Epithelial and Mesenchymal-like Pancreatic Cancer Cells Exhibit Different Stem Cell Phenotypes Associated with Different Metastatic Propensities
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines and Cell Culture
2.2. Single-Cell Cloning and Clone Expansion
2.3. Colony-Formation Assay (CFA)
2.4. RNA Isolation and RT-qPCR
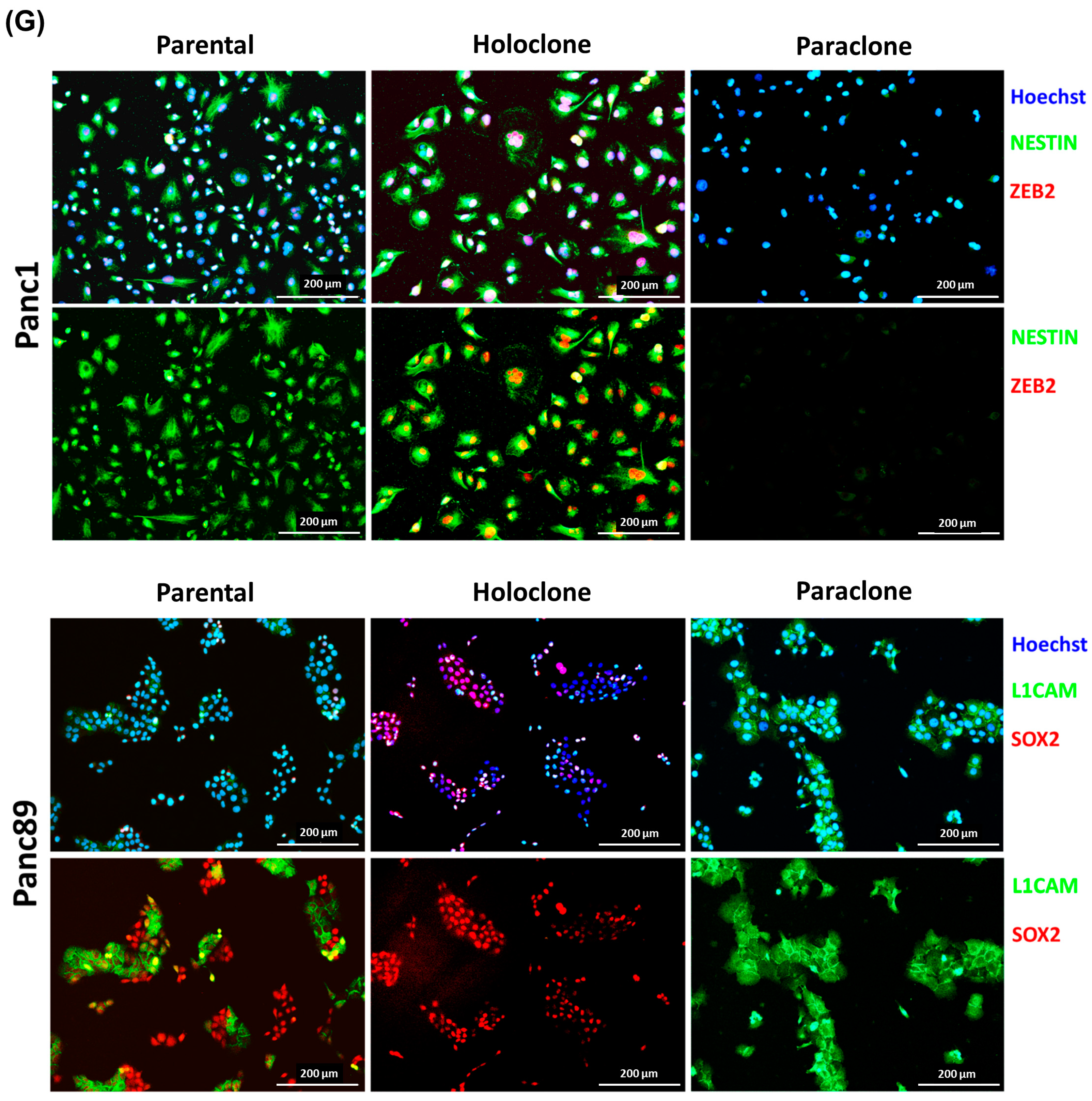
2.5. Analysis of CSC and EMT Markers via Immunofluorescence Staining
2.5.1. Concomitant Double IFS of Nestin and ZEB2 in Panc1 Cells
2.5.2. Sequential IFS of SOX2 and L1CAM in Panc89
2.6. RNA Sequencing and Transcriptomic Analysis
2.7. Cell Growth Analysis
2.8. Treatment Response Analysis
2.9. Migration Assay
2.10. Invasion Assay
2.11. Adhesion Assay
2.12. Tumorigenicity and Metastasis Assay In Vivo
2.13. Immunohistochemical Staining of Paraffin-Embedded Tissue Sections
2.14. Statistical Analysis
3. Results
3.1. In Vitro Analysis of Panc1 and Panc89 Cell Variants
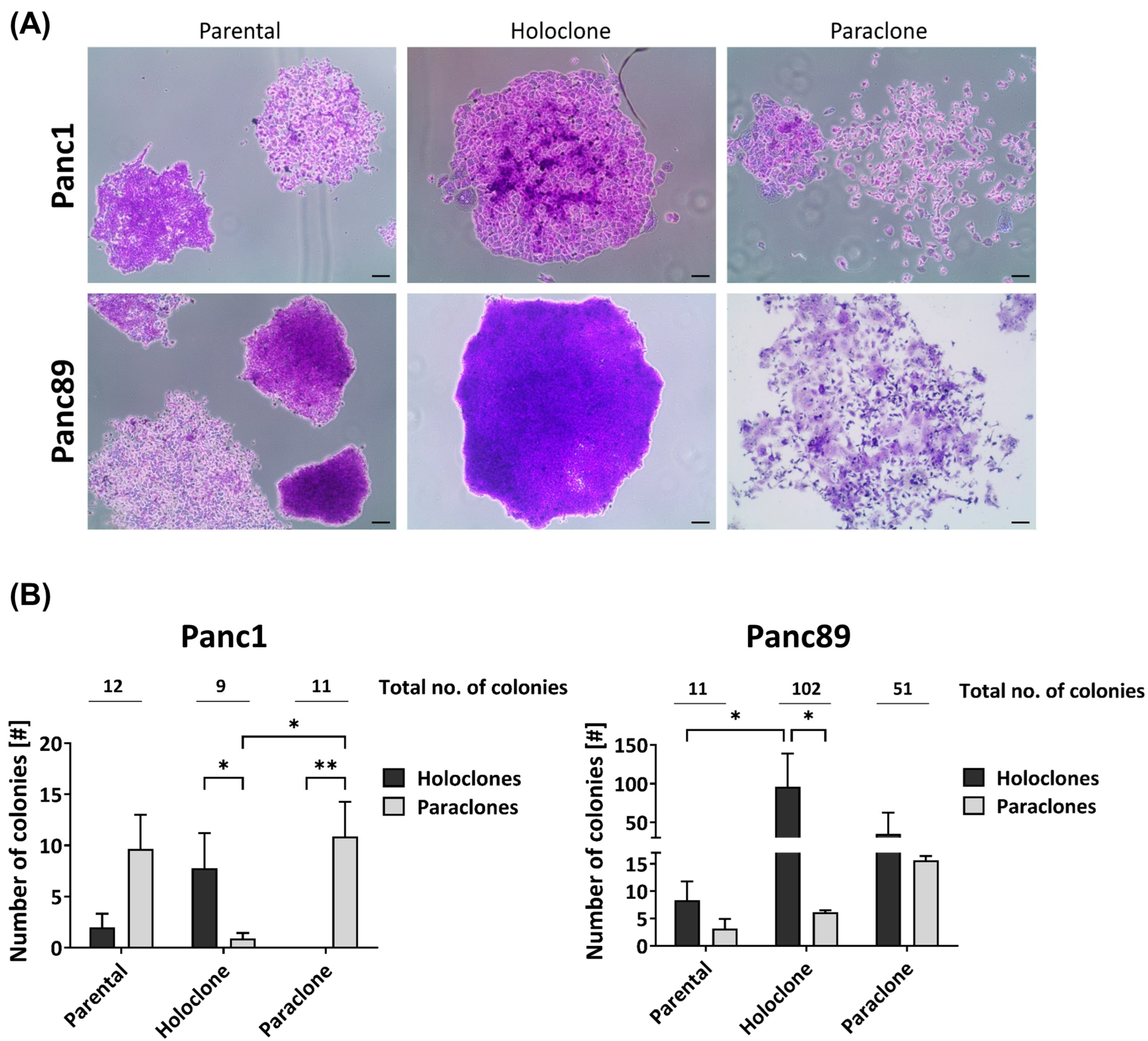
3.1.1. Panc1 and Panc89 Cell Variants Exhibit Differences in Colony-Formation Ability
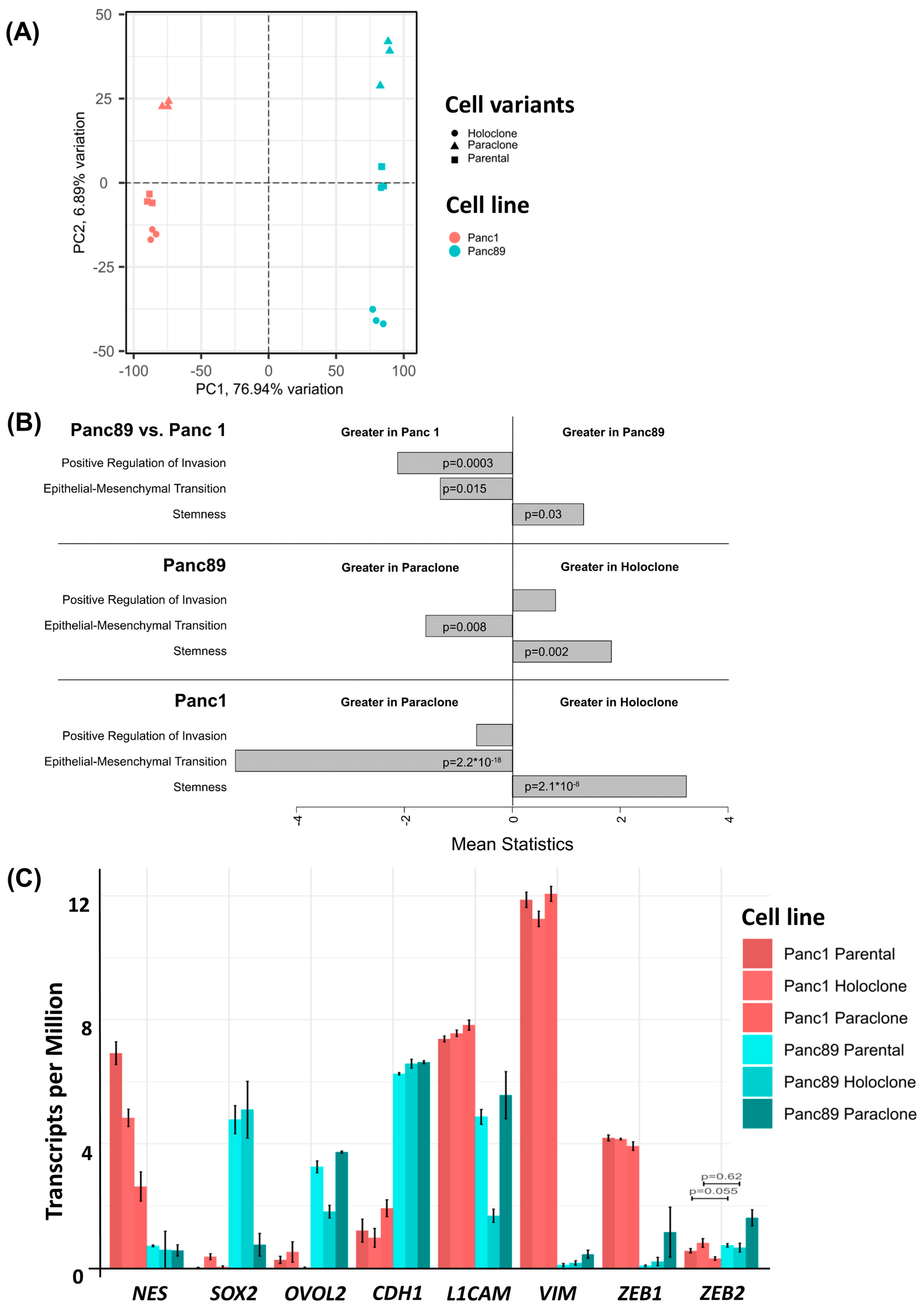
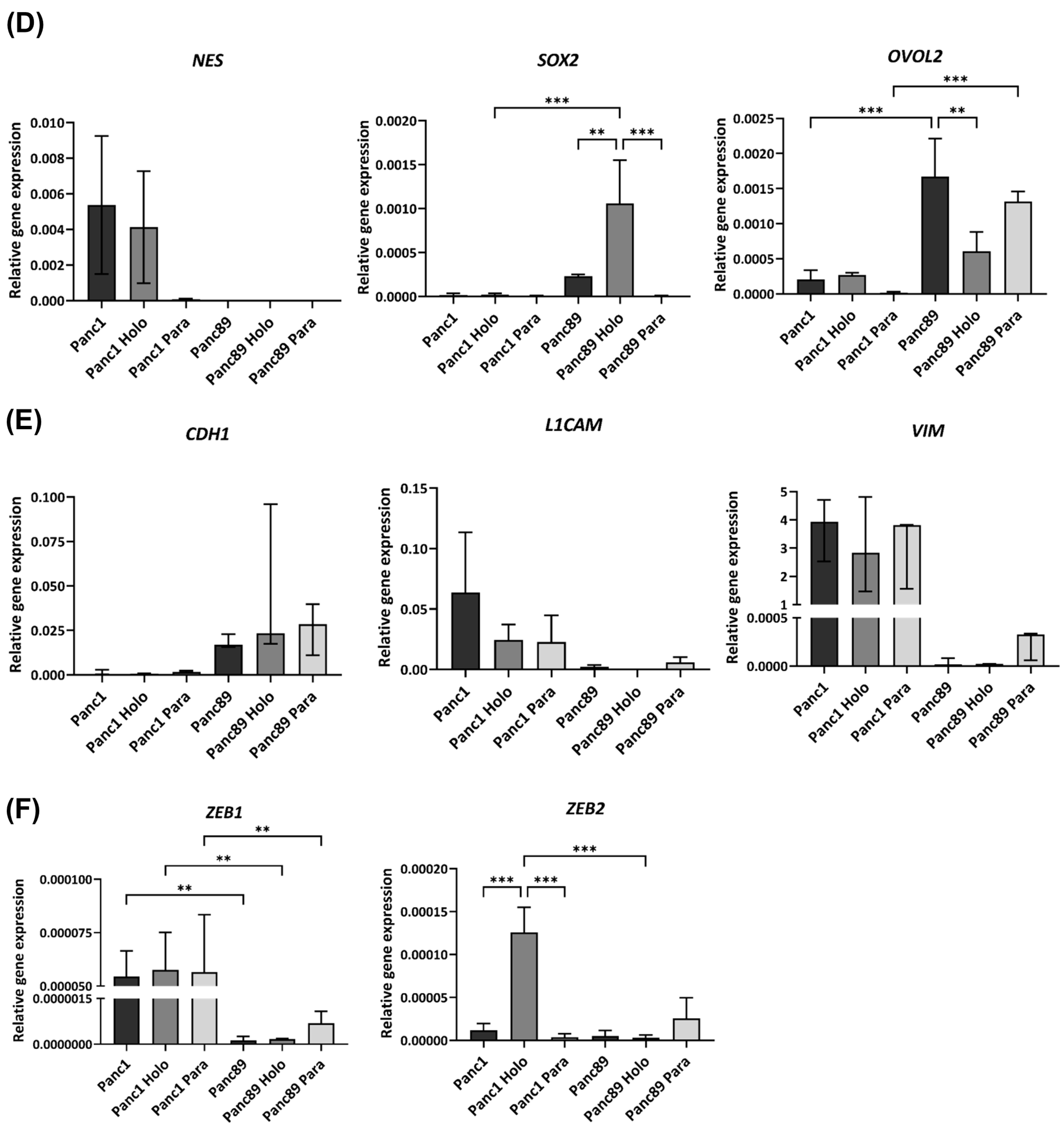
3.1.2. Parental Panc1 and Panc89 as Well as Their Derived Holo- and Paraclone Cells Show Distinct Transcriptional CSC and EMT Signatures
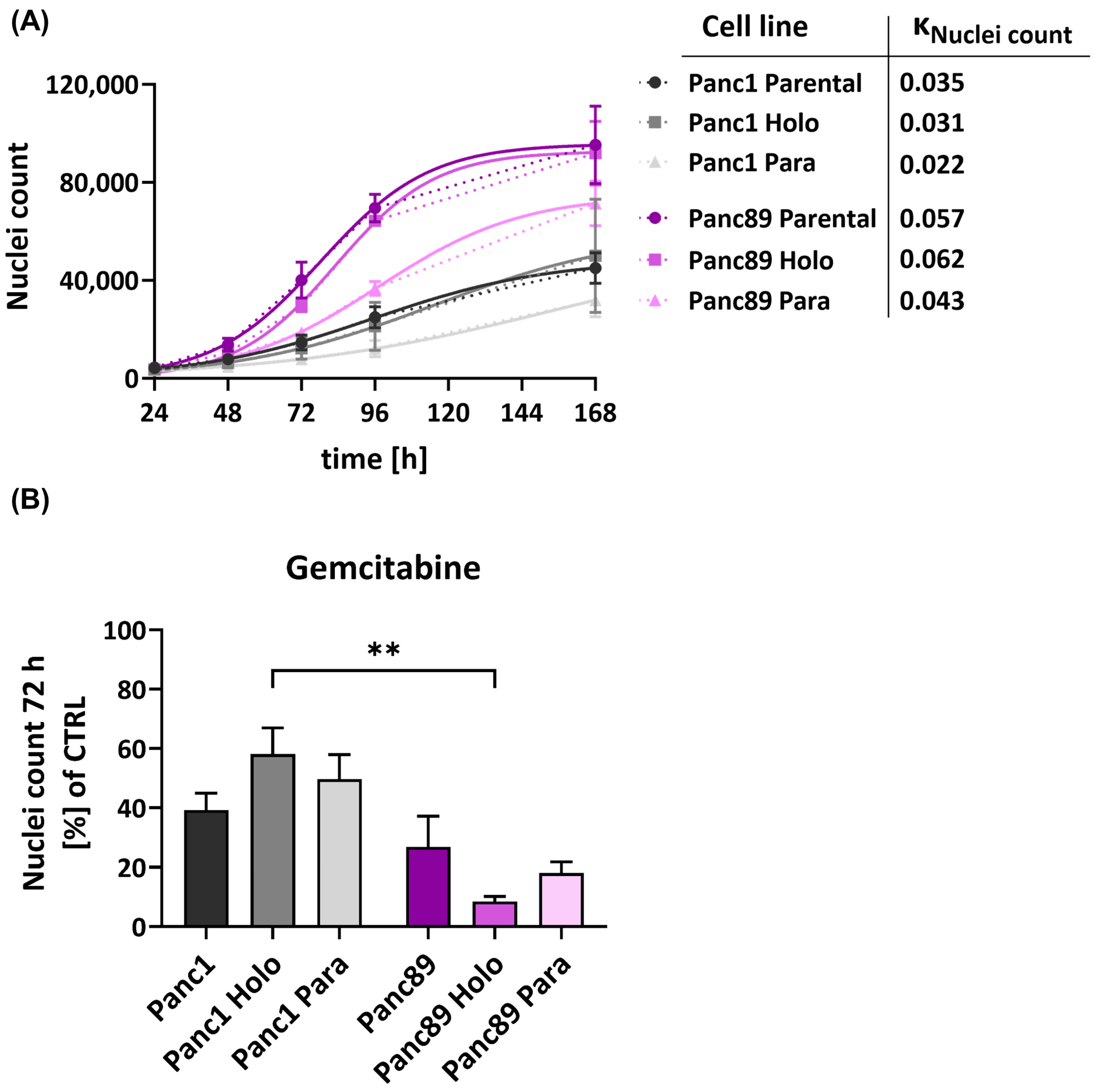
3.1.3. Panc89 Cell Variants Show Enhanced Cell Growth Rates Compared to Panc1 Cell Populations and Differ with Respect to Responses to Chemotherapeutic Treatments
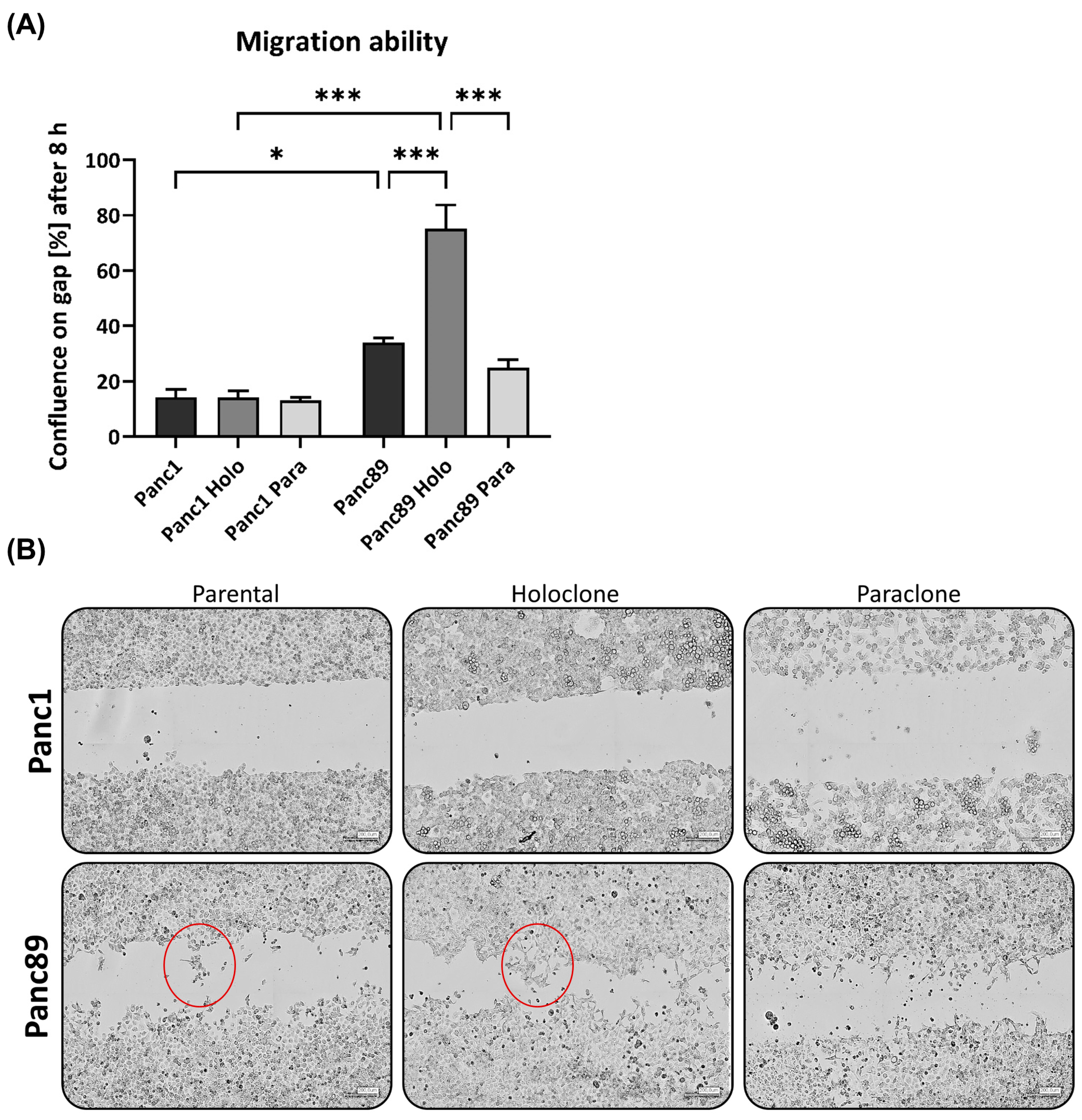
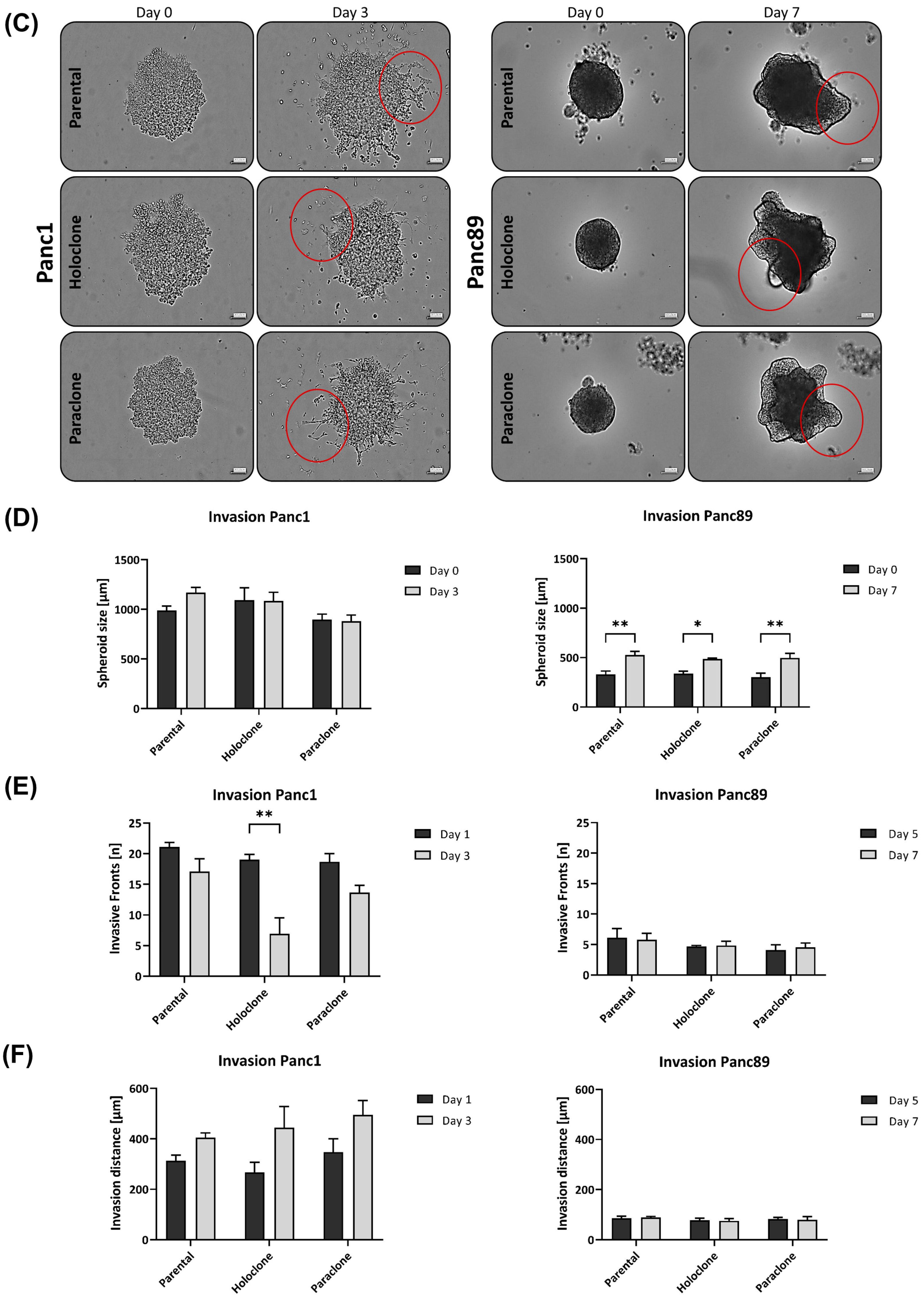
3.1.4. Panc1 Holoclone Cells Are Less Migratory but Highly Invasive in a Mesenchymal-like Invasion Manner, While Panc89 Holoclone Cells Show Pronounced Cell Migration but Slow Invasion in Clusters
3.2. Tumorigenicity and Metastasis Analysis of Panc1 and Panc89 Holo- or Paraclone Cells In Vivo
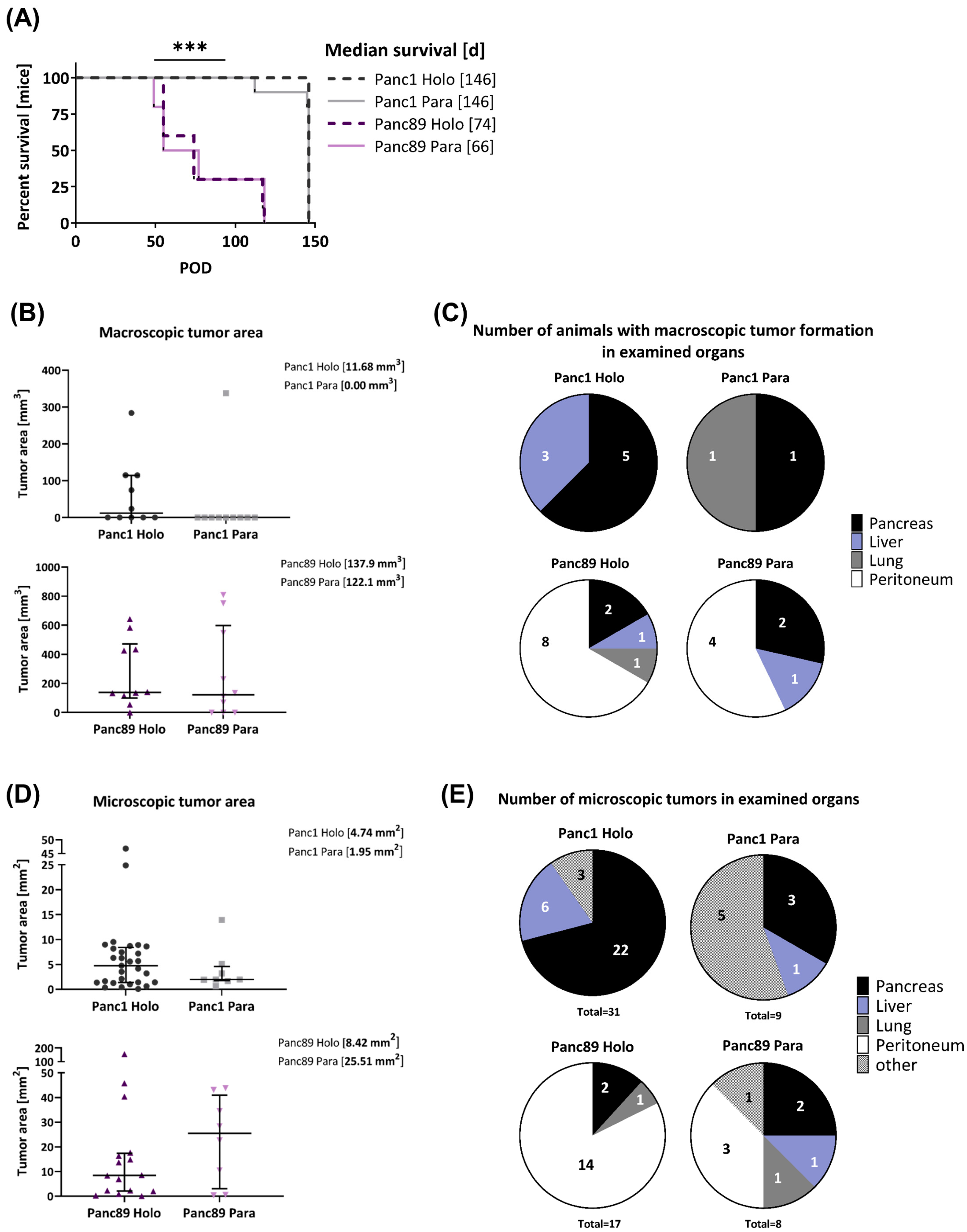
3.2.1. Panc1 and Panc89 Cell Variants Essentially Differ with Respect to Their Metastatic Capacity In Vivo
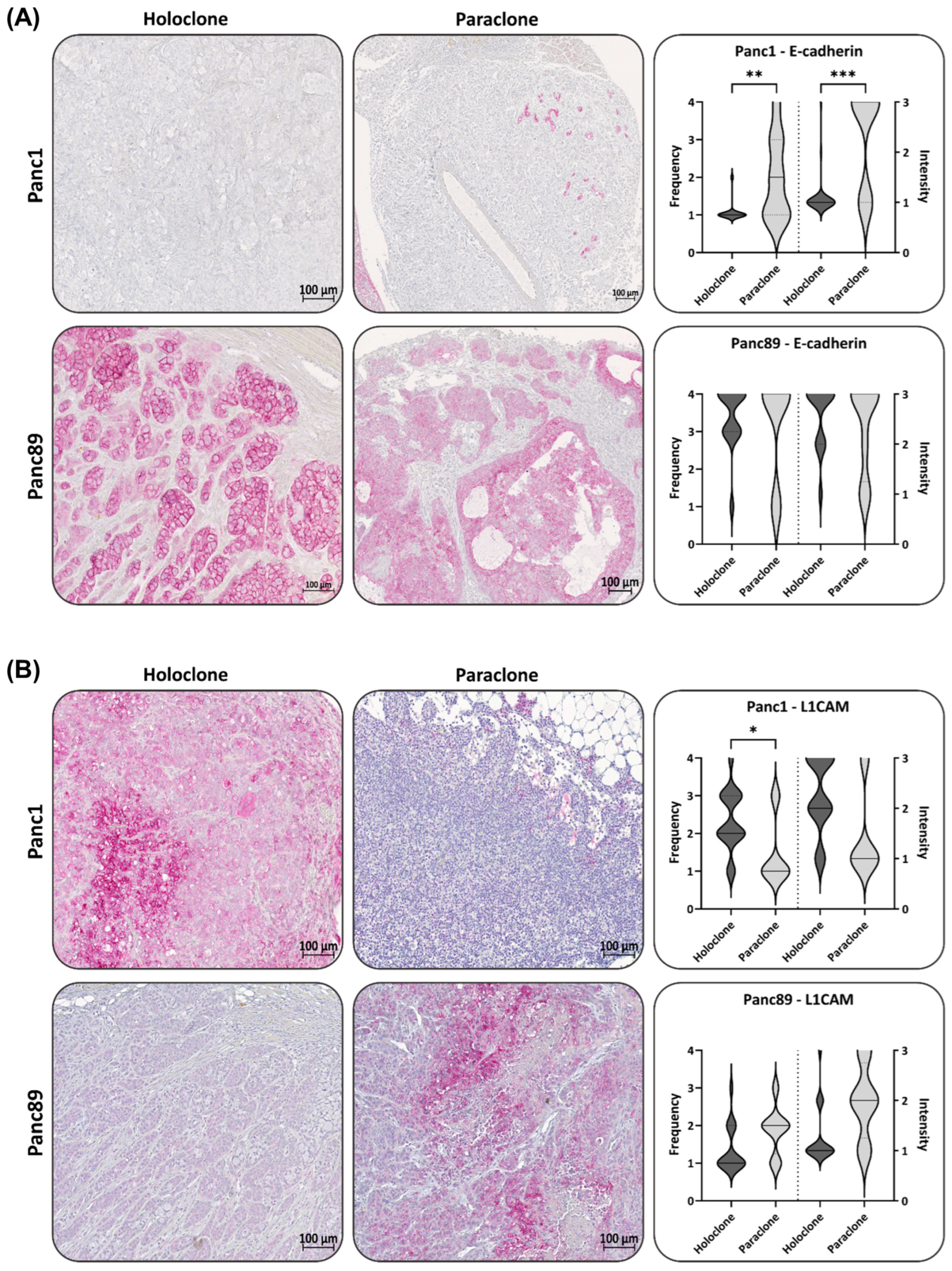
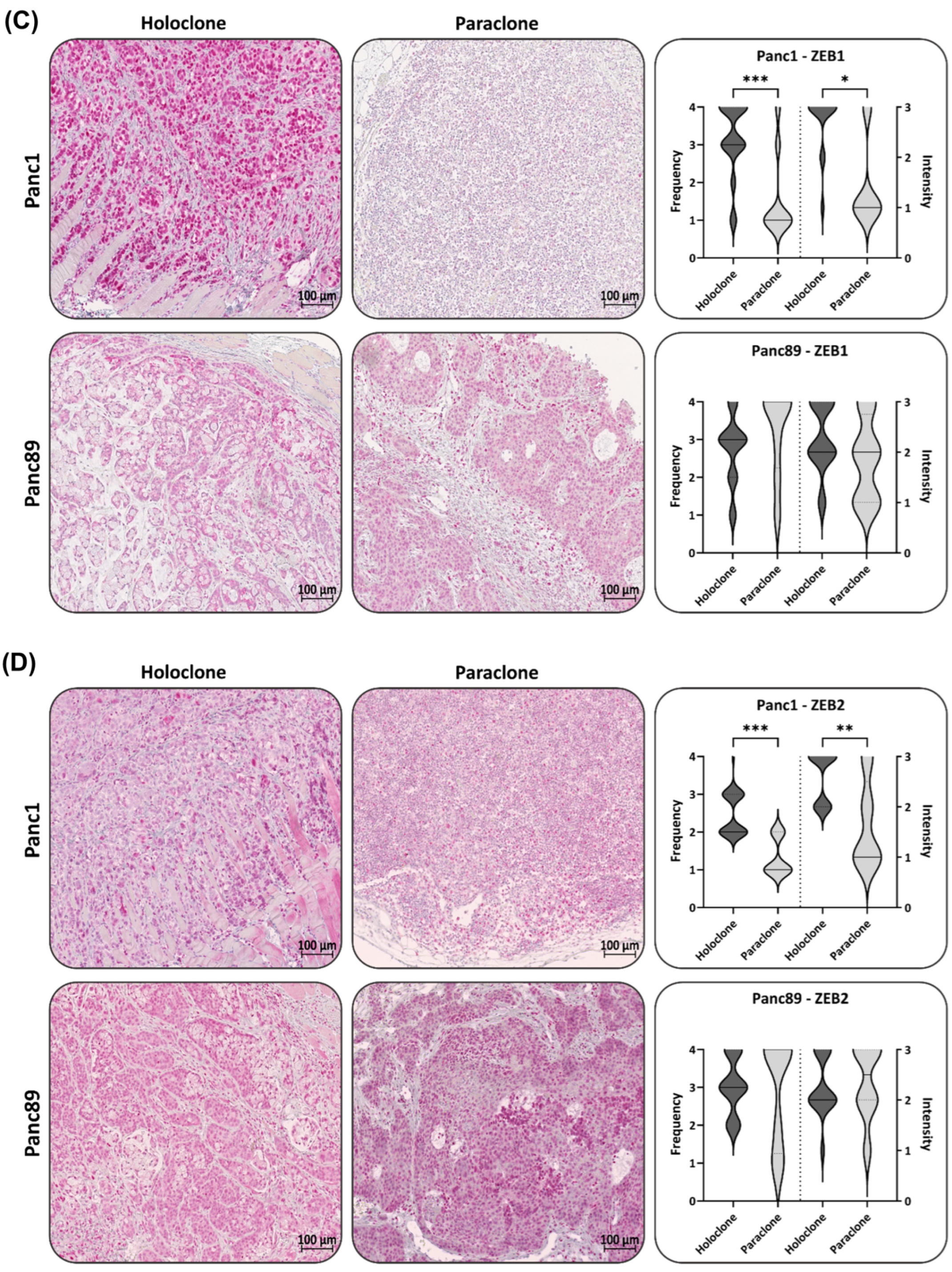
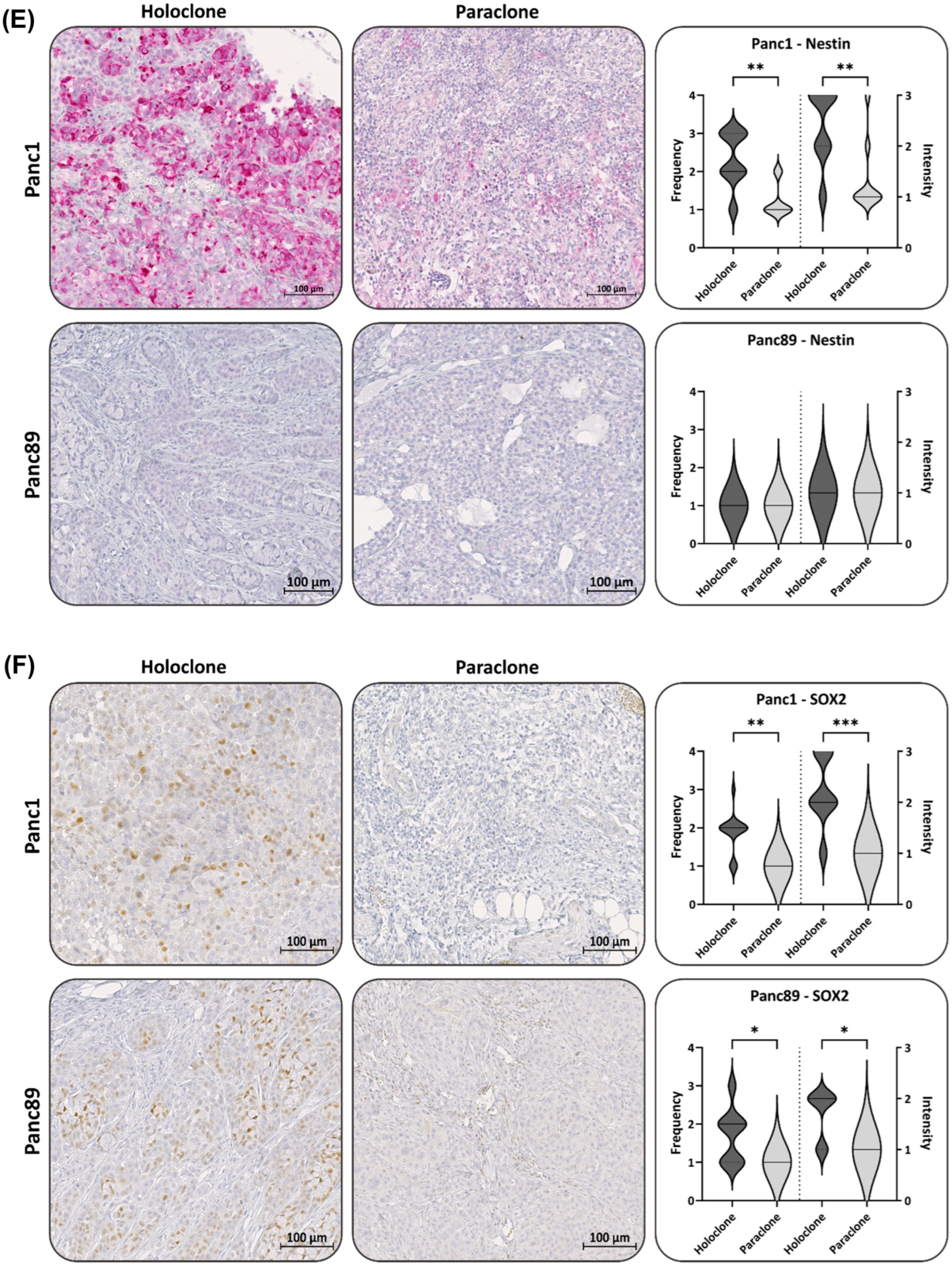
3.2.2. Panc1 and Panc89 Holo- and Paraclone Tumors Exhibit Differences in EMT and CSC Marker Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Rahib, L.; Smith, B.D.; Aizenberg, R.; Rosenzweig, A.B.; Fleshman, J.M.; Matrisian, L.M. Projecting cancer incidence and deaths to 2030: The unexpected burden of thyroid, liver, and pancreas cancers in the united states. Cancer Res. 2014, 74, 2913–2921. [Google Scholar] [CrossRef] [PubMed]
- Meslar, E. Pancreatic adenocarcinoma. JAAPA 2020, 33, 50–51. [Google Scholar] [CrossRef] [PubMed]
- Sperti, C.; Pasquali, C.; Piccoli, A.; Pedrazzoli, S. Recurrence after resection for ductal adenocarcinoma of the pancreas. World J. Surg. 1997, 21, 195–200. [Google Scholar] [CrossRef] [PubMed]
- Van den Broeck, A.; Sergeant, G.; Ectors, N.; Van Steenbergen, W.; Aerts, R.; Topal, B. Patterns of recurrence after curative resection of pancreatic ductal adenocarcinoma. Eur. J. Surg. Oncol. 2009, 35, 600–604. [Google Scholar] [CrossRef]
- Groot, V.P.; Rezaee, N.; Wu, W.; Cameron, J.L.; Fishman, E.K.; Hruban, R.H.; Weiss, M.J.; Zheng, L.; Wolfgang, C.L.; He, J. Patterns, Timing, and Predictors of Recurrence Following Pancreatectomy for Pancreatic Ductal Adenocarcinoma. Ann. Surg. 2018, 267, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Itchins, M.; Arena, J.; Nahm, C.; Pavlakis, N.; Clarke, S.; Gill, A.; Samra, J.S.; Mittal, A. Patterns and Determinants of Recurrence for Pancreatic Ductal Adenocarcinoma after Resection. J. Pancreas 2017, 18, 458–464. [Google Scholar]
- Valastyan, S.; Weinberg, R.A. Tumor metastasis: Molecular insights and evolving paradigms. Cell 2011, 147, 275–292. [Google Scholar] [CrossRef]
- Toh, B.; Wang, X.; Keeble, J.; Sim, W.J.; Khoo, K.; Wong, W.C.; Kato, M.; Prevost-Blondel, A.; Thiery, J.P.; Abastado, J.P. Mesenchymal transition and dissemination of cancer cells is driven by myeloid-derived suppressor cells infiltrating the primary tumor. PLoS Biol. 2011, 9, e1001162. [Google Scholar] [CrossRef]
