Understanding Cancer’s Defense against Topoisomerase-Active Drugs: A Comprehensive Review
Simple Summary
Abstract
1. Introduction
2. Topoisomerases
3. Mechanisms of Action of Topo-Active Drugs
4. Tumor Heterogeneity and Topo-Active Drugs
5. Mutation of Target Enzymes
6. Altered Drug Metabolisms and Topo-Active Drugs
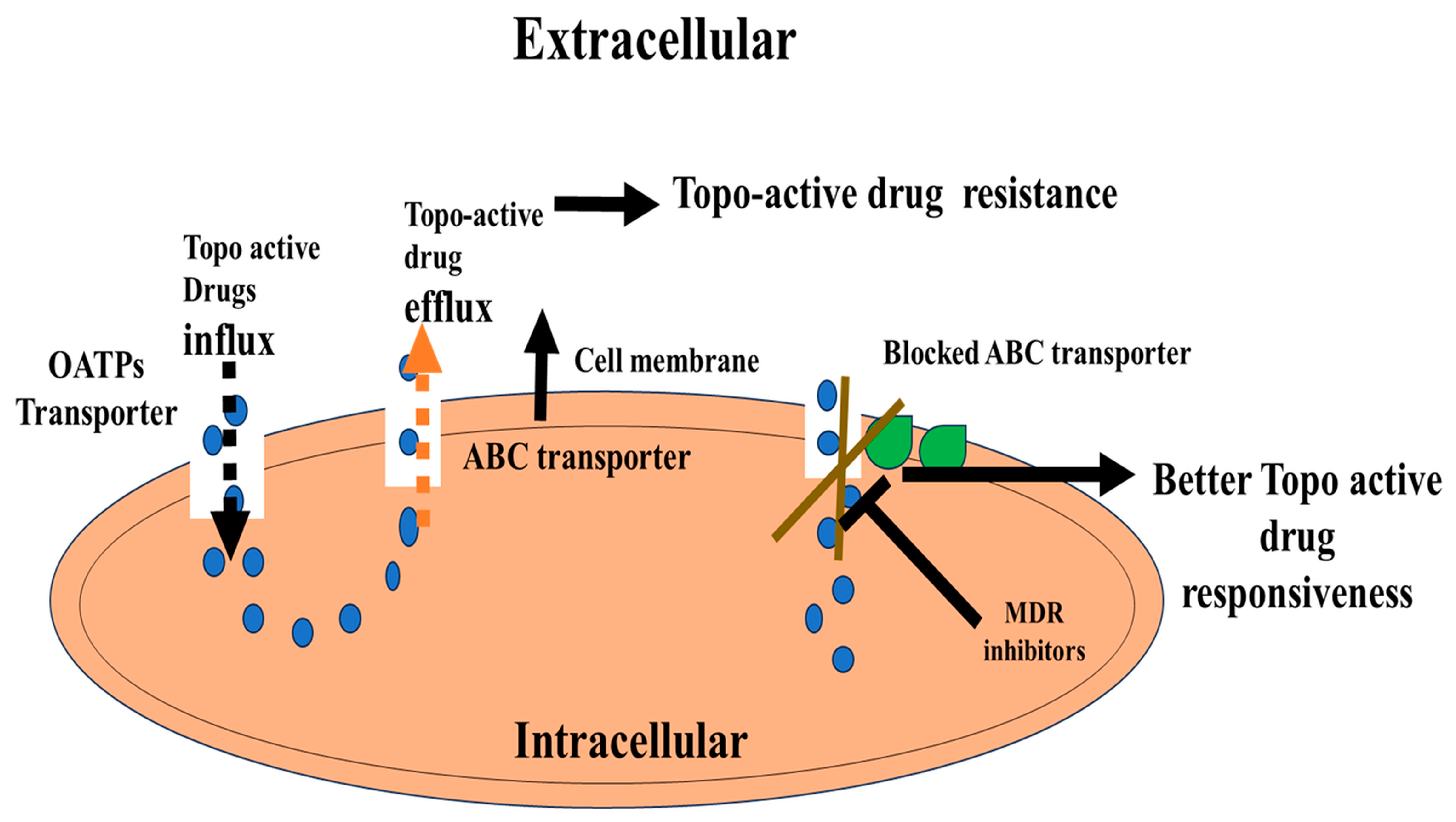
7. ABC Transporter-Mediated Resistance of Topo Drugs
8. GSH Depletion and Topo-Drug Resistance
9. DNA Damage Response Pathways and Topo-Active Drug Resistance
10. Metabolic Reprogramming and Topo-Active Drug Resistance
11. Immune Checkpoint Inhibitors and Topo-Active Drugs
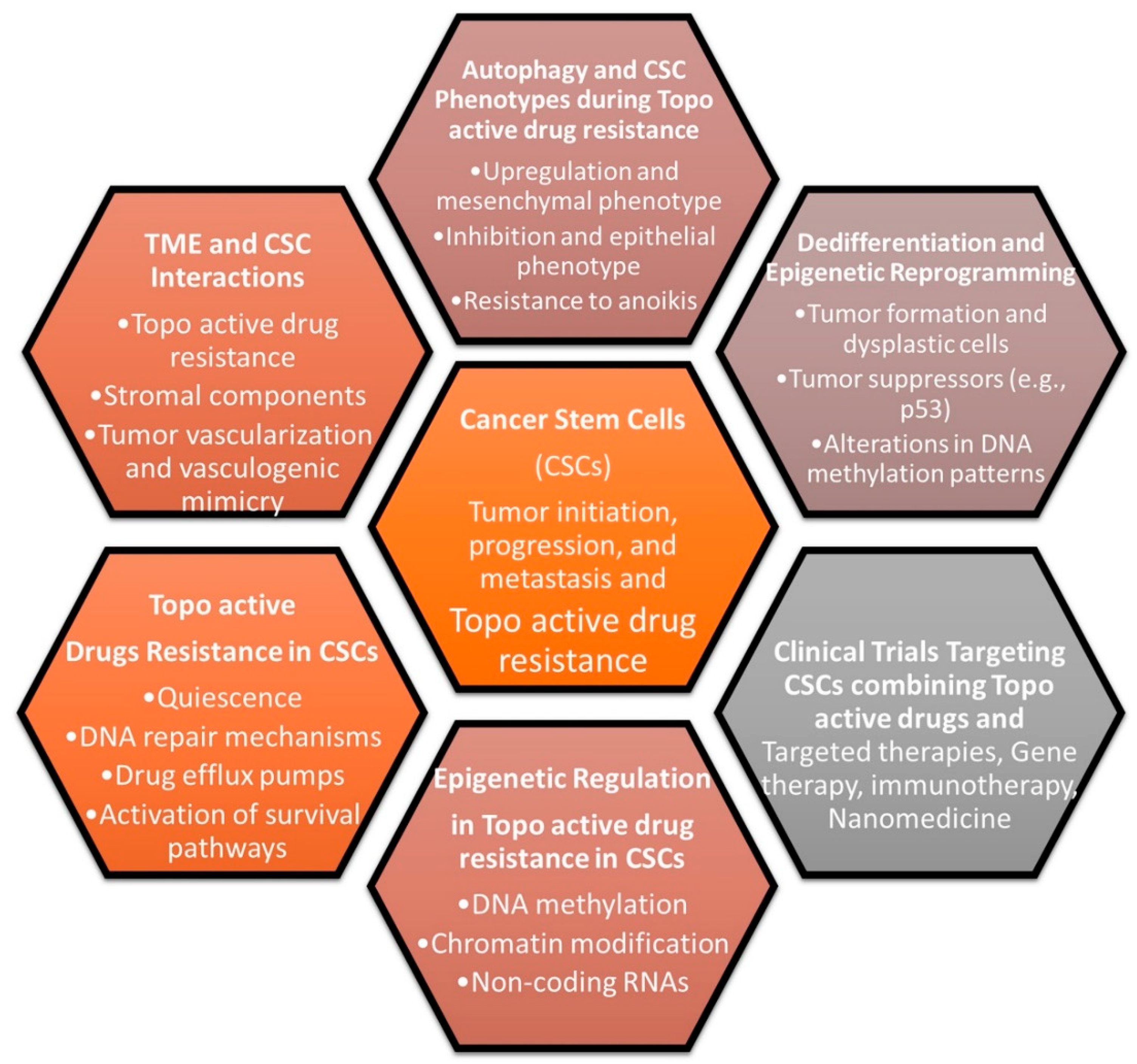
12. Topo-Active Drugs and CSCs
13. Epigenetic Changes and Topo-Active Drugs
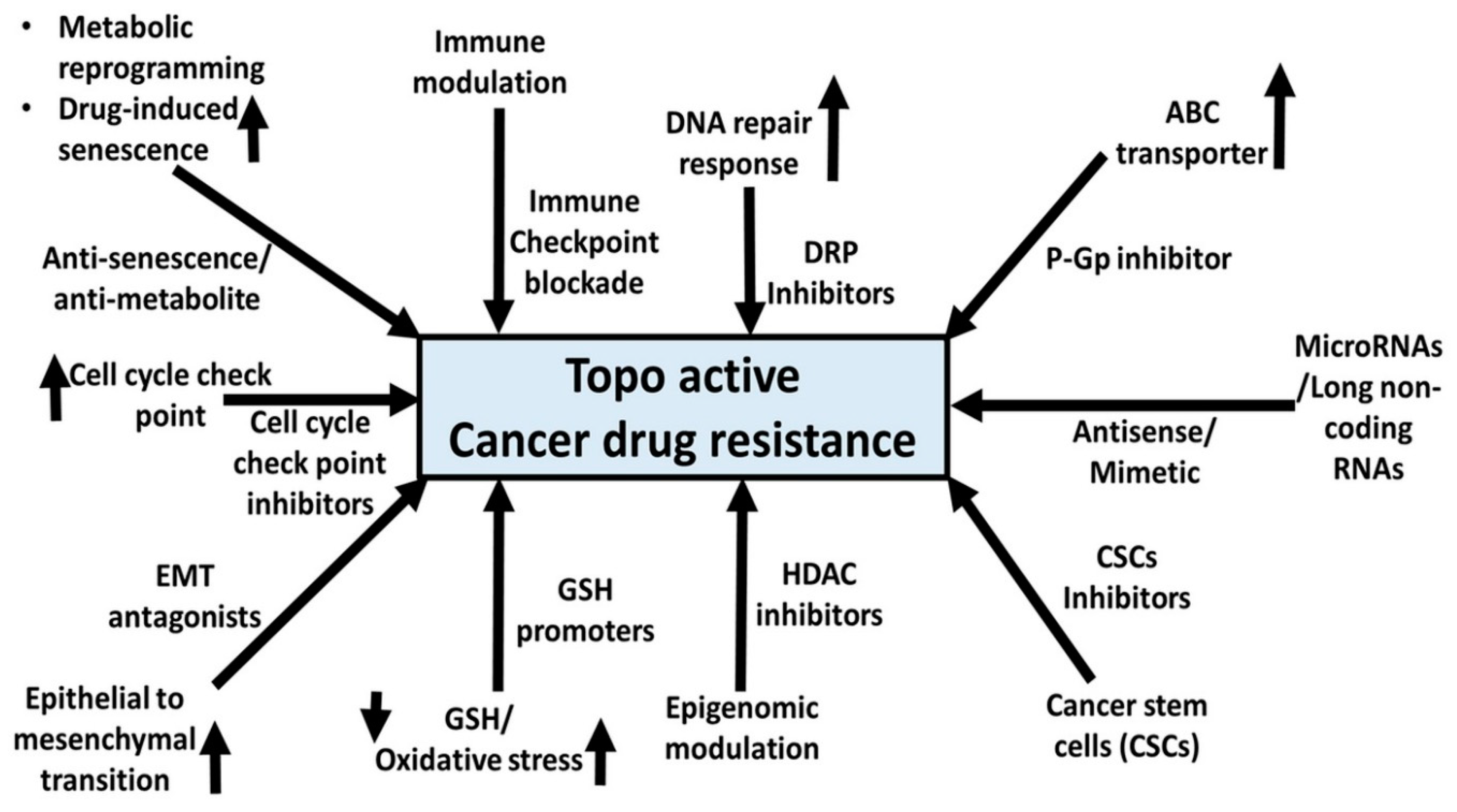
14. Miscellaneous Mechanisms of Resistance and Topo-Active Drug-Induced Senescence and Drug Resistance
14.1. Regulatory RNAs and Topo-Active Drugs
14.2. EMT and Topo-Active Drugs
15. Conclusion and Future Developments
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| ABC transporters | ATP-binding cassette transporters |
| ATM | Ataxia Telangiectasia Mutated |
| BBB | Blood–brain barrier |
| BCL-XL | B-cell lymphoma-extra large |
| CAF | Cancer-associated fibroblast |
| CAR-T | Chimeric antigen receptor T-cell therapy |
| CAT | Catalase |
| CDH2 | Cadherin 2 |
| cGAS | Cyclic GMP-AMP synthase |
| Chk2 | Checkpoint kinase 2 |
| CPT | Camptothecin |
| CRPC | Castration-resistant prostate cancer |
| CSC | Cancer stem cell |
| DOX | Doxorubicin |
| DSB | Double-strand break |
| dsDNA | Double-stranded DNA |
| ECM | Extracellular matrix |
| EV | Extracellular Vesicles |
| GSH | Glutathione |
| HDAC | Histone deacetylase |
| HGF | Hepatocyte growth factor |
| HR | Homologous recombination |
| ICIT | Immune checkpoint inhibitor therapies |
| MCH-I | Major histocompatibility class I |
| MDR1 | Multi-Drug Resistance Protein 1 |
| miRNA | microRNA |
| MMDX | Nemorubicin |
| MXT | Mitoxantrone |
| NADPH | Nicotinamide adenine dinucleotide phosphate |
| NGS | Next-generation sequencing |
| NHEJ | Non-homologous end joining |
| PARP | Poly(ADP-ribose) polymerase |
| PD-1/PD-L1 [PD-(L)1] | Programmed cell death protein 1/Programmed death-ligand 1 |
| PEG | Polyethylene glycol |
| ROS | Reactive oxygen species |
| RPA | Replication Protein A |
| SNP | Single nucleotide polymorphism |
| SOD | Superoxide dismutase |
| ssDNA | Single-stranded DNA |
| TGF | Tumor growth factor |
| TGF-β | Transforming growth factor beta |
| TME | Tumor microenvironment |
| TNBC | Triple-negative breast cancer |
| TOP1cc | Topoisomerase I cleavage complex |
| TOP3α | Topoisomerase III alpha |
| TOP3β | Topoisomerase III beta |
| TOPI | DNA topoisomerase I |
| TOPII | DNA topoisomerase II |
| TOPmt | Mitochondrial DNA topoisomerase |
| VP-16 | Etoposide |
| ZEB1 | Zinc finger E-box-binding homeobox 1 |
References
- Chhikara, B.S.; Parang, K. Global Cancer Statistics 2022: The Trends Projection Analysis. Chem. Biol. Lett. 2023, 10, 451. [Google Scholar]
- Morgan, E.; Arnold, M.; Gini, A.; Lorenzoni, V.; Cabasag, C.J.; Laversanne, M.; Vignat, J.; Ferlay, J.; Murphy, N.; Bray, F. Global Burden of Colorectal Cancer in 2020 and 2040: Incidence and Mortality Estimates from GLOBOCAN. Gut 2023, 72, 338–344. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, H.; Chen, X. Drug Resistance and Combating Drug Resistance in Cancer. Cancer Drug Resist. 2019, 2, 141–160. [Google Scholar] [CrossRef]
- Emran, T.B.; Shahriar, A.; Mahmud, A.R.; Rahman, T.; Abir, M.H.; Siddiquee, M.F.; Ahmed, H.; Rahman, N.; Nainu, F.; Wahyudin, E.; et al. Multidrug Resistance in Cancer: Understanding Molecular Mechanisms, Immunoprevention and Therapeutic Approaches. Front. Oncol. 2022, 12, 891652. [Google Scholar] [CrossRef] [PubMed]
- Vaidya, F.U.; Sufiyan Chhipa, A.; Mishra, V.; Gupta, V.K.; Rawat, S.G.; Kumar, A.; Pathak, C. Molecular and Cellular Paradigms of Multidrug Resistance in Cancer. Cancer Rep. 2022, 5, e1291. [Google Scholar] [CrossRef]
- Nikolaou, M.; Pavlopoulou, A.; Georgakilas, A.G.; Kyrodimos, E. The Challenge of Drug Resistance in Cancer Treatment: A Current Overview. Clin. Exp. Metastasis 2018, 35, 309–318. [Google Scholar] [CrossRef]
- Ramos, A.; Sadeghi, S.; Tabatabaeian, H. Battling Chemoresistance in Cancer: Root Causes and Strategies to Uproot Them. Int. J. Mol. Sci. 2021, 22, 9451. [Google Scholar] [CrossRef] [PubMed]
- You, F.; Gao, C. Topoisomerase Inhibitors and Targeted Delivery in Cancer Therapy. Curr. Top. Med. Chem. 2019, 19, 713–729. [Google Scholar] [CrossRef]
- Nitiss, J.L.; Kiianitsa, K.; Sun, Y.; Nitiss, K.C.; Maizels, N. Topoisomerase Assays. Curr. Protoc. 2021, 1, e250. [Google Scholar] [CrossRef]
- Singh, S.; Pandey, V.P.; Yadav, K.; Yadav, A.; Dwivedi, U.N. Natural Products as Anti-Cancerous Therapeutic Molecules Targeted towards Topoisomerases. Curr. Protein. Pept. Sci. 2020, 21, 1103–1142. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.Y.; Kim, D.; Kim, N.D. Recent Developments in Combination Chemotherapy for Colorectal and Breast Cancers with Topoisomerase Inhibitors. Int. J. Mol. Sci. 2023, 24, 8457. [Google Scholar] [CrossRef] [PubMed]
- Madkour, M.M.; Ramadan, W.S.; Saleh, E.; El-Awady, R. Epigenetic Modulations in Cancer: Predictive Biomarkers and Potential Targets for Overcoming the Resistance to Topoisomerase I Inhibitors. Ann. Med. 2023, 55, 2203946. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Patel, A.; Kumar, P.; Chen, Z.S. Role of ABC Transporters in Cancer Chemotherapy. Chin. J. Cancer 2012, 31, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Wtorek, K.; Długosz, A.; Janecka, A. Drug Resistance in Topoisomerase-Targeting Therapy. Postępy Hig. I Med. Doświadczalnej 2018, 72, 1073–1083. [Google Scholar] [CrossRef]
- Liu, J.; Qu, L.; Meng, L.; Shou, C. Topoisomerase Inhibitors Promote Cancer Cell Motility via ROS-Mediated Activation of JAK2-STAT1-CXCL1 Pathway. J. Exp. Clin. Cancer Res. 2019, 38, 370. [Google Scholar] [CrossRef] [PubMed]
- Nwabufo, C.K. Relevance of ABC Transporters in Drug Development. Curr. Drug. Metab. 2022, 23, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Simůnek, T.; Stérba, M.; Popelová, O.; Adamcová, M.; Hrdina, R.; Gersl, V. Anthracycline-Induced Cardiotoxicity: Overview of Studies Examining the Roles of Oxidative Stress and Free Cellular Iron. Pharmacol. Rep. 2009, 61, 154–171. [Google Scholar] [CrossRef]
- Cai, F.; Luis, M.A.F.; Lin, X.; Wang, M.; Cai, L.; Cen, C.; Biskup, E. Anthracycline-Induced Cardiotoxicity in the Chemotherapy Treatment of Breast Cancer: Preventive Strategies and Treatment. Mol. Clin. Oncol. 2019, 11, 15–23. [Google Scholar] [CrossRef]
- Kennedy, L.; Sandhu, J.K.; Harper, M.E.; Cuperlovic-Culf, M. Role of Glutathione in Cancer: From Mechanisms to Therapies. Biomolecules 2020, 10, 1429. [Google Scholar] [CrossRef]
- Sekhar, R.V. GlyNAC Supplementation Improves Glutathione Deficiency, Oxidative Stress, Mitochondrial Dysfunction, Inflammation, Aging Hallmarks, Metabolic Defects, Muscle Strength, Cognitive Decline, and Body Composition: Implications for Healthy Aging. J. Nutr. 2021, 151, 3606–3616. [Google Scholar] [CrossRef]
- Carrasco, R.; Castillo, R.L.; Gormaz, J.G.; Carrillo, M.; Thavendiranathan, P. Role of Oxidative Stress in the Mechanisms of Anthracycline-Induced Cardiotoxicity: Effects of Preventive Strategies. Oxid. Med. Cell. Longev. 2021, 2021, 8863789. [Google Scholar] [CrossRef]
- Hickman, J.A.; Potten, C.S.; Merritt, A.J.; Fisher, T.C. Apoptosis and Cancer Chemotherapy. Philos. Trans. R Soc. Lond. B Biol. Sci. 1994, 345, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S.; Pommier, Y. DNA Cleavage Assay for the Identification of Topoisomerase I Inhibitors. Nat. Protoc. 2008, 3, 1736–1750. [Google Scholar] [CrossRef] [PubMed]
- Gokduman, K. Strategies Targeting DNA Topoisomerase I in Cancer Chemotherapy: Camptothecins, Nanocarriers for Camptothecins, Organic Non-Camptothecin Compounds and Metal Complexes. Curr. Drug. Targets 2016, 17, 1928–1939. [Google Scholar] [CrossRef]
- Jurkovicova, D.; Neophytou, C.M.; Gašparović, A.Č.; Gonçalves, A.C. DNA Damage Response in Cancer Therapy and Resistance: Challenges and Opportunities. Int. J. Mol. Sci. 2022, 23, 14672. [Google Scholar] [CrossRef]
- Sharma, P.; Hu-Lieskovan, S.; Wargo, J.A.; Ribas, A. Primary, Adaptive, and Acquired Resistance to Cancer Immunotherapy. Cell 2017, 168, 707–723. [Google Scholar] [CrossRef] [PubMed]
- Jackson, C.M.; Choi, J.; Lim, M. Mechanisms of Immunotherapy Resistance: Lessons from Glioblastoma. Nat. Immunol. 2019, 20, 1100–1109. [Google Scholar] [CrossRef]
- van Elsas, M.J.; van Hall, T.; van der Burg, S.H. Future Challenges in Cancer Resistance to Immunotherapy. Cancers 2020, 12, 935. [Google Scholar] [CrossRef]
- Dragu, D.L.; Necula, L.G.; Bleotu, C.; Diaconu, C.C.; Chivu-Economescu, M. Therapies Targeting Cancer Stem Cells: Current Trends and Future Challenges. World J. Stem. Cells 2015, 7, 1185–1201. [Google Scholar] [CrossRef]
- Harris, K.S.; Shi, L.; Foster, B.M.; Mobley, M.E.; Elliott, P.L.; Song, C.J.; Watabe, K.; Langefeld, C.D.; Kerr, B.A. CD117/c-kit Defines a Prostate CSC-Like Subpopulation Driving Progression and TKI Resistance. Sci. Rep. 2021, 11, 1465. [Google Scholar] [CrossRef]
