Simple Summary
Heat shock proteins (HSPs) are extensively distributed throughout cells. They play a crucial role as molecular chaperones and regulate various cellular processes, such as metabolism, growth, differentiation, cell signaling, and programmed cell death. However, in cancers, HSPs are frequently overexpressed and associated with tumor advancement and metastasis, as well as in acquiring drug resistance against chemotherapeutic agents, leading to poor prognosis. Thus, the expression of HSPs can be modulated to imitate the cellular response against cancer cells by targeting the tumor microenvironment through different mechanisms. For instance, HSPs bind to antigens (peptides) associated with tumors, forming a complex that is easily recognized as an antigen-presenting cell (APC), leading to specific antitumor responses. This review summarizes and discusses recent advances, perspectives, and involvement of HSPs, including small and large HSPs, as well as the molecular mechanisms underlying tumor progression and metastasis. This study may offer new insights into the development of safer and more effective anticancer therapeutics.
Abstract
Heat shock proteins (HSPs) are developmentally conserved families of protein found in both prokaryotic and eukaryotic organisms. HSPs are engaged in a diverse range of physiological processes, including molecular chaperone activity to assist the initial protein folding or promote the unfolding and refolding of misfolded intermediates to acquire the normal or native conformation and its translocation and prevent protein aggregation as well as in immunity, apoptosis, and autophagy. These molecular chaperonins are classified into various families according to their molecular size or weight, encompassing small HSPs (e.g., HSP10 and HSP27), HSP40, HSP60, HSP70, HSP90, and the category of large HSPs that include HSP100 and ClpB proteins. The overexpression of HSPs is induced to counteract cell stress at elevated levels in a variety of solid tumors, including anticancer chemotherapy, and is closely related to a worse prognosis and therapeutic resistance to cancer cells. HSPs are also involved in anti-apoptotic properties and are associated with processes of cancer progression and development, such as metastasis, invasion, and cell proliferation. This review outlines the previously mentioned HSPs and their significant involvement in diverse mechanisms of tumor advancement and metastasis, as well as their contribution to identifying potential targets for therapeutic interventions.
1. Introduction
Heat shock proteins (HSPs) are ubiquitous polypeptide proteins found mostly in every cell from prokaryotes to eukaryotes [1]. HSPs are essential cellular components contributing to the maintenance of cellular protein homeostasis and internal conditions under both moderate and damaged growth conditions [2,3]. It is commonly known that HSPs are involved in a variety of cellular functions, including protein synthesis, folding and assembly, translocation, conformational maintenance, and degradation during cellular processes. These proteins also contribute to the normal growth and development of cells [4,5]. They play essential roles in a myriad of cellular activities: assist in protein synthesis, prevent aggregation of unfolded or misfolded proteins that aid in proper folding and assembly to form functional structures, aid in the translocation of proteins for their proper localization within the cell, maintain proper conformation, thus preventing proteins to acquire potentially harmful structures, help in the degradation of misfolded proteins through proteolytic pathways such as the ubiquitin-proteasome system, and finally assist in the regular processes of cellular growth and development by playing regulatory roles in signaling pathways that control cell proliferation and differentiation. HSPs also have a crucial role in activating client proteins within cells [6]. They have also been found to help in membrane stabilization and protein refolding under stressed conditions [7]. They also help to eliminate damaged or old cells under stress conditions to restore cellular homeostasis [8,9].
Cellular stress includes elevated temperature, reduced oxygen availability, ischemic conditions, and exposure to harmful substances, pathogens, ultraviolet (UV) radiation, or inflammatory signaling molecules. Exposure of organisms to pathophysiological, metabolic, or environmental stress conditions leads to the selective upregulation of HSPs as a natural cellular defense response [10]. This elevated expression of HSPs helps cells resist and thus acts via cytoprotective mechanisms [11].
2. Major HSPs and Their Role in the Various Biological Pathways of Cells
Classified by their molecular weights, HSPs fall into six primary families: HSPH denoted as HSP110, HSPC denoted as HSP90, HSPA denoted as HSP70, DNAJ denoted as HSP40, HSPB- small HSPs, and chaperonins, HSP60. The HSPB gene family generates small HSPs, including 11 ubiquitous molecular chaperones that function independently of ATP. The small HSPs have a size that varies between 12 and 43 kDa. Their size is determined by a β-sandwich structure of the α-crystallin domain, which is conserved. This structure is surrounded by N- and C-terminal sections that can vary greatly [12]. Several high-molecular-weight HSPs are well documented as ATP-dependent chaperones; that is, their function depends on ATPase activity [13]. For example, the HSP40 family is encoded by DNAJ genes and, being the largest among the various HSPs, has 49 members. The chaperonin family of HSP60 in humans consists of 14 members encoded by CCT, HSPD/E, and various other genes [11].
Members of the HSP70 family encode 13 HSPA genes. HSPC contains five homologous members of the HSP90 family. Large HSP families have two members, HSP110 and GRP170 (glucose-regulated protein 170), which are encoded by the HSPH gene family [11,14]. HSP expression is regulated by multiple factors, including cellular stress factors, generally known as the heat shock response (HSR) [15], and is regulated by heat shock factors (HSFs) at the transcriptional level [16]. In vertebrates, HSFs such as HSF1, HSF2, HSF3, HSF4, and HSFY mutually utilize identical structures, with the most highly conserved N-terminal helix-turn-helix DNA-binding domain and C-terminal transactivation domain [17]. Heat Shock Factor 1, initially identified as the primary regulator of HSR and the production of HSP genes, functions via cis-regulatory elements located in the promoter region upstream of HSP genes, which are known as heat shock elements (HSEs) [18]. In contrast to the traditional HSR, certain HSP members, such as HSC70, GRP78, MTP70, and HSP90β, and almost none of the types of HSPs require stress or heat shock for their expression; however, they are constitutively expressed under normal conditions [19]. Table 1 describes the various types of HSP families and their relevant members, along with their cellular locations and proposed functions.

Table 1.
Summary of various families of HSPs and their members with details of cellular location and key functions.
