MicroRNAs Associated with Androgen Receptor and Metastasis in Triple-Negative Breast Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Selection
2.2. AR Detection by Immunohistochemistry (IHC)
2.3. Preparation and Relative Quantification of miRNA in Tissue Samples
2.4. Bioinformatics Analyses
2.5. Statistical Analyses
3. Results
3.1. Included Patients
3.2. Identification of DE miRNAs
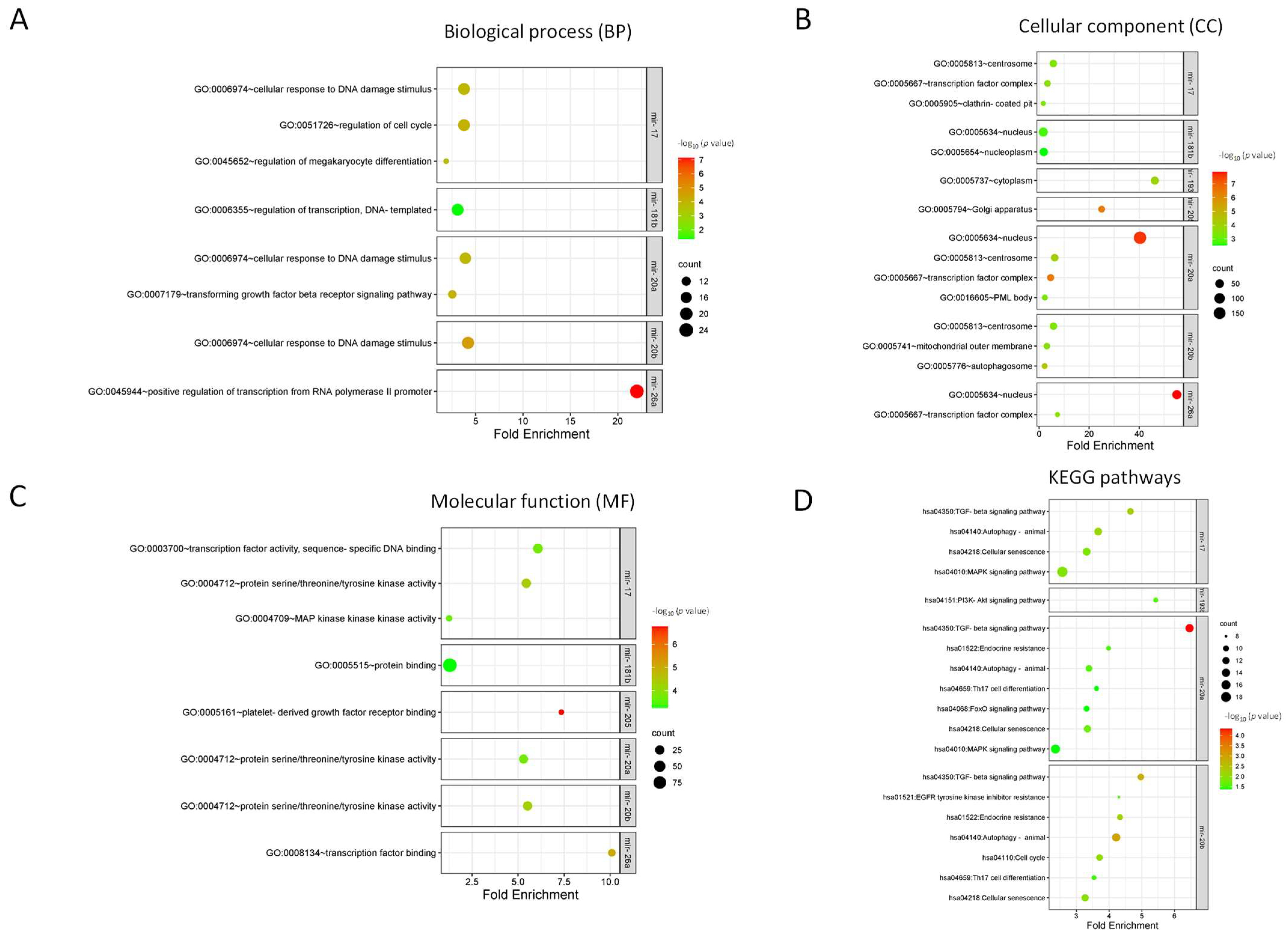
3.3. Identification of Target Genes and Enrichment Analysis
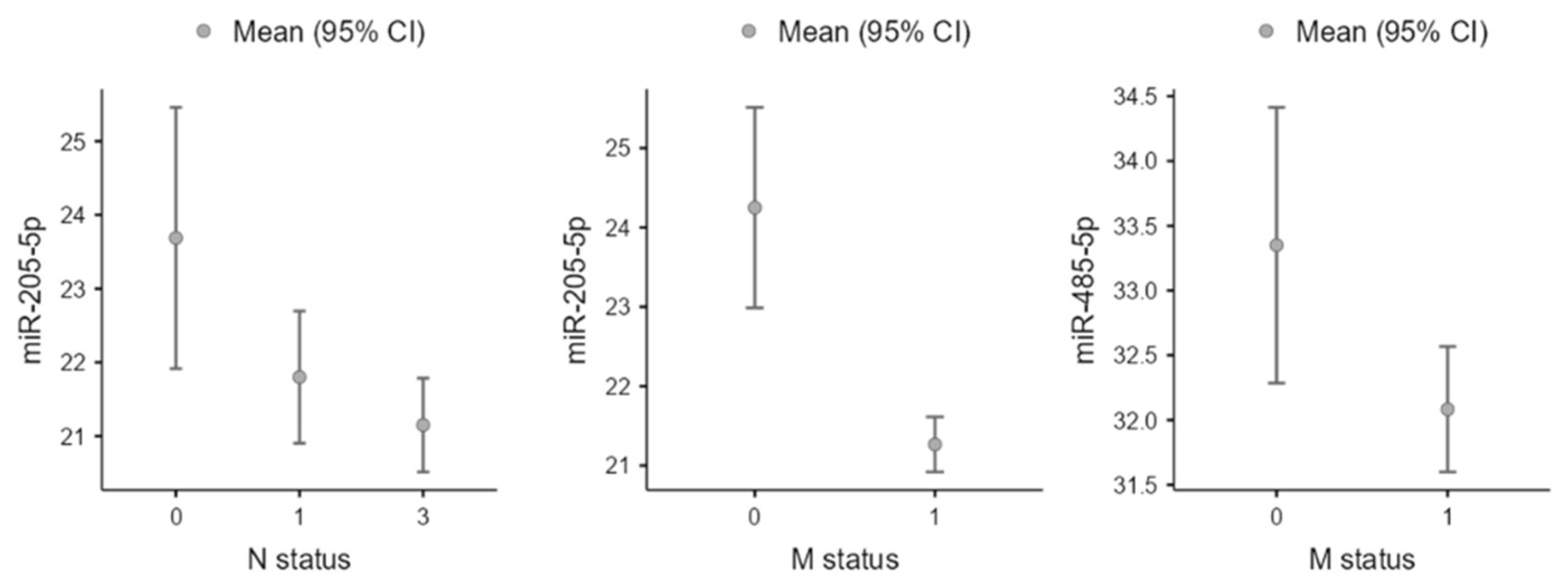
3.4. Clinicopathological Associations
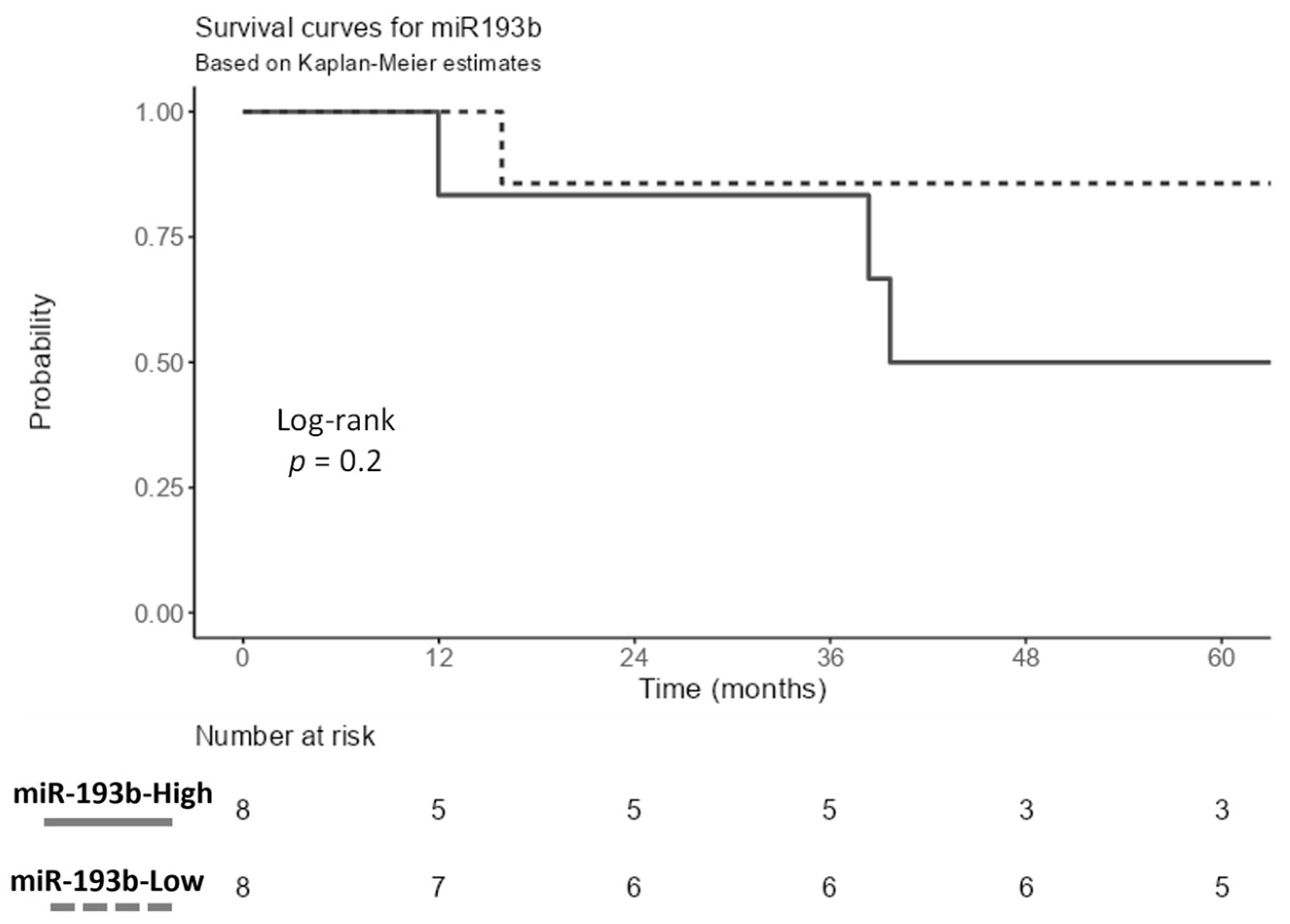
3.5. Survival Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Al-Othman, N.; Ahram, M.; Alqaraleh, M. Role of androgen and microRNA in triple-negative breast cancer. Breast Dis. 2020, 39, 15–27. [Google Scholar] [CrossRef]
- Kargutkar, N.; Hariharan, P.; Nadkarni, A. Dynamic interplay of microRNA in diseases and therapeutic. Clin. Genet. 2023, 103, 268–276. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.U.; Saeed, S.; Sheikh, A.N.; Arbi, F.M.; Shahzad, A.; Faryal, U.; Lu, K. Crafting a Blueprint for MicroRNA in Cardiovascular Diseases (CVDs). Curr. Probl. Cardiol. 2023, 48, 102010. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, A.E.; Gil-Jaramillo, N.; Tapias, M.A.; González-Giraldo, Y.; Pinzón, A.; Puentes-Rozo, P.J.; Aristizábal-Pachón, A.F.; González, J. MicroRNA: A Linking between Astrocyte Dysfunction, Mild Cognitive Impairment, and Neurodegenerative Diseases. Life 2022, 12, 1439. [Google Scholar] [CrossRef] [PubMed]
- Szczepanek, J.; Skorupa, M.; Tretyn, A. MicroRNA as a Potential Therapeutic Molecule in Cancer. Cells 2022, 11, 1008. [Google Scholar] [CrossRef] [PubMed]
- Barbato, S.; Solaini, G.; Fabbri, M. MicroRNAs in Oncogenesis and Tumor Suppression. Int. Rev. Cell Mol. Biol. 2017, 333, 229–268. [Google Scholar] [CrossRef] [PubMed]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Abdelaal, A.M.; Sohal, I.S.; Iyer, S.; Sudarshan, K.; Kothandaraman, H.; Lanman, N.A.; Low, P.S.; Kasinski, A.L. A first-in-class fully modified version of miR-34a with outstanding stability, activity, and anti-tumor efficacy. Oncogene 2023, 42, 2985–2999. [Google Scholar] [CrossRef]
- Gong, C.; Tian, J.; Wang, Z.; Gao, Y.; Wu, X.; Ding, X.; Qiang, L.; Li, G.; Han, Z.; Yuan, Y.; et al. Functional exosome-mediated co-delivery of doxorubicin and hydrophobically modified microRNA 159 for triple-negative breast cancer therapy. J. Nanobiotechnol. 2019, 17, 93. [Google Scholar] [CrossRef]
- Yin, H.; Xiong, G.; Guo, S.; Xu, C.; Xu, R.; Guo, P.; Shu, D. Delivery of Anti-miRNA for Triple-Negative Breast Cancer Therapy Using RNA Nanoparticles Targeting Stem Cell Marker CD133. Mol. Ther. 2019, 27, 1252–1261. [Google Scholar] [CrossRef]