- Nguyen, D.X.; Bos, P.D.; Massagué, J. Metastasis: From dissemination to organ-specific colonization. Nat. Rev. Cancer 2009, 9, 274–284. [Google Scholar] [CrossRef]
- Sökeland, G.; Schumacher, U. The functional role of integrins during intra- and extravasation within the metastatic cascade. Mol. Cancer 2019, 18, 12. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.J.; Nieto, M.A. Epithelial-Mesenchymal Transitions in Development and Disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Nieto, M.A.; Huang, R.Y.-J.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Roca, H.; Hernandez, J.; Weidner, S.; McEachin, R.C.; Fuller, D.; Sud, S.; Schumann, T.; Wilkinson, J.E.; Zaslavsky, A.; Li, H.; et al. Transcription Factors OVOL1 and OVOL2 Induce the Mesenchymal to Epithelial Transition in Human Cancer. PLoS ONE 2013, 8, e76773. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Liu, Y.; Noguchi, S.; Murray, M.; Chang, J.C.; Kishima, M.; Nishimura, H.; Hashimoto, K.; Minoda, A.; Suzuki, H. OVOL2 induces mesenchymal-to-epithelial transition in fibroblasts and enhances cell-state reprogramming towards epithelial lineages. Sci. Rep. 2019, 9, 6490. [Google Scholar] [CrossRef] [PubMed]
- Xia, L.; Gao, J.; Ma, K.; Lin, H.; Chen, Y.; Luo, Q.; Lian, J. OVOL2 attenuates the expression of MAP3K8 to suppress epithelial mesenchymal transition in colorectal cancer. Pathol. Res. Pract. 2021, 224, 153493. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef] [PubMed]
- Andriani, F.; Bertolini, G.; Facchinetti, F.; Baldoli, E.; Moro, M.; Casalini, P.; Caserini, R.; Milione, M.; Leone, G.; Pelosi, G.; et al. Conversion to stem-cell state in response to microenvironmental cues is regulated by balance between epithelial and mesenchymal features in lung cancer cells. Mol. Oncol. 2016, 10, 253–271. [Google Scholar] [CrossRef]
- Schwitalla, S.; Fingerle, A.A.; Cammareri, P.; Nebelsiek, T.; Göktuna, S.I.; Ziegler, P.K.; Canli, O.; Heijmans, J.; Huels, D.J.; Moreaux, G.; et al. Intestinal tumorigenesis initiated by dedifferentiation and acquisition of stem-cell-like properties. Cell 2013, 152, 25–38. [Google Scholar] [CrossRef]
- Matsuda, Y.; Yoshimura, H.; Ueda, J.; Naito, Z.; Korc, M.; Ishiwata, T. Nestin delineates pancreatic cancer stem cells in metastatic foci of NOD/Shi-scid IL2R??null (NOG) mice. Am. J. Pathol. 2014, 184, 674–685. [Google Scholar] [CrossRef]
- Heeschen, C.; Herrler, T.; Ellwart, J.W.; Guba, M.; Huber, S.L.; Bruns, C.J.; Aicher, A.; Hermann, P.C. Distinct Populations of Cancer Stem Cells Determine Tumor Growth and Metastatic Activity in Human Pancreatic Cancer. Cell Stem Cell 2007, 1, 313–323. [Google Scholar]
- Li, C.; Heidt, D.G.; Dalerba, P.; Burant, C.F.; Zhang, L.; Adsay, V.; Wicha, M.; Clarke, M.F.; Simeone, D.M. Identification of pancreatic cancer stem cells. Cancer Res. 2007, 67, 1030–1037. [Google Scholar] [CrossRef]
- Chaffer, C.L.; Brueckmann, I.; Scheel, C.; Kaestli, A.J.; Wiggins, P.A.; Rodrigues, L.O.; Brooks, M.; Reinhardt, F.; Su, Y.; Polyak, K.; et al. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc. Natl. Acad. Sci. USA 2011, 108, 7950–7955. [Google Scholar] [CrossRef]
- Dalla Pozza, E.; Dando, I.; Biondani, G.; Brandi, J.; Costanzo, C.; Zoratti, E.; Fassan, M.; Boschi, F.; Melisi, D.; Cecconi, D.; et al. Pancreatic ductal adenocarcinoma cell lines display a plastic ability to bi-directionally convert into cancer stem cells. Int. J. Oncol. 2015, 46, 1099–1108. [Google Scholar] [CrossRef]
- O’Leary, D.P.; O’Leary, E.; Foley, N.; Cotter, T.G.; Wang, J.H.; Redmond, H.P. Effects of surgery on the cancer stem cell niche. Eur. J. Surg. Oncol. 2016, 42, 319–325. [Google Scholar] [CrossRef]
- Mukherjee, S.; Manna, A.; Bhattacharjee, P.; Mazumdar, M.; Saha, S.; Chakraborty, S.; Guha, D.; Adhikary, A.; Jana, D.; Gorain, M.; et al. Non-migratory tumorigenic intrinsic cancer stem cells ensure breast cancer metastasis by generation of CXCR4+ migrating cancer stem cells. Oncogene 2016, 35, 4937–4948. [Google Scholar] [CrossRef]