Moreover, HSP expression may be constitutive (cognate) or inductively regulated. Hence, the HSP family can be classified based on their expression as constitutive or inducible HSPs. For instance, HSP73 exhibits continuous expression, whereas the closely related protein HSP72 is responsive to stress for its induction. Both HSP72 and HSP73 function as molecular chaperones, engaging in brief interactions with newly formed polypeptides [33]. A study indicated the dispersion patterns of the constant HSC70 (heat shock cognate 70) and stress triggered HSP70 within the 70 kDa HSP category. The distribution of HSP70s is related to the metabolic needs of the rat eye, which are required for the detection and absorption of light [34]. HSP70 expression is usually strongly induced by stress, but immune cells express these proteins under normal conditions. However, HSC70 is constitutively expressed in mammalian cells [34]. First, the trimerization of HSF occurs before the affinity of the heat shock element (HSE) to the DNA. Phosphorylation triggered by stress is pivotal in regulating the activation and interaction of HSF1 with DNA. The association between HSBP1 and HSF1 monomers hinders the activation of HSF1 [35]. These results suggest that HSP70 can trigger the transition from a fully activated HSF trimer to an inactive monomer [36]. This represents a critical element in reducing the transcriptional response to heat shock. In summary, stress induces HSP70 expression, constitutive expression of HSC70 is observed, and the regulation of HSP expression involves HSFs, trimerization, and stress-dependent phosphorylation, with HSP70 influencing the transition of active HSF trimers to inactive monomers.
Recent studies have reported that molecular chaperones are essential for the translocation of proteins from one cellular compartment to another. For example, mitochondria and chloroplasts import over 95% of their protein constituents from the cytosol, which is further transported into a particular cell organelle via a specific protein import machinery called ATP-driven HSP70 (cpHSP70) [37]. In another study, Sin et al. reported that sHSP20 (small heat shock protein 20) is essential for the nuclear translocation of PKD1 (protein kinase D1) [38].
HSPs participate in various cellular functions such as protein folding, acting as chaperones, regulating apoptosis, managing autophagy, and contributing to immunity. Broadly, these proteins safeguard cells against stressful situations, including elevated temperatures. Additionally, diverse functions of HSPs have been associated with their role in the immune system. These encompass intracellular roles, such as antigen expression and presentation facilitated by innate immunity, as well as extracellular functions, such as participating in tumor immunosurveillance and autoimmunity [39]. Extracellular HSPs also occur in extracellular sites, plasma membrane, cytosol, extracellular vesicles, and biological fluids. Extracellular HSPs interact with the immune system and serve as regulators and stimulants [39]. Based on their location in intracellular or extracellular sites, HSPs have been demonstrated to have dual roles, as reported in multiple studies. Extracellular HSP90 (for example HSP90α) is located on the cell surface of tumors facing the extracellular space and plays an important role in tumor metastasis [40]. Similarly, extracellular HSP70 has been detected in the medium of antigen-presenting cells (APCs), inducing T-cell activation. Extracellular HSP60 has been identified in the plasma of some individuals, but its source remains unknown [41]. Extracellular HSPs such as HSP27, αBC, and HSP20 are secreted into the extracellular medium via non-classical pathways and play important roles in cell-to-cell communication, cell signaling, immunity, stress tolerance, inflammation, and anti-apoptotic activities [42]. Internally located HSPs perform a vital function in safeguarding cells through various mechanisms, providing essential cytoprotection. HSP27 and various molecular machinery involved in programmed cell death are directly involved in its interaction with HSP70 [43]. The biological functions of different HSPs in various cellular processes are shown in Figure 1.
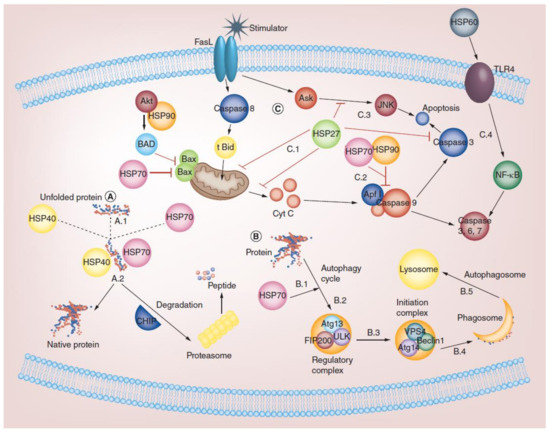
Figure 1.
Role of HSP in biochemical functions of cells. (A) Chaperone activity: misfolded proteins that accumulate in the cytosol bind to HSPs to maintain homeostatic balance (A.1). For example, the HSP70/HSP40 complex guides the degradation of misfolded proteins into short peptides via proteasome (A.2). (B) Role of HSP in the autophagy pathway: the autophagy cycle begins by binding HSP70 to the protein (B.1). Regulation of Atg13, FIP200, and ULK1 complex induced by HSPs (B.2). The initiation of the VPS4, Atg14, and Beclin1 complexes is regulated by HSPs (B.3). Ultimately, phagosomes are formed using phagosomal markers (B.4). Finally, autophagosomes or amphisomes fuse with lysosomes (B.5). (C) Role of HSP in different apoptotic pathways: HSPs help in modulating pro-apoptotic signals through FasL at the mitochondrial and post-mitochondrial levels. HSP70 and HSP27 appear to suppress the release of mitochondrial pro-apoptotic proteins by activating Bax and tBid (truncated Bid), respectively (C.1). HSP27, HSP70, and HSP90 can interact with and recruit Apaf-1 (apoptosis protease-activating factor 1) by directly sequestering cytochrome C to prevent apoptosome oligomerization and activation (C.2). HSP27 can also inhibit apoptosis via inactivation of caspase 3 and ASK (apoptosis signal-regulated kinase) (C.3). Furthermore, HSP60 and TLR-4 interactions mediate the NF-kappaB (NF-κB) signaling pathway, resulting in the activation of caspases 3, 6, and 7 and DNase (C.4). Reprinted (adapted) with permission from Ref. [44]. Copyright 2019, Future Medicine Ltd., London, UK.
3. HSPs and Cancer
The rapid growth of cancer cells demands constant protein synthesis to support replication, leading to significant alterations in signal transduction pathways and metabolic stress. As a result, cancer cells are more protein-dependent. Hence, cancer cells are increasingly dependent on proteins, such as stress-inducible HSPs with molecular chaperone activity, for protein folding to the native form. Furthermore, the cytoprotective functions of HSPs are essential for the maintenance and survival of cancer cells. Undoubtedly, HSP27, HSP70, and HSP90 have been associated with cancer cells, which also indicates that HSPs control cellular processes and functions during carcinogenesis [14].
Numerous studies have documented an increase in the expression levels of HSPs in cancer cells when exposed to different stimuli, such as chemotherapy, inhibition of tyrosine kinases, oxidative stress, elevated temperature (hyperthermia), and activation of the Fas (Apo-1/CD95) apoptotic receptor [16]. This confirmed a notable interaction between the expression levels of HSPs and the resistance of cancer cells during chemotherapy. Various studies have shown that inhibition or depletion of HSPs significantly reduces tumor size and hence cancer progression, as in the case of HSP70. Therefore, targeting different HSPs has been proposed as a potential innovative approach for cancer therapy.
