1 polymorphisms do not affect the rate of TELCYTA™ (TLK286) prodrug activation. Cancer Res. 2004, 64 (Suppl. S7), 883. [Google Scholar]
- Chen, K.; Arnold, F.H. Tuning the activity of an enzyme for unusual environments: Sequential random mutagenesis of subtilisin E for catalysis in dimethylformamide. Proc. Natl. Acad. Sci. USA 1993, 90, 5618–5622. [Google Scholar] [CrossRef] [PubMed]
- Mendonça, L.M.F.; Marana, S.R. Single mutations outside the active site affect the substrate specificity in a β-glycosidase. Biochim. Biophys. Acta 2011, 1814, 1616–1623. [Google Scholar] [CrossRef]
- Prade, L.; Huber, R.; Manoharan, T.H.; Fahl, W.E.; Reuter, W. Structures of class pi glutathione S-transferase from human placenta in complex with substrate, transition-state analogue and inhibitor. Structure 1997, 5, 1287–1295. [Google Scholar] [CrossRef] [PubMed]
- Risso, V.A.; Romero-Rivera, A.; Gutierrez-Rus, L.I.; Ortega-Muñoz, M.; Santoyo-Gonzalez, F.; Gavira, J.A.; Sanchez-Ruiz, J.M.; Kamerlin, S.C.L. Enhancing a de novo enzyme activity by computationally-focused ultra-low-throughput screening. Chem. Sci. 2020, 11, 6134–6148. [Google Scholar] [CrossRef] [PubMed]
- Bartlett, G.J.; Porter, C.T.; Borkakoti, N.; Thornton, J.M. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 2002, 324, 105–121. [Google Scholar] [CrossRef]