- García-Vazquez, R.; Ruiz-García, E.; García, A.M.; Astudillo-De La Vega, H.; Lara-Medina, F.; Alvarado-Miranda, A.; Maldonado-Martínez, H.; González-Barrios, J.A.; Campos-Parra, A.D.; Cuevas, S.R.; et al. A microRNA signature associated with pathological complete response to novel neoadjuvant therapy regimen in triple-negative breast cancer. Tumour Biol. J. Int. Soc. Oncodev. Biol. Med. 2017, 39, 1010428317702899. [Google Scholar] [CrossRef]
- Lowery, A.J.; Miller, N.; Devaney, A.; McNeill, R.E.; Davoren, P.A.; Lemetre, C.; Benes, V.; Schmidt, S.; Blake, J.; Ball, G.; et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009, 11, R27. [Google Scholar] [CrossRef]
- Mattie, M.D.; Benz, C.C.; Bowers, J.; Sensinger, K.; Wong, L.; Scott, G.K.; Fedele, V.; Ginzinger, D.; Getts, R.; Haqq, C. Optimized high-throughput microRNA expression profiling provides novel biomarker assessment of clinical prostate and breast cancer biopsies. Mol. Cancer 2006, 5, 24. [Google Scholar] [CrossRef] [PubMed]
- Ouyang, M.; Li, Y.; Ye, S.; Ma, J.; Lu, L.; Lv, W.; Chang, G.; Li, X.; Li, Q.; Wang, S.; et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS ONE 2014, 9, e96228. [Google Scholar] [CrossRef] [PubMed]
- Rothé, F.; Ignatiadis, M.; Chaboteaux, C.; Haibe-Kains, B.; Kheddoumi, N.; Majjaj, S.; Badran, B.; Fayyad-Kazan, H.; Desmedt, C.; Harris, A.L.; et al. Global microRNA expression profiling identifies MiR-210 associated with tumor proliferation, invasion and poor clinical outcome in breast cancer. PLoS ONE 2011, 6, e20980. [Google Scholar] [CrossRef] [PubMed]
- Kurozumi, S.; Yamaguchi, Y.; Kurosumi, M.; Ohira, M.; Matsumoto, H.; Horiguchi, J. Recent trends in microRNA research into breast cancer with particular focus on the associations between microRNAs and intrinsic subtypes. J. Hum. Genet. 2017, 62, 15–24. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Ni, J.; Beretov, J.; Graham, P.; Li, Y. Triple-negative breast cancer therapeutic resistance: Where is the Achilles’ heel? Cancer Lett. 2021, 497, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Szymiczek, A.; Lone, A.; Akbari, M.R. Molecular intrinsic versus clinical subtyping in breast cancer: A comprehensive review. Clin. Genet. 2021, 99, 613–637. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Bauer, J.A.; Chen, X.; Sanders, M.E.; Chakravarthy, A.B.; Shyr, Y.; Pietenpol, J.A. Identification of human triple-negative breast cancer subtypes and preclinical models for selection of targeted therapies. J. Clin. Investig. 2011, 121, 2750–2767. [Google Scholar] [CrossRef]
- D’Amato, N.C.; Gordon, M.A.; Babbs, B.; Spoelstra, N.S.; Carson Butterfield, K.T.; Torkko, K.C.; Phan, V.T.; Barton, V.N.; Rogers, T.J.; Sartorius, C.A.; et al. Cooperative dynamics of AR and ER activity in breast cancer. Mol. Cancer Res. 2016, 14, 1054–1067. [Google Scholar] [CrossRef]