3.1. Role of HSP27 in Cancer Development and Progression
3.1.1. Structure, Function, and Interaction of HSP27
HSP27, an important chaperone, belongs to the sHSP family with a common highly conserved alpha-crystallin domain, as shown in Figure 2A [45]. It plays an essential role in cell proliferation, invasion, metastasis, differentiation, and death. The molecular chaperone HSP27 is ubiquitously distributed in the body and is regulated by phosphorylation at three specific serine residues (Ser82, Ser78, and Ser15), resulting in the formation of an oligomer. The process of phosphorylation described is reversible and primarily facilitated by the enzyme mitogen-activated protein kinase-activated protein kinase 2/3 (MAPKAPK 2/3 kinase). The activation of these MAPKAPK occurs when upstream kinases phosphorylate the p38 mitogen-activated protein kinase (Figure 2B) [46]. The oligomeric status of HSP27 regulates its function as a molecular chaperone [47]. For instance, dephosphorylation provokes large oligomer formation and shows intrinsic chaperonin activity, whereas phosphorylation favors small oligomer formation, which binds to the microfilaments to stabilize HSP27 as an extrinsic function, as shown in Figure 2B [48]. However, under in vivo conditions, the oligomerization of HSP27 is entangled with cell–cell interactions, and the phosphorylation status is independent. Client proteins involved in various stages of the apoptotic pathway, such as p53, NF-kB, and HSP27, control apoptosis by regulating Akt. Overexpression of HSP27 reduces apoptotic cell death in response to different cellular stressors, such as cytotoxic drugs, hyperthermia, and oxidative stress. The anti-apoptotic functions of HSP27 have also been identified through the formation of large oligomers and their tumorigenic properties.
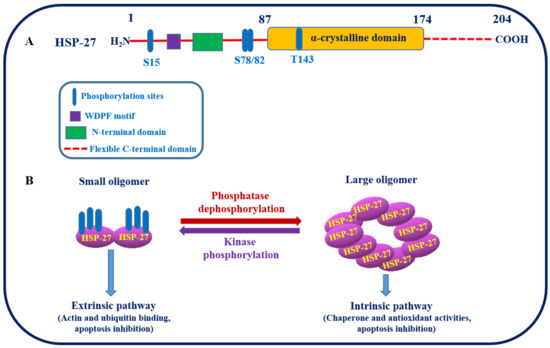
Figure 2.
(A) Structure of HSP27 and its putative phosphorylation sites. (B) Schematic representation showing the two different conformational states of HSP27 induced by phosphorylation, that is, large oligomers when unphosphorylated and small oligomers when phosphorylation is catalyzed by MAPKAPK 2/3 kinase at specific serine residues of HSP27. These conformational, structural, and functional changes actively contribute to the maintenance of cellular proteostasis [49].
3.1.2. HSP27 as Target for Cancers Therapeutics
In several studies, HSP27 was found to be overexpressed, playing critical roles in tumorigenesis, metastasis, and invasiveness, as well as in the poor prognosis of various cancers. Moreover, the phosphorylation patterns of HSP27 in cancer cells are unique and align with those in primary non-transformed cells [50]. Hence, the distinct phosphorylation patterns of HSP27 may be utilized as a useful tumor biomarker [51]. A high level of HSP27 in the tissues of meningioma patients, confirmed by immunohistochemical and Western blot studies, correlated with poor prognostic factors in cancer patients [52]. The overexpression and heat shock accumulation of HSP27 are associated with several characteristics of carcinogenesis, including multi-drug resistance, increased cytoprotection, and apoptosis inhibition with several pro-apoptotic proteins [53,54]. Thus, HSP27 is considered a promising therapeutic target in cancers because of its important role in drug resistance and tumor progression [55,56].
3.1.3. HSP27 as a Therapeutic Target
Three distinct strategies for cancer therapies based on HSP27, as shown in Figure 3, are described as follows:
- ❖
- First, the inhibition of HSP27 is associated with the binding of small molecules [57,58].
- ❖
- Second, protein aptamers are intended to bind to certain proteins of interest, consequently disrupting and inhibiting their functions and preventing dimerization or oligomerization [59].
- ❖
- Finally, antisense oligonucleotide (ASO) binds to HSP27 mRNA and inhibits HSP27 translation [60].
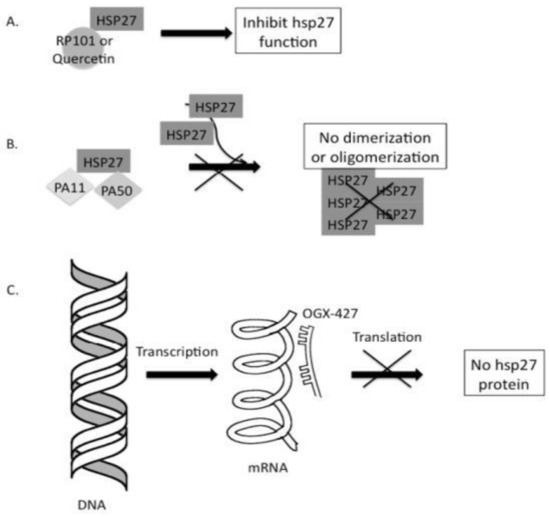
Figure 3.
Strategies targeting cancer cells based on HSP27 inhibition. (A) Small molecules, such as quercetin and RP101 (brivudine), can directly bind to HSP27 and suppress its function. (B) Peptide aptamers such as PA11 and PA50 can specifically bind to HSP27, inhibiting HSP27 dimerization or oligomerization, thereby disabling its function. (C) The final strategy applies Apatorsen (OGX-427), an antisense oligonucleotide that binds to HSP27 mRNA and blocks HSP27 translation, thereby HSP 27 protein production inhibited. Consequently, the amount of HSP27 is reduced, causing cell death, which protects the cell through its anti-apoptotic activity. Reprinted (adapted) with permission from Ref. [61]. Copyright 2013, Elsevier.
HSP27 inhibitors, such as quercetin and RP101, are two important molecules that have already been studied. RP101 (Brivudine) also known as bromovinyldeoxyuridine (BVDU) is an antiviral nucleoside that binds with HSP27 via binding π-stacking with Phe-29 and Phe-33 of HSP27 to inhibit its function (Figure 3A) [58,62]. When RP101 binds to HSP27, there is less binding of HSP27 to pro-caspase3, cytochrome C, and Akt1, which affects apoptosis [58,63]. Hence, the development of RP101 as a cancer drug is a unique approach to enhance chemosensitivity and prevent chemoresistance. In clinical studies involving stage 3 and 4 pancreatic cancer patients, the median overall survival rate increased by more than 8.5 months due to the RP101vs historical control group (NCT00550004) [58]. Additionally, during a phase II clinical trial targeting pancreatic cancer, the combination of RP101 with gemcitabine resulted in an increase in survival period by around 2.17 months. Despite the termination of the clinical study due to adverse side effects of gemcitabine, the RP101-based trial reported no such adverse effects. Consequently, the advancement of second-generation RP101 candidates is presently in the developmental stages.