- Lo Bello, M.; Oakley, A.J.; Battistoni, A.; Mazzetti, A.P.; Nuccetelli, M.; Mazzarese, G.; Rossjohn, J.; Parker, M.W.; Ricci, G. Multifunctional role of Tyr 108 in the catalytic mechanism of human glutathione transferase P1-1. Crystallographic and kinetic studies on the Y108F mutant enzyme. Biochemistry 1997, 36, 6207–6217. [Google Scholar] [CrossRef] [PubMed]
- Nuccetelli, M.; Mazzetti, A.P.; Rossjohn, J.; Parker, M.W.; Board, P.; Caccuri, A.M.; Federici, G.; Ricci, G.; Lo Bello, M. Shifting substrate specificity of human glutathione transferase (from class Pi to class alpha) by a single point mutation. Biochem. Biophys. Res. Commun. 1998, 252, 184–189. [Google Scholar] [CrossRef]
- Shishido, Y.; Tomoike, F.; Kuwata, K.; Fujikawa, H.; Sekido, Y.; Murakami-Tonami, Y.; Kameda, T.; Abe, N.; Kimura, Y.; Shuta, S.; et al. A covalent inhibitor for glutathione S-transferase Pi (GSTP1-1) in human cells. ChemBioChem 2019, 20, 900–905. [Google Scholar] [CrossRef]
- Dourado, D.F.A.R.; Fernandes, P.A.; Mannervik, B.; Ramos, M.J. Glutathione transferase: New model for glutathione activation. Chem. Eur. J. 2008, 14, 9591–9598. [Google Scholar] [CrossRef]
- Kolm, R.H.; Sroga, G.E.; Mannervik, B. Participation of the phenolic hydroxyl group of Tyr-8 in the catalytic mechanism of human glutathione transferase P1-1. Biochem. J. 1992, 285 Pt 2, 537–540. [Google Scholar] [CrossRef]
