Hypoxia-Derived Exosomes Promote Lung Adenocarcinoma by Regulating HS3ST1-GPC4-Mediated Glycolysis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Gene Expression Profiling Interactive Analysis (GEPIA)
2.2. Cell Culture
2.3. Co-Immunoprecipitation (Co-IP)
2.4. Immunofluorescence Staining
2.5. Mitochondrial Stress Test
2.6. Glycolysis Stress Test
2.7. Glucose Uptake and Lactate Product Assay
2.8. Western Blotting Assay
2.9. Exosome Isolation and Detection
2.10. Cell Transfection
2.11. Reverse Transcription-Quantitative PCR (RT-qPCR)
2.12. Cell Viability Analysis
2.13. Colony Formation Assays
2.14. Cell Cycle and Apoptosis Analyses
2.15. Dual-Luciferase Reporter Assay
2.16. Xenograft Tumor Experiments
2.17. Samples Collection
3. Statistical Analysis
4. Results
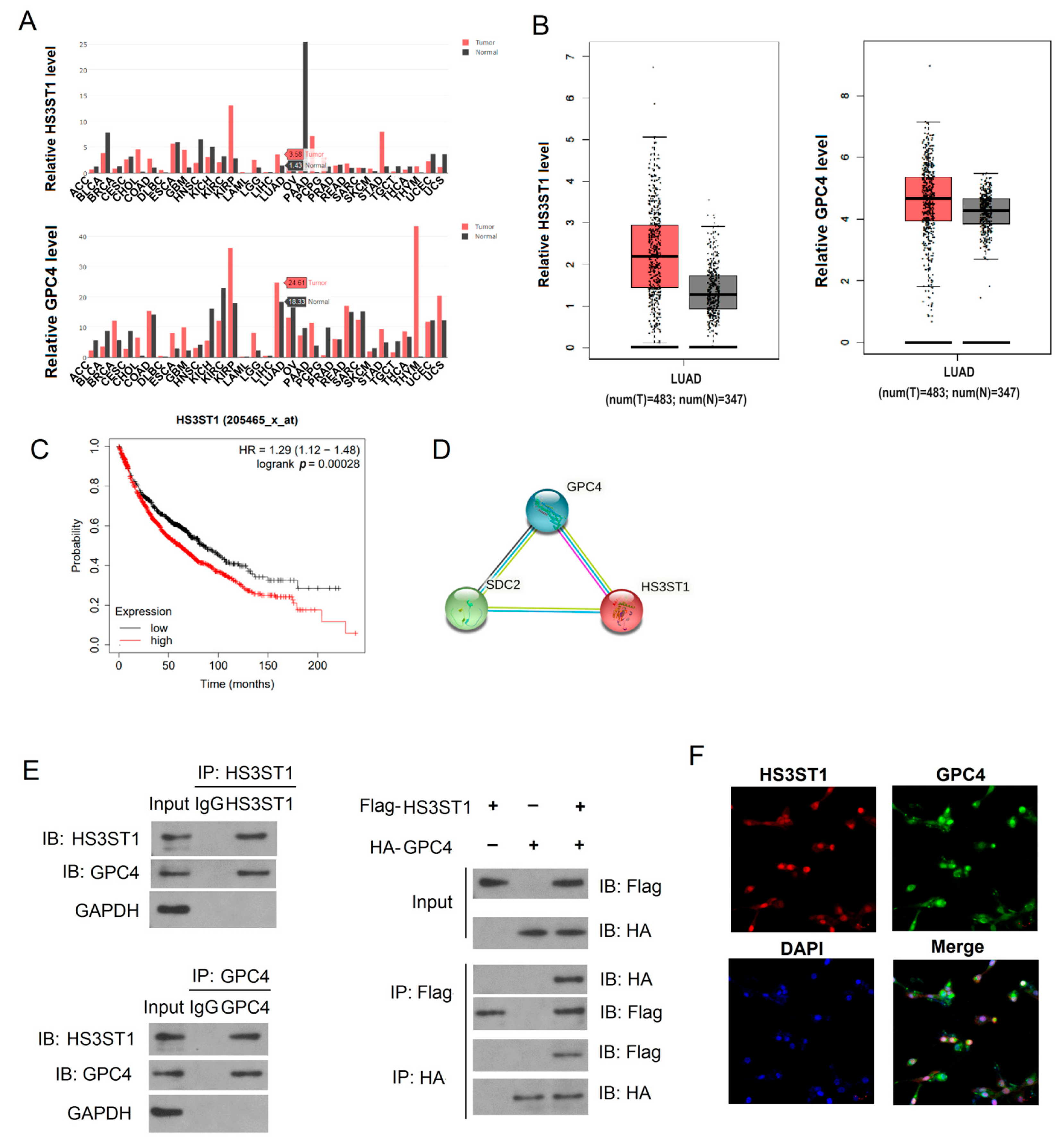
4.1. HS3ST1 Promotes Glycolysis in LUAD Cells
4.2. GPC4 Promotes HS3ST1-Mediated Glycolysis in LUAD Cells
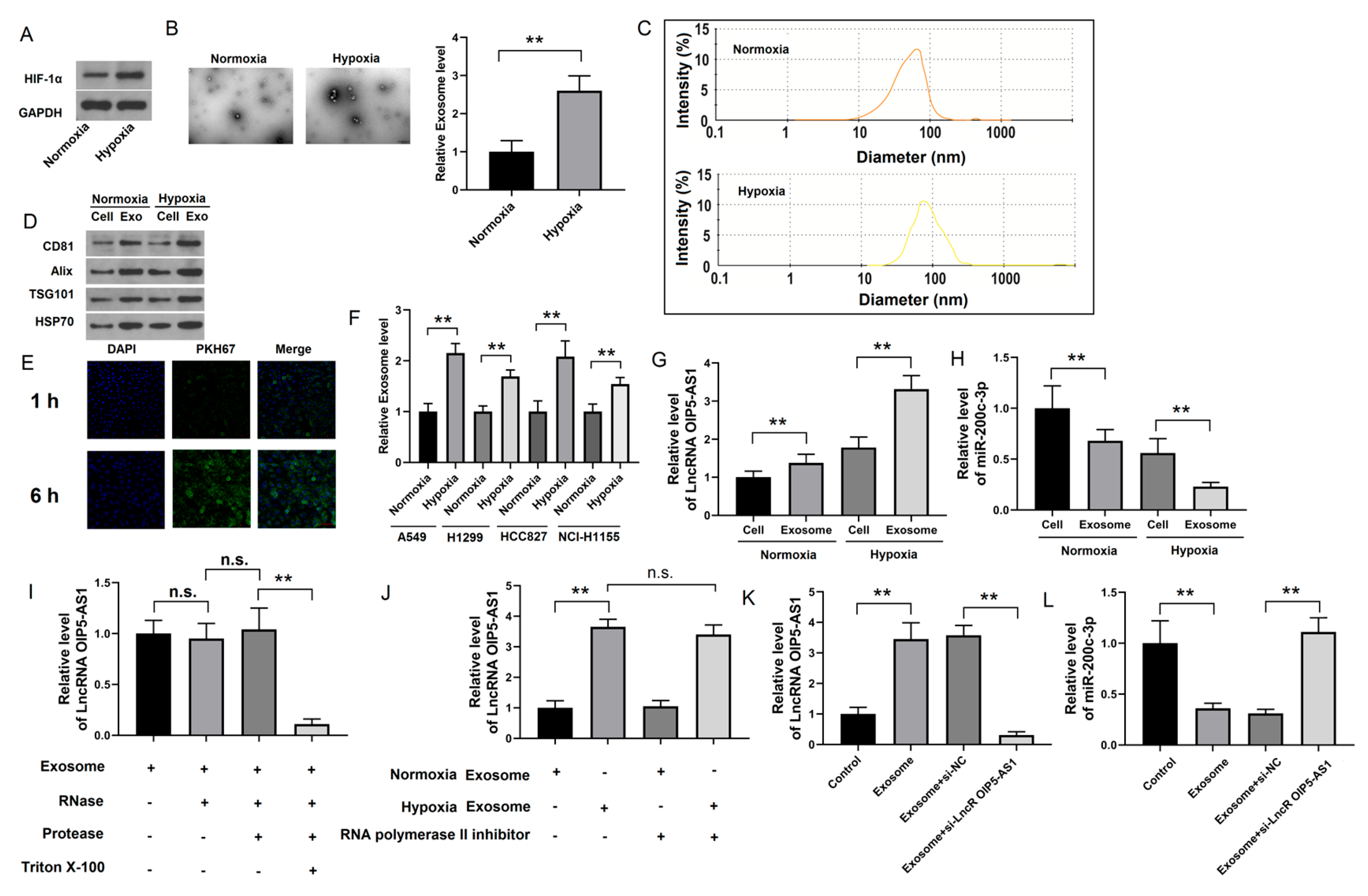
4.3. Identification of lncRNA OIP5-AS1 in Hypoxic LUAD Cells
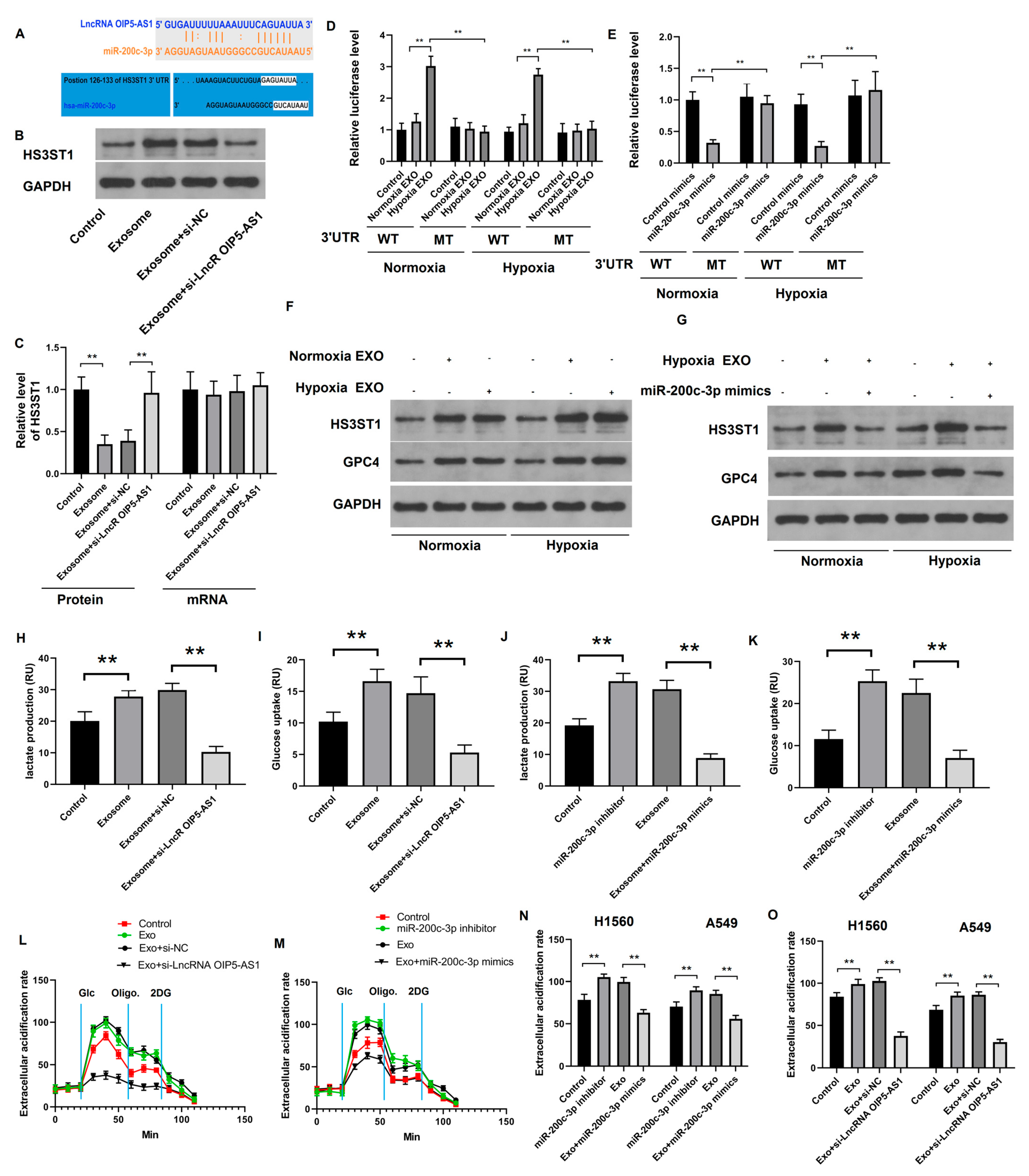
4.4. Hypoxia-Derived Exosomal lncRNA OIP5-AS1 Targets HS3ST1-GPC4 and miR-200c-3p in LUAD Cells
4.5. Hypoxia-Derived Exosomal lncRNA OIP5-AS1 Promotes Glycolysis via miR-200c-3p Axis in LUAD Cells
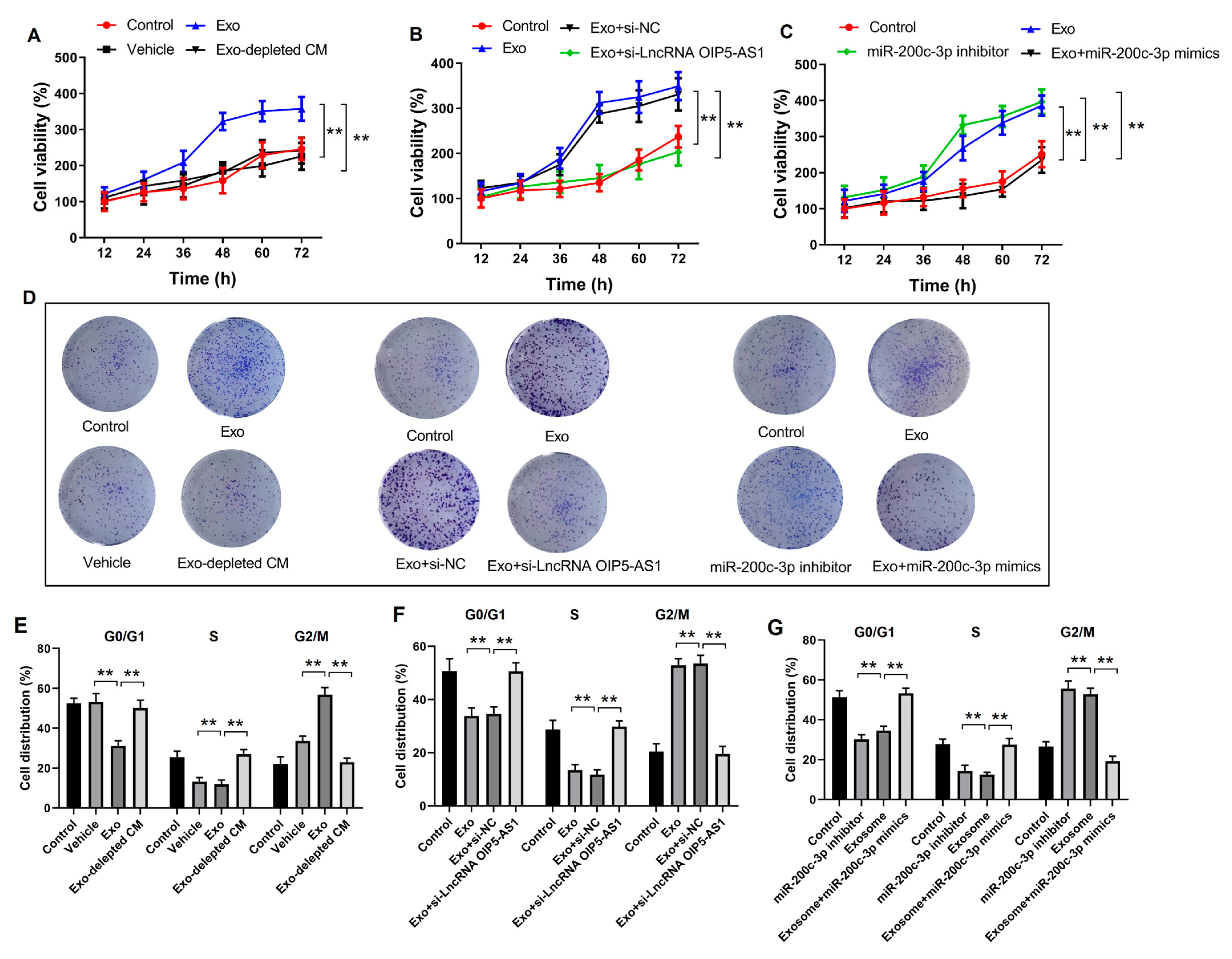
4.6. Hypoxia-Derived Exosomal lncRNA OIP5-AS1 Promotes LUAD Proliferation and Metastasis via miR-200c-3p In Vitro
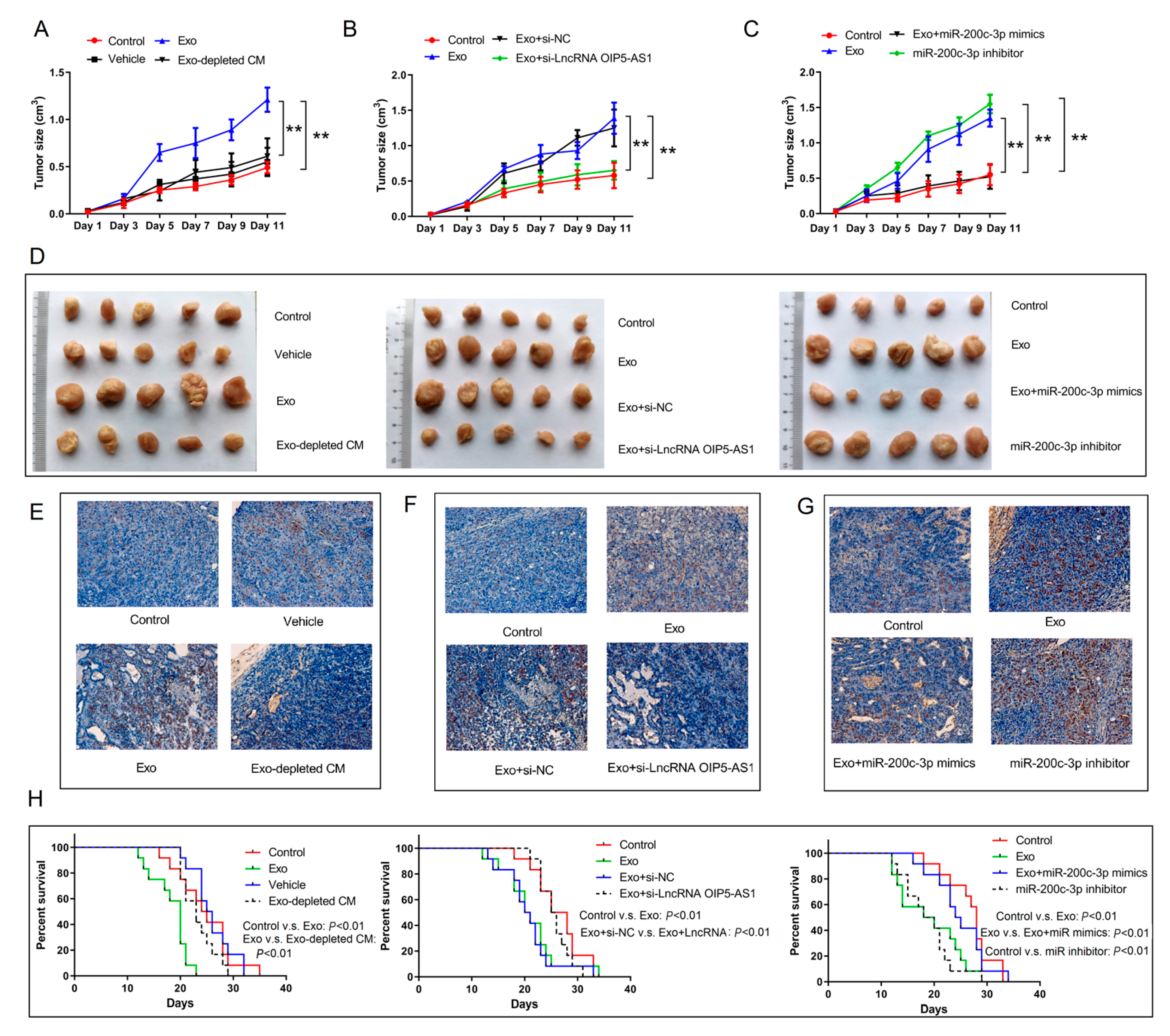
4.7. Hypoxia-Derived Exosomal lncRNA OIP5-AS1 Promotes LUAD Tumor Growth and Metastasis via miR-200c-3p Axis In Vivo
4.8. Long Non-Coding RNA OIP5-AS1 Is Upregulated in LUAD Tissues, Serum, and Serum Exosomes
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Miller, K.D.; Nogueira, L.; Devasia, T.; Mariotto, A.B.; Yabroff, K.R.; Jemal, A.; Kramer, J.; Siegel, R.L. Cancer treatment and survivorship statistics. CA Cancer J. Clin. 2022, 72, 409–436. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer statistics. CA Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef]