Quercetin is a bioflavonoid widely distributed in ferns. This compound is also a popular natural compound with anticancer activity via HSF1 (heat shock transcriptional factor1)-dependent HSPs in several cancer cell lines [64]. Most non-small-cell lung cancer (NSCLC) cell lines, such as A549, have been reported to inhibit HSP-27, thereby reducing cell survival. Furthermore, quercetin also serves as a chemosensitizer when used in combination with chemotherapeutics, such as cisplatin or gemcitabine, as it exhibits more potent cytotoxic activity in A549 cells than when applied alone [65]. However, the exact mechanism of action of quercetin is not adequately understood, and a recent study showed that the inhibition of cellular expression of CK2 (casein kinase 2) by quercetin facilitates the proteasome-mediated degradation of HSP27 [66,67]. Casein kinase 2 plays an essential role in increasing the stability of HSP27 protein [68], indicating that quercetin acts as an important regulator of HSP27 protein stability by inhibiting CK2. It is a widely recognized natural compound that exhibits anticancer effects through the regulation of heat shock transcriptional factor 1 (HSF1)-dependent HSPs in numerous cancer cell lines [18]. In several NSCLC cell lines, including A549, it has been observed that HSP27 inhibition occurs, leading to a decrease in cell viability.
The second approach involves the application of specific peptides that inhibit HSP27 anti-apoptotic activity by binding to a particular protein domain (Figure 3B) [69]. Currently, PA11 and PA50 are two important peptide aptamers that specifically bind to HSP-27, thereby interfering with its oligomerization or dimerization, and disrupting its function. PA11 inhibits HSP27 oligomerization, which ultimately perturbs early cellular proteostasis and induces proteotoxic stress and cell death ensues [59]. The mechanism of PA50 is to inhibit the dimerization of HSP27. By inhibiting HSP27 dimerization, PA50 interrupts HSP27 to participate in cell-signaling events that are essential for cell survival.
Moreover, like molecular inhibitors, peptide aptamers are not capable on their own but function as chemo-sensitizing agents in combination with other therapies to sensitize cancers. For instance, both PA11 and PA50 accelerate the cellular radiosensitivity of HNSCC (head and neck squamous cell carcinoma) cell lines by 47% and 32%, respectively, against control. Although peptide aptamers have demonstrated promising results in preclinical models and trials for cancer therapy, they encounter specific limitations. These constraints include size restrictions on the studied proteins, challenges in addressing membrane protein complexes, and difficulties in operating in an RNase-free environment. To address these issues, overcoming obstacles and enhancing the understanding of the structure–function relationship of HSP27 is crucial, leading to the discovery of new anticancer drugs.
The third approach involves the application of second-generation antisense oligonucleotides (ASOs) that target HSP27 mRNA. The sequence of Apatorsen or OGX-427 (OncoGenex Pharmaceuticals, Bothell, WA, USA) is an antisense oligonucleotide that inhibits the expression of HSP27 (Figure 3C) by preventing HSP27 mRNA translation through binding to the initiation site (5′-GGGACGCGGCGCTCGGTCAT-3′) [70]. In a xenograft model of prostate cancer (PC-3), the co-administration of OGX-427 and chloroquine resulted in a 50% reduction in tumor volume after a seven-week treatment period, compared to the effect observed with chloroquine treatment alone [71].
Inhibition of HSP27 significantly enhanced radiation-induced cell death and apoptosis and promoted Akt inactivation. In SQ20B-cancer-cell-bearing mice (radiation-resistant lung cancer), knockdown of HSP27 using OGX-427 enhanced the efficiency of radiation therapy by increasing the cytotoxic efficacy and decreasing glutathione antioxidant defenses and cell survival [72]. Radiation and OGX-427 decreased angiogenesis, which was related to a reduction in Akt pathway activity. This combination treatment also enhances cell survival and antitumor effects in SQ20B-tumor-bearing mice, with no effect on acute or delayed tumor growth [72]. Furthermore, a clinical study demonstrated that the survival rate of patients with metastatic pancreatic cancer and the duration of progression-free survival were extended when standard chemotherapy was combined with OGX-427.
In summary, HSP27 plays an important role in cancer treatment and has become a novel therapeutic target. However, HSP27, with its greater structural complexity, has become a significant challenge in the search and design of therapeutic molecules that can be easily neutralized. Both the large and small oligomers of HSP27 are in dynamic equilibrium, which is regulated by HSP27 phosphorylation and functions in its oligomeric form. Therefore, it is preferable to target only the phosphorylated or unphosphorylated forms of HSP27 using various molecules. Moreover, HSP27 is involved in the prevention of protein aggregation, thereby inhibiting several neurodegenerative diseases. Hence, HSP27 may be considered for the prevention of neurodegenerative diseases along with its use as a target in cancer therapy.
3.2. Role of HSP40 in Cancer Progression and Potential Therapeutic Target
3.2.1. HSP40 Structural Studies and Functional Implications
The HSP40 family plays a crucial role in the translation, folding, refolding, translocation, degradation, and assembly/disassembly of proteins. The DNAJ protein family encodes 49 proteins in humans and is primarily categorized into three subtypes, types I, II, and III (also indicated as DNAJA, DNAJB, and DNAJC, respectively), depending on the presence and location of the domain structure [73]. Type I HSP40 (DNAJA) comprises four protein domains: an N-terminal JD (J-domain), a GF-motif (a glycine/phenylalanine-rich region), a Cys-repeat (a cysteine repeat), and a largely uncharacterized C-terminus [74]. Type II (DNAJB) comprises 13 distinct members, characterized by the absence of the Cys-repeat region and an elongated GF-motif region. In contrast, Type III (DNAJC) encompasses 32 members, exhibiting substantial distinctions from both Type I and Type II DNAJs. Notably, Type III lacks both GF-motif and Cys-repeat regions, and the J-domain may be positioned anywhere within the protein domain. The structure of the J-domain consists of approximately 70 amino acids folded into four α-helices (helices I–IV), with a loop region between helices II and III containing a highly conserved tripeptide, the HPD (His-Pro-Asp) motif [75]. The helical wheel representation of helix II is crucial for its interactions with the ATPase domain of HSP70 [76].