- Zhu, Y.-Y.; Yuan, Z. Pancreatic cancer stem cells. Am. J. Cancer Res. 2015, 5, 894–906. [Google Scholar] [PubMed]
- Neradil, J.; Veselska, R. Nestin as a marker of cancer stem cells. Cancer Sci. 2015, 106, 803–811. [Google Scholar] [CrossRef] [PubMed]
- Bhagwandin, V.J.; Bishop, J.M.; Wright, W.E.; Shay, J.W. The metastatic potential and chemoresistance of human pancreatic cancer stem cells. PLoS ONE 2016, 11, e0148807. [Google Scholar] [CrossRef] [PubMed]
- Jeter, C.R.; Yang, T.; Wang, J.; Chao, H.P.; Tang, D.G. Concise review: NANOG in cancer stem cells and tumor development: An update and outstanding questions. Stem Cells 2016, 33, 2381–2390. [Google Scholar] [CrossRef]
- Herreros-Villanueva, M.; Bujanda, L.; Billadeau, D.D.; Zhang, J.S. Embryonic stem cell factors and pancreatic cancer. World J. Gastroenterol. 2014, 20, 2247–2254. [Google Scholar] [CrossRef]
- Narita, K.; Matsuda, Y.; Seike, M.; Naito, Z.; Gemma, A.; Ishiwata, T. Nestin regulates proliferation, migration, invasion and stemness of lung adenocarcinoma. Int. J. Oncol. 2014, 44, 1118–1130. [Google Scholar] [CrossRef]
- Gawlik-Rzemieniewska, N.; Bednarek, I. The role of NANOG transcriptional factor in the development of malignant phenotype of cancer cells. Cancer Biol. Ther. 2016, 17, 1–10. [Google Scholar] [CrossRef]
- Lu, Y.; Zhu, H.; Shan, H.; Lu, J.; Chang, X.; Li, X.; Lu, J.; Fan, X.; Zhu, S.; Wang, Y.; et al. Knockdown of Oct4 and Nanog expression inhibits the stemness of pancreatic cancer cells. Cancer Lett. 2013, 340, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Su, H.-T.; Weng, C.-C.; Hsiao, P.-J.; Chen, L.-H.; Kuo, T.-L.; Chen, Y.-W.; Kuo, K.-K.; Cheng, K.-H. Stem cell marker nestin is critical for TGF-beta1-mediated tumor progression in pancreatic cancer. Mol. Cancer Res. 2013, 11, 768–779. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, Y.; Naito, Z.; Kawahara, K.; Nakazawa, N.; Korc, M.; Ishiwata, T. Nestin is a novel target for suppressing pancreatic cancer cell migration, invasion and metastasis. Cancer Biol. Ther. 2011, 11, 512–523. [Google Scholar] [CrossRef] [PubMed]
- Arai, T.; Yamashita, S.; Matsuda, Y.; Yoshimura, H.; Ushijima, T.; Ishiwata, T. Systemic Administration of Small Interfering RNA Targeting Human Nestin Inhibits Pancreatic Cancer Cell Proliferation and Metastasis. Pancreas 2015, 45, 93–100. [Google Scholar]
- Sanada, Y.; Yoshida, K.; Ohara, M.; Oeda, M.; Konishi, K.; Tsutani, Y. Histopathologic evaluation of stepwise progression of pancreatic carcinoma with immunohistochemical analysis of gastric epithelial transcription factor SOX2: Comparison of expression patterns between invasive components and cancerous or nonneoplastic intr. Pancreas 2006, 32, 164–170. [Google Scholar] [CrossRef]
- Han, X.; Fang, X.; Lou, X.; Hua, D.; Ding, W.; Foltz, G.; Hood, L.; Yuan, Y.; Lin, B. Silencing SOX2 induced mesenchymal-epithelial transition and its expression predicts liver and lymph node metastasis of CRC patients. PLoS ONE 2012, 7, e41335. [Google Scholar] [CrossRef]
- Knaack, H.; Lenk, L.; Philipp, L.-M.; Miarka, L.; Rahn, S.; Viol, F.; Hauser, C.; Egberts, J.-H.; Gundlach, J.-P.; Will, O.; et al. Liver metastasis of pancreatic cancer: The hepatic microenvironment impacts differentiation and self-renewal capacity of pancreatic ductal epithelial cells. Oncotarget 2018, 9, 31771–31786. [Google Scholar] [CrossRef]
- Beaver, C.M.; Ahmed, A.; Masters, J.R. Clonogenicity: Holoclones and meroclones contain stem cells. PLoS ONE 2014, 9, e89834. [Google Scholar] [CrossRef] [PubMed]
- Harper, L.J.; Piper, K.; Common, J.; Fortune, F.; Mackenzie, I.C. Stem cell patterns in cell lines derived from head and neck squamous cell carcinoma. J. Oral Pathol. Med. Off. Publ. Int. Assoc. Oral Pathol. Am. Acad. Oral Pathol. 2007, 36, 594–603. [Google Scholar] [CrossRef] [PubMed]