- Hitchens, T.K.; Mannervik, B.; Rule, S.G. Disorder-to-order transition of the active site of human class pi glutathione transferase, GST P1-1. Biochemistry 2001, 40, 11660–11669. [Google Scholar] [CrossRef]
- Stella, L.; Caccuri, A.M.; Rosato, N.; Nicotra, M.; Lo Bello, M.; De Matteis, F.; Mazzetti, A.P.; Federici, G.; Ricci, G. Flexibility of helix 2 in the human glutathione transferase P1-1: Time-resolved fluorescence spectroscopy. J. Biol. Chem. 1998, 273, 23267–23273. [Google Scholar] [CrossRef][Green Version]
- Mishra, S.; Tiwari, A.K.M.; Singh, R.B.; Mahdi, A.A. A review on conventional and modern techniques of protein engineering and their applications. Am. J. Biochem. Mol. Biol. 2019, 9, 17–28. [Google Scholar] [CrossRef][Green Version]
- Musdal, Y.; Govindarajan, S.; Mannervik, B. Exploring sequence-function space of a poplar glutathione transferase using designed information-rich gene variants. Protein Eng. Des. Sel. 2017, 30, 543–549. [Google Scholar] [CrossRef]
- Eriksson, A.E.; Baase, W.A.; Zhang, X.J.; Heinz, D.W.; Blaber, M.; Baldwin, E.P.; Matthews, B.W. Response of a protein structure to cavity-creating mutations and its relation to the hydrophobic effect. Science 1992, 255, 178–183. [Google Scholar] [CrossRef]