DNAJ stabilizes the interaction between HSP70 and its substrates. The highly conserved HPD motif of the J-domain plays an essential role in HSP40/HSP70 interactions, and HSP40 stimulates HSP70 ATPase activity [77]. The ATPase domain of HSP70 results in ATP hydrolysis to convert HSP70 into an ADP-bound state, thus maintaining its interaction with client polypeptides [78]. The ATP-binding site of HSP70 resides in the N-terminal ATPase domain and has low affinity for client substrate proteins. Currently, two isoforms of DNAJ directly interact with HSP90: DNAJC7 (Tpr2) and DNAJB2 isoform b (HSJ1b) [79]. DNAJC7 contains various highly conserved tetratricopeptide repeat (TPR) motifs that connect other co-chaperones to HSP90 and HSP70, such as HSP90–HSP70 sorting-out proteins (HOP) [80]. The tetratricopeptide-repeat-containing (TPR-containing) chaperone is associated with HSP70 and HSP90 through the C-terminal EEVD motif [80,81].
Another principal function of HSP40 is translocation. For instance, DNAJC6/auxilin and DNAJC26/auxilin-2 play important roles in endocytosis by uncoating clathrin-coated vesicles in association with HSC70 [82]. DNAJs/HSP40s also play a vital role in targeting denatured and damaged proteins for proteasomal degradation in the cytosol by preventing conglomeration.
3.2.2. Role of HSP40 in Carcinogenesis
The overexpression of HSP40 plays a potential role in malignancy progression in many human cancers, such as liver, cervical, gastric, lung, prostate, and colorectal cancers. The dual roles of HSP40 family members are due to their contribution to both pro-malignant and anti-malignant effects [83]. DNAJA3, recognized as Tid1, binds to HSP70 and directly attaches to p53. This interaction results in the translocation of mitochondria, triggering a crucial apoptotic process in MCF-7 breast cancer cells. It functions as a regulator of apoptosis mediated by p53 [84,85]. Decreased Tid1 levels facilitate the promotion of cell growth and a significant reduction in apoptosis in osteosarcoma cells, contributing to Tid1’s role as a tumor suppressor. Conversely, an increase in Tid1 expression is associated with the initiation of colorectal cancer in certain cases [86].
DNAJB11 (ERdj3) is a stress-inducible endoplasmic reticulum (ER) DNAJ homolog that binds to the Kaposi sarcoma-related herpes virus (KSHV) K1 protein, enhances its expression, and shows anti-apoptotic function that promotes cancer [87]. Different members of the HSP40 family exhibit oncogenic activities. The expression of DNAJB1 destabilizes PDCD5 (programmed cell death protein) to inhibit p53-associated apoptosis in multiple cancer cells [88]. The overexpression of DNAJC6 and DNAJB8 has been reported in the development of tumors in hepatocellular carcinoma and renal cancer cells, respectively [89,90].
3.2.3. HSP40 as a Therapeutic Target
HSP40 members regulate the effects of chemotherapeutic agents. An immunological methodology to target HSP40 involves immunization against the DNAJB8 isoform. DNAJ (HSP40) homolog subfamily DNAJB8 belongs to the HSP40 family, and its expression was detected in the development and metastasis of cancer. Enhanced expression of DNAJB8 has also been observed in stem cells derived from cancer (CSCs) in patients diagnosed with renal cell carcinoma. This suggests that DNAJB8 plays a special role in the onset of malignant growth and can be used to target CSCs for therapy [89,91].
In the pursuit of diverse strategies to combat cancer, a range of inhibitors have surfaced, each uniquely targeting and influencing the mechanisms associated with HSP40. N-formyl-3,4-methylenedioxy-benzylidene-γ-butyrolactam (KNK437), a pan-HSP inhibitor, inhibits the expression of various HSPs, including HSP40, in human colon carcinoma cells [92,93]. BMS-690514 is a potent inhibitor of the epidermal growth factor (EGF) receptor (HER)/vascular endothelial growth factor (VEGFR) receptor and exerts anti-apoptotic effects by decreasing the expression of HSP40 and other HSPs in non-small-cell lung cancer (NSCLC) cells [94,95]. Farnesyltransferase inhibitor (FTI) R115777 (tipifamib) is an active inhibitor that restricts tumor development, cell survival, and angiogenesis (HDJ2, VEGF, and MMP1) pathways in breast carcinoma cells [96,97]. In addition, R115777 inhibitor targets HDJ-2 to enhance the radiosensitization of glioblastoma multiforme (GBM) cells [98]. A derivative of phenoxy-N-arylacetamides is known to inhibit the expression of HSP40 but is still in the initial stages of development [99].
Knockdown of DNAJB1 increased the effect of gefitinib (a selective EGF receptor inhibitor) by stimulating mitogen-inducible gene 6 (MIG6) in lung adenocarcinoma A549 human cells [100]. Prenylation is a post-translational modification of type 1 DNAJs/HSP40s. In vivo studies have recently demonstrated that simvastatin treatment inhibits geranylgeranylation in murine mononuclear cells (MNCs) from patients with acute myeloid leukemia (AML) [101]. Enhanced chemosensitivity of AML cells following simvastatin treatment was also shown. It was concluded that simvastatin caused increased chemosensitization in a subset of AML patients by inhibiting the prenylation of DNAJs/HSP-40s [102].
3.3. Role of HSP60 in Cancer and Its Potential Therapeutic Target
3.3.1. Structure and Function of HSP60
The molecular chaperone HSP60 has emerged as a potential target in cancer therapy. It is widely distributed in the mitochondria and assists in the correct folding of proteins. HSP60 and its co-chaperone HSP10 form a complex that mediates the folding of proteins imported into mitochondria [103]. Human HSP60 and its bacterial homolog are found mainly in the form of a tetradecameric structure and a homo-oligomeric ring of seven subunits [104]. The human HSP60 protein has three cysteine residues (Cys237, Cys442, and Cys447) but not in the bacterial homolog (GroEL) [105]. HSP60 has three structural domains: equatorial (ATP binding), intermediate (connecting the equatorial and apical domains), and apical (substrate binding) [106].
In addition to mitochondrial protein folding, some evidence indicates that intracellular HSP60 interacts with survivin, mortalin (mitochondrial HSP70), and p53 to mediate apoptosis [107,108]. Thus, increasing the expression of surface HSP60s activates cellular immunity and dendritic cell maturation; consequently, an antitumor response of T-cells is created [109]. Extracellular HSP60 can engage with multiple cell-surface receptors, such as CD14, CD40, and Toll-like receptors (TLRs), causing either a pro- or an inflammatory response [110,111] and presenting tumor-associated peptide antigens to the immune system. The pro-inflammatory effect of HSP60, including cytokines secreted by APCs, induces T-cell activation [112]. In addition, HSP60 induces maturation and activation of dendritic cells through the TLR-2 and TLR-4 signal-transduction pathways, synergizing the proinflammatory action of IFNγ [113].