- Locke, M.; Heywood, M.; Fawell, S.; Mackenzie, I.C. Retention of intrinsic stem cell hierarchies in carcinoma-derived cell lines. Cancer Res. 2005, 65, 8944–8950. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.J.; Sun, B.C.; Zhao, X.L.; Zhao, X.M.; Sun, T.; Gu, Q.; Yao, Z.; Dong, X.Y.; Zhao, N.; Liu, N. CD133+ cells with cancer stem cell characteristics associates with vasculogenic mimicry in triple-negative breast cancer. Oncogene 2013, 32, 544–553. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.; Sui, X.; Deng, H.; Ding, M. Holoclone forming cells from pancreatic cancer cells enrich tumor initiating cells and represent a novel model for study of cancer stem cells. PLoS ONE 2011, 6, e23383. [Google Scholar] [CrossRef] [PubMed]
- Rochat, A.; Grasset, N.; Gorostidi, F.; Lathion, S.; Barrandon, Y. Regeneration of Epidermis from Adult Human Keratinocyte Stem Cells. In Handbook of Stem Cells, 2nd ed.; Academic Press: Cambridge, MA, USA, 2012; Volumes 1–2, pp. 767–780. [Google Scholar]
- Barrandon, Y.; Green, H. Three clonal types of keratinocyte with different capacities for multiplication. Proc. Natl. Acad. Sci. USA 1987, 84, 2302–2306. [Google Scholar] [CrossRef]
- Brabletz, S.; Schuhwerk, H.; Brabletz, T.; Stemmler, M.P. Dynamic EMT: A multi-tool for tumor progression. EMBO J. 2021, 40, e108647. [Google Scholar] [CrossRef]
- Mehta, P.K.; Karls, R.K.; White, E.H.; Ades, E.W.; Quinn, F.D. Entry and intracellular replication of Mycobacterium tuberculosis in cultured human microvascular endothelial cells. Microb. Pathog. 2006, 41, 119–124. [Google Scholar] [CrossRef]
- Matsumura, T.; Takesue, M.; Westerman, K.A.; Okitsu, T.; Sakaguchi, M.; Fukazawa, T.; Totsugawa, T.; Noguchi, H.; Yamamoto, S.; Stolz, D.B.; et al. Establishment of an immortalized human-liver endothelial cell line with SV40T and hTERT. Transplantation 2004, 77, 1357–1365. [Google Scholar] [CrossRef]
- Lechner, J.F.; Tokiwa, T.; LaVeck, M.; Benedict, W.F.; Banks-Schlegel, S.; Yeager, H.; Banerjee, A.; Harris, C.C. Asbestos-associated chromosomal changes in human mesothelial cells. Proc. Natl. Acad. Sci. USA 1985, 82, 3884–3888. [Google Scholar] [CrossRef]
- Reddel, R.R.; Yang, K.; Rhim, J.S.; Brash, D.; Su, R.T.; Lechner, J.F.; Gerwin, B.I.; Harris, C.C.; Amstad, P. Immortalized Human Bronchial Epitherial Mesothelial Cell Lines. U.S. Patent 4,885,238, 5 December 1989. [Google Scholar]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Soneson, C.; Love, M.I.; Robinson, M.D. Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000Research 2015, 4, 1521. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Zhu, A.; Ibrahim, J.G.; Love, M.I. Heavy-Tailed prior distributions for sequence count data: Removing the noise and preserving large differences. Bioinformatics 2019, 35, 2084–2092. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Friedman, M.S.; Shedden, K.; Hankenson, K.D.; Woolf, P.J. GAGE: Generally applicable gene set enrichment for pathway analysis. BMC Bioinform. 2009, 10, 161. [Google Scholar] [CrossRef]
- Müller, F.J.; Laurent, L.C.; Kostka, D.; Ulitsky, I.; Williams, R.; Lu, C.; Park, I.H.; Rao, M.S.; Shamir, R.; Schwartz, P.H.; et al. Regulatory networks define phenotypic classes of human stem cell lines. Nature 2008, 455, 401–405. [Google Scholar] [CrossRef] [PubMed]
- Moffitt, R.A.; Marayati, R.; Flate, E.L.; Volmar, K.E.; Loeza, S.G.H.; Hoadley, K.A.; Rashid, N.U.; Williams, L.A.; Eaton, S.C.; Chung, A.H.; et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat. Genet. 2015, 47, 1168–1178. [Google Scholar] [CrossRef] [PubMed]
- Galván, J.A.; Zlobec, I.; Wartenberg, M.; Lugli, A.; Gloor, B.; Perren, A.; Karamitopoulou, E. Expression of E-cadherin repressors SNAIL, ZEB1 and ZEB2 by tumour and stromal cells influences tumour-budding phenotype and suggests heterogeneity of stromal cells in pancreatic cancer. Br. J. Cancer 2015, 112, 1944–1950. [Google Scholar] [CrossRef]
- Gallmeier, E.; Gress, T.M. Pancreatic ductal adenocarcinoma. Internist 2018, 59, 805–822. [Google Scholar] [CrossRef]
- Fulawka, L.; Donizy, P.; Halon, A. Cancer stem cells--the current status of an old concept: Literature review and clinical approaches. Biol. Res. 2014, 47, 66. [Google Scholar] [CrossRef]