- Aceto, A.; Caccuri, A.M.; Sacchetta, P.; Bucciarelli, T.; Dragani, B.; Rosato, N.; Federici, G.; Di Ilio, C. Dissociation and unfolding of Pi-class glutathione transferase. Evidence for a monomeric inactive intermediate. Biochem. J. 1992, 285 Pt 1, 241–245. [Google Scholar] [CrossRef]
- Rahman, R.N.Z.A.; Fujiwara, S.; Nakamura, H.; Takagi, M.; Imanaka, T. Ion Pairs Involved in maintaining a thermostable structure of glutamate dehydrogenase from a hyperthermophilic archaeon. Biochem. Biophys. Res. Commun. 1998, 248, 920–926. [Google Scholar] [CrossRef]
- Sharma, S.K.; Bagshawe, K.D. Antibody directed enzyme prodrug therapy (ADEPT): Trials and tribulations. Adv. Drug. Deliv. Rev. 2017, 118, 2–7. [Google Scholar] [CrossRef] [PubMed]
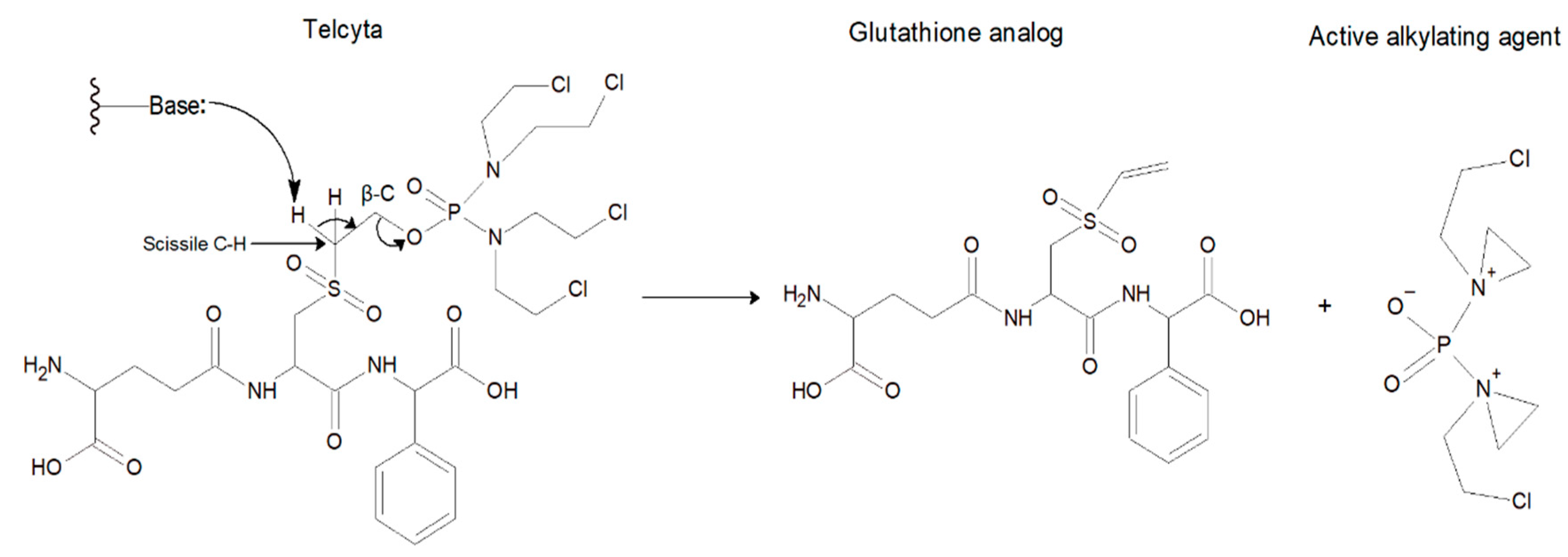

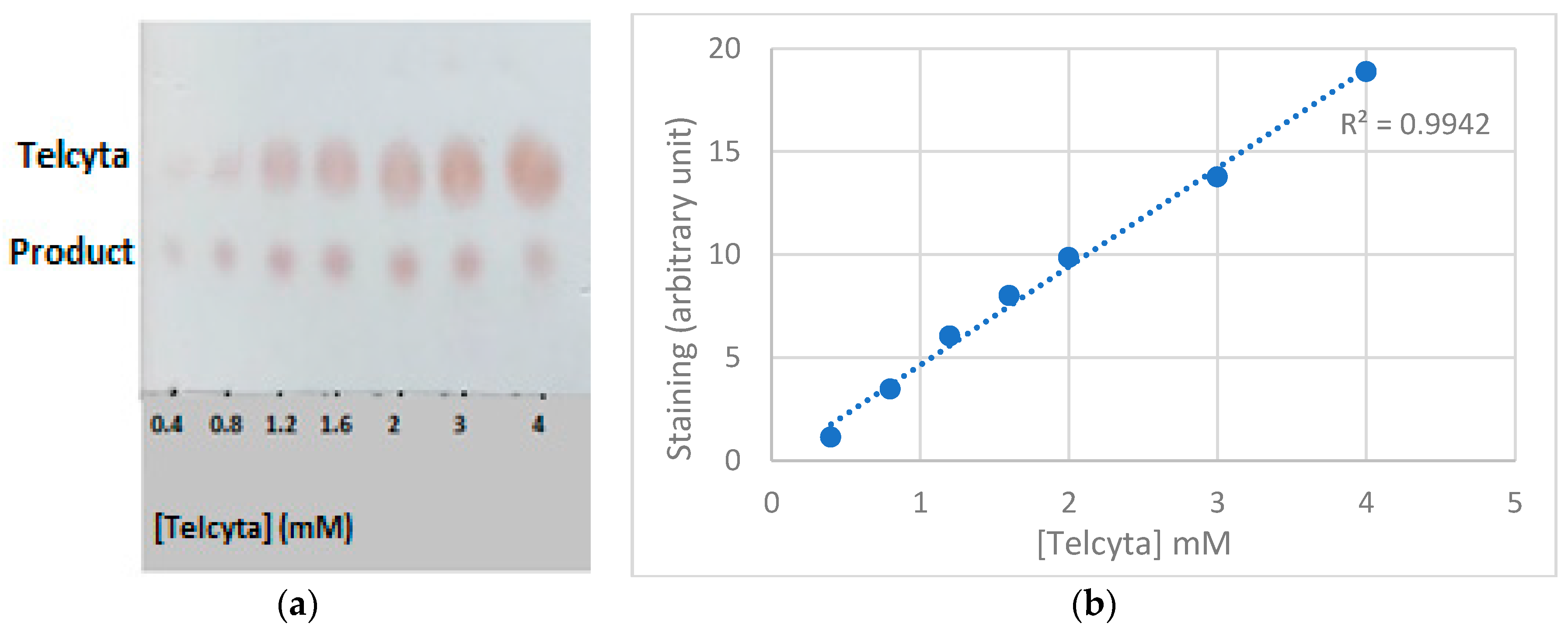
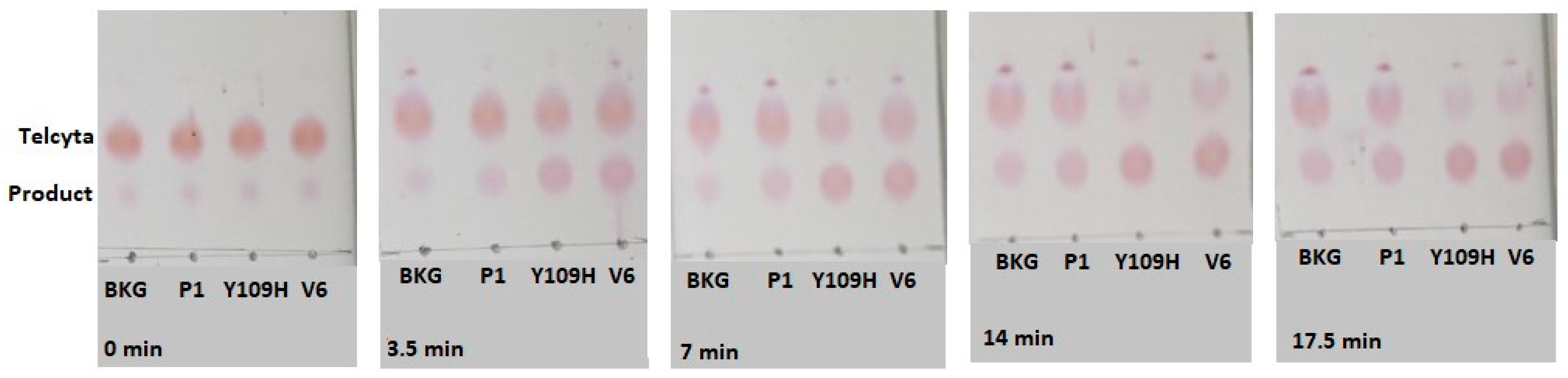
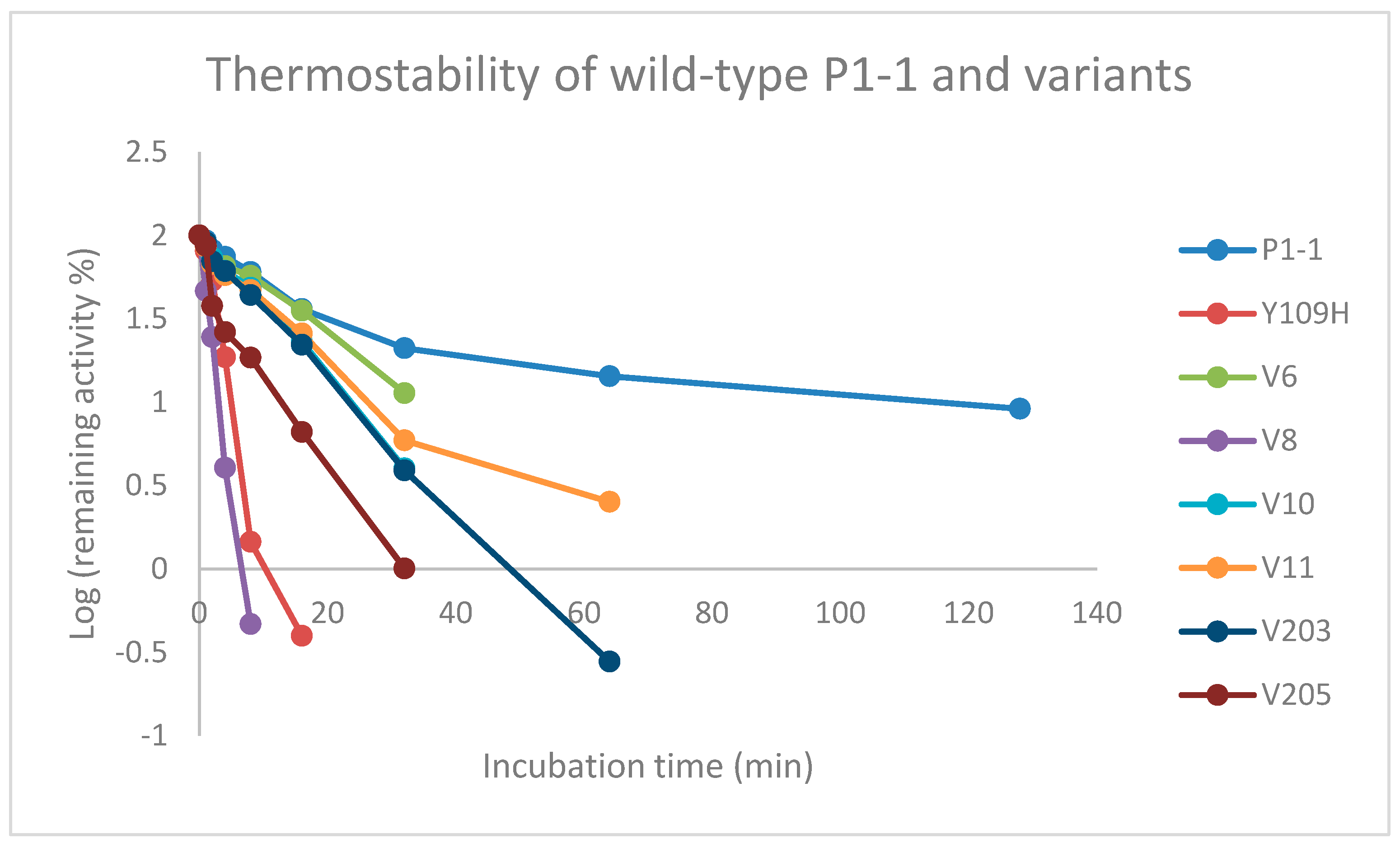



| Activity Relative to Wildtype GST P1-1 | ||
|---|---|---|
| Variants | Telcyta (Fold) | CDNB (µmol min−1 mg−1) |
| Human P1-1 | 1 | 106 ± 4 |
| Mouse P1-1 | 0.20 ± 0.06 | 76 ± 2.7 |
| Mouse P2-2 | ND | 0.10 ± 0.007 |
| Rat P1-1 | + | 17 ± 1.0 |
| Dog P1-1 1 | + | 23 ± 1.3 |
| Human P1-1 variant Y8H | ND | 0.08 ± 0.004 |
| Human P1-1 variant Y8E | ND | ND |
| Human P1-1 variant Y109H | 2.9 ± 0.6 | 20.9 ± 0.7 |
| Human P1-1 variant V6 (Q85R-C102S-S106T-Y109H-V200L) | 3.1 ± 0.9 | 28.2 ± 0.3 |