- Guedj, M.; Marisa, L.; De Reynies, A.; Orsetti, B.; Schiappa, R.; Bibeau, F.; MacGrogan, G.; Lerebours, F.; Finetti, P.; Longy, M.; et al. A refined molecular taxonomy of breast cancer. Oncogene 2012, 31, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Astvatsaturyan, K.; Yue, Y.; Walts, A.E.; Bose, S. Androgen receptor positive triple negative breast cancer: Clinicopathologic, prognostic, and predictive features. PLoS ONE 2018, 13, e0197827. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Yang, F.; Zhang, W.; Song, W.; Liu, Y.; Guan, X. The Androgen Receptor Promotes Cellular Proliferation by Suppression of G-Protein Coupled Estrogen Receptor Signaling in Triple-Negative Breast Cancer. Cell. Physiol. Biochem. 2017, 43, 2047–2061. [Google Scholar] [CrossRef] [PubMed]
- Barton, V.N.; D’Amato, N.C.; Gordon, M.A.; Lind, H.T.; Spoelstra, N.S.; Babbs, B.L.; Heinz, R.E.; Elias, A.; Jedlicka, P.; Jacobsen, B.M.; et al. Multiple molecular subtypes of triple-negative breast cancer critically rely on androgen receptor and respond to enzalutamide in vivo. Mol. Cancer Ther. 2015, 14, 769–778. [Google Scholar] [CrossRef] [PubMed]
- Traina, T.A.; Miller, K.; Yardley, D.A.; Eakle, J.; Schwartzberg, L.S.; O’Shaughnessy, J.; Gradishar, W.; Schmid, P.; Winer, E.; Kelly, C.; et al. Enzalutamide for the treatment of androgen receptor-expressing triple-negative breast cancer. J. Clin. Oncol. 2018, 36, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.M.; Oh, M.H.; Go, J.H.; Han, K.; Choi, S.Y. Molecular subtypes of triple-negative breast cancer: Understanding of subtype categories and clinical implication. Genes Genom. 2020, 42, 1381–1387. [Google Scholar] [CrossRef] [PubMed]
- Cascione, L.; Gasparini, P.; Lovat, F.; Carasi, S.; Pulvirenti, A.; Ferro, A.; Alder, H.; He, G.; Vecchione, A.; Croce, C.M.; et al. Integrated microRNA and mRNA signatures associated with survival in triple negative breast cancer. PLoS ONE 2013, 8, e55910. [Google Scholar] [CrossRef]
- Nakano, K.; Miki, Y.; Hata, S.; Ebata, A.; Takagi, K.; Mcnamara, K.M.; Sakurai, M.; Masuda, M.; Hirakawa, H.; Ishida, T.; et al. Identification of Androgen-responsive microRNAs and Androgen-related Genes in Breast Cancer. Anticancer. Res. 2013, 33, 4811–4819. [Google Scholar]
- Ahram, M.; Mustafa, E.; Zaza, R.; Abu Hammad, S.; Alhudhud, M.; Bawadi, R.; Zihlif, M. Differential expression and androgen regulation of microRNAs and metalloprotease 13 in breast cancer cells. Cell Biol. Int. 2017, 41, 1345–1355. [Google Scholar] [CrossRef]
- Al-Othman, N.; Hammad, H.; Ahram, M. Dihydrotestosterone regulates expression of CD44 via miR-328-3p in triple-negative breast cancer cells. Gene 2018, 675, 128–135. [Google Scholar] [CrossRef]
- Ahram, M.; Amarin, J.Z.; Suradi, H.H.; Abdelhamid, S.S.; Makhamreh, M.M.; Bawadi, R.M.; Al-Hussaini, M. Association of MicroRNAs with the Clinicopathologic Characteristics of Ependymoma. J. Mol. Neurosci. 2018, 66, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.W.; Chen, Y.; Chen, S.; Wang, X. OncomiR: An online resource for exploring pan-cancer microRNA dysregulation. Bioinformatics 2018, 34, 713–715. [Google Scholar] [CrossRef] [PubMed]
- Al-Othman, N.; Hammad, H.; Ahram, M. Type of Serum as a Cell Culture Supplement Influences Regulation of MicroRNA Expression in Breast MDA-MB-231 Cancer Cells. Jordan Med. J. 2017, 51, 1–7. [Google Scholar]