3.3.2. HSP60 and Cancer Development
HSP60 promotes the survival and progression of certain types of cancer cells. For instance, cytosolic HSP60 prevents the activity of IkB kinase (IKK), resulting in the survival of human cervical carcinoma (HeLa) cells [114]. Other studies have shown that HSP60 stabilizes the mitochondrial pool of survivin [115]. The complete exclusion of HSP60 leads to mitochondrial dysfunction, which in turn triggers apoptosis. This process involves the disintegration of the HSP60 and p53 complex, which results in the stabilization of p53, upregulation of BAX expression, and onset of apoptosis dependent on BAX in breast and colon carcinoma cells [108]. HSP60 regulates apoptosis by interacting with cyclophilin D (a protein within the mitochondrial permeability transition pore). Furthermore, the activation of mitochondrial HSP60 triggers CypD-dependent mitochondrial permeability transition, caspase-dependent apoptosis, and the suppression of tumor growth [116]. HSP60 directly interacts with β-catenin and promotes metastasis of malignant cells [117]. In both early and advanced prostate tumors, there is a considerable increase in the expression of HSP10 and HSP60 [118].
3.3.3. HSP60 as Therapeutic Target
HSP60 plays a vital role in the persistence, advancement, and development of resistance in cancer cells, cytoprotection against cell stressors, and anti-apoptotic potential [119]. For example, 5-FU-resistant SW480 colorectal cancer (CRC) cells showed a reversal of drug resistance after the inhibition of HSP-60 expression [120]. Mizoribine, an imidazole nucleoside antibiotic isolated from Eupenicillium brefeldianum derivative, was the first HSP60 inhibitor to be reported. On associating with HSP-60, Mizoribine suppresses its ATPase activity and destroys the HSP60-HSP10 complex [121]. The antitumor effects of bortezomib (a proteasome inhibitor) elevated the expression of HSP60 and HSP90 on cancer cell surfaces and promoted phagocytosis by dendritic cells (DCs) in a murine model of ovarian cancer [122]. Synthetic HSP60 inhibitors, such as o-carboranyl-phenoxy-acetanilide and gold (III) porphyrin complexes, have been found to be highly effective against malignant growth in numerous cancer cell lines [123,124]. Small interfering RNA (siRNA) used to silence the HSP60 gene results in mitochondrial damage, disruption of the Hsp60-p53 complex, Bax expression, and Bax-dependent (caspase-dependent) apoptosis in breast and colon cancers [125]. siRNA-mediated gene silencing of HSP60 in healthy cells does not induce apoptosis [125]. Hence, this approach could be used as a target-specific therapy for breast and colon cancer.
3.4. The Role of HSP70 in Cancer Development and Its Application as a Therapeutic Target
3.4.1. Molecular Structure and Cellular Functions of HSP70
Human HSP70s comprise a family of 13 members that perform their functions depending on their cellular localization, among which five members that are most concerned with cancer development are stress-inducible HSP70 (HSP72 or HSPA1) and HSPA6 (HSP70B) and constitutively expressed GRP78 (HSPA5), HSC70 (HSPA8), and mortalin (HSPA9) [11]. Under healthy conditions, HSP70 uses ATP to assist in the folding and assembly of newly synthesized polypeptides, maintain proteostasis, and enhance the survival of cells under or after stress [126]. HSP70 contains an interdomain linker that links two distinct highly conserved functional domain structures, namely an N-terminal ATPase domain (45 kDa nucleotide-binding domain, NBD) and a C-terminal polypeptide-binding domain (25 kDa protein substrate-binding domain, SBD) [127]. Furthermore, a short, highly conserved linker links these two domains, which is only 12 residues long, as shown in Figure 4B [127]. Amongst the HSP70 family members, the least conserved region is the C-terminal to the “lid,” which terminates with an EEVD (amino acid designation) motif and is able to interact with the tetra-trico-peptide repeat (TPR) domain. The interaction and sequence variation of HSP70 is believed to allow family members to have distinct co-chaperones, thereby regulating its chaperone function. The co-chaperones of HSP70 are HSP40, Bcl-2-associated athanogene1 (BAG-1), and the C-terminus of HSP70 interacting protein (CHIP).
These can be classified into three categories. They are as follows:
- The J-domain co-chaperones, a family of HSP40, bind to the NDB of HSP70 and stimulate the ATPase activity of this protein.
- Nucleotide exchange factor (NEF) co-chaperone members including Bag-1, HSP110, and HSPBP1 (HSP70 binding protein1) stimulate ADP release and complete the chaperone cycle.
- The tetratricopeptide repeat (TPR) domain co-chaperones members include Hop and CHIP binding to the C-terminal EEVD motif present in both HSP70 and HSP90 chaperones, which is essential for the assembly and intermolecular properties of the HSP70/HSP90 multichaperone complex. Thus, CHIP with ubiquitin ligase activity has also been implicated in the ubiquitination and degradation of HSP70 client proteins [128].
3.4.2. Role of HSP70 in Cancer Development and Survival
HSP70 plays a crucial role in occurrence, progression, and cancer metastasis and is frequently expressed at extreme levels, especially the stress-inducible isoform of HSP70 (also known as HSP-701A1, HSP70-1, or HSP72). Usually, these are undetectable in cells; however, HSP70 can be detected in cells using flow cytometry or confocal microscopy, and their elevated expression helps cancer cells rebalance proteostasis under high-stress conditions [126]. HSP70 overexpression in cancer is associated with the promotion of metastasis, differentiation, apoptosis, tumor growth, drug resistance, and poor prognosis [129]. HSP70 is also involved in most molecular mechanisms, and several checkpoints are involved in cancer development. Evidently, HSP70 is thought to inhibit both the intrinsic and extrinsic apoptotic pathways. In the intrinsic pathway, HSP70 inhibits Bax activation, thereby preventing the permeabilization of the mitochondrial outer membrane and the release of apoptosis-inducing factors [130]. In the extrinsic pathway, HSP70 blocks death-inducing signaling complex (DISC) assembly [131]. Senescence can also act as a powerful antitumor mechanism by preventing the proliferation of cancer cells. Reduced HSP72 expression triggers cell senescence in specific cancer cell lines through pathways leading to senescence that involve both p53-dependent and p53-independent mechanisms [132]. Bag3, serving as a nucleotide exchange factor for HSP-70, is regulated by different transcription factors such as Hif1α, NF-κB, and FoxM1; translation regulators such as HuR; and cell-cycle regulators such as p21 and survivin [133]. Furthermore, BAG3 is also involved in autophagy and cell death. Additionally, HSP70 plays a crucial role in maintaining the genomic integrity of DNA by binding to poly (ADP-ribose) polymerase-1 (PARP-1) in the base pair excision system. DNA-damaging agents have been used extensively in cancer chemotherapy. Hence, the use of HSP70 inhibitors alone or in combination with other drugs may be a better therapeutic strategy for cancer as shown in Figure 4 [134].