- Hu, J.; Sun, T.; Wang, H.; Chen, Z.; Wang, S.; Yuan, L.; Liu, T.; Li, H.R.; Wang, P.; Feng, Y.; et al. MiR-215 is induced post-transcriptionally via HIF-Drosha complex and mediates glioma-initiating cell adaptation to hypoxia by targeting KDM1B. Cancer Cell 2016, 29, 49–60. [Google Scholar] [CrossRef]
- Wang, T.; Gilkes, D.M.; Takano, N.; Xiang, L.; Luo, W.; Bishop, C.J.; Chaturvedi, P.; Green, J.J.; Semenza, G.L. Hypoxia-inducible factors and RAB22A mediate formation of microvesicles that stimulate breast cancer invasion and metastasis. Proc. Natl. Acad. Sci. USA 2014, 111, E3234–E3242. [Google Scholar] [CrossRef]
- Pientka, F.K.; Hu, J.; Schindler, S.G.; Brix, B.; Thiel, A.; Jöhren, O.; Fandrey, J.; Berchner-Pfannschmidt, U.; Depping, R. Oxygen sensing by the prolyl-4-hydroxylase PHD2 within the nuclear compartment and the influence of compartmentalisation on HIF-1 signalling. J. Cell Sci. 2012, 125, 5168–5176. [Google Scholar]
- Wang, Q.; Hu, D.F.; Rui, Y.; Jiang, A.B.; Liu, Z.L.; Huang, L.N. Prognosis value of HIF-1α expression in patients with non-small cell lung cancer. Gene 2014, 541, 69–74. [Google Scholar] [CrossRef]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef]
- Zhou, R.; Chen, K.K.; Zhang, J.; Xiao, B.; Huang, Z.; Ju, C.; Sun, J.; Zhang, F.; Lv, X.B.; Huang, G. The decade of exosomal long RNA species: An emerging cancer antagonist. Mol. Cancer 2018, 17, 75. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringnér, M.; Mörgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Luo, G.; Zhang, K.; Cao, J.; Huang, C.; Jiang, T.; Liu, B.; Su, L.; Qiu, Z. Hypoxic tumor-derived exosomal miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to promote pancreatic cancer metastasis. Cancer Res. 2018, 78, 4586–4598. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Liao, T.; Liu, H.; Yuan, H.; Ouyang, T.; Wang, J.; Chai, S.; Li, J.; Chen, J.; Li, X.; et al. Hypoxic glioma stem cell-derived exosomes containing Linc01060 promote progression of glioma by regulating the MZF1/c-Myc/HIF1α axis. Cancer Res. 2021, 81, 114–128. [Google Scholar] [CrossRef] [PubMed]