- Petrović, N.; Todorović, L.; Nedeljković, M.; Božović, A.; Bukumirić, Z.; Tanić, N.D.; Jovanović-Ćupić, S.; Šami, A.; Mandušić, V. Dual function miR-205 is positively associated with ER and negatively with five-year survival in breast cancer patients. Pathol. Res. Pract. 2022, 238, 154080. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Y.; Li, Y.; Tao, H.; Humphries, B.; Li, A.; Jiang, Y.; Yang, C.; Luo, R.; Wang, Z. Integrin α5 down-regulation by miR-205 suppresses triple negative breast cancer stemness and metastasis by inhibiting the Src/Vav2/Rac1 pathway. Cancer Lett. 2018, 433, 199–209. [Google Scholar] [CrossRef] [PubMed]
- De Cola, A.; Volpe, S.; Budani, M.C.; Ferracin, M.; Lattanzio, R.; Turdo, A.; D’Agostino, D.; Capone, E.; Stassi, G.; Todaro, M.; et al. MIR-205-5p-mediated downregulation of ERBB/HER receptors in breast cancer stem cells results in targeted therapy resistance. Cell Death Dis. 2015, 6, e1823. [Google Scholar] [CrossRef] [PubMed]
- Lin, L.F.; Li, Y.T.; Han, H.; Lin, S.G. MicroRNA-205-5p targets the HOXD9-Snail1 axis to inhibit triple negative breast cancer cell proliferation and chemoresistance. Aging 2021, 13, 3945–3956. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Zhou, Z.; Guo, Y.; Du, Q.; Li, L. CircCDK1 knockdown reduces CDK1 expression by targeting miR-489-3p to suppress the development of breast cancer and strengthen the sensitivity of Tamoxifen. Anti Cancer Drugs 2022, 33, 286–299. [Google Scholar] [CrossRef]
- Liu, Q.; Yang, G.; Qian, Y. Loss of MicroRNA-489-3p promotes osteosarcoma metastasis by activating PAX3-MET pathway. Mol. Carcinog. 2017, 56, 1312–1321. [Google Scholar] [CrossRef]
- Zhang, P.; Li, L.; Wang, B.; Ran, X.; Yang, S.; Luo, Y.; Li, Y.; Wang, Z.; Liu, Y.; Zhu, B. miR-489-3p promotes malignant progression of non-small cell lung cancer through the inactivation of Wnt/β-catenin signaling pathway via regulating USP48. Respir. Res. 2022, 23, 93. [Google Scholar] [CrossRef]
- Li, J.; Qu, W.; Jiang, Y.; Sun, Y.; Cheng, Y.; Zou, T.; Du, S. miR-489 Suppresses Proliferation and Invasion of Human Bladder Cancer Cells. Oncol. Res. 2016, 24, 391–398. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.; Chen, T. MiR-489-3p inhibits cell proliferation, migration, and invasion, and induces apoptosis, by targeting the BDNF-mediated PI3K/AKT pathway in glioblastoma. Open Life Sci. 2020, 15, 274–283. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhu, X.; Yan, Z.; Li, C.; Zhao, H.; Ma, L.; Zhang, D.; Liu, J.; Liu, Z.; Du, N.; et al. miR-489-3p/SIX1 Axis Regulates Melanoma Proliferation and Glycolytic Potential. Mol. Ther. Oncolytics 2019, 16, 30–40. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.; Wang, G.; Zhu, W.; Luo, C.; Guo, Z. LEF1-AS1 accelerates tumorigenesis in glioma by sponging miR-489-3p to enhance HIGD1A. Cell Death Dis. 2020, 11, 690. [Google Scholar] [CrossRef]