- Karamitopoulou, E. Tumor budding cells, cancer stem cells and epithelial-mesenchymal transition-type cells in pancreatic cancer. Front. Oncol. 2013, 2, 2009–2013. [Google Scholar] [CrossRef]
- Castellanos, J.A.; Merchant, N.B.; Nagathihalli, N.S. Emerging targets in pancreatic cancer: Epithelial-mesenchymal transition and cancer stem cells. Onco. Targets. Ther. 2013, 6, 1261–1267. [Google Scholar]
- Zhan, H.X.; Xu, J.W.; Wu, D.; Zhang, T.P.; Hu, S.Y. Pancreatic cancer stem cells: New insight into a stubborn disease. Cancer Lett. 2015, 357, 429–437. [Google Scholar] [CrossRef]
- Neesse, A.; Bauer, C.A.; Öhlund, D.; Lauth, M.; Buchholz, M.; Michl, P.; Tuveson, D.A.; Gress, T.M. Stromal biology and therapy in pancreatic cancer: Ready for clinical translation? Gut 2019, 68, 159–171. [Google Scholar] [CrossRef]
- Yuan, S.; Norgard, R.J.; Stanger, B.Z. Cellular plasticity in cancer. Cancer Discov. 2019, 9, 837–851. [Google Scholar] [CrossRef] [PubMed]
- Elaskalani, O.; Razak, N.B.A.; Falasca, M.; Metharom, P. Epithelial-mesenchymal transition as a therapeutic target for overcoming chemoresistance in pancreatic cancer. World J. Gastrointest. Oncol. 2017, 9, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Gaianigo, N.; Melisi, D.; Carbone, C. EMT and treatment resistance in pancreatic cancer. Cancers 2017, 9, 122. [Google Scholar] [CrossRef]
- Kyuno, D.; Yamaguchi, H.; Ito, T.; Kono, T.; Kimura, Y.; Imamura, M.; Konno, T.; Hirata, K.; Sawada, N.; Kojima, T. Targeting tight junctions during epithelial to mesenchymal transition in human pancreatic cancer. World J. Gastroenterol. 2014, 20, 10813–10824. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, T. Cancer stem cells and epithelial-mesenchymal transition: Novel therapeutic targets for cancer. Pathol. Int. 2016, 66, 601–608. [Google Scholar] [CrossRef]
- Shibue, T.; Weinberg, R.A. EMT, CSCs, and drug resistance: The mechanistic link and clinical implications. Nat. Rev. Clin. Oncol. 2017, 14, 611–629. [Google Scholar] [CrossRef]
- Rhim, A.D. Epithelial to mesenchymal transition and the generation of stem-like cells in pancreatic cancer. Pancreatology 2013, 13, 114–117. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Paul, R.; Dorsey, J.F.; Fan, Y. Cell plasticity, senescence, and quiescence in cancer stem cells: Biological and therapeutic implications. Pharmacol. Ther. 2022, 231, 107985. [Google Scholar] [CrossRef] [PubMed]
- Kreso, A.; Dick, J.E. Evolution of the cancer stem cell model. Cell Stem Cell 2014, 14, 275–291. [Google Scholar] [CrossRef] [PubMed]
- Valle, S.; Martin-Hijano, L.; Alcalá, S.; Alonso-Nocelo, M.; Sainz, B. The ever-evolving concept of the cancer stem cell in pancreatic cancer. Cancers 2018, 10, 33. [Google Scholar] [CrossRef]
- Burdziak, C.; Alonso-Curbelo, D.; Walle, T.; Reyes, J.; Barriga, F.M.; Haviv, D.; Xie, Y.; Zhao, Z.; Zhao, C.J.; Chen, H.-A.; et al. Epigenetic plasticity cooperates with cell-cell interactions to direct pancreatic tumorigenesis. Science 2023, 380, eadd5327. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Clarke, M.F. Self-renewal and solid tumor stem cells. Oncogene 2004, 23, 7274–7282. [Google Scholar] [CrossRef]
- Yu, Q.R. Stem cells and cancer stem cells. J. Clin. Rehabil. Tissue Eng. Res. 2007, 11, 2948–2951. [Google Scholar]
- Aponte, P.M.; Caicedo, A. Stemness in Cancer: Stem Cells, Cancer Stem Cells, and Their Microenvironment. Stem Cells Int. 2017, 2017, 5619472. [Google Scholar] [CrossRef]
- Sipos, B.; Möser, S.; Kalthoff, H.; Török, V.; Löhr, M.; Klöppel, G. A comprehensive characterization of pancreatic ductal carcinoma cell lines: Towards the establishment of an in vitro research platform. Virchows Arch. 2003, 442, 444–452. [Google Scholar] [CrossRef]