- Lei, X.; He, Q.; Li, Z.; Zou, Q.; Xu, P.; Yu, H.; Ding, Y.; Zhu, W. Cancer Stem Cells in Colorectal Cancer and the Association with Chemotherapy Resistance. Med. Oncol. 2021, 38, 43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, G.; Qin, M.; Zhang, X.; Yang, J.; Yu, H. Topotecan Induces Hepatocellular Injury via ASCT2 Mediated Oxidative Stress. Gastroenterol. Hepatol. 2021, 44, 1–12. [Google Scholar] [CrossRef] [PubMed]
- El-Far, A.H.; Tantawy, M.A.; Al Jaouni, S.K.; Mousa, S.A. Thymoquinone-Chemotherapeutic Combinations: New Regimen to Combat Cancer and Cancer Stem Cells. Naunyn Schmiedebergs Arch. Pharmacol. 2020, 393, 1581–1598. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mayea, Y.; Mir, C.; Masson, F.; Paciucci, R.; LLeonart, M.E. Insights into New Mechanisms and Models of Cancer Stem Cell Multidrug Resistance. Semin. Cancer Biol. 2020, 60, 166–180. [Google Scholar] [CrossRef] [PubMed]
- Phan, L.M.; Yeung, S.C.; Lee, M.H. Cancer Metabolic Reprogramming: Importance, Main Features, and Potentials for Precise Targeted Anti-Cancer Therapies. Cancer Biol. Med. 2014, 11, 1–19. [Google Scholar] [CrossRef] [PubMed]
- Szwed, A.; Kim, E.; Jacinto, E. Regulation and Metabolic Functions of mTORC1 and mTORC2. Physiol. Rev. 2021, 101, 1371–1426. [Google Scholar] [CrossRef]
- Faubert, B.; Solmonson, A.; DeBerardinis, R.J. Metabolic Reprogramming and Cancer Progression. Science 2020, 368, eaaw5473. [Google Scholar] [CrossRef] [PubMed]
- Wagner, A.; Kosnacova, H.; Chovanec, M.; Jurkovicova, D. Mitochondrial Genetic and Epigenetic Regulations in Cancer: Therapeutic Potential. Int. J. Mol. Sci. 2022, 23, 7897. [Google Scholar] [CrossRef]
- Karlstaedt, A.; Moslehi, J.; de Boer, R.A. Cardio-Onco-Metabolism: Metabolic Remodeling in Cardiovascular Disease and Cancer. Nat. Rev. Cardiol. 2022, 19, 414–425. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer Metabolism: Looking Forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef]
- Pommier, Y.; Kerrigan, D.; Hartman, K.D.; Glazer, R.I. Phosphorylation of Mammalian DNA Topoisomerase I and Activation by Protein Kinase C. J. Biol. Chem. 1990, 265, 9418–9422. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.F.; Rowe, T.C.; Yang, L.; Tewey, K.M.; Chen, G.L. Cleavage of DNA by Mammalian DNA Topoisomerase II. J. Biol. Chem. 1983, 258, 15365–15370. [Google Scholar] [CrossRef] [PubMed]
- Champoux, J.J. DNA topoisomerases: Structure, function, and mechanism. Annu. Rev. Biochem. 2001, 70, 369–413. [Google Scholar] [CrossRef] [PubMed]
- Nitiss, J.L. DNA topoisomerase II and its growing repertoire of biological functions. Nat. Rev. Cancer. 2009, 9, 327–337. [Google Scholar] [CrossRef] [PubMed]
- Buzun, K.; Bielawska, A.; Bielawski, K.; Gornowicz, A. DNA Topoisomerases as Molecular Targets for Anticancer Drugs. J. Enzym. Inhib. Med. Chem. 2020, 35, 1781–1799. [Google Scholar] [CrossRef]
- Sinha, B.K. Topoisomerase Inhibitors. A Review of Their Therapeutic Potential in Cancer. Drugs 1995, 49, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Robert, J.; Rivory, L. Pharmacology of Irinotecan. Drugs Today 1998, 34, 777–803. [Google Scholar] [CrossRef] [PubMed]
- Takagi, T.; Saotome, T. Chemotherapy with Irinotecan (CPT-11), a Topoisomerase-I Inhibitor, for Refractory and Relapsed Non-Hodgkin’s Lymphoma. Leuk. Lymphoma 2001, 42, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Staker, B.L.; Hjerrild, K.; Feese, M.D.; Behnke, C.A.; Burgin, A.B., Jr.; Stewart, L. The Mechanism of Topoisomerase I Poisoning by a Camptothecin Analog. Proc. Natl. Acad. Sci. USA 2002, 99, 15387–15392. [Google Scholar] [CrossRef]
- Pommier, Y.; Barcelo, J.M.; Rao, V.A.; Sordet, O.; Jobson, A.G.; Thibaut, L.; Miao, Z.H.; Seiler, J.A.; Zhang, H.; Marchand, C.; et al. Repair of Topoisomerase I-Mediated DNA Damage. Prog. Nucleic Acid. Res. Mol. Biol. 2006, 81, 179–229. [Google Scholar] [CrossRef]
- Zhang, F.L.; Wang, P.; Liu, Y.H.; Liu, L.B.; Liu, X.B.; Li, Z.; Xue, Y.X. Topoisomerase I Inhibitors, Shikonin and Topotecan, Inhibit Growth and Induce Apoptosis of Glioma Cells and Glioma Stem Cells. PLoS ONE 2013, 8, e81815. [Google Scholar] [CrossRef] [PubMed]
- Sakasai, R.; Iwabuchi, K. The Distinctive Cellular Responses to DNA Strand Breaks Caused by a DNA Topoisomerase I Poison in Conjunction with DNA Replication and RNA Transcription. Genes Genet. Syst. 2016, 90, 187–194. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Pommier, Y.; Orr, A.; Kohn, K.W.; Riou, J.F. Differential Effects of Amsacrine and Epipodophyllotoxins on Topoisomerase II Cleavage in the Human c-myc Protooncogene. Cancer Res. 1992, 52, 3125–3130. [Google Scholar]
- Danesi, R.; Agen, C.; Grandi, M.; Nardini, V.; Bevilacqua, G.; Del Tacca, M. 3’-Deamino-3’-(2-methoxy-4-morpholinyl)-doxorubicin (FCE 23762): A New Anthracycline Derivative with Enhanced Cytotoxicity and Reduced Cardiotoxicity. Eur. J. Cancer 1993, 29, 1560–1565. [Google Scholar] [CrossRef]
- Burden, D.A.; Kingma, P.S.; Froelich-Ammon, S.J.; Bjornsti, M.A.; Patchan, M.W.; Thompson, R.B.; Osheroff, N. Topoisomerase II-Etoposide Interactions Direct the Formation of Drug-Induced Enzyme-DNA Cleavage Complexes. J. Biol. Chem. 1996, 271, 29238–29244. [Google Scholar] [CrossRef] [PubMed]
- Quintieri, L.; Geroni, C.; Fantin, M.; Battaglia, R.; Rosato, A.; Speed, W.C.; Zanovello, P.; Floreani, M. Formation and Antitumor Activity of PNU-159682, A Major Metabolite of Nemorubicin in Human Liver Microsomes. Am. Assoc. Cancer Res. 2005, 11, 1608–1617. [Google Scholar] [CrossRef] [PubMed]
- McClendon, A.K.; Osheroff, N. DNA Topoisomerase II, Genotoxicity, and Cancer. Mutat. Res. 2007, 623, 83–97. [Google Scholar] [CrossRef]
- Scalabrin, M.; Quintieri, L.; Palumbo, M.; Riccardi Sirtori, F.; Gatto, B. Virtual Cross-Linking of the Active Nemorubicin Metabolite PNU-159682 to Double-Stranded DNA. Chem. Res. Toxicol. 2017, 30, 614–624. [Google Scholar] [CrossRef]
- Selas, A.; Martin-Encinas, E.; Fuertes, M.; Masdeu, C.; Rubiales, G.; Palacios, F.; Alonso, C. A Patent Review of Topoisomerase I Inhibitors (2016–Present). Expert Opin. Ther. Pat. 2021, 31, 473–508. [Google Scholar] [CrossRef]
- Sinha, B.K.; Kumar, A.; Bhattacharjee, S.; Espey, M.G.; Mason, R.P. Effect of Nitric Oxide on the Anticancer Activity of the Topoisomerase-Active Drugs Etoposide and Adriamycin in Human Melanoma Cells. J. Pharmacol. Exp. Ther. 2013, 347, 607–614. [Google Scholar] [CrossRef]
- Sharma, N.K.; Kumar, A.; Kumari, A.; Tokar, E.J.; Waalkes, M.P.; Bortner, C.D.; Williams, J.; Ehrenshaft, M.; Mason, R.P.; Sinha, B.K. Nitric Oxide Down-Regulates Topoisomerase I and Induces Camptothecin Resistance in Human Breast MCF-7 Tumor Cells. PLoS ONE 2015, 10, e0141897. [Google Scholar] [CrossRef]
- Baikar, S.; Malpathak, N. Secondary Metabolites as DNA Topoisomerase Inhibitors: A New Era towards Designing of Anticancer Drugs. Pharmacogn. Rev. 2010, 4, 12–26. [Google Scholar] [CrossRef]
- Muhammad, N.; Usmani, D.; Tarique, M.; Naz, H.; Ashraf, M.; Raliya, R.; Tabrez, S.; Zughaibi, T.A.; Alsaieedi, A.; Hakeem, I.J.; et al. The Role of Natural Products and Their Multitargeted Approach to Treat Solid Cancer. Cells 2022, 11, 2209. [Google Scholar] [CrossRef]
- Khalil, O.M.; Gedawy, E.M.; El-Malah, A.A.; Adly, M.E. Novel nalidixic acid derivatives targeting topoisomerase II enzyme; Design, synthesis, anticancer activity and effect on cell cycle profile. Bioorg. Chem. 2019, 83, 262–276. [Google Scholar] [CrossRef]
- Kizek, R.; Adam, V.; Hrabeta, J.; Eckschlager, T.; Smutny, S.; Burda, J.V.; Frei, E.; Stiborova, M. Anthracyclines and ellipticines as DNA-damaging anticancer drugs: Recent advances. Pharmacol. Ther. 2012, 133, 26–39. [Google Scholar] [CrossRef]
- Okoro, C.O.; Fatoki, T.H. A Mini Review of Novel Topoisomerase II Inhibitors as Future Anticancer Agents. Int. J. Mol. Sci. 2023, 24, 2532. [Google Scholar] [CrossRef]
- Iacopetta, D.; Rosano, C.; Puoci, F.; Parisi, O.I.; Saturnino, C.; Caruso, A.; Longo, P.; Ceramella, J.; Malzert-Fréon, A.; Dallemagne, P.; et al. Multifaceted properties of 1,4-dimethylcarbazoles: Focus on trimethoxybenzamide and trimethoxyphenylurea derivatives as novel human topoisomerase II inhibitors. Eur. J. Pharm. Sci. 2017, 96, 263–272. [Google Scholar] [CrossRef] [PubMed]
- Saturnino, C.; Caruso, A.; Iacopetta, D.; Rosano, C.; Ceramella, J.; Muià, N.; Mariconda, A.; Bonomo, M.G.; Ponassi, M.; Rosace, G.; et al. Inhibition of Human Topoisomerase II by N,N,N-Trimethylethanammonium Iodide Alkylcarbazole Derivatives. ChemMedChem 2018, 13, 2635–2643. [Google Scholar] [CrossRef] [PubMed]
- Ceramella, J.; Iacopetta, D.; Caruso, A.; Mariconda, A.; Petrou, A.; Geronikaki, A.; Rosano, C.; Saturnino, C.; Catalano, A.; Longo, P.; et al. 5,8-Dimethyl-9H-Carbazole Derivatives Blocking hTopo I Activity and Actin Dynamics. Pharmaceuticals 2023, 16, 353. [Google Scholar] [CrossRef]
- Kumar, S.; Kushwaha, P.P.; Gupta, S. Emerging Targets in Cancer Drug Resistance. Cancer Drug. Resist. 2019, 2, 161–177. [Google Scholar] [CrossRef] [PubMed]
- Vasan, N.; Baselga, J.; Hyman, D.M. A View on Drug Resistance in Cancer. Nature 2019, 575, 299–309. [Google Scholar] [CrossRef]
- Lim, Z.F.; Ma, P.C. Emerging Insights of Tumor Heterogeneity and Drug Resistance Mechanisms in Lung Cancer Targeted Therapy. J. Hematol. Oncol. 2019, 12, 134. [Google Scholar] [CrossRef]
- Galmarini, C.M.; Galmarini, F.C. Multidrug Resistance in Cancer Therapy: Role of the Microenvironment. Curr. Opin. Investig. Drugs 2003, 4, 1416–1421. [Google Scholar]