- Feng, W.; Li, B.; Wang, J.; Zhang, H.; Liu, Y.; Xu, D.; Cheng, K.; Zhuang, J. Long Non-coding RNA LINC00115 Contributes to the Progression of Colorectal Cancer by Targeting miR-489-3p via the PI3K/AKT/mTOR Pathway. Front. Genet. 2020, 11, 567630. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Gao, Q.; Wang, M.; Xin, H. LncRNA SNHG1 contributes to the regulation of acute myeloid leukemia cell growth by modulating miR-489-3p/SOX12/Wnt/β-catenin signaling. J. Cell. Physiol. 2021, 236, 653–663. [Google Scholar] [CrossRef]
- Wang, J.; Zhang, J.Q.; Zhao, X.L.; Lu, J.Y.; Weng, Z.M.; Ding, Z.M.; Yang, F.Q. Circular RNA DHX33 promotes malignant behavior in ccRCC by targeting miR-489-3p/MEK1 axis. Aging 2020, 12, 14885–14896. [Google Scholar] [CrossRef]
- Delmas, E.; Jah, N.; Pirou, C.; Bouleau, S.; Le Floch, N.; Vayssière, J.L.; Mignotte, B.; Renaud, F. FGF1 C-terminal domain and phosphorylation regulate intracrine FGF1 signaling for its neurotrophic and anti-apoptotic activities. Cell Death Dis. 2016, 7, e2079. [Google Scholar] [CrossRef]
- Huang, R.-S.; Zheng, Y.-L.; Li, C.; Ding, C.; Xu, C.; Zhao, J. MicroRNA-485-5p suppresses growth and metastasis in non-small cell lung cancer cells by targeting IGF2BP2. Life Sci. 2018, 199, 104–111. [Google Scholar] [CrossRef]
- Jang, T.H.; Huang, W.C.; Tung, S.L.; Lin, S.C.; Chen, P.M.; Cho, C.Y.; Yang, Y.Y.; Yen, T.C.; Lo, G.H.; Chuang, S.E.; et al. MicroRNA-485-5p targets keratin 17 to regulate oral cancer stemness and chemoresistance via the integrin/FAK/Src/ERK/β-catenin pathway. J. Biomed. Sci. 2022, 29, 42. [Google Scholar] [CrossRef]
- Zhao, R.; Shan, Y.; Zhou, X.; Zhang, C.; Zhao, R.; Zhao, L.; Shan, B. MicroRNA-485-5p suppresses the progression of esophageal squamous cell carcinoma by targeting flotillin-1 and inhibits the epithelial-mesenchymal transition. Oncol. Rep. 2021, 45, 93. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Dai, W.; Wu, A.; Li, Y. CircCDC45 promotes the malignant progression of glioblastoma by modulating the miR-485-5p/CSF-1 axis. BMC Cancer 2021, 21, 1090. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Zhou, S.; Cheng, G.; Ruan, Y.; Tian, Y.; Lv, K.; Han, S.; Zhou, X. CircLMTK2 Silencing Attenuates Gemcitabine Resistance in Pancreatic Cancer by Sponging miR-485-5p and to Target PAK1. J. Oncol. 2022, 2022, 1911592. [Google Scholar] [CrossRef] [PubMed]
- Tie, W.; Ge, F. MALAT1 Inhibits Proliferation of HPV16-Positive Cervical Cancer by Sponging miR-485-5p to Promote Expression of MAT2A. DNA Cell Biol. 2021, 40, 1407–1417. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Chen, X.; Li, J.; Wang, F. CircRUNX1 functions as an oncogene in colorectal cancer by regulating circRUNX1/miR-485-5p/SLC38A1 axis. Eur. J. Clin. Investig. 2021, 51, e13540. [Google Scholar] [CrossRef] [PubMed]
- Zeng, F.; Yao, M.; Wang, Y.; Zheng, W.; Liu, S.; Hou, Z.; Cheng, X.; Sun, S.; Li, T.; Zhao, H.; et al. Fatty acid β-oxidation promotes breast cancer stemness and metastasis via the miRNA-328-3p-CPT1A pathway. Cancer Gene Ther. 2022, 29, 383–395. [Google Scholar] [CrossRef] [PubMed]