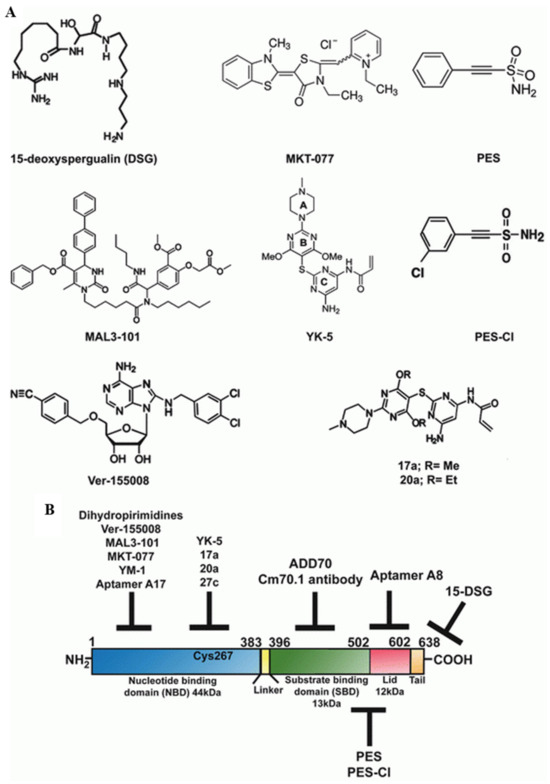
Figure 4.
(A) Structures of HSP70 inhibitors. (B) Schematic diagram of the domain architecture of HSP70 and the possible sites targeted by various inhibitors. Reprinted (adapted) with permission from Ref. [135]. Copyright 2015, Springer Nature.
3.4.3. HSP70 as an Emerging Drug Target
Three distinct strategies for cancer therapy based on HSP70 are shown in Figure 4:
- Targeting C-terminal peptide or substrate-binding domain (SBD).
- Targeting the N-terminal ATP-binding domain (ABD) or N-terminal domain of the nucleotide.
- Targeting HSP70 co-chaperones.
3.4.4. Targeting C-Terminal Peptide or Substrate-Binding Domain (SBD)
HSP70 has a prominent cytoprotective ability to inhibit apoptosis by neutralizing its function and interacting with apoptosis-inducing factor (AIF) and apoptotic protease activating factor-1 (APAF-1) [136]. Garrido and co-workers rationally designed decay targets of HSP70, which are derived from AIF that neutralized HSP70 function, and hence inducted the sensitization of cancer cells and apoptosis [137]. The blocking peptide derived from AIF corresponds to amino acids 150–228 in the AIF region, which is a crucial segment for binding to the SBD of HSP70, referred to as peptide ADD70 (AIF-Derived Decay). ADD70 disrupts the interaction between HSP70 and AIF, along with other proteins designated as client proteins [137]. 2-phenylethynesulfonamide (PES), a small-molecule inhibitor of HSP70, specifically interacts with its C-terminal substrate-binding domain of HSP70 (Figure 4). Consequently, this triggers the aggregation of improperly folded proteins, disrupts autophagic processes, and destabilizes the usual functions of lysozyme, ultimately prompting autophagic cell death [138]. The outcome of PES induction of either autophagic or apoptotic cell death is contingent on the cell type.
3.4.5. Targeting the N-Terminal ATPase Domain (ABD) or N-Terminal Nucleotide-Binding Domain (NBD)
Compounds that replace ATP from HSP70 are anticipated to be powerful tools for regulating HSP90 inhibitors. VER-155008 an adenosine-based HSP70 inhibitor binds to the ATPase site of the HSC70/BAG-1 crystal structure. It has also been reported that VER-155008 targets the degradation of various HSP90 client proteins such as CDK4, HER2, and RAF-1 [139]. Interestingly, MKT-077 is a cationic rhodacyanine dye analog that binds to a negatively charged pocket close to the ABD of HSP70 and thus disrupts the HSP70 folding cycle [140]. MKT-077 has been explored as an anticancer agent in mouse xenografts, and the trial showed severe renal impairment and toxicity, thus limiting further study of MKT-077 [141]. Recently, YM-1, a derivative of MKT-077, was shown to exhibit selective toxicity against diverse tumor cell lines, highlighting its potential as a promising therapeutic candidate [142]. Reports indicate that VER-155008 is effective in disrupting the degradation of several HSP90 client proteins, including CDK4, HER2, and RAF-1 [35]. Notably, MKT-077, a cationic rhodacyanine dye analog, binds to a negatively charged pocket near the ABD of HSP70, thereby disrupting its folding cycle of HSP70.
Screening for HSP70 ATPase activity revealed that 15-deoxyspergualin (DSG) inhibits HSP70 steady-state ATPase activity by interacting with the C-terminus and hindering its function [143]. However, the efficacy of DSG as an anticancer drug has been limited, which has subsequently led to the search for compounds with structural similarity, resulting in the identification of MAL3-101, a second-generation dihydropyrimidine [144]. MAL3-101 inhibits the ability of HSP40 co-chaperone, accelerates the ATPase activity of HSP70, and prevents its function [145]. Recently, smaller peptide aptamers, A17, were developed using peptide aptamer libraries of Escherichia coli thioredoxin scaffolds. A17 binds to the ATP-binding domains of HSP70 and specifically inhibits chaperone activity [146].
Multhoff and Hightower reported that the presence of HSP70 on the surface of tumor cells stabilizes the lysosomal membrane of tumor cells to escape from cell death as a potential therapeutic target [147,148]. HSP70 is expressed only on the surface of tumor cells and stains positively for the IgG1 mouse monoclonal antibody (mAb) cmHSP70.1 [149]. Furthermore, mAb CmHSP70.1 recognizes conserved epitopes in the C-terminal amino acids (aa 450–461) of inducible HSP70 on both viable human tumors (in vitro) and mouse tumors (in vivo), as shown in Figure 4B [150]. The injection of the Cm70.1 antibody into mice-bearing tumors significantly inhibited tumor growth and improved overall survival. This finding indicated that cmHSP70.1 could be used as a therapeutic agent.
3.4.6. Targeting HSP70 Co-Chaperones
The best way to inhibit HSP70 function involves determining specific molecules that precisely interrupt the interaction between HSP70 and its specific co-chaperone; however, this has not been explored in detail. Using a high-throughput screening approach, pyrimidotriazinediones were designed to interfere with Hop/HSP70 interaction and were found to be toxic to WST-1 cells in vitro [151]. It was also recently reported that targeting Hip impairs the proteostatic activity of the HSP70 chaperone and stimulates neurodegeneration [152].