- Bhan, A.; Soleimani, M.; Mandal, S.S. Long noncoding RNA and cancer: A new paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef]
- Wang, Z.; Yang, B.; Zhang, M.; Guo, W.; Wu, Z.; Wang, Y.; Jia, L.; Li, S.; Cancer Genome Atlas Research Network; Xie, W.; et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell 2018, 33, 706–720. [Google Scholar] [CrossRef]
- Hu, W.L.; Jin, L.; Xu, A.; Wang, Y.F.; Thorne, R.F.; Zhang, X.D.; Wu, M. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat. Cell Biol. 2018, 20, 492–502. [Google Scholar] [CrossRef]
- Hosono, Y.; Niknafs, Y.S.; Prensner, J.R.; Iyer, M.K.; Dhanasekaran, S.M.; Mehra, R.; Pitchiaya, S.; Tien, J.; Escara-Wilke, J.; Poliakov, A.; et al. Oncogenic role of THOR, a conserved cancer/testis long non-coding RNA. Cell 2023, 186, 4254–4255. [Google Scholar] [CrossRef]
- Schmitt, A.M.; Chang, H.Y. Long noncoding RNAs in cancer pathways. Cancer Cell 2016, 29, 452–463. [Google Scholar] [CrossRef]
- Liu, B.; Sun, L.; Liu, Q.; Gong, C.; Yao, Y.; Lv, X.; Lin, L.; Yao, H.; Su, F.; Li, D.; et al. A cytoplasmic NF-κB interacting long noncoding RNA blocks IκB phosphorylation and suppresses breast cancer metastasis. Cancer Cell 2015, 27, 370–381. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Tan, D.S.W.; Chong, F.T.; Leong, H.S.; Toh, S.Y.; Lau, D.P.; Kwang, X.L.; Zhang, X.; Sundaram, G.M.; Tan, G.S.; Chang, M.M.; et al. Long noncoding RNA EGFR-AS1 mediates epidermal growth factor receptor addiction and modulates treatment response in squamous cell carcinoma. Nat. Med. 2017, 23, 1167–1175. [Google Scholar] [CrossRef]
- Wang, Y.; Zhou, J.; Xu, Y.J.; Hu, H.B. Long non-coding RNA LINC00968 acts as oncogene in NSCLC by activating the Wnt signaling pathway. J. Cell. Physiol. 2018, 233, 3397–3406. [Google Scholar] [CrossRef] [PubMed]