- Liu, Y.; Zhang, J.; Li, L.; Yin, G.; Zhang, J.; Zheng, S.; Cheung, H.; Wu, N.; Lu, N.; Mao, X.; et al. Genomic Heterogeneity of Multiple Synchronous Lung Cancer. Nat. Commun. 2016, 7, 13200. [Google Scholar] [CrossRef]
- Haider, T.; Pandey, V.; Banjare, N.; Gupta, P.N.; Soni, V. Drug Resistance in Cancer: Mechanisms and Tackling Strategies. Phamacol. Rep. 2020, 72, 1125–1151. [Google Scholar] [CrossRef] [PubMed]
- Maleki, E.H.; Bahrami, A.R.; Matin, M.M. Cancer Cell Cycle Heterogeneity as a Critical Determinant of Therapeutic Resistance. Genes Dis. 2023, 11, 189–204. [Google Scholar] [CrossRef]
- Asif, P.J.; Longobardi, C.; Hahne, M.; Medema, J.P. The Role of Cancer-Associated Fibroblasts in Cancer Invasion and Metastasis. Cancers 2021, 13, 4720. [Google Scholar] [CrossRef]
- Wright, K.; Ly, T.; Kriet, M.; Czirok, A.; Thomas, S.M. Cancer-Associated Fibroblasts: Master Tumor Microenvironment Modifiers. Cancers 2023, 15, 1899. [Google Scholar] [CrossRef]
- Li, M.; Li, M.; Yin, T.; Shi, H.; Wen, Y.; Zhang, B.; Chen, M.; Xu, G.; Ren, K.; Wei, Y. Targeting of Cancer-Associated Fibroblasts Enhances the Efficacy of Cancer Chemotherapy by Regulating the Tumor Microenvironment. Mol. Med. Rep. 2016, 13, 2476–2484. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between Cancer-Associated Fibroblasts and Immune Cells in the Tumor Microenvironment: New Findings and Future Perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef]
- Rimal, R.; Desai, P.; Daware, R.; Hosseinnejad, A.; Prakash, J.; Lammers, T.; Singh, S. Cancer-Associated Fibroblasts: Origin, Function, Imaging, and Therapeutic Targeting. Adv. Drug. Deliv. Rev. 2022, 189, 114504. [Google Scholar] [CrossRef] [PubMed]
- Tie, Y.; Tang, F.; Peng, D.; Zhang, Y.; Shi, H. TGF-β Signal Transduction: Biology, Function and Therapy for Diseases. Mol. Biomed. 2022, 3, 45. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Fei, Y.; Wang, H.; Hu, S.; Liu, C.; Hu, R.; Du, Q. CAFs Orchestrates Tumor Immune Microenvironment-A New Target in Cancer Therapy? Front. Pharmacol. 2023, 14, 1113378. [Google Scholar] [CrossRef]
- Beca, F.; Polyak, K. Intratumor Heterogeneity in Breast Cancer. Adv. Exp. Med. Biol. 2016, 882, 169–189. [Google Scholar] [CrossRef]
- Tong, M.; Deng, Z.; Zhang, X.; He, B.; Yang, M.; Cheng, W.; Liu, Q. New Insights from the Widening Homogeneity Perspective to Target Intratumor Heterogeneity. Cancer Commun. 2018, 38, 17. [Google Scholar] [CrossRef]
- Turajlic, S.; Sottoriva, A.; Graham, T.; Swanton, C. Resolving Genetic Heterogeneity in Cancer. Nat. Rev. Genet. 2019, 20, 404–416. [Google Scholar] [CrossRef]
- Marusyk, A.; Janiszewska, M.; Polyak, K. Intratumor Heterogeneity: The Rosetta Stone of Therapy Resistance. Cancer Cell 2020, 37, 471–484. [Google Scholar] [CrossRef] [PubMed]
- Comaills, V.; Castellano-Pozo, M. Chromosomal Instability in Genome Evolution: From Cancer to Macroevolution. Biology 2023, 12, 671. [Google Scholar] [CrossRef]
- Guo, M.; Peng, Y.; Gao, A.; Du, C.; Herman, J.G. Epigenetic Heterogeneity in Cancer. Biomark. Res. 2019, 7, 23. [Google Scholar] [CrossRef]
- Mattos, E.C.; Silva, L.P.; Valero, C.; de Castro, P.A.; Dos Reis, T.F.; Ribeiro, L.F.C.; Marten, M.R.; Silva-Rocha, R.; Westmann, C.; da Silva, C.H.T.P.; et al. The Aspergillus fumigatus Phosphoproteome Reveals Roles of High-Osmolarity Glycerol Mitogen-Activated Protein Kinases in Promoting Cell Wall Damage and Caspofungin Tolerance. mBio 2020, 11, e02962-19. [Google Scholar] [CrossRef]
- Beyes, S.; Bediaga, N.G.; Zippo, A. An Epigenetic Perspective on Intra-Tumour Heterogeneity: Novel Insights and New Challenges from Multiple Fields. Cancers 2021, 13, 4969. [Google Scholar] [CrossRef]
- Vessoni, A.T.; Filippi-Chiela, E.C.; Lenz, G.; Batista, L.F.Z. Tumor Propagating Cells: Drivers of Tumor Plasticity, Heterogeneity, and Recurrence. Oncogene 2020, 39, 2055–2068. [Google Scholar] [CrossRef]
- Biswas, A.; De, S. Drivers of Dynamic Intratumor Heterogeneity and Phenotypic Plasticity. Am. J. Physiol. Cell Physiol. 2021, 320, C750–C760. [Google Scholar] [CrossRef] [PubMed]
- Jones, P.A.; Baylin, S.B. The Fundamental Role of Epigenetic Events in Cancer. Nat. Rev. Genet. 2002, 3, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Garinis, G.A.; Patrinos, G.P.; Spanakis, N.E.; Menounos, P.G. DNA Hypermethylation: When Tumour Suppressor Genes Go Silent. Hum. Genet. 2002, 111, 115–127. [Google Scholar] [CrossRef] [PubMed]
- Vaissière, T.; Sawan, C.; Herceg, Z. Epigenetic Interplay between Histone Modifications and DNA Methylation in Gene Silencing. Mutat. Res. 2008, 659, 40–48. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Allis, C.D.; Wang, G.G. The Language of Chromatin Modification in Human Cancers. Nat. Rev. Cancer 2021, 21, 413–430. [Google Scholar] [CrossRef] [PubMed]
- McLeod, H.L.; Douglas, F.; Oates, M.; Symonds, R.P.; Prakash, D.; van der Zee, A.G.; Kaye, S.B.; Brown, R.; Keith, W.N. Topoisomerase I and II Activity in Human Breast, Cervix, Lung and Colon Cancer. Int. J. Cancer 1994, 59, 607–611. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yao, G.; Zhang, J.; Bian, J.; Li, G.; Xu, J. An Integrated Multi-Omics Analysis of Topoisomerase Family in Pan-Cancer: Friend or Foe? PLoS ONE 2022, 17, e0274546. [Google Scholar] [CrossRef]
- Gongora, C.; Vezzio-Vie, N.; Tuduri, S.; Denis, V.; Causse, A.; Auzanneau, C.; Collod-Beroud, G.; Coquelle, A.; Pasero, P.; Pourquier, P.; et al. New Topoisomerase I Mutations Are Associated with Resistance to Camptothecin. Mol. Cancer 2011, 10, 64. [Google Scholar] [CrossRef]
- Sinha, B.K.; Haim, N.; Dusre, L.; Kerrigan, D.; Pommier, Y. DNA Strand Breaks Produced by Etoposide (VP-16,213) in Sensitive and Resistant Human Breast Tumor Cells: Implications for the Mechanism of Action. Cancer Res. 1988, 48, 5096–5100. [Google Scholar]
- Beck, W.T.; Danks, M.K. Mechanisms of Resistance to Drugs That Inhibit DNA Topoisomerases. Semin. Cancer Biol. 1991, 2, 235–244. [Google Scholar]
- Pommier, Y.; Leo, E.; Zhang, H.; Marchand, C. DNA Topoisomerases and Their Poisoning by Anticancer and Antibacterial Drugs. Chem. Biol. 2010, 17, 421–433. [Google Scholar] [CrossRef] [PubMed]
- Vos, S.M.; Tretter, E.M.; Schmidt, B.H.; Berger, J.M. All Tangled Up: How Cells Direct, Manage and Exploit Topoisomerase Function. Nat. Rev. Mol. Cell Biol. 2011, 12, 827–841. [Google Scholar] [CrossRef] [PubMed]
- Pommier, Y.; Sun, Y.; Huang, S.N.; Nitiss, J.L. Roles of Eukaryotic Topoisomerases in Transcription, Replication and Genomic Stability. Nat. Rev. Mol. Cell Biol. 2016, 17, 703–721. [Google Scholar] [CrossRef]
- Boot, A.; Liu, M.; Stantial, N.; Shah, V.; Yu, W.; Nitiss, K.C.; Nitiss, J.L.; Jinks-Robertson, S.; Rozen, S.G. Recurrent Mutations in Topoisomerase IIα Cause a Previously Undescribed Mutator Phenotype in Human Cancers. Proc. Natl. Acad. Sci. USA 2022, 119, e2114024119. [Google Scholar] [CrossRef] [PubMed]
- Bandak, A.F.; Blower, T.R.; Nitiss, K.C.; Shah, V.; Nitiss, J.L.; Berger, J.M. Using Energy to Go Downhill—A Genoprotective Role for ATPase Activity in DNA Topoisomerase II. Nucleic Acids Res. 2023, gkad1157. [Google Scholar] [CrossRef] [PubMed]
- Hinds, M.; Deisseroth, K.; Mayes, J.; Altschuler, E.; Jansen, R.; Ledley, F.D.; Zwelling, L.A. Identification of a Point Mutation in the Topoisomerase II Gene from a Human Leukemia Cell Line Containing an Amsacrine-Resistant Form of Topoisomerase II. Cancer Res. 1991, 51, 4729–4731. [Google Scholar]
- Campain, J.A.; Gottesman, M.M.; Pastan, I. A Novel Mutant Topoisomerase II Alpha Present in VP-16-Resistant Human Melanoma Cell Lines Has a Deletion of Alanine 429. Biochemistry 1994, 33, 11327–11332. [Google Scholar] [CrossRef]
- Robert, J.; Larsen, A.K. Drug Resistance to Topoisomerase II Inhibitors. Biochimie 1998, 80, 247–254. [Google Scholar] [CrossRef]
- Borst, P. Genetic Mechanisms of Drug Resistance. A Review. Acta. Oncol. 1991, 30, 87–105. [Google Scholar] [CrossRef]
- Ganapathi, R.N.; Ganapathi, M.K. Mechanisms Regulating Resistance to Inhibitors of Topoisomerase II. Front. Pharmacol. 2013, 4, 89. [Google Scholar] [CrossRef]
- Licata, S.; Saponiero, A.; Mordente, A.; Minotti, G. Doxorubicin Metabolism and Toxicity in Human Myocardium: Role of Cytoplasmic Deglycosidation and Carbonyl Reduction. Chem. Res. Toxicol. 2000, 13, 414–420. [Google Scholar] [CrossRef]
- Chen, A.Y.; Chou, R.; Shih, S.J.; Lau, D.; Gandara, D. Enhancement of Radiotherapy with DNA Topoisomerase I-Targeted Drugs. Crit. Rev. Oncol. Hematol. 2004, 50, 111–119. [Google Scholar] [CrossRef]
- Kumar, A.; Patel, S.; Bhatkar, D.; Sarode, S.C.; Sharma, N.K. A Novel Method to Detect Intracellular Metabolite Alterations in MCF-7 Cells by Doxorubicin Induced Cell Death. Metabolomics 2021, 17, 3. [Google Scholar] [CrossRef]
- Potęga, A. Glutathione-Mediated Conjugation of Anticancer Drugs: An Overview of Reaction Mechanisms and Biological Significance for Drug Detoxification and Bioactivation. Molecules 2022, 27, 5252. [Google Scholar] [CrossRef]
- Sinha, B.K.; Mimnaugh, E.G. Free Radicals and Anticancer Drug Resistance: Oxygen Free Radicals in the Mechanisms of Drug Cytotoxicity and Resistance by Certain Tumors. Free Radic. Biol. Med. 1990, 8, 567–581. [Google Scholar] [CrossRef] [PubMed]
- Bartoszek, A.; Wolf, C.R. Enhancement of Doxorubicin Toxicity Following Activation by NADPH Cytochrome P450 Reductase. Biochem. Pharmacol. 1992, 43, 1449–1457. [Google Scholar] [CrossRef] [PubMed]