- Jolly, M.K.; Boareto, M.; Huang, B.; Jia, D.; Lu, M.; Onuchic, J.N.; Levine, H.; Ben-Jacob, E. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front. Oncol. 2015, 5, 155. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Ware, K.E.; Gilja, S.; Somarelli, J.A.; Levine, H. EMT and MET: Necessary or permissive for metastasis? Mol. Oncol. 2017, 11, 755–769. [Google Scholar] [CrossRef]
- Carstens, J.L.; Yang, S.; Correa de Sampaio, P.; Zheng, X.; Barua, S.; McAndrews, K.M.; Rao, A.; Burks, J.K.; Rhim, A.D.; Kalluri, R. Stabilized epithelial phenotype of cancer cells in primary tumors leads to increased colonization of liver metastasis in pancreatic cancer. Cell Rep. 2021, 35, 108990. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Tripathi, S.C.; Somarelli, J.A.; Hanash, S.M.; Levine, H. Epithelial/mesenchymal plasticity: How have quantitative mathematical models helped improve our understanding? Mol. Oncol. 2017, 11, 739–754. [Google Scholar] [CrossRef] [PubMed]
- Beckinger, S.; Daunke, T.; Aldag, L.; Krüger, S.; Heckl, S.; Wesch, D.; Schäfer, H.; Röcken, C.; Rahn, S.; Sebens, S. Hepatic myofibroblasts exert immunosuppressive effects independent of the immune checkpoint regulator PD-L1 in liver metastasis of pancreatic ductal adenocarcinoma. Front. Oncol. 2023, 13, 1160824. [Google Scholar] [CrossRef] [PubMed]
- Aldag, L.; Beckinger, S.; Daunke, T.; Philipp, L.M.; Surrow, A.; Yesilyurt, U.U.; Wandmacher, A.M.; Mehdorn, A.S.; Sebens, S. The heterogeneity of the tumor microenvironment as essential determinant of development, progression and therapy response of pancreatic cancer. Cancers 2021, 13, 4932. [Google Scholar]
- Geismann, C.; Morscheck, M.; Koch, D.; Bergmann, F.; Ungefroren, H.; Arlt, A.; Tsao, M.S.; Bachem, M.G.; Altevogt, P.; Sipos, B.; et al. Up-regulation of L1CAM in pancreatic duct cells is transforming growth factor β1- and slug-dependent: Role in malignant transformation of pancreatic cancer. Cancer Res. 2009, 69, 4517–4526. [Google Scholar] [CrossRef]
- Helm, O.; Held-Feindt, J.; Grage-Griebenow, E.; Reiling, N.; Ungefroren, H.; Vogel, I.; Krüger, U.; Becker, T.; Ebsen, M.; Röcken, C.; et al. Tumor-associated macrophages exhibit pro- and anti-inflammatory properties by which they impact on pancreatic tumorigenesis. Int. J. Cancer 2014, 135, 843–861. [Google Scholar] [CrossRef]
- Yang, J.; Liao, D.; Chen, C.; Liu, Y.; Chuang, T.H.; Xiang, R.; Markowitz, D.; Reisfeld, R.A.; Luo, Y. Tumor-associated macrophages regulate murine breast cancer stem cells through a novel paracrine egfr/stat3/sox-2 signaling pathway. Stem Cells 2013, 31, 248–258. [Google Scholar] [CrossRef]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef]
- Collisson, E.A.; Sadanandam, A.; Olson, P.; Gibb, W.J.; Truitt, M.; Gu, S.; Cooc, J.; Weinkle, J.; Kim, G.E.; Jakkula, L.; et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat. Med. 2011, 17, 500–503. [Google Scholar] [CrossRef]
- Tsujii, M. Cancer therapy targeting cancer stem cell. Nihon Rinsho. 2014, 72, 35–41. [Google Scholar]
- Benjamin, B. Cytotoxic Drugs. In Introduction to Basics of Pharmacology and Toxicology: Volume 2: Essentials of Systemic Pharmacology: From Principles to Practice; Paul, A., Anandabaskar, N., Mathaiyan, J., Raj, G.M., Eds.; Springer Nature: Singapore, 2021; pp. 1077–1090. [Google Scholar] [CrossRef]
- Sazonova, E.V.; Chesnokov, M.S.; Zhivotovsky, B.; Kopeina, G.S. Drug toxicity assessment: Cell proliferation versus cell death. Cell Death Discov. 2022, 8, 417. [Google Scholar] [CrossRef]
- Liotta, L.A.; Tryggvason, K.; Garbisa, S.; Hart, I.; Foltz, C.M.; Shafie, S. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature 1980, 284, 67–68. [Google Scholar] [CrossRef]
- Maatta, M.; Soini, Y.; Liakka, A.; Autio-Harmainen, H. Differential expression of matrix metalloproteinase (MMP)-2, MMP-9, and membrane type 1-MMP in hepatocellular and pancreatic adenocarcinoma: Implications for tumor progression and clinical prognosis. Clin. Cancer Res. 2000, 6, 2726–2734. [Google Scholar]
- Kidd, M.E.; Shumaker, D.K.; Ridge, K.M. The role of Vimentin intermediate filaments in the progression of lung cancer. Am. J. Respir. Cell Mol. Biol. 2014, 50, 1–6. [Google Scholar] [CrossRef]