- Lu, Q.C.; Rui, Z.H.; Guo, Z.L.; Xie, W.; Shan, S.; Ren, T. LncRNA-DANCR contributes to lung adenocarcinoma progression by sponging miR-496 to modulate mTOR expression. J. Cell. Mol. Med. 2018, 22, 1527–1537. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.L.; Lin, S.; Cao, C.; Gimbrone, N.T.; Yang, R.; Fu, D.A.; Carper, M.B.; Haura, E.B.; Schabath, M.B.; et al. cAMP/CREB-regulated LINC00473 marks LKB1-inactivated lung cancer and mediates tumor growth. J. Clin. Investig. 2016, 126, 2267–2279. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Li, C.; Kang, B.; Gao, G.; Li, C.; Zhang, Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017, 45, W98–W102. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.; Huang, C.; Sun, J.; Bollt, O.; Wang, X.; Martine, E.; Kang, J.; Taylor, M.D.; Fang, B.; Singh, P.K.; et al. The mitochondrial deoxyguanosine kinase is required for cancer cell stemness in lung adenocarcinoma. EMBO Mol. Med. 2019, 11, e10849. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zheng, Q.; Bao, C.; Li, S.; Guo, W.; Zhao, J.; Chen, D.; Gu, J.; He, X.; Huang, S. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. 2015, 25, 981–984. [Google Scholar] [CrossRef] [PubMed]
- Nasim, F.; Sabath, B.F.; Eapen, G.A. Lung Cancer. Med. Clin. N. Am. 2019, 103, 463–473. [Google Scholar] [CrossRef]
- Jain, D.; Roy-Chowdhuri, S. Advances in cytology of lung cancer. Semin. Diagn. Pathol. 2021, 38, 109–115. [Google Scholar] [CrossRef]
- Denisenko, T.V.; Budkevich, I.N.; Zhivotovsky, B. Cell death-based treatment of lung adenocarcinoma. Cell Death Dis. 2018, 9, 117. [Google Scholar] [CrossRef] [PubMed]
- Jones, G.S.; Baldwin, D.R. Recent advances in the management of lung cancer. Clin. Med. 2018, 18 (Suppl. 2), s41–s46. [Google Scholar] [CrossRef] [PubMed]
- The Lancet. Lung cancer: Some progress, but still a lot more to do. Lancet 2019, 394, 1880. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.B.; Xu, G.L.; Jia, W.D.; Li, J.S.; Ma, J.L.; Chen, K.; Wang, Z.H.; Ge, Y.S.; Ren, W.H.; Yu, J.H.; et al. Prognostic significance and mechanisms of patterned matrix vasculogenic mimicry in hepatocellular carcinoma. Med. Oncol. 2011, 28 (Suppl. 1), S228–S238. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Shen, Z.Y.; Zhang, H.; Fan, Y.Z.; Zhang, W.Z.; Zhang, J.T.; Lu, X.S.; Ye, C. Overexpression of HIF-1α in primary gallbladder carcinoma and its relation to vasculogenic mimicry and unfavourable prognosis. Oncol. Rep. 2012, 27, 1990–2002. [Google Scholar] [PubMed]