| Estimated Activity Relative to Wildtype GST P1-1 | ||
|---|---|---|
| Variants | Telcyta (Score) | CDNB (µmol min−1 mg−1) |
| Human P1-1 | +++ | 106 ± 4 |
| Y109H | ++++ | 20.9 ± 0.7 |
| Y8H | ND | 0.08 ± 0.004 |
| Y8E | ND | N/D |
| F9H-Y109H | ND | 0.71 ± 0.01 |
| V11H-Y109H | ND | 0.01 ± 0.001 |
| V11A-Y109H | + | 17.5 ± 0.4 |
| V11S-Y109H | ND | 1.82 ± 0.02 |
| V11T-Y109H | + | 5.2 ± 0.2 |
| V11E-Y109H | ND | 0.027 ± 0.002 |
| V36R-Y109H | +++ | 20.6 ± 1.2 |
| V36M-Y109H | ++++ | 24.4 ± 1.1 |
| V36G-Y109H | +++ | 12.4 ± 0.3 |
| V36L-Y109H | ++++ | 22.0 ± 0.3 |
| V36K-Y109H | ++++ | 27.0 ± 2.5 |
| V36I-Y109H | ++++ | 18.5 ± 0.2 |
| V36T-Y109H | ++++ | 15.6 ± 0.2 |
| Substrate: CDNB | |
|---|---|
| P1-1 Variants | t½ (min) |
| Human P1-1 | 9.1 |
| Y109H | 2.4 |
| V36T-Y109H | 2.8 |
| V36L-Y109H | 2.3 |
| V1 (T35S-Q40L-A46S-Q85R-Y109H) | 2.9 |
| V2 (Q40M-E41Q-A46S-Y109H-V200L) | 1.1 |
| V3 (Q40L-S43P-Q85K-Y109H-V200L) | 1.7 |
| V4 (T35S-E41Q-Q85K-S106T-Y109H) | 2.9 |
| V5 (Q40M-S43P-Q85R-Y109H-S185C) | 5.9 |
| V6 (Q85R-C102S-S106T-Y109H-V200L) | 10.9 |
| V7 (A46S-S106T-Y109H-S185C-V200A) | 4.1 |
| V8 (Q40L-E41Q-Q84P-Y109H-V200A) | 0.94 |
| V9 (T35S-S43P-C102S-Y109H-V200A) | 1.3 |
| V10 (Q40M-Q84P-Q85K-C102S-Y109H) | 7.3 |
| V11 (T35S-Q84P-Y109H-S185C-V200L) | 6.7 |
| V201 (T35S-Q40L-E41Q-Q84P-Q85K-S106T-Y109H) | 1.4 |