- Domagala, W.; Lasota, J.; Dukowicz, A.; Markiewski, M.; Striker, G.; Weber, K.; Osborn, M. Vimentin expression appears to be associated with poor prognosis in node-negative ductal NOS breast carcinomas. Am. J. Pathol. 1990, 137, 1299–1304. [Google Scholar] [PubMed]
- Burch, T.C.; Watson, M.T.; Nyalwidhe, J.O. Variable Metastatic Potentials Correlate with Differential Plectin and Vimentin Expression in Syngeneic Androgen Independent Prostate Cancer Cells. PLoS ONE 2013, 8, e65005. [Google Scholar] [CrossRef] [PubMed]
- Dauphin, M.; Barbe, C.; Lemaire, S.; Nawrocki-Raby, B.; Lagonotte, E.; Delepine, G.; Birembaut, P.; Gilles, C.; Polette, M. Vimentin expression predicts the occurrence of metastases in non small cell lung carcinomas. Lung Cancer 2013, 81, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Divanyan, A.; Jourd’heuil, F.L.; Goldman, R.D.; Ridge, K.M.; Jourd’heuil, D.; Lopez-Soler, R.I. Vimentin expression is required for the development of EMT-related renal fibrosis following unilateral ureteral obstruction in mice. Am. J. Physiol. Ren. Physiol. 2018, 315, F769–F780. [Google Scholar] [CrossRef] [PubMed]
- Ridge, K.M.; Eriksson, J.E.; Pekny, M.; Goldman, R.D. Roles of vimentin in health and disease. Genes Dev. 2022, 36, 391–407. [Google Scholar] [CrossRef] [PubMed]
- Grage-Griebenow, E.; Jerg, E.; Gorys, A.; Wicklein, D.; Wesch, D.; Freitag-Wolf, S.; Goebel, L.; Vogel, I.; Becker, T.; Ebsen, M.; et al. L1CAM promotes enrichment of immunosuppressive T cells in human pancreatic cancer correlating with malignant progression. Mol. Oncol. 2014, 8, 982–997. [Google Scholar] [CrossRef] [PubMed]
- Sebens Müerköster, S.; Werbing, V.; Sipos, B.; Debus, M.A.; Witt, M.; Großmann, M.; Leisner, D.; Kötteritzsch, J.; Kappes, H.; Klöppel, G.; et al. Drug-induced expression of the cellular adhesion molecule L1CAM confers anti-apoptotic protection and chemoresistance in pancreatic ductal adenocarcinoma cells. Oncogene 2007, 26, 2759–2768. [Google Scholar] [CrossRef] [PubMed]
- Schäfer, H.; Geismann, C.; Heneweer, C.; Egberts, J.H.; Korniienko, O.; Kiefel, H.; Moldenhauer, G.; Bachem, M.G.; Kalthoff, H.; Altevogt, P.; et al. Myofibroblast-induced tumorigenicity of pancreatic ductal epithelial cells is L1CAM dependent. Carcinogenesis 2012, 33, 84–93. [Google Scholar] [CrossRef][Green Version]
- Kasashima, H.; Duran, A.; Martinez-Ordoñez, A.; Nakanishi, Y.; Kinoshita, H.; Linares, J.F.; Reina-Campos, M.; Kudo, Y.; L’Hermitte, A.; Yashiro, M.; et al. Stromal SOX2 Upregulation Promotes Tumorigenesis through the Generation of a SFRP1/2-Expressing Cancer-Associated Fibroblast Population. Dev. Cell 2021, 56, 95–110.e10. [Google Scholar] [CrossRef]
- Wuebben, E.L.; Wilder, P.J.; Cox, J.L.; Grunkemeyer, J.A.; Caffrey, T.; Hollingsworth, M.A.; Rizzino, A. SOX2 functions as a molecular rheostat to control the growth, tumorigenicity and drug responses of pancreatic ductal adenocarcinoma cells. Oncotarget 2016, 7, 34890–34906. [Google Scholar] [CrossRef]
- Bylund, M.; Andersson, E.; Novitch, B.G.; Muhr, J. Vertebrate neurogenesis is counteracted by Sox1-3 activity. Nat. Neurosci. 2003, 6, 1162–1168. [Google Scholar] [CrossRef]
| Target | 5′-3′ Sequence | Annealing [C°] |
|---|---|---|
| CDH1 (E-cadherin) ** | fw-TGCTCTTGCTGTTTCTTCGG rv-TGCCCCATTCGTTCAAGTAG | 55 |
| GAPDH * |
fw-TCCATGACAACTTTGGTATCGTGG rv-GACGCCTGCTTCACCACCTTCT | 58 |
| L1CAM ** |
fw-GAACTGGATGTGGTGGAGAG rv-GAGGGTGGTAGAGGTCTGGT | 58 |
| NES (Nestin) * |
fw-GAAACAGCCATAGAGGGCAAA rv-TGGTTTTCCAGAGTCTTCAGTGA | 58 |
| OVOL2 (ZNF339) ** |
fw-GGGACAAGCTCTACGTCTGC rv-GTCTGTCCTCCCCTTCCTTC | 58 |
| SOX2 * |
fw-TCCCATCACCCACAGCAAATGA rv-TTTCTTGTCGGCATCGCGGTTT | 58 |
| VIM (Vimentin) ** |
fw-TCCAAGTTTGCTGACCTCTC rv-TCAACGGCAAAGTTCTCTTC | 58 |
| ZEB1 * |
fw-TCCATGCTTAAGAGCGCTAGCT rv-ACCGTAGTTGAGTAGGTGTATGCCA | 61 |
| ZEB2 ** |
fw-CACATCAGCAGCAAGAAATG rv-AAACCCGTGTGTAGCCATAA | 58 |
| Antigen | Buffer (pH) | Temperature, Time |
|---|---|---|
| anti-E-cadherin | S1699 (pH 6.1) | Steaming 121 °C, 10 min |
| anti-L1CAM | EDTA (pH 8.0) |