- Kierans, S.J.; Taylor, C.T. Regulation of glycolysis by the hypoxia-inducible factor (HIF): Implications for cellular physiology. J. Physiol. 2021, 599, 23–37. [Google Scholar] [CrossRef]
- Scharping, N.E.; Rivadeneira, D.B.; Menk, A.V.; Vignali, P.D.A.; Ford, B.R.; Rittenhouse, N.L.; Peralta, R.; Wang, Y.; Wang, Y.; DePeaux, K.; et al. Mitochondrial stress induced by continuous stimulation under hypoxia rapidly drives T cell exhaustion. Nat. Immunol. 2021, 22, 205–215. [Google Scholar] [CrossRef]
- Zhang, Y.; Kurupati, R.; Liu, L.; Zhou, X.Y.; Zhang, G.; Hudaihed, A.; Filisio, F.; Giles-Davis, W.; Xu, X.; Karakousis, G.C.; et al. Enhancing CD8+ T cell fatty acid catabolism within a metabolically challenging tumor microenvironment increases the efficacy of melanoma immunotherapy. Cancer Cell 2017, 32, 377–391. [Google Scholar] [CrossRef]
- Peng, H.; Weng, L.; Lei, S.; Hou, S.; Yang, S.; Li, M.; Zhao, D. Hypoxia-hindered methylation of PTGIS in endometrial stromal cells accelerates endometriosis progression by inducing CD16- NK-cell differentiation. Exp. Mol. Med. 2022, 54, 890–905. [Google Scholar] [CrossRef]
- Lin, J.; Wang, X.; Zhai, S.; Shi, M.; Peng, C.; Deng, X.; Fu, D.; Wang, J.; Shen, B. Hypoxia-induced exosomal circPDK1 promotes pancreatic cancer glycolysis via c-myc activation by modulating miR-628-3p/BPTF axis and degrading BIN1. J. Hematol. Oncol. 2022, 15, 128. [Google Scholar] [CrossRef]
- Lin, J.; Wang, X.; Zhai, S.; Shi, M.; Peng, C.; Deng, X.; Fu, D.; Wang, J.; Shen, B. Hypoxia-responsive circRNAs: A novel but important participant in non-coding RNAs ushered toward tumor hypoxia. Cell Death Dis. 2022, 13, 666. [Google Scholar]
- Ward, P.S.; Thompson, C.B. Metabolic reprogramming: A cancer hallmark even warburg did not anticipate. Cancer Cell 2012, 21, 297–308. [Google Scholar] [CrossRef]
- Birkeland, E.S.; Koch, L.M.; Dechant, R. Another consequence of the Warburg effect? Metabolic regulation of Na+/H+ exchangers may link aerobic glycolysis to cell growth. Front. Oncol. 2020, 10, 1561. [Google Scholar] [CrossRef]