- Laroche-Clary, A.; Le Morvan, V.; Yamori, T.; Robert, J. Cytochrome P450 1B1 Gene Polymorphisms as Predictors of Anticancer Drug Activity: Studies with In Vitro Models. Mol. Cancer Ther. 2010, 9, 3315–3321. [Google Scholar] [CrossRef] [PubMed]
- Pathania, S.; Bhatia, R.; Baldi, A.; Singh, R.; Rawal, R.K. Drug Metabolizing Enzymes and Their Inhibitors’ Role in Cancer Resistance. Biomed. Pharmacother. 2018, 105, 53–65. [Google Scholar] [CrossRef] [PubMed]
- Mathijssen, R.H.; de Jong, F.A.; van Schaik, R.H.; Lepper, E.R.; Friberg, L.E.; Rietveld, T.; de Bruijn, P.; Graveland, W.J.; Figg, W.D.; Verweij, J.; et al. Prediction of Irinotecan Pharmacokinetics by Use of Cytochrome P450 3A4 Phenotyping Probes. J. Natl. Cancer Inst. 2004, 96, 1585–1592. [Google Scholar] [CrossRef] [PubMed]
- de Man, F.M.; Goey, A.K.L.; van Schaik, R.H.N.; Mathijssen, R.H.J.; Bins, S. Individualization of Irinotecan Treatment: A Review of Pharmacokinetics, Pharmacodynamics, and Pharmacogenetics. Clin. Pharmacokinet. 2018, 57, 1229–1254. [Google Scholar] [CrossRef]
- Riordan, J.R.; Ling, V. Purification of P-Glycoprotein from Plasma Membrane Vesicles of Chinese Hamster Ovary Cell Mutants with Reduced Colchicine Permeability. J. Biol. Chem. 1979, 254, 12701–12705. [Google Scholar] [CrossRef]
- Alpsoy, A.; Yasa, S.; Gündüz, U. Etoposide Resistance in MCF-7 Breast Cancer Cell Line Is Marked by Multiple Mechanisms. Biomed. Pharmacother. 2014, 68, 351–355. [Google Scholar] [CrossRef]
- Goldwirt, L.; Beccaria, K.; Carpentier, A.; Farinotti, R.; Fernandez, C. Irinotecan and Temozolomide Brain Distribution: A Focus on ABCB1. Cancer Chemother. Pharmacol. 2014, 74, 185–193. [Google Scholar] [CrossRef]
- Choi, Y.H.; Yu, A.M. ABC Transporters in Multidrug Resistance and Pharmacokinetics, and Strategies for Drug Development. Curr. Pharm. Des. 2014, 20, 793–807. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.; Tampé, R. Structural and Mechanistic Principles of ABC Transporters. Ann. Rev. Biochem. 2020, 89, 605–636. [Google Scholar] [CrossRef] [PubMed]
- Wei, T.; Bi, G.; Bian, Y.; Ruan, S.; Yuan, G.; Xie, H.; Zhao, M.; Shen, R.; Zhu, Y.; Wang, Q.; et al. The Significance of Secreted Phosphoprotein 1 in Multiple Human Cancers. Front. Mol. Biosci. 2020, 7, 565383. [Google Scholar] [CrossRef]
- Długosz, A.; Gach-Janczak, K.; Szymański, J.; Deredas, D.; Janecki, T.; Janecka, A. Involvement of α-Methylene-γ- and δ-Lactones in the Suppression of Multidrug Resistance in MCF-7 Cells. Pharmacol. Rep. 2018, 70, 631–638. [Google Scholar] [CrossRef]
- Zander, S.A.; Kersbergen, A.; Sol, W.; Gonggrijp, M.; van de Wetering, K.; Jonkers, J.; Borst, P.; Rottenberg, S. Lack of ABCG2 Shortens Latency of BRCA1-Deficient Mammary Tumors and This Is Not Affected by Genistein or Resveratrol. Cancer Prev. Res. 2012, 5, 1053–1060. [Google Scholar] [CrossRef]
- Natarajan, K.; Xie, Y.; Baer, M.R.; Ross, D.D. Role of Breast Cancer Resistance Protein (BCRP/ABCG2) in Cancer Drug Resistance. Biochem. Pharmacol. 2012, 83, 1084–1103. [Google Scholar] [CrossRef]
- Omori, M.; Noro, R.; Seike, M.; Matsuda, K.; Hirao, M.; Fukuizumi, A.; Takano, N.; Miyanaga, A.; Gemma, A. Inhibitors of ABCB1 and ABCG2 Overcame Resistance to Topoisomerase Inhibitors in Small Cell Lung Cancer. Thorac. Cancer. 2022, 13, 2142–2151. [Google Scholar] [CrossRef]
- Kawabata, S.; Oka, M.; Shiozawa, K.; Tsukamoto, K.; Nakatomi, K.; Soda, H.; Fukuda, M.; Ikegami, Y.; Sugahara, K.; Yamada, Y.; et al. Breast Cancer Resistance Protein Directly Confers SN-38 Resistance of Lung Cancer Cells. Biochem. Biophys. Res. Commun. 2012, 80, 1216–1223. [Google Scholar] [CrossRef]
- Nakanishi, T.; Ross, D.D. Breast Cancer Resistance Protein (BCRP/ABCG2): Its Role in Multidrug Resistance and Regulation of Its Gene Expression. Chin. J. Cancer 2012, 31, 73–99. [Google Scholar] [CrossRef]
- Bukowski, K.; Kciuk, M.; Kontek, R. Mechanisms of Multidrug Resistance in Cancer Chemotherapy. Int. J. Mol. Sci. 2020, 21, 3233. [Google Scholar] [CrossRef]
- Robey, R.W.; Pluchino, K.M.; Hall, M.D.; Fojo, A.T.; Bates, S.E.; Gottesman, M.M. Revisiting the Role of ABC Transporters in Multidrug-Resistant Cancer. Nat. Rev. Cancer 2018, 18, 452–464. [Google Scholar] [CrossRef] [PubMed]
- Bonitto, E.P.; McKeown, B.T.; Goralski, K.B. Jadomycins: A Potential Chemotherapy for Multi-Drug Resistant Metastatic Breast Cancer. Pharmacol. Res. Perspect. 2021, 9, e00886. [Google Scholar] [CrossRef] [PubMed]
- Alferiev, I.S.; Guerrero, D.T.; Guan, P.; Nguyen, F.; Kolla, V.; Soberman, D.; Pressly, B.B.; Fishbein, I.; Brodeur, G.M.; Chorny, M. Poloxamer-Linked Prodrug of a Topoisomerase I Inhibitor SN22 Shows Efficacy in Models of High-Risk Neuroblastoma with Primary and Acquired Chemoresistance. FASEB J. 2022, 36, e22213. [Google Scholar] [CrossRef] [PubMed]
- Sies, H. Glutathione and Its Role in Cellular Functions. Free Radic. Biol. Med. 1999, 27, 916–921. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Roh, J.L. Unleashing Ferroptosis in Human Cancers: Targeting Ferroptosis Suppressor Protein 1 for Overcoming Therapy Resistance. Antioxidants 2023, 12, 1218. [Google Scholar] [CrossRef] [PubMed]
- Ortega, A.L.; Mena, S.; Estrela, J.M. Glutathione in Cancer Cell Death. Cancers 2011, 3, 1285–1310. [Google Scholar] [CrossRef]
- Daga, M.; Ullio, C.; Argenziano, M.; Dianzani, C.; Cavalli, R.; Trotta, F.; Ferretti, C.; Zara, G.P.; Gigliotti, C.L.; Ciamporcero, E.S.; et al. GSH-Targeted Nanosponges Increase Doxorubicin-Induced Toxicity “In Vitro” and “In Vivo” in Cancer Cells with High Antioxidant Defenses. Free Radic. Biol. Med. 2016, 97, 24–37. [Google Scholar] [CrossRef]
- Colla, R.; Izzotti, A.; De Ciucis, C.; Fenoglio, D.; Ravera, S.; Speciale, A.; Ricciarelli, R.; Furfaro, A.L.; Pulliero, A.; Passalacqua, M.; et al. Glutathione-Mediated Antioxidant Response and Aerobic Metabolism: Two Crucial Factors Involved in Determining the Multi-Drug Resistance of High-Risk Neuroblastoma. Oncotarget 2016, 7, 70715–70737. [Google Scholar] [CrossRef]
- Cai, Y.; Zhao, B.; Liang, Q.; Zhang, Y.; Cai, J.; Li, G. The Selective Effect of Glycyrrhizin and Glycyrrhetinic Acid on Topoisomerase IIα and Apoptosis in Combination with Etoposide on Triple Negative Breast Cancer MDA-MB-231 Cells. Eur. J. Pharmacol. 2017, 809, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Deshpande, N.U.; Jayakannan, M. Cisplatin-Stitched Polysaccharide Vesicles for Synergistic Cancer Therapy of Triple Antagonistic Drugs. Biomacromolecules 2017, 18, 113–126. [Google Scholar] [CrossRef]
- Rashmi, R.; Nedungadi, D.; Podder, A.; Mishra, N.; Bhuniya, S. Monitoring of Topoisomerase (I) Inhibitor Camptothecin Release from Endogenous Redox-Stimulated GO-Polymer Hybrid Carrier. J. Photochem. Photobiol. B 2018, 189, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, S.; Gupta, P.; Nazim, U.; Ali, M.; Karadkhelkar, N.; Ahmad, M.; Chen, Z.S. Anti-Cancer Effect of Indanone-Based Thiazolyl Hydrazone Derivative on Colon Cancer Cell Lines. Int. J. Biochem. Cell Biol. 2019, 110, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Dilshara, M.G.; Jayasooriya, R.G.P.T.; Choi, Y.H.; Kim, G.Y. Camptothecin Induces c-Myc- and Sp1-Mediated hTERT Expression in LNCaP Cells: Involvement of Reactive Oxygen Species and PI3K/Akt. Food Chem. Toxicol. 2019, 127, 53–60. [Google Scholar] [CrossRef]
- Sinha, B.K.; van ‘t Erve, T.J.; Kumar, A.; Bortner, C.D.; Motten, A.G.; Mason, R.P. Synergistic Enhancement of Topotecan-Induced Cell Death by Ascorbic Acid in Human Breast MCF-7 Tumor Cells. Free Radic. Biol. Med. 2017, 113, 406–412. [Google Scholar] [CrossRef]
- Sinha, B.K.; Tokar, E.J.; Bushel, P.R. Elucidation of Mechanisms of Topotecan-Induced Cell Death in Human Breast MCF-7 Cancer Cells by Gene Expression Analysis. Front. Genet. 2020, 11, 775. [Google Scholar] [CrossRef]
- Maszczyk, M.; Rzepka, Z.; Rok, J.; Beberok, A.; Wrześniok, D. Neobavaisoflavone May Modulate the Activity of Topoisomerase Inhibitors towards U-87 MG Cells: An In Vitro Study. Molecules 2021, 26, 4516. [Google Scholar] [CrossRef]
- Shi, C.; Zhang, Z.; Xu, R.; Zhang, Y.; Wang, Z. Contribution of HIF-1α/BNIP3-Mediated Autophagy to Lipid Accumulation during Irinotecan-Induced Liver Injury. Sci. Rep. 2023, 13, 6528. [Google Scholar] [CrossRef]
- Al-Karmalawy, A.A.; Rashed, M.; Sharaky, M.; Abulkhair, H.S.; Hammouda, M.M.; Tawfik, H.O.; Shaldam, M.A. Novel Fused Imidazotriazines Acting as Promising Top. II Inhibitors and Apoptotic Inducers with Greater Selectivity Against Head and Neck Tumors: Design, Synthesis, and Biological Assessments. Eur. J. Med. Chem. 2023, 259, 115661. [Google Scholar] [CrossRef]
- Nayak, J.; Seshu Vardhan, P.; Sahoo, S.K.; Kumar, M.; Vashistha, V.K.; Kumar, R. Computational Insight of Antioxidant and Doxorubicin Combination for Effective Cancer Therapy. J. Biomol. Struct. Dyn. 2023, 41, 2242507. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Costa, M. The Role of NUPR1 in Response to Stress and Cancer Development. Toxicol. Appl. Pharmacol. 2022, 454, 116244. [Google Scholar] [CrossRef] [PubMed]
- Balaji, S.; Neupane, R.; Malla, S.; Khupse, R.; Amawi, H.; Kumari, S.; Tukaramrao, D.B.; Chattopadhyay, S.; Ashby, C.R., Jr.; Boddu, S.H.S.; et al. IND-2, a Quinoline Derivative, Inhibits the Proliferation of Prostate Cancer Cells by Inducing Oxidative Stress, Apoptosis and Inhibiting Topoisomerase II. Life 2022, 12, 1879. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, J.; Du, W.; Mickelsen, D.M.; Shi, H.; Yu, H.; Kumar, S.; Yan, C. PDE10A Inactivation Prevents Doxorubicin-Induced Cardiotoxicity and Tumor Growth. Circ. Res. 2023, 133, 138–157. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fan, Z.; Wang, F.; Yin, L.; Wu, J.; Li, D.; Song, S.; Wang, X.; Tang, Y.; Huang, C. Tubeimoside I Ameliorates Doxorubicin-Induced Cardiotoxicity by Upregulating SIRT3. Oxid. Med. Cell Longev. 2023, 2023, 9966355. [Google Scholar] [CrossRef] [PubMed]