1. microwave broiling 800 watts 2. microwave 560 watts, 3 × 5 min |
| anti-Nestin | Citrate buffer (pH 6.0) | Steaming 121 °C, 10 min |
| anti-PanCK |
Target Retrieval Solution, Citrate (pH 6.1, 10×), S1699 (DAKO Agilent, Santa Clara, CA, USA) |
1. Boiling 2. Waterbath, 95 °C, 20 min |
| anti-SOX2 |
Target Retrieval Solution, Citrate (pH 6.1, 10×), S1699 (DAKO Agilent, Santa Clara, CA, USA) | Steaming 125 °C, 4 min |
| anti-ZEB1 | Citrate buffer (pH 6.0) | Steaming 121 °C, 10 min |
| anti-ZEB2 | Citrate buffer (pH 6.0) | Steaming 121 °C, 10 min |
| Primary Antibody | Secondary Antibody | Isotype |
|---|---|---|
|
E-cadherin (1:100; clone NHC-38; DAKO Agilent, Santa Clara, CA, USA) |
Dako REAL™ Detection System (Biotinylated goat anti-mouse/anti-rabbit immunoglobulins) |
Mouse IgG1 (1:1484; 02-6100; Invitrogen via Thermo Fisher Scientific, Darmstadt, Germany) |
|
L1CAM (1:1000; clone L1-11A, Peter Altevogt, German Cancer Research Center, Heidelberg, Germany) |
Goat anti-mouse-biotin (1:200; LS-C149505; LS-Bio, Shirley, MA, USA) |
Mouse IgG1 (1:228; 02-6100; Invitrogen via Thermo Fisher Scientific, Darmstadt, Germany) |
|
Nestin (1:100; clone 10C2; Merck, Darmstadt, Germany) |
Goat anti-mouse biotin (1:200; LS-C149505; LS-Bio, Shirley, MA, USA) |
Mouse IgG1 (1:100; 02-6100; Invitrogen via Thermo Fisher Scientific, Darmstadt, Germany) |
|
Pan-cytokeratin (PanCK) (12.7 µg/mL; clone AE1/AE3; DAKO Agilent, Santa Clara, CA, USA) |
Goat anti-mouse biotin (1:200; LS-C149505; LS-Bio, Shirley, MA, USA) |
Mouse IgG1 (1:80; 02-6100; Invitrogen via Thermo Fisher Scientific, Darmstadt, Germany) |
|
SOX2 (1:20, polyclonal; Atlas Antibodies, Bromma, Sweden) |
Goat anti-rabbit biotin (1:200; LS-C350860; LS-Bio, Shirley, MA, USA) |
Rabbit polyclonal IgG (1:2000; ab37415; Abcam, Cambridge, UK) |
|
ZEB1 (1:100; polyclonal; Atlas Antibodies, Bromma, Sweden) |
Goat anti-rabbit biotin (1:200; LS-C350860; LS-Bio, Shirley, MA, USA) |
Rabbit polyclonal IgG (1:2500; ab37415; Abcam, Cambridge, UK) |
|
ZEB2 (1:100; polyclonal; Atlas Antibodies, Bromma, Sweden) |
Dako REAL™ Detection System (Biotinylated goat anti-mouse/anti-rabbit immunoglobulins) |
Rabbit polyclonal IgG (1:5000; ab37415; Abcam, Cambridge, UK) |
| Frequency Score | Intensity Score |
|---|---|
| 1—Negative/low (0–10%) | 1—Negative/low |
| 2—Intermediate (11–50%) | 2—Intermediate |
| 3—High (51–90%) | 3—Strong |
| 4—Strong (>90%) | – |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Philipp, L.-M.; Yesilyurt, U.-U.; Surrow, A.; Künstner, A.; Mehdorn, A.-S.; Hauser, C.; Gundlach, J.-P.; Will, O.; Hoffmann, P.; Stahmer, L.; et al. Epithelial and Mesenchymal-like Pancreatic Cancer Cells Exhibit Different Stem Cell Phenotypes Associated with Different Metastatic Propensities. Cancers 2024, 16, 686. https://doi.org/10.3390/cancers16040686
Philipp L-M, Yesilyurt U-U, Surrow A, Künstner A, Mehdorn A-S, Hauser C, Gundlach J-P, Will O, Hoffmann P, Stahmer L, et al. Epithelial and Mesenchymal-like Pancreatic Cancer Cells Exhibit Different Stem Cell Phenotypes Associated with Different Metastatic Propensities. Cancers. 2024; 16(4):686. https://doi.org/10.3390/cancers16040686
Chicago/Turabian StylePhilipp, Lisa-Marie, Umut-Ulas Yesilyurt, Arne Surrow, Axel Künstner, Anne-Sophie Mehdorn, Charlotte Hauser, Jan-Paul Gundlach, Olga Will, Patrick Hoffmann, Lea Stahmer, and et al. 2024. "Epithelial and Mesenchymal-like Pancreatic Cancer Cells Exhibit Different Stem Cell Phenotypes Associated with Different Metastatic Propensities" Cancers 16, no. 4: 686. https://doi.org/10.3390/cancers16040686
APA StylePhilipp, L.-M., Yesilyurt, U.-U., Surrow, A., Künstner, A., Mehdorn, A.-S., Hauser, C., Gundlach, J.-P., Will, O., Hoffmann, P., Stahmer, L., Franzenburg, S., Knaack, H., Schumacher, U., Busch, H., & Sebens, S. (2024). Epithelial and Mesenchymal-like Pancreatic Cancer Cells Exhibit Different Stem Cell Phenotypes Associated with Different Metastatic Propensities. Cancers, 16(4), 686. https://doi.org/10.3390/cancers16040686