- Reuss, A.M.; Groos, D.; Buchfelder, M.; Savaskan, N. The acidic brain-glycolytic switch in the microenvironment of malignant glioma. Int. J. Mol. Sci. 2021, 22, 5518. [Google Scholar] [CrossRef]
- Mertens, R.T.; Parkin, S.; Awuah, S.G. Cancer cell-selective modulation of mitochondrial respiration and metabolism by potent organogold(iii) dithiocarbamates. Chem. Sci. 2020, 11, 10465–10482. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Chen, C.; Ding, Q.; Zhao, Y.; Wang, Z.; Chen, J.; Jiang, Z.; Zhang, Y.; Xu, G.; Zhang, J.; et al. METTL3-mediated m(6)A modification of HDGF mRNA promotes gastric cancer progression and has prognostic significance. Gut 2020, 69, 1193–1205. [Google Scholar] [CrossRef]
- Jia, R.; Chai, P.; Wang, S.; Sun, B.; Xu, Y.; Yang, Y.; Ge, S.; Jia, R.; Yang, Y.G.; Fan, X. M(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol. Cancer 2019, 18, 161. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Xuan, B.; Yan, T.; Ma, Y.; Xu, P.; Tian, X.; Zhang, X.; Cao, Y.; Ma, D.; Zhu, X.; et al. M(6)A-dependent glycolysis enhances colorectal cancer progression. Mol. Cancer 2020, 19, 72. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor micro-environment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Pefanis, E.; Wang, J.; Rothschild, G.; Lim, J.; Kazadi, D.; Sun, J.; Federation, A.; Chao, J.; Elliott, O.; Liu, Z.P.; et al. RNA exosome-regulated long non-coding RNA transcription controls super-enhancer activity. Cell 2015, 161, 774–789. [Google Scholar] [CrossRef]
- Hosseini-Beheshti, E.; Choi, W.; Weiswald, L.B.; Kharmate, G.; Ghaffari, M.; Roshan-Moniri, M.; Hassona, M.D.; Chan, L.; Chin, M.Y.; Tai, I.T.; et al. Exosomes confer pro-survival signals to alter the phenotype of prostate cells in their surrounding environment. Oncotarget 2016, 7, 14639–14658. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Li, C.; Wang, S.; Wang, Z.; Jiang, J.; Wang, W.; Li, X.; Chen, J.; Liu, K.; Li, C.; et al. Exosomes derived from hypoxic oral squamous cell carcinoma cells deliver miR-21 to normoxic cells to elicit a prometastatic phenotype. Cancer Res. 2016, 76, 1770–1780. [Google Scholar] [CrossRef]