| V202 (T35S-Q40L-E41Q-Q85K-S106T-Y109H-S185C) | 2.1 |
| V203 (T35S-E41Q-Q84P-Q85K-S106T-Y109H-S185C) | 6.5 |
| V204 (T35S-Q40L-E41Q-Q84P-Q85K-S106T-Y109H-S185C) | 1.8 |
| V205 (E41Q-Q84P-Q85K-S106T-Y109H-S185C) | 6.8 |
| V206 (Q40L-E41Q-Q84P-Q85P-S106T-Y109H-S185C) | 2.3 |
| V401 (Q85R-Y109H) | 5.6 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ismail, A.; Govindarajan, S.; Mannervik, B. Human GST P1-1 Redesigned for Enhanced Catalytic Activity with the Anticancer Prodrug Telcyta and Improved Thermostability. Cancers 2024, 16, 762. https://doi.org/10.3390/cancers16040762
Ismail A, Govindarajan S, Mannervik B. Human GST P1-1 Redesigned for Enhanced Catalytic Activity with the Anticancer Prodrug Telcyta and Improved Thermostability. Cancers. 2024; 16(4):762. https://doi.org/10.3390/cancers16040762
Chicago/Turabian StyleIsmail, Aram, Sridhar Govindarajan, and Bengt Mannervik. 2024. "Human GST P1-1 Redesigned for Enhanced Catalytic Activity with the Anticancer Prodrug Telcyta and Improved Thermostability" Cancers 16, no. 4: 762. https://doi.org/10.3390/cancers16040762
APA StyleIsmail, A., Govindarajan, S., & Mannervik, B. (2024). Human GST P1-1 Redesigned for Enhanced Catalytic Activity with the Anticancer Prodrug Telcyta and Improved Thermostability. Cancers, 16(4), 762. https://doi.org/10.3390/cancers16040762