- Selvakumaran, M.; Pisarcik, D.A.; Bao, R.; Yeung, A.T.; Hamilton, T.C. Enhanced Cisplatin Cytotoxicity by Disturbing the Nucleotide Excision Repair Pathway in Ovarian Cancer Cell Lines. Cancer Res. 2003, 63, 1311–1316. [Google Scholar]
- Mountzios, G.; Dimopoulos, M.A.; Papadimitriou, C. Excision Repair Cross-Complementation Group 1 Enzyme as a Molecular Determinant of Responsiveness to Platinum-Based Chemotherapy for Non Small-Cell Lung Cancer. Biomark. Insights 2008, 3, 219–226. [Google Scholar] [CrossRef]
- Curtin, N.J. DNA Repair Dysregulation from Cancer Driver to Therapeutic Target. Nat. Rev. Cancer 2012, 12, 801–817. [Google Scholar] [CrossRef]
- Awwad, S.W.; Serrano-Benitez, A.; Thomas, J.C.; Gupta, V.; Jackson, S.P. Revolutionizing DNA Repair Research and Cancer Therapy with CRISPR-Cas Screens. Nat. Rev. Mol. Cell. Biol. 2023, 24, 477–494. [Google Scholar] [CrossRef]
- Rocha, J.C.; Busatto, F.F.; Guecheva, T.N.; Saffi, J. Role of Nucleotide Excision Repair Proteins in Response to DNA Damage Induced by Topoisomerase II Inhibitors. Mutat. Res. Rev. Mutat. Res. 2016, 768, 68–77. [Google Scholar] [CrossRef]
- Carlsen, L.; El-Deiry, W.S. Anti-Cancer Immune Responses to DNA Damage Response Inhibitors: Molecular Mechanisms and Progress Toward Clinical Translation. Front. Oncol. 2022, 12, 998388. [Google Scholar] [CrossRef] [PubMed]
- Foo, T.K.; Xia, B. BRCA1-Dependent and Independent Recruitment of PALB2-BRCA2-RAD51 in the DNA Damage Response and Cancer. Cancer Res. 2022, 82, 3191–3197. [Google Scholar] [CrossRef]
- da Costa, A.A.B.A.; Chowdhury, D.; Shapiro, G.I.; D’Andrea, A.D.; Konstantinopoulos, P.A. Targeting Replication Stress in Cancer Therapy. Nat. Rev. Drug. Discov. 2023, 22, 38–58. [Google Scholar] [CrossRef]
- Groelly, F.J.; Fawkes, M.; Dagg, R.A.; Blackford, A.N.; Tarsounas, M. Targeting DNA Damage Response Pathways in Cancer. Nat. Rev. Cancer 2023, 23, 78–94. [Google Scholar] [CrossRef]
- Saha, L.K.; Saha, S.; Yang, X.; Huang, S.N.; Sun, Y.; Jo, U.; Pommier, Y. Replication-Associated Formation and Repair of Human Topoisomerase IIIα Cleavage Complexes. Nat. Commun. 2023, 14, 1925. [Google Scholar] [CrossRef] [PubMed]
- Dexheimer, T.S.; Antony, S.; Marchand, C.; Pommier, Y. Tyrosyl-DNA Phosphodiesterase as a Target for Anticancer Therapy. Anticancer. Agents Med. Chem. 2008, 8, 381–389. [Google Scholar] [CrossRef] [PubMed]
- Caldecott, K.W. DNA Single-Strand Break Repair and Human Genetic Disease. Trends Cell Biol. 2022, 32, 733–745. [Google Scholar] [CrossRef]
- Okamoto, R.; Takano, H.; Okamura, T.; Park, J.S.; Tanimoto, K.; Sekikawa, T.; Yamamoto, W.; Sparreboom, A.; Verweij, J.; Nishiyama, M. O(6)-Methylguanine-DNA Methyltransferase (MGMT) as a Determinant of Resistance to Camptothecin Derivatives. Jpn. J. Cancer Res. 2002, 93, 93–102. [Google Scholar] [CrossRef]
- Xu, Y.; Villalona-Calero, M.A. Irinotecan: Mechanisms of Tumor Resistance and Novel Strategies for Modulating Its Activity. Ann. Oncol. 2002, 13, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Okafuji, M.; Traganos, F.; Luther, E.; Holden, E.; Darzynkiewicz, Z. Assessment of Histone H2AX Phosphorylation Induced by DNA Topoisomerase I and II Inhibitors Topotecan and Mitoxantrone and by the DNA Cross-Linking Agent Cisplatin. Cytom. A 2004, 58, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Di Leo, A.; Claudino, W.M.; Pestrin, M.; Licitra, S.; Biganzoli, L. Using Specific Cytotoxics with a Targeted Mind. Breast 2007, 16 (Suppl. S2), S120–S126. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, J.; Feng, S.; Zhao, T.; Li, Z.; Wang, L.; Wang, P.; Du, H.; Yuan, S.; Sun, L. A Novel Camptothecin Derivative 3j Inhibits NSCLC Proliferation via Induction of Cell Cycle Arrest by Topo I-Mediated DNA Damage. Anticancer. Agents Med. Chem. 2019, 19, 365–374. [Google Scholar] [CrossRef]
- Huang, X.; Kurose, A.; Tanaka, T.; Traganos, F.; Dai, W.; Darzynkiewicz, Z. Activation of ATM and Histone H2AX Phosphorylation Induced by Mitoxantrone but Not by Topotecan Is Prevented by the Antioxidant N-Acetyl-L-Cysteine. Cancer Biol. Ther. 2006, 5, 959–964. [Google Scholar] [CrossRef]
- Zhao, H.; Traganos, F.; Darzynkiewicz, Z. Phosphorylation of p53 on Ser15 during Cell Cycle Caused by Topo I and Topo II Inhibitors in Relation to ATM and Chk2 Activation. Cell Cycle 2008, 7, 3048–3055. [Google Scholar] [CrossRef]
- Peleg, R.; Bobilev, D.; Priel, E. Topoisomerase I as a Target of Erlotinib and Gefitinib: Efficacy of Combined Treatments with Camptothecin. Int. J. Oncol. 2014, 44, 934–942. [Google Scholar] [CrossRef]
- Ranganathan, P.; Kashyap, T.; Yu, X.; Meng, X.; Lai, T.H.; McNeil, B.; Bhatnagar, B.; Shacham, S.; Kauffman, M.; Dorrance, A.M.; et al. XPO1 Inhibition using Selinexor Synergizes with Chemotherapy in Acute Myeloid Leukemia by Targeting DNA Repair and Restoring Topoisomerase IIα to the Nucleus. Clin. Cancer Res. 2016, 22, 6142–6152. [Google Scholar] [CrossRef]
- Nateewattana, J.; Dutta, S.; Reabroi, S.; Saeeng, R.; Kasemsook, S.; Chairoungdua, A.; Weerachayaphorn, J.; Wongkham, S.; Piyachaturawat, P. Induction of Apoptosis in Cholangiocarcinoma by an Andrographolide Analogue Is Mediated through Topoisomerase II Alpha Inhibition. Eur. J. Pharmacol. 2014, 723, 148–155. [Google Scholar] [CrossRef]
- Yuan, Z.; Chen, S.; Chen, C.; Chen, J.; Chen, C.; Dai, Q.; Gao, C.; Jiang, Y. Design, Synthesis and Biological Evaluation of 4-Amidobenzimidazole Acridine Derivatives as Dual PARP and Topo Inhibitors for Cancer Therapy. Eur. J. Med. Chem. 2017, 138, 1135–1146. [Google Scholar] [CrossRef]
- de Camargo, M.S.; De Grandis, R.A.; da Silva, M.M.; da Silva, P.B.; Santoni, M.M.; Eismann, C.E.; Menegário, A.A.; Cominetti, M.R.; Zanelli, C.F.; Pavan, F.R.; et al. Determination of in Vitro Absorption in Caco-2 Monolayers of Anticancer Ru(II)-Based Complexes Acting as Dual Human Topoisomerase and PARP Inhibitors. Biometals 2019, 32, 89–100. [Google Scholar] [CrossRef]
- Baglini, E.; Salerno, S.; Barresi, E.; Robello, M.; Da Settimo, F.; Taliani, S.; Marini, A.M. Multiple Topoisomerase I (TopoI), Topoisomerase II (TopoII) and Tyrosyl-DNA Phosphodiesterase (TDP) Inhibitors in the Development of Anticancer Drugs. Eur. J. Pharm. Sci. 2021, 156, 105594. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, Y.; Wang, Y.; Han, Q.; Yan, H.; Yang, T.; Guo, Z.; Wang, X. Platinum Complexes as Inhibitors of DNA Repair Protein Ku70 and Topoisomerase IIα in Cancer Cells. Dalton Trans. 2022, 51, 3188–3197. [Google Scholar] [CrossRef]
- Boichuk, S.; Bikinieva, F.; Mustafin, I.; Zykova, S.; Ryzkin, S.; Galembikova, A. 2-Amino-Pyrrole-Carboxylate Attenuates Homology-Mediated DNA Repair and Sensitizes Cancer Cells to Doxorubicin. Biochemistry 2022, 87, 391–399. [Google Scholar] [CrossRef]
- Collins, A.; Møller, P.; Gajski, G.; Vodenková, S.; Abdulwahed, A.; Anderson, D.; Bankoglu, E.E.; Bonassi, S.; Boutet-Robinet, E.; Brunborg, G.; et al. Measuring DNA Modifications with the Comet Assay: A Compendium of Protocols. Nat. Protoc. 2023, 18, 929–989. [Google Scholar] [CrossRef] [PubMed]
- Petrella, G.; Corsi, F.; Ciufolini, G.; Germini, S.; Capradossi, F.; Pelliccia, A.; Torino, F.; Ghibelli, L.; Cicero, D.O. Metabolic Reprogramming of Castration-Resistant Prostate Cancer Cells as a Response to Chemotherapy. Metabolites 2022, 13, 65. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Balan, M.; Sabarwal, A.; Choueiri, T.K.; Pal, S. Metabolic Reprogramming in Renal Cancer: Events of a Metabolic Disease. Biochim. Biophys. Acta. Rev. Cancer 2021, 1876, 188559. [Google Scholar] [CrossRef]
- Chen, M.; Lan, H.; Yao, S.; Jin, K.; Chen, Y. Metabolic Interventions in Tumor Immunity: Focus on Dual Pathway Inhibitors. Cancers 2023, 15, 2043. [Google Scholar] [CrossRef] [PubMed]
- Xiang, H.; Yang, R.; Tu, J.; Xi, Y.; Yang, S.; Lv, L.; Zhai, X.; Zhu, Y.; Dong, D.; Tao, X. Metabolic Reprogramming of Immune Cells in Pancreatic Cancer Progression. Biomedicines 2023, 13, 2043. [Google Scholar] [CrossRef] [PubMed]
- Kang, H.; Kim, H.; Lee, S.; Youn, H.; Youn, B. Role of Metabolic Reprogramming in Epithelial–Mesenchymal Transition (EMT). Int. J. Mol. Sci. 2019, 20, 2042. [Google Scholar] [CrossRef]
- Naidoo, J.; Li, B.T.; Schindler, K.; Page, D.B. What Does the Future Hold for Immunotherapy in Cancer? Ann. Transl. Med. 2016, 4, 177. [Google Scholar] [CrossRef]
- Vermeulen, J.F.; Van Hecke, W.; Adriaansen, E.J.M.; Jansen, M.K.; Bouma, R.G.; Villacorta Hidalgo, J.; Fisch, P.; Broekhuizen, R.; Spliet, W.G.M.; Kool, M.; et al. Prognostic Relevance of Tumor-Infiltrating Lymphocytes and Immune Checkpoints in Pediatric Medulloblastoma. Oncoimmunology 2017, 7, e1398877. [Google Scholar] [CrossRef]
- Toor, S.M.; Nair, V.S.; Decock, J.; Elkord, E. Immune Checkpoints in the Tumor Microenvironment. Semin. Cancer Biol. 2020, 65, 140–149. [Google Scholar] [CrossRef]