- Sánchez, C.A.; Andahur, E.I.; Valenzuela, R.; Castellón, E.A.; Fullá, J.A.; Ramos, C.G.; Triviño, J.C. Exosomes from bulk and stem cells from human prostate cancer have a differential microRNA content that contributes cooperatively over local and pre-metastatic niche. Oncotarget 2016, 7, 3993–4008. [Google Scholar] [CrossRef] [PubMed]
- Anderson, J.D.; Johansson, H.J.; Graham, C.S.; Vesterlund, M.; Pham, M.T.; Bramlett, C.S.; Montgomery, E.N.; Mellema, M.S.; Bardini, R.L.; Contreras, Z.; et al. Comprehensive proteomic analysis of mesenchymal stem cell exosomes reveals modulation of angiogenesis via nuclear factor-kappaB signaling. Stem Cells 2016, 34, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Das, P.K.; Siddika, M.A.; Asha, S.Y.; Aktar, S.; Rakib, M.A.; Khanam, J.A.; Pillai, S.; Islam, F. MicroRNAs, a promising target for breast cancer stem cells. Mol. Diagn. Ther. 2020, 24, 69–83. [Google Scholar] [CrossRef]
- Das, P.K.; Asha, S.Y.; Abe, I.; Islam, F.; Lam, A.K. Roles of non-coding RNAs on anaplastic thyroid carcinomas. Cancers 2020, 12, 3159. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J.T. Functional classification and experimental dissection of long noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef]
- Islam, F.; Zhou, Y.; Lam, A.K. Long non-coding RNAs profiling using microarray in papillary thyroid carcinoma. In Papillary Thyroid Carcinoma: Methods and Protocols; Methods in Molecular Biology; Humana: New York, NY, USA, 2022; Volume 2534, pp. 135–148. [Google Scholar]
- Trimarchi, T.; Bilal, E.; Ntziachristos, P.; Fabbri, G.; Dalla-Favera, R.; Tsirigos, A.; Aifantis, I. Genome-wide mapping and characterization of notch-regulated long noncoding RNAs in acute leukemia. Cell 2014, 158, 593–606. [Google Scholar] [CrossRef]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Prensner, J.R.; Chinnaiyan, A.M. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011, 1, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Suhovskih, A.V.; Tsidulko, A.Y.; Kutsenko, O.S.; Kovner, A.V.; Aidagulova, S.V.; Ernberg, I.; Grigorieva, E.V. Transcriptional activity of heparan sulfate biosynthetic machinery is specifically impaired in benign prostate hyperplasia and prostate cancer. Front. Oncol. 2014, 4, 79. [Google Scholar] [CrossRef] [PubMed]
- Knelson, E.H.; Nee, J.C.; Blobe, G.C. Heparan sulfate signaling in cancer. Trends Biochem. Sci. 2014, 39, 277–288. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, H.; Zhu, K.; Yang, Y.; Li, J.; Jiang, H.; Liu, Z. Investigation of candidate molecular biomarkers for expression profile analysis of the Gene expression omnibus (GEO) in acute lymphocytic leukemia (ALL). Biomed. Pharmacother. 2019, 120, 109530. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Qu, Y.; Xia, P.; Chen, Y.; Zhu, X.; Zhang, J.; Wang, G.; Tian, Y.; Ying, J.; Fan, Z. Transdifferentiation of tumor infiltrating innate lymphoid cells during progression of colorectal cancer. Cell Res. 2020, 30, 610–622, Correction in Cell Res. 2020, 30, 630. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ji, X.; Zhu, R.; Gao, C.; Xie, H.; Gong, X.; Luo, J. Hypoxia-Derived Exosomes Promote Lung Adenocarcinoma by Regulating HS3ST1-GPC4-Mediated Glycolysis. Cancers 2024, 16, 695. https://doi.org/10.3390/cancers16040695
Ji X, Zhu R, Gao C, Xie H, Gong X, Luo J. Hypoxia-Derived Exosomes Promote Lung Adenocarcinoma by Regulating HS3ST1-GPC4-Mediated Glycolysis. Cancers. 2024; 16(4):695. https://doi.org/10.3390/cancers16040695
Chicago/Turabian StyleJi, Xianxiu, Ren Zhu, Caixia Gao, Huikang Xie, Xiaomei Gong, and Jie Luo. 2024. "Hypoxia-Derived Exosomes Promote Lung Adenocarcinoma by Regulating HS3ST1-GPC4-Mediated Glycolysis" Cancers 16, no. 4: 695. https://doi.org/10.3390/cancers16040695
APA StyleJi, X., Zhu, R., Gao, C., Xie, H., Gong, X., & Luo, J. (2024). Hypoxia-Derived Exosomes Promote Lung Adenocarcinoma by Regulating HS3ST1-GPC4-Mediated Glycolysis. Cancers, 16(4), 695. https://doi.org/10.3390/cancers16040695