- Al-Momany, B.; Hammad, H.; Ahram, M. Dihydrotestosterone Induces Chemo-Resistance of Triple-Negative Breast MDA-MB-231 Cancer Cells Towards Doxorubicin Independent of ABCG2 and miR-328-3p. Curr. Mol. Pharmacol. 2021, 14, 860–870. [Google Scholar] [CrossRef] [PubMed]
- Thomson, D.W.; Dinger, M.E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Ma, S.; Wei, H.; Wang, C.; Han, J.; Chen, X.; Li, Y. MiR-26b-5p inhibits cell proliferation and EMT by targeting MYCBP in triple-negative breast cancer. Cell. Mol. Biol. Lett. 2021, 26, 52. [Google Scholar] [CrossRef]
- Cai, Y.; Zhang, T.; Chen, G.; Liu, C. MiR-26a-5p Heightens Breast Cancer Cell Sensitivity to Paclitaxel via Targeting Flap Endonuclease 1. Ann. Clin. Lab. Sci. 2023, 53, 116–125. [Google Scholar]
- Du, Q.; Yuan, Z.; Huang, X.; Huang, Y.; Zhang, J.; Li, R. miR-26b-5p suppresses chemoresistance in breast cancer by targeting serglycin. Anti Cancer Drugs 2022, 33, 308–319. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Mei, Y.; Li, K.; Huang, X.; Yang, H. Downregulation of miR-17-92a cluster promotes autophagy induction in response to celastrol treatment in prostate cancer cells. Biochem. Biophys. Res. Commun. 2016, 478, 804–810. [Google Scholar] [CrossRef] [PubMed]
- Pidíková, P.; Herichová, I. miRNA Clusters with Up-Regulated Expression in Colorectal Cancer. Cancers 2021, 13, 2979. [Google Scholar] [CrossRef] [PubMed]
- Kalecky, K.; Modisette, R.; Pena, S.; Cho, Y.R.; Taube, J. Integrative analysis of breast cancer profiles in TCGA by TNBC subgrouping reveals novel microRNA-specific clusters, including miR-17-92a, distinguishing basal-like 1 and basal-like 2 TNBC subtypes. BMC Cancer 2020, 20, 141. [Google Scholar] [CrossRef] [PubMed]
- Rosas, E.; Roberts, J.T.; O’Neill, K.I.; Christenson, J.L.; Williams, M.M.; Hanamura, T.; Spoelstra, N.S.; Vahrenkamp, J.M.; Gertz, J.; Richer, J.K. A Positive Feedback Loop Between TGFβ and Androgen Receptor Supports Triple-negative Breast Cancer Anoikis Resistance. Endocrinology 2021, 162, bqaa226. [Google Scholar] [CrossRef] [PubMed]
- Dews, M.; Fox, J.L.; Hultine, S.; Sundaram, P.; Wang, W.; Liu, Y.Y.; Furth, E.; Enders, G.H.; El-Deiry, W.; Schelter, J.M.; et al. The myc-miR-17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}-dependent antiangiogenic factors. Cancer Res. 2010, 70, 8233–8246. [Google Scholar] [CrossRef] [PubMed]
- Leung, C.M.; Chen, T.W.; Li, S.C.; Ho, M.R.; Hu, L.Y.; Liu, W.S.; Wu, T.T.; Hsu, P.C.; Chang, H.T.; Tsai, K.W. MicroRNA expression profiles in human breast cancer cells after multifraction and single-dose radiation treatment. Oncol. Rep. 2014, 31, 2147–2156. [Google Scholar] [CrossRef]
- Alsawalha, L.; Ahram, M.; Abdullah, M.S.; Dalmizrak, O. Enzalutamide Overcomes Dihydrotestosterone-Induced Chemoresistance in Triple- Negative Breast Cancer Cells via Apoptosis. Anti Cancer Agents Med. Chem. 2022, 22, 3038–3048. [Google Scholar] [CrossRef]