- Byun, D.J.; Wolchok, J.D.; Rosenberg, L.M.; Girotra, M. Cancer Immunotherapy—Immune Checkpoint Blockade and Associated Endocrinopathies. Nat. Rev. Endocrinol. 2017, 13, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Beyersdorf, N.; Kerkau, T.; Hünig, T. CD28 Co-Stimulation in T-Cell Homeostasis: Recent Perspective. Immunotargets Ther. 2015, 4, 111–122. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Sznol, M.; McDermott, D.F.; Kluger, H.M.; Carvajal, R.D.; Sharfman, W.H.; Brahmer, J.R.; Lawrence, D.P.; Atkins, M.B.; Powderly, J.D.; et al. Survival, Durable Tumor Remission, and Long-Term Safety in Patients with Advanced Melanoma Receiving Nivolumab. J. Clin. Oncol. 2014, 32, 1020–1030. [Google Scholar] [CrossRef]
- Latchman, Y.; Wood, C.R.; Chernova, T.; Chaudhary, D.; Borde, M.; Chernova, I.; Iwai, Y.; Long, A.J.; Brown, J.A.; Nunes, R.; et al. PD-L2 Is a Second Ligand for PD-1 and Inhibits T Cell Activation. Nat. Immunol. 2001, 2, 261–268. [Google Scholar] [CrossRef] [PubMed]
- Topalian, S.L.; Drake, C.G.; Pardoll, D.M. Targeting the PD-1/B7-H1(PD-L1) Pathway to Activate Anti-Tumor Immunity. Curr. Opin. Immunol. 2012, 24, 207–212. [Google Scholar] [CrossRef]
- Ramsay, A.G. Immune Checkpoint Blockade Immunotherapy to Activate Anti-Tumour T-Cell Immunity. Br. J. Haematol. 2013, 162, 313–325. [Google Scholar] [CrossRef]
- Han, Y.; Liu, D.; Li, L. PD-1/PD-L1 Pathway: Current Researches in Cancer. Am. J. Cancer Res. 2020, 10, 727–742. [Google Scholar] [PubMed]
- Mishima, S.; Shitara, K. Trastuzumab Deruxtecan for the Treatment of HER2-Positive Gastric Cancer. Expert Opin. Biol. Ther. 2021, 21, 825–830. [Google Scholar] [CrossRef] [PubMed]
- Cai, S.; Chen, Z.; Wang, Y.; Wang, M.; Wu, J.; Tong, Y.; Chen, L.; Lu, C.; Yang, H. Reducing PD-L1 Expression with a Self-Assembled Nanodrug: An Alternative to PD-L1 Antibody for Enhanced Chemo-Immunotherapy. Theranostics 2021, 11, 1970–1981. [Google Scholar] [CrossRef] [PubMed]
- Chung, Y.M.; Khan, P.P.; Wang, H.; Tsai, W.B.; Qiao, Y.; Yu, B.; Larrick, J.W.; Hu, M.C. Sensitizing Tumors to Anti-PD-1 Therapy by Promoting NK and CD8+ T Cells via Pharmacological Activation of FOXO3. J. Immunother. Cancer 2021, 9, e002772. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Hua, S.; Wang, X.; Chen, F.; Gou, S. The Introduction of Immunosuppresso (TDO Inhibitor) Significantly Improved the Efficacy of Irinotecan in Treating Hepatocellular Carcinoma. Cancer Immunol. Immunother. 2021, 70, 497–508. [Google Scholar] [CrossRef] [PubMed]
- McKenzie, J.A.; Mbofung, R.M.; Malu, S.; Zhang, M.; Ashkin, E.; Devi, S.; Williams, L.; Tieu, T.; Peng, W.; Pradeep, S.; et al. The Effect of Topoisomerase I Inhibitors on the Efficacy of T-Cell-Based Cancer Immunotherapy. J. Natl. Cancer Inst. 2018, 110, 777–786. [Google Scholar] [CrossRef] [PubMed]
- Goldenberg, D.M.; Sharkey, R.M. Sacituzumab Govitecan, a Novel, Third-Generation, Antibody-Drug Conjugate (ADC) for Cancer Therapy. Expert Opin. Biol. Ther. 2020, 20, 871–885. [Google Scholar] [CrossRef]
- Bardia, A.; Hurvitz, S.A.; Tolaney, S.M.; Loirat, D.; Punie, K.; Oliveira, M.; Brufsky, A.; Sardesai, S.D.; Kalinsky, K.; Zelnak, A.B.; et al. ASCENT Clinical Trial Investigators. Sacituzumab Govitecan in Metastatic Triple-Negative Breast Cancer. N. Engl. J. Med. 2021, 384, 1529–1541. [Google Scholar] [CrossRef]
- Zhou, Y.; Bastian, I.N.; Long, M.D.; Dow, M.; Li, W.; Liu, T.; Ngu, R.K.; Antonucci, L.; Huang, J.Y.; Phung, Q.T.; et al. Activation of NF-κB and p300/CBP Potentiates Cancer Chemoimmunotherapy through Induction of MHC-I Antigen Presentation. Proc. Natl. Acad. Sci. USA 2021, 118, e2025840118. [Google Scholar] [CrossRef]
- Hassan, M.; Trung, V.; Bedi, D.; Shaddox, S.; Gunturu, D.; Yates, C.; Datta, P.; Samuel, T. Interference with Pathways Activated by Topoisomerase Inhibition Alters the Surface Expression of PD-L1 and MHC I in Colon Cancer Cells. Oncol. Lett. 2022, 25, 41. [Google Scholar] [CrossRef]
- Hao, X.; Zhao, B.; Zhou, W.; Liu, H.; Fukumoto, T.; Gabrilovich, D.; Zhang, R. Sensitization of Ovarian Tumor to Immune Checkpoint Blockade by Boosting Senescence-Associated Secretory Phenotype. iScience 2020, 24, 102016. [Google Scholar] [CrossRef]
- Vitiello, A.; Ferrara, F.; Lasala, R.; Zovi, A. Precision Medicine in the Treatment of Locally Advanced or Metastatic Urothelial Cancer: New Molecular Targets and Pharmacological Therapies. Cancers 2022, 14, 5167. [Google Scholar] [CrossRef]
- Bedi, D.; Henderson, H.J.; Manne, U.; Samuel, T. Camptothecin Induces PD-L1 and Immunomodulatory Cytokines in Colon Cancer Cells. Medicines 2019, 6, 51. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Hu, J.; Zhang, H.; Xu, F.; He, W.; Wang, X.; Li, M.; Lu, W.; Zeng, G.; et al. cGAS/STING Axis Mediates a Topoisomerase II Inhibitor-Induced Tumor Immunogenicity. J. Clin. Investig. 2019, 129, 4850–4862. [Google Scholar] [CrossRef]
- Li, X.; Li, Y.; Zhao, Z.; Miao, N.; Liu, G.; Deng, L.; Wei, S.; Hou, J. Immunogenicity of Small-Cell Lung Cancer Associates with STING Pathway Activation and Is Enhanced by AT and TOP1 Inhibition. Cancer Med. 2023, 12, 4864–4881. [Google Scholar] [CrossRef] [PubMed]
- Mohan, N.; Jiang, J.; Dokmanovic, M.; Wu, W.J. Trastuzumab-Mediated Cardiotoxicity: Current Understanding, Challenges, and Frontiers. Antib. Ther. 2018, 1, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Tan, X.; Fang, P.; Li, K.; You, M.; Cao, Y.; Xu, H.; Zhu, X.; Wang, L.; Wei, X.; Wen, H.; et al. A HER2-Targeted Antibody-Novel DNA Topoisomerase I Inhibitor Conjugate Induces Durable Adaptive Antitumor Immunity by Activating Dendritic Cells. MAbs 2023, 15, 2220466. [Google Scholar] [CrossRef]
- Liu, L.; Kshirsagar, P.G.; Gautam, S.K.; Gulati, M.; Wafa, E.I.; Christiansen, J.C.; White, B.M.; Mallapragada, S.K.; Wannemuehler, M.J.; Kumar, S.; et al. Nanocarriers for Pancreatic Cancer Imaging, Treatments, and Immunotherapies. Theranostics 2022, 12, 1030–1060. [Google Scholar] [CrossRef]
- Cao, J.; Huang, D.; Peppas, N.A. Advanced Engineered Nanoparticulate Platforms to Address Key Biological Barriers for Delivering Chemotherapeutic Agents to Target Sites. Adv. Drug Deliv. Rev. 2020, 167, 170–188. [Google Scholar] [CrossRef]
- Bahmani, B.; Uehara, M.; Ordikhani, F.; Li, X.F.; Jiang, L.W.; Banouni, N.; Ichimura, T.; Kasinath, V.; Eskandari, S.K.; Annabi, N.; et al. Ectopic High Endothelial Venules in Pancreatic Ductal Adenocarcinoma: A Unique Site for Targeted Delivery. EBioMedicine 2018, 38, 79–88. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Li, F.; Li, Y.; Wang, H.; Ren, H.; Chen, J.; Nie, G.; Hao, J. Co-Delivery of HIF1α siRNA and Gemcitabine via Biocompatible Lipid-Polymer Hybrid Nanoparticles for Effective Treatment of Pancreatic Cancer. Biomaterials 2015, 46, 13–25. [Google Scholar] [CrossRef]
- Mittal, A.; Chitkara, D.; Behrman, S.W.; Mahato, R.I. Efficacy of Gemcitabine Conjugated and miRNA-205 Complexed Micelles for Treatment of Advanced Pancreatic Cancer. Biomaterials 2014, 35, 7077–7087. [Google Scholar] [CrossRef]
- Li, Y.; Wang, Z.; Ajani, J.A.; Song, S. Drug Resistance and Cancer Stem Cells. Cell Commun. Signal 2021, 19, 19. [Google Scholar] [CrossRef]
- Hsu, J.M.; Xia, W.; Hsu, Y.H.; Chan, L.C.; Yu, W.H.; Cha, J.H.; Chen, C.T.; Liao, H.W.; Kuo, C.W.; Khoo, K.H.; et al. STT3-Dependent PD-L1 Accumulation on Cancer Stem Cells Promotes Immune Evasion. Nat. Commun. 2018, 9, 1908. [Google Scholar] [CrossRef] [PubMed]
- Schulten, H.J.; Hussein, D. Array Expression Meta-Analysis of Cancer Stem Cell Genes Identifies Upregulation of PODXL Especially in DCC Low Expression Meningiomas. PLoS ONE 2019, 14, e0215452. [Google Scholar] [CrossRef]
- Lee, P.J.; Ho, C.C.; Ho, H.; Chen, W.J.; Lin, C.H.; Lai, Y.H.; Juan, Y.C.; Chu, W.C.; Lee, J.H.; Su, S.F.; et al. Tumor Microenvironment-Based Screening Repurposes Drugs Targeting Cancer Stem Cells and Cancer-Associated Fibroblasts. Theranostics 2021, 11, 9667–9686. [Google Scholar] [CrossRef] [PubMed]
- Maroufi, N.F.; Amiri, M.; Dizaji, B.F.; Vahedian, V.; Akbarzadeh, M.; Roshanravan, N.; Haiaty, S.; Nouri, M.; Rashidi, M.R. Inhibitory Effect of Melatonin on Hypoxia-Induced Vasculogenic Mimicry via Suppressing Epithelial-Mesenchymal Transition (EMT) in Breast Cancer Stem Cells. Eur. J. Pharmacol. 2020, 881, 173282. [Google Scholar] [CrossRef]
- Nicolay, N.H.; Rühle, A.; Perez, R.L.; Trinh, T.; Sisombath, S.; Weber, K.J.; Schmezer, P.; Ho, A.D.; Debus, J.; Saffrich, R.; et al. Mesenchymal Stem Cells Exhibit Resistance to Topoisomerase Inhibition. Cancer Lett. 2016, 374, 75–84. [Google Scholar] [CrossRef]
- Hong, Y.; Sang, M.; Shang, C.; Xue, Y.X.; Liu, Y.H. Quantitative Analysis of Topoisomerase II Alpha and Evaluation of Its Effects on Cell Proliferation and Apoptosis in Glioblastoma Cancer Stem Cells. Neurosci. Lett. 2012, 518, 138–143. [Google Scholar] [CrossRef] [PubMed]
- Peleg, R.; Romzova, M.; Kogan-Zviagin, I.; Apte, R.N.; Priel, E. Modification of Topoisomerases in Mammospheres Derived from Breast Cancer Cell Line: Clinical Implications for Combined Treatments with Tyrosine Kinase Inhibitors. BMC Cancer 2014, 14, 910. [Google Scholar] [CrossRef] [PubMed]
- Conley, S.J.; Baker, T.L.; Burnett, J.P.; Theisen, R.L.; Lazarus, D.; Peters, C.G.; Clouthier, S.G.; Eliasof, S.; Wicha, M.S. CRLX101, an Investigational Camptothecin-Containing Nanoparticle-Drug Conjugate, Targets Cancer Stem Cells and Impedes Resistance to Antiangiogenic Therapy in Mouse Models of Breast Cancer. Breast Cancer Res. Treat 2015, 150, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Boichuk, S.; Dunaev, P.; Mustafin, I.; Mani, S.; Syuzov, K.; Valeeva, E.; Bikinieva, F.; Galembikova, A. Infigratinib (BGJ 398), a Pan-FGFR Inhibitor, Targets P-Glycoprotein and Increases Chemotherapeutic-Induced Mortality of Multidrug-Resistant Tumor Cells. Biomedicines 2022, 10, 601. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Agarwal, P.; Zhao, S.; Xu, R.X.; Yu, J.; Lu, X.; He, X. Hyaluronic Acid-Decorated Dual Responsive Nanoparticles of Pluronic F127, PLGA, and Chitosan for Targeted Co-Delivery of Doxorubicin and Irinotecan to Eliminate Cancer Stem-Like Cells. Biomaterials 2015, 72, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Ngo, S.N.T.; Williams, D.B. Protective Effect of Isothiocyanates from Cruciferous Vegetables on Breast Cancer: Epidemiological and Preclinical Perspectives. Anticancer. Agents Med. Chem. 2021, 21, 1413–1430. [Google Scholar] [CrossRef] [PubMed]
- Capelôa, T.; Benyahia, Z.; Zampieri, L.X.; Blackman, M.C.N.M.; Sonveaux, P. Metabolic and Non-Metabolic Pathways That Control Cancer Resistance to Anthracyclines. Semin. Cell Dev. Biol. 2020, 98, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Perez-Plasencia, C.; Duenas-Gonzalez, A. Can the State of Cancer Chemotherapy Resistance Be Reverted by Epigenetic Therapy? Mol. Cancer 2006, 5, 27. [Google Scholar] [CrossRef]
- Guo, L.; Lee, Y.T.; Zhou, Y.; Huang, Y. Targeting Epigenetic Regulatory Machinery to Overcome Cancer Therapy Resistance. Semin. Cancer Biol. 2022, 83, 487–502. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Peng, Q.; Jiang, X.; Tan, S.; Yang, Y.; Yang, W.; Han, Y.; Chen, Y.; Oyang, L.; Lin, J.; et al. Metabolic Reprogramming and Epigenetic Modifications in Cancer: From the Impacts and Mechanisms to the Treatment Potential. Exp. Mol. Med. 2023, 55, 1357–1370. [Google Scholar] [CrossRef]
- Das, C.; Adhikari, S.; Bhattacharya, A.; Chakraborty, S.; Mondal, P.; Yadav, S.S.; Adhikary, S.; Hunt, C.R.; Yadav, K.K.; Pandita, S.; et al. Epigenetic-Metabolic Interplay in the DNA Damage Response and Therapeutic Resistance of Breast Cancer. Cancer Res. 2023, 83, 657–666. [Google Scholar] [CrossRef]
- Oura, K.; Morishita, A.; Hamaya, S.; Fujita, K.; Masaki, T. The Roles of Epigenetic Regulation and the Tumor Microenvironment in the Mechanism of Resistance to Systemic Therapy in Hepatocellular Carcinoma. Int. J. Mol. Sci. 2023, 24, 2805. [Google Scholar] [CrossRef]
- Arts, J.; De Schepper, S.; Van Emelen, K. Histone Deacetylase Inhibitors: From Chromatin Remodeling to Experimental Cancer Therapeutics. Curr. Med. Chem. 2003, 10, 2343–2350. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6, a018713. [Google Scholar] [CrossRef]
- Kim, H.J.; Bae, S.C. Histone Deacetylase Inhibitors: Molecular Mechanisms of Action and Clinical Trials as Anti-Cancer Drugs. Am. J. Transl. Res. 2011, 3, 166–179. [Google Scholar]
- Tiffon, C.; Adams, J.; Van Der Fits, L.; Wen, S.; Townsend, P.; Ganesan, A.; Hodges, E.; Vermeer, M.; Packham, G. The Histone Deacetylase Inhibitors Vorinostat and Romidepsin Downmodulate IL-10 Expression in Cutaneous T-Cell Lymphoma Cells. Br. J. Pharmacol. 2011, 162, 1590–1602. [Google Scholar] [CrossRef]
- Kaewpiboon, C.; Srisuttee, R.; Malilas, W.; Moon, J.; Oh, S.; Jeong, H.G.; Johnston, R.N.; Assavalapsakul, W.; Chung, Y.H. Upregulation of Stat1-HDAC4 Confers Resistance to Etoposide through Enhanced Multidrug Resistance 1 Expression in Human A549 Lung Cancer Cells. Mol. Med. Rep. 2015, 11, 2315–2321. [Google Scholar] [CrossRef]
- Wasim, L.; Chopra, M. Panobinostat Induces Apoptosis via Production of Reactive Oxygen Species and Synergizes with Topoisomerase Inhibitors in Cervical Cancer Cells. Biomed. Pharmacother. 2016, 84, 1393–1405. [Google Scholar] [CrossRef]
- Sampson, V.B.; Vetter, N.S.; Kamara, D.F.; Collier, A.B.; Gresh, R.C.; Kolb, E.A. Vorinostat Enhances Cytotoxicity of SN-38 and Temozolomide in Ewing Sarcoma Cells and Activates STAT3/AKT/MAPK Pathways. PLoS ONE 2015, 10, e0142704. [Google Scholar] [CrossRef]
- Meisenberg, C.; Ashour, M.E.; El-Shafie, L.; Liao, C.; Hodgson, A.; Pilborough, A.; Khurram, S.A.; Downs, J.A.; Ward, S.E.; El-Khamisy, S.F. Epigenetic Changes in Histone Acetylation Underpin Resistance to the Topoisomerase I Inhibitor Irinotecan. Nucleic Acids Res. 2017, 45, 1159–1176. [Google Scholar] [CrossRef] [PubMed]
- He, S.; Dong, G.; Wang, Z.; Chen, W.; Huang, Y.; Li, Z.; Jiang, Y.; Liu, N.; Yao, J.; Miao, Z.; et al. Discovery of Novel Multiacting Topoisomerase I/II and Histone Deacetylase Inhibitors. ACS Med. Chem. Lett. 2015, 6, 239–243. [Google Scholar] [CrossRef] [PubMed]
- Cincinelli, R.; Musso, L.; Artali, R.; Guglielmi, M.; Bianchino, E.; Cardile, F.; Colelli, F.; Pisano, C.; Dallavalle, S. Camptothecin-psammaplin A hybrids as topoisomerase I and HDAC dual-action inhibitors. Eur. J. Med. Chem. 2018, 143, 2005–2014. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, M.; Tahara, T.; Hayakawa, S.; Matsumoto, H.; Wada, S.I.; Tomioka, K.; Iida, A. Synthesis and Biological Evaluation of Histone Deacetylase and DNA Topoisomerase II-Targeted Inhibitors. Bioorg. Med. Chem. 2018, 26, 1920–1928. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Li, D.; Li, W.; Yin, J.; Zhang, Y.; Yuan, Z.; Gao, C.; Liu, F.; Jiang, Y. Design, Synthesis and Anticancer Evaluation of Acridine Hydroxamic Acid Derivatives as Dual Topo and HDAC Inhibitors. Bioorg. Med. Chem. 2018, 26, 3958–3966. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Wan, Y.; Xiao, Y.; Xia, C.; Duan, G. Dual-Target Inhibitors Based on HDACs: Novel Antitumor Agents for Cancer Therapy. J. Med. Chem. 2020, 63, 8977–9002. [Google Scholar] [CrossRef] [PubMed]
- Marx, C.; Sonnemann, J.; Beyer, M.; Maddocks, O.D.K.; Lilla, S.; Hauzenberger, I.; Piée-Staffa, A.; Siniuk, K.; Nunna, S.; Marx-Blümel, L.; et al. Mechanistic Insights into p53-Regulated Cytotoxicity of Combined Entinostat and Irinotecan against Colorectal Cancer Cells. Mol. Oncol. 2021, 15, 3404–3429. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Huang, Y.; Wang, T.; Li, K.; Lu, J.; Huang, M.; Dong, G.; Sheng, C. Evodiamine-Inspired Topoisomerase-Histone Deacetylase Dual Inhibitors: Novel Orally Active Antitumor Agents for Leukemia Therapy. J. Med. Chem. 2022, 65, 4818–4831. [Google Scholar] [CrossRef] [PubMed]
- Zhu, F.; Meng, X.; Liang, H.; Sheng, C.; Dong, G.; Liu, D.; Wu, S. Design, Synthesis, and Structure-Activity Relationships of Evodiamine-Based Topoisomerase (Top)/Histone Deacetylase (HDAC) Dual Inhibitors. Bioorg. Chem. 2022, 122, 105702. [Google Scholar] [CrossRef] [PubMed]
- El-Kalyoubi, S.; Elbaramawi, S.S.; Eissa, A.G.; Al-Ageeli, E.; Hobani, Y.H.; El-Sharkawy, A.A.; Mohamed, H.T.; Al-Karmalawy, A.A.; Abulkhair, H.S. Design and Synthesis of Novel Uracil-Linked Schiff Bases as Dual Histone Deacetylase Type II/Topoisomerase Type I Inhibitors with Apoptotic Potential. Future Med. Chem. 2023, 15, 937–958. [Google Scholar] [CrossRef]
- Kim, H.Y.; Choi, S.A.; Koh, E.J.; Kim, K.H.; Phi, J.H.; Lee, J.Y.; Kim, S.K. Combination Treatment of CI-994 With Etoposide Potentiates Anticancer Effects Through a Topoisomerase II-Dependent Mechanism in Atypical Teratoid/Rhabdoid Tumor (AT/RT). Front. Oncol. 2021, 11, 648023. [Google Scholar] [CrossRef]
- Zhao, M.; Yang, K.; Zhu, X.; Gao, T.; Yu, W.; Liu, H.; You, Z.; Liu, Z.; Qiao, X.; Song, Y. Design, Synthesis and Biological Evaluation of Dual Topo II/HDAC Inhibitors Bearing Pyrimido[5,4-b]indole and Pyrazolo[3,4-d]pyrimidine Motifs. Eur. J. Med. Chem. 2023, 252, 115303. [Google Scholar] [CrossRef]
- Flor, A.C.; Doshi, A.P.; Kron, S.J. Modulation of Therapy-Induced Senescence by Reactive Lipid Aldehydes. Cell Death Discov. 2016, 2, 16045. [Google Scholar] [CrossRef]
- Ogiso, Y.; Tomida, A.; Lei, S.; Omura, S.; Tsuruo, T. Proteasome Inhibition Circumvents Solid Tumor Resistance to Topoisomerase II-Directed Drugs. Cancer Res. 2000, 60, 2429–2434. [Google Scholar]
- Drummond, C.J.; Finlay, G.J.; Broome, L.; Marshall, E.S.; Richardson, E.; Baguley, B.C. Action of SN 28049, a New DNA Binding Topoisomerase II-Directed Antitumour Drug: Comparison with Doxorubicin and Etoposide. Investig. New Drugs 2011, 29, 1102–1110. [Google Scholar] [CrossRef]
- Polewska, J.; Skwarska, A.; Augustin, E.; Konaopa, J. DNA-Damaging Imidazoacridinone C-1311 Induces Autophagy Followed by Irreversible Growth Arrest and Senescence in Human Lung Cancer Cells. J. Pharmacol. Exp. Ther. 2013, 346, 393–405. [Google Scholar] [CrossRef]
- Huang, R.X.; Zhou, P.K. DNA Damage Response Signaling Pathways and Targets for Radiotherapy Sensitization in Cancer. Signal Transduct. Target Ther. 2020, 5, 60. [Google Scholar] [CrossRef]
- Courapied, S.; Cherier, J.; Vigneron, A.; Troadec, M.B.; Giraud, S.; Valo, I.; Prigent, C.; Gamelin, E.; Coqueret, O.; Barré, B. Regulation of the Aurora-A Gene Following Topoisomerase I Inhibition: Implication of the Myc Transcription Factor. Mol. Cancer 2010, 9, 205. [Google Scholar] [CrossRef]
- Taschner-Mandl, S.; Schwarz, M.; Blaha, J.; Kauer, M.; Kromp, F.; Frank, N.; Rifatbegovic, F.; Weiss, T.; Ladenstein, R.; Hohenegger, M.; et al. Metronomic Topotecan Impedes Tumor Growth of MYCN-Amplified Neuroblastoma Cells In Vitro and In Vivo by Therapy Induced Senescence. Oncotarget 2016, 7, 3571–3586. [Google Scholar] [CrossRef]
- Hao, L.; Rohani, N.; Zhao, R.T.; Pulver, E.M.; Mak, H.; Kelada, O.J.; Ko, H.; Fleming, H.E.; Gertler, F.B.; Bhatia, S.N. Microenvironment-Triggered Multimodal Precision Diagnostics. Nat. Mater. 2021, 20, 1440–1448. [Google Scholar] [CrossRef]