- Ebrahimi, A.; Bakhshaei Shahrebabaki, P.; Fouladi, H.; Mansoori Derakhshan, S. The impact of microRNAs on the resistance of breast cancer subtypes to chemotherapy. Pathol. Res. Pract. 2023, 249, 154702. [Google Scholar] [CrossRef]
- Chamandi, G.; El-Hajjar, L.; El Kurdi, A.; Le Bras, M.; Nasr, R.; Lehmann-Che, J. ER Negative Breast Cancer and miRNA: There Is More to Decipher Than What the Pathologist Can See! Biomedicines 2023, 11, 2300. [Google Scholar] [CrossRef]
- Jordan-Alejandre, E.; Campos-Parra, A.D.; Castro-López, D.L.; Silva-Cázares, M.B. Potential miRNA Use as a Biomarker: From Breast Cancer Diagnosis to Metastasis. Cells 2023, 12, 525. [Google Scholar] [CrossRef]
- Pekarek, L.; Sánchez Cendra, A.; Roberts Cervantes, E.D.; Sánchez Cendra, C.; Fraile-Martinez, O.; García-Montero, C.; Diaz-Pedrero, R.; Torres-Carranza, D.; Lopez-Gonzalez, L.; Aguado-Henche, S.; et al. Clinical and Translational Applications of Serological and Histopathological Biomarkers in Metastatic Breast Cancer: A Comprehensive Review. Int. J. Mol. Sci. 2023, 24, 8396. [Google Scholar] [CrossRef]
| Criteria | Mean (SD)/Count (%) |
|---|---|
| Age at diagnosis (years) | 52.4 (11.1) |
| Laterality | |
| Right | 11 (68.8) |
| Left | 5 (31.3) |
| Nationality | |
| Jordanian | 13 (81.3) |
| Others | 3 (18.7) |
| Histology | |
| Not otherwise specified | 14 (88.5) |
| Apocrine | 2 (12.5) |
| Grade | |
| High (3) | 14 (87.5) |
| Low (1 and 2) | 2 (12.5) |
| Lymphovascular invasion | |
| Present | 11 (68.8) |
| Absent | 5 (31.3) |
| T status | |
| 1 | 3 (21.4) |
| 2 | 8 (57.1) |
| 3 | 3 (21.4) |
| N status | |
| 0 | 8 (61.5) |
| 1 | 3 (23.1) |
| 2 | 0 (0) |
| 3 | 2 (15.4) |
| M status | |
| 0 | 12 (50) |
| 1 | 12 (50) |
| Vital status: Alive | 12 (75) |
| Median overall survival (months) | 58.2 |
| miRNA | Expression Fold | p-Value | Notes |
|---|---|---|---|
| |||
| miR-17-5p | −3.6 | 0.031 | Down-regulated |
| miR-193b-3p | 2.6 | 0.007 | Up-regulated |
| miR-20a-5p | −3.93 | 0.012 | Down-regulated |
| miR-20b-5p | −5.12 | 0.028 | Down-regulated |
| miR-328-3p # | 3.11 | 0.031 | Up-regulated |
| miR-485-5p | 1.73 | 0.027 | Up-regulated |
| miR-489-3p | 3.29 | 0.009 | Up-regulated |
| |||
| miR-205-5p | 4.41 | 0.029 | Up-regulated |
| |||
| miR-223-3p | 2.33 | 0.04 | Up-regulated |
| miR-26a-5p | 2.1 | 0.04 | Up-regulated |
| miR-26b-5p | 2.77 | 0.03 | Up-regulated |
| miR-489-3p | 3.11 | 0.01 | Up-regulated |
| |||
| miR-181d-5p | 2.52 | 0.04 | Up-regulated |
| miR-193b-3p | 3.21 | 0.03 | Up-regulated |
| miR-328-3p | 2.61 | 0.03 | Up-regulated |
| |||
| miR-205-5p | 9.08 | <0.01 | Up-regulated |
| |||
| None | |||
| |||
| miR-193b-3p | 2.8 | 0.04 | Up-regulated |
| miR-205-5p | 5.86 | 0.01 | Up-regulated |
| miR-20b-5p | −7.82 | 0.03 | Down-regulated |
| miR-328-3p | 2.38 | 0.04 | Up-regulated |
| |||
| miR-489-3p | 2.67 | 0.04 | Down-regulated |
| miRNA Name | Clinical Parameter | ANOVA p-Value | ANOVA FDR # | Multivariate Log Rank p-Value | Multivariate Log Rank FDR |
|---|---|---|---|---|---|
| miR-17-5p | Pathologic N Status | 4.43 × 10−4 | 2.58 × 10−2 | 3.07 × 10−1 | 9.11 × 10−1 |