- Marx, O.M.; Mankarious, M.M.; Eshelman, M.A.; Ding, W.; Koltun, W.A.; Yochum, G.S. Transcriptome Analyses Identify Deregulated MYC in Early Onset Colorectal Cancer. Biomolecules 2022, 12, 1223. [Google Scholar] [CrossRef] [PubMed]
- Saleh, T.; Carpenter, V.J.; Tyutyunyk-Massey, L.; Murray, G.; Leverson, J.D.; Souers, A.J.; Alotaibi, M.R.; Faber, A.C.; Reed, J.; Harada, H.; et al. Clearance of Therapy-Induced Senescent Tumor Cells by the Senolytic ABT-263 via Interference with BCL-XL–BAX Interaction. Mol. Oncol. 2023, 14, 2504–2519. [Google Scholar] [CrossRef] [PubMed]
- Meredith, A.M.; Dass, C.R. Increasing Role of the Cancer Chemotherapeutic Doxorubicin in Cellular Metabolism. J. Pharm. Pharmacol. 2016, 68, 729–741. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Flores, R.R.; Jang, I.H.; Saathoff, A.; Robbins, P.D. Immune Senescence, Immunosenescence and Aging. Front. Aging 2022, 3, 900028. [Google Scholar] [CrossRef] [PubMed]
- Vulpis, E.; Cuollo, L.; Borrelli, C.; Antonangeli, F.; Masuelli, L.; Cippitelli, M.; Fionda, C.; Caracciolo, G.; Petrucci, M.T.; Santoni, A.; et al. Doxorubicin-Mediated miR-433 Expression on Exosomes Promotes Bystander Senescence in Multiple Myeloma Cells in a DDR-Independent Manner. Int. J. Mol. Sci. 2023, 24, 6862. [Google Scholar] [CrossRef] [PubMed]
- Takigawa, N.; Vaziri, S.A.; Grabowski, D.R.; Chikamori, K.; Rybicki, L.R.; Bukowski, R.M.; Ganapathi, M.K.; Ganapathi, R.; Mekhail, T. Proteasome Inhibition with Bortezomib Enhances Activity of Topoisomerase I-Targeting Drugs by NF-kappaB-Independent Mechanisms. Anticancer Res. 2006, 26, 1869–1876. [Google Scholar] [PubMed]
- Rudolf, E.; John, S.; Cervinka, M. Irinotecan Induces Senescence and Apoptosis in Colonic Cells In Vitro. Toxicol. Lett. 2012, 214, 192–199. [Google Scholar] [CrossRef]
- Gewirtz, D.A. Growth Arrest and Cell Death in Breast Tumor Cells in Response to Ionizing Radiation and Chemotherapeutic Agents Inducing DNA Damage. Breast Cancer Res. Treat 2000, 62, 223–235. [Google Scholar] [CrossRef]
- Han, H.; Bearss, D.J.; Browne, L.W.; Calaluce, R.; Nagle, R.B.; Von Hoff, D.D. Identification of differentially expressed genes in pancreatic cancer cells using cDNA microarray. Cancer Res. 2002, 62, 2890–2896. [Google Scholar]
- Elmore, J.G.; Armstrong, K.; Lehman, C.D.; Fletcher, S.W. Screening for breast cancer. JAMA 2005, 293, 1245–1256. [Google Scholar] [CrossRef] [PubMed]
- Guichard, G.; Larré, S.; Gallina, A.; Lazar, A.; Faucon, H.; Chemama, S.; Allory, Y.; Patar, J.; Vordos, D.; Hoznek, A.; et al. Extended 21-sample needle biopsy protocol for diagnosis of prostate cancer in 1000 consecutive patients. Eur. Urol. 2007, 52, 430–435. [Google Scholar] [CrossRef]
- Gleyzer, N.; Scarpulla, R.C. Activation of a PGC-1-related coactivator (PRC)-dependent inflammatory stress program linked to apoptosis and premature senescence. J. Biol. Chem. 2013, 288, 8004–8015. [Google Scholar] [CrossRef]
- Yeo, S.K.; Wen, J.; Chen, S.; Guan, J. Autophagy differentially regulates distinct breast cancer stem-like cells in murine models via EGFR/STAT3 and TGFΒ/SMAD signaling. Am. Assoc. Cancer Res. 2016, 76, 3397–3410. [Google Scholar] [CrossRef]
- Chen, S.; Zhao, Y.; Liu, S.; Zhang, J.; Assaraf, Y.G.; Cui, W.; Wang, L. Epigenetic enzyme mutations as mediators of anti-cancer drug resistance. Drug. Resist. Updat. 2022, 61, 100821. [Google Scholar] [CrossRef]
- Zhang, X.; Dang, C.V. Time to hit pause on mitochondria-targeting cancer therapies. Nat. Med. 2023, 29, 29–30. [Google Scholar] [CrossRef]
- Çalışkan, M.; Güler, H.; Bozok Çetintaş, V. Current updates on microRNAs a regulators of chemoresistance. Biomed. Pharmacother. 2017, 95, 1000–1012. [Google Scholar] [CrossRef]
- Torki, Z.; Ghavi, D.; Hashemi, S.; Rahmati, Y.; Rahmanpour, D.; Pornour, M.; Alivand, M.R. The related miRNAs involved in doxorubicin resistance or sensitivity of various cancers: An update. Cancer Chemother. Pharmacol. 2021, 88, 771–793. [Google Scholar] [CrossRef]
- Taheri, M.; Mahmud Hussen, B.; Tondro Anamag, F.; Shoorei, H.; Dinger, M.E.; Ghafouri-Fard, S. The role of miRNAs and lncRNAs in conferring resistance to doxorubicin. J. Drug Target 2022, 30, 1–21. [Google Scholar] [CrossRef]
- Sritharan, S.; Guha, S.; Hazarika, S.; Sivalingam, N. Meta analysis of bioactive compounds, miRNA, siRNA and cell death regulators as sensitizers to doxorubicin induced chemoresistance. Apoptosis 2022, 27, 622–646. [Google Scholar] [CrossRef]
- Sun, M.; Zhang, Q.; Yang, X.; Qian, S.Y.; Guo, B. Vitamin D Enhances the Efficacy of Irinotecan through miR-627-Mediated Inhibition of Intratumoral Drug Metabolism. Mol. Cancer Ther. 2016, 15, 2086–2095. [Google Scholar] [CrossRef]
- Carvajal-Moreno, J.; Hernandez, V.A.; Wang, X.; Li, J.; Yalowich, J.C.; Elton, T.S. Effects of hsa-miR-9-3p and hsa-miR-9-5p on Topoisomerase IIβ Expression in Human Leukemia K562 Cells with Acquired Resistance to Etoposide. J. Pharmacol. Exp. Ther. 2023, 384, 265–276. [Google Scholar] [CrossRef] [PubMed]
- Kubiliūtė, R.; Šulskytė, I.; Daniūnaitė, K.; Daugelavičius, R.; Jarmalaitė, S. Molecular features of doxorubicin-resistance development in colorectal cancer CX-1 cell line. Medicina 2016, 52, 298–306. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, S.; Abadi, A.J.; Gholami, M.H.; Hashemi, F.; Zabolian, A.; Hushmandi, K.; Zarrabi, A.; Entezari, M.; Aref, A.R.; Khan, H.; et al. The involvement of epithelial-to-mesenchymal transition in doxorubicin resistance: Possible molecular targets. Eur. J. Pharmacol. 2021, 908, 174344. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Abraham, A.D.; Li, L.; Babalmorad, A.; Bagby, S.; Arcaroli, J.J.; Hansen, R.J.; Valeriote, F.A.; Gustafson, D.L.; Schaack, J.; et al. Topoisomerase IIα mediates TCF-dependent epithelial-mesenchymal transition in colon cancer. Oncogene 2016, 35, 4990–4999. [Google Scholar] [CrossRef]
- Abraham, A.D.; Esquer, H.; Zhou, Q.; Tomlinson, N.; Hamill, B.D.; Abbott, J.M.; Li, L.; Pike, L.A.; Rinaldetti, S.; Ramirez, D.A.; et al. Drug Design Targeting T-Cell Factor-Driven Epithelial-Mesenchymal Transition as a Therapeutic Strategy for Colorectal Cancer. J. Med. Chem. 2019, 62, 10182–10203. [Google Scholar] [CrossRef]
- Davis, J.T.; Ghosh, T.M.; Mazumder, S.; Mitra, A.; Bird, R.C.; Arnold, R.D. Extended Exposure Topotecan Significantly Improves Long-Term Drug Sensitivity by Decreasing Malignant Cell Heterogeneity and by Preventing Epithelial-Mesenchymal Transition. Int. J. Mol. Sci. 2023, 24, 8490. [Google Scholar] [CrossRef]
- Ohtsuka, K.; Ohnìshì, H.; Morii, T.; Fujiwara, M.; Kishino, T.; Ogura, W.; Chiba, M.; Matsushima, S.; Goya, T.; Watanabe, T. Downregulated ABCG2 enhances sensitivity to topoisomerase I inhibitor in epidermal growth factor receptor tyrosine kinase Inhibitor-Resistant non-small cell lung cancer. J. Thorac. Oncol. 2010, 5, 1726–1733. [Google Scholar] [CrossRef]
- Goswami, S.; Sharma-Walia, N. Osteoprotegerin secreted by inflammatory and invasive breast cancer cells induces aneuploidy, cell proliferation and angiogenesis. BMC Cancer 2015, 15, 935. [Google Scholar] [CrossRef] [PubMed]
- Bentires-Alj, M.; Barbu, V.; Fillet, M.; Chariot, A.; Relic, B.; Jacobs, N.; Gielen, J.; Merville, M.P.; Bours, V. NF-kappaB transcription factor induces drug resistance through MDR1 expression in cancer cells. Oncogene 2003, 22, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Linder, S.; Bazzaro, M. Drug Development Targeting the Ubiquitin-Proteasome System (UPS) for the Treatment of Human Cancers. Cancers 2020, 12, 902. [Google Scholar] [CrossRef]
- Żabka, A.; Winnicki, K.; Polit, J.T.; Maszewski, J. The effects of anti-DNA topoisomerase II drugs, etoposide and ellipticine, are modified in root meristem cells of Allium cepa by MG132, an inhibitor of 26S proteasomes. Plant Physiol. Biochem. 2015, 96, 72–82. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.K.; Perera, L.; Cannon, R.E. Reversal of drug resistance by JS-K and nitric oxide in ABCB1- and ABCG2-expressing multi-drug resistant human tumor cells. Biomed. Pharmacother. 2019, 120, 109468. [Google Scholar] [CrossRef] [PubMed]
- Sinha, B.K.; Perera, L.; Cannon, R.E. NCX-4040, a Unique Nitric Oxide Donor, Induces Reversal of Drug-Resistance in Both ABCB1- and ABCG2-Expressing Multidrug Human Cancer Cells. Cancers 2021, 13, 1680. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sharma, N.K.; Bahot, A.; Sekar, G.; Bansode, M.; Khunteta, K.; Sonar, P.V.; Hebale, A.; Salokhe, V.; Sinha, B.K. Understanding Cancer’s Defense against Topoisomerase-Active Drugs: A Comprehensive Review. Cancers 2024, 16, 680. https://doi.org/10.3390/cancers16040680
Sharma NK, Bahot A, Sekar G, Bansode M, Khunteta K, Sonar PV, Hebale A, Salokhe V, Sinha BK. Understanding Cancer’s Defense against Topoisomerase-Active Drugs: A Comprehensive Review. Cancers. 2024; 16(4):680. https://doi.org/10.3390/cancers16040680
Chicago/Turabian StyleSharma, Nilesh Kumar, Anjali Bahot, Gopinath Sekar, Mahima Bansode, Kratika Khunteta, Priyanka Vijay Sonar, Ameya Hebale, Vaishnavi Salokhe, and Birandra Kumar Sinha. 2024. "Understanding Cancer’s Defense against Topoisomerase-Active Drugs: A Comprehensive Review" Cancers 16, no. 4: 680. https://doi.org/10.3390/cancers16040680
APA StyleSharma, N. K., Bahot, A., Sekar, G., Bansode, M., Khunteta, K., Sonar, P. V., Hebale, A., Salokhe, V., & Sinha, B. K. (2024). Understanding Cancer’s Defense against Topoisomerase-Active Drugs: A Comprehensive Review. Cancers, 16(4), 680. https://doi.org/10.3390/cancers16040680