| Pathologic Stage | 2.28 × 10−2 | 2.24 × 10−1 | 4.59 × 10−1 | 9.98 × 10−1 | |
| Pathologic T Status | 3.70 × 10−4 | 5.48 × 10−3 | 4.70 × 10−1 | 9.98 × 10−1 | |
| miR-193b-3p | Pathologic M Status | 2.25 × 10−3 | 1.50 × 10−2 | 3.19 × 10−1 | 9.48 × 10−1 |
| Pathologic T Status | 1.47 × 10−2 | 8.85 × 10−2 | 1.43 × 10−1 | 7.29 × 10−1 | |
| miR-20a-5p | Pathologic M Status | 4.88 × 10−2 | 1.70 × 10−1 | 8.44 × 10−1 | 9.97 × 10−1 |
| Pathologic N Status | 4.82 × 10−3 | 7.67 × 10−2 | 8.35 × 10−1 | 9.98 × 10−1 | |
| Pathologic T Status | 1.08 × 10−3 | 1.25 × 10−2 | 9.84 × 10−1 | 9.98 × 10−1 | |
| miR-205-5p | Pathologic M Status | 1.80 × 10−3 | 1.27 × 10−2 | 1.41 × 10−1 | 7.17 × 10−1 |
| Pathologic N Status | 1.15 × 10−2 | 1.04 × 10−1 | 1.14 × 10−1 | 6.66 × 10−1 | |
| Pathologic Stage | 5.75 × 10−4 | 2.41 × 10−2 | 1.15 × 10−1 | 6.88 × 10−1 | |
| Pathologic T Status | 1.26 × 10−6 | 1.13 × 10−04 | 1.04 × 10−1 | 6.51 × 10−1 | |
| miR-223-3p | Pathologic N Status | 4.71 × 10−2 | 2.19 × 10−1 | 1.49 × 10−1 | 7.31 × 10−1 |
| Pathologic Stage | 4.56 × 10−2 | 3.13 × 10−1 | 1.64 × 10−1 | 7.75 × 10−1 | |
| Pathologic T Status | 6.68 × 10−3 | 5.01 × 10−2 | 1.40 × 10−1 | 7.29 × 10−1 | |
| miR-26a-5p | Pathologic M Status | 8.70 × 10−1 | 9.41 × 10−1 | 4.75 × 10−2 | 5.27 × 10−1 |
| Pathologic N Status | 4.80 × 10−1 | 7.91 × 10−1 | 2.50 × 10−2 | 4.12 × 10−1 | |
| Pathologic Stage | 9.15 × 10−2 | 4.37 × 10−1 | 1.81 × 10−2 | 4.02 × 10−1 | |
| Pathologic T Status | 4.51 × 10−4 | 6.30 × 10−3 | 2.75 × 10−2 | 3.96 × 10−1 | |
| miR-26b-5p | Pathologic M Status | 9.24 × 10−3 | 5.08 × 10−2 | 1.33 × 10−1 | 7.11 × 10−1 |
| miR-328-3p | Pathologic M Status | 1.89 × 10−2 | 8.63 × 10−2 | 1.14 × 10−1 | 6.84 × 10−1 |
| Pathologic N Status | 1.25 × 10−2 | 1.10 × 10−1 | 9.13 × 10−2 | 6.27 × 10−1 | |
| Pathologic T Status | 2.11 × 10−2 | 1.11 × 10−1 | 7.64 × 10−2 | 6.13 × 10−1 | |
| miR-489-3p | Pathologic M Status | 2.02 × 10−2 | 9.06 × 10−2 | 8.49 × 10−1 | 9.97 × 10−1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahram, M.; Abu Alragheb, B.; Abushukair, H.; Bawadi, R.; Al-Hussaini, M. MicroRNAs Associated with Androgen Receptor and Metastasis in Triple-Negative Breast Cancer. Cancers 2024, 16, 665. https://doi.org/10.3390/cancers16030665
Ahram M, Abu Alragheb B, Abushukair H, Bawadi R, Al-Hussaini M. MicroRNAs Associated with Androgen Receptor and Metastasis in Triple-Negative Breast Cancer. Cancers. 2024; 16(3):665. https://doi.org/10.3390/cancers16030665
Chicago/Turabian StyleAhram, Mamoun, Bayan Abu Alragheb, Hassan Abushukair, Randa Bawadi, and Maysa Al-Hussaini. 2024. "MicroRNAs Associated with Androgen Receptor and Metastasis in Triple-Negative Breast Cancer" Cancers 16, no. 3: 665. https://doi.org/10.3390/cancers16030665
APA StyleAhram, M., Abu Alragheb, B., Abushukair, H., Bawadi, R., & Al-Hussaini, M. (2024). MicroRNAs Associated with Androgen Receptor and Metastasis in Triple-Negative Breast Cancer. Cancers, 16(3), 665. https://doi.org/10.3390/cancers16030665