3.5. HSP90 as a Target for Cancer Development
HSP90 is a vital chaperone and client protein of the HSPs family that is involved in cancer progression. HSP90 is the most abundant, highly conserved, and expressed molecular chaperone in eukaryotic cells, representing 1–2% of the total cellular proteome under physiological homeostatic conditions. Under stress, HSP90 expression rises to approximately 4–6% of cellular proteins, stabilizing itself and boosting cell survival [153]. HSP90 is essential for promoting the development, resilience, and effectiveness of cancer-causing client proteins [154]. HSP90 performs housekeeping functions such as proper folding of newly synthesized proteins, restoration of denatured proteins to their functional states, directing proteins to specific cellular compartments, averting the aggregation of proteins, facilitating the maturation of signaling proteins, and orchestrating the regulated degradation of proteins [155]. HSP90 functions in a multicomponent machine of chaperone proteins, including p23, p60/Hop, p50Cdc37, HSP40/HDJ2, HSP70, and various immunophilins [156]. HSP90 is unique among molecular chaperones due to its preference for protein kinases and steroid hormone receptors as client proteins. These proteins play essential roles in signal transduction pathways [156].
Many of these client proteins, including erythroblastic oncogene B (ERBB2), tumor suppressor protein (p53), breakpoint cluster region-Abelson (Bcr-Abl), epidermal growth factor receptor 2 (HER2), B-Raf proto-oncogene serine/threonine kinase (B-Raf), C-Raf proto-oncogene serine/threonine kinase (C-Raf), protein kinase B (AKT), mesenchymal-epithelial transition factor (MET), vascular endothelial growth factor (VEGF), fms-like tyrosine kinase 3 (FLT3), hypoxia-inducible factor (HIF)-1, telomerase, androgen, and estrogen receptors, have been shown to promote cell growth, proliferation, and cell survival. Many of these clients are frequently mutated and/or overexpressed in cancer cells [157,158,159]. HSP90 frequently exhibits higher levels of expression in comparison to its healthy counterparts, and it plays a vital role in promoting the proliferation and survival of cancer cells. Additionally, there have been reports indicating that the suppression of HSP90 results in the breakdown of client proteins through the ubiquitin/proteasome pathway. HSP90 is essential for correctly folding client proteins and has become an attractive target for the development of anticancer medications [159].
3.5.1. Structure and Functional Relationship of HSP90
HSP90 is a large and flexible homodimer protein that functions under physiological conditions. Each monomeric unit is composed of three highly conserved functional domains: the N-terminal domain (NTD) or ATP-binding domain, the middle domain (MD) or catalytic domain, and the C-terminal domain (CTD) or dimerization domain [160]. In addition, it has specific sites for its binding partners, including co-chaperones and client proteins. The N-terminal domain contains a nucleotide-binding pocket for binding to ATP/ADP. Hence, the binding of ATP and its subsequent hydrolysis provide the energy required for protein folding. The C-terminal domain provides dimerization for HSP90 and has a putative ATP-binding pocket that serves as an allosteric regulator for the function of HSP90. The middle domain provides a site for the interactions of client proteins and co-chaperones with HSP90 to form the HSP90–co-chaperone–client protein complex. Hence, the dynamic nature of the HSP90 chaperone cycle provides multiple targeting sites for the modulation of chaperone activity using small-molecule ligands. Moreover, some members of the HSP90 family, such as cytosolic eukaryotic HSP90 and GRP94 (94 kDa glucose-regulated protein), have disordered regions, termed the charged linker, which separates NTD and MD. Furthermore, cytosolic eukaryotic HSP90 contains an MEEVD motif at the C-terminal extension. The NTD- or ATP-binding domain of HSP90 is unique and distinct from the ATP-binding cleft of HSP70 and other protein kinases. The p23 (yeast homolog Sba1) and p50 (yeast homolog Cdc37) co-chaperones are also connected to the NTD binding site [161].
The MD of HSP90 comprises a binding site for co-chaperone Aha1 (activator of HSP90 ATPase activity1) and client proteins such as Cdk4, Akt, eNOS, phycobilisome-linker polypeptides, and staphylococcal nuclease. The dimerization of HSP90 is crucially influenced by its carboxy-terminal domain, which is also responsible for interacting with client proteins such as the tumor suppressor protein p53 [162]. The N-terminal ATP binding site leads to the dimerization of HSP90 (open state) and facilitates co-chaperone and client protein binding. The ATP hydrolysis conformational cycle of HSP90 (closed state) was enhanced by the co-chaperone, as shown in Figure 5 [163]. Several co-chaperones, Hop (HSP organizing protein-yeast homolog Sti1), protein phosphatase 5 (PP5-Yeast homolog Ppt1), and FK506-binding protein FKBP52, which contains multiple TPR domains, have the ability to recruit the HSP90 chaperone and simultaneously bind to the C-terminal MEEVD motif of HSP90 [164].
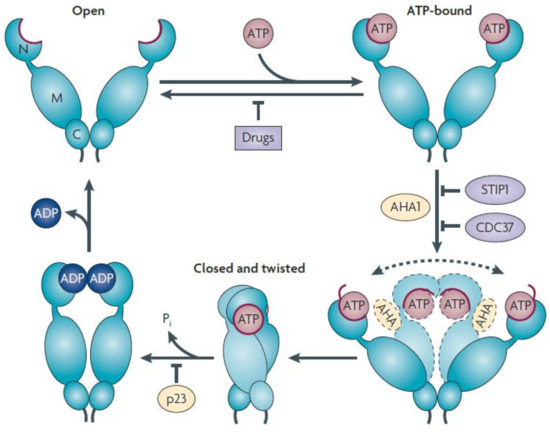
Figure 5.
The chaperone, HSP90, undergoes multiple conformational states in the absence of nucleotides or other factors. ATP binding and hydrolysis shift conformational equilibrium by lowering the energy barrier between specific conformations, thus providing a conformational cycle for HSP90 [165]. ATP binds to the undimerized NTD (open state) of HSP90 and leads to N-terminal dimerization caused by the “lid” segment (red) repositioning. The subsequent structural rearrangement establishes a (closed and twisted) conformation of HSP90, which is suitable for ATP hydrolysis. The co-chaperone AHA1 increases HSP90 ATPase 1 activity by promoting conformational changes required to achieve ATPase competence. In the absence of AHA1, it was very difficult for HSP90 to achieve ATPase-competent conformation (dotted arrow). A number of co-chaperones, including Sti1p (p60HOP), Cdc37 homolog (cell division cycle 37), and N-domain-binding HSP90 inhibitors, prevent dimerization of the N-terminal domain. PTGES3 (prostaglandin E synthase 3 cytosolic, so-called p23 protein) deregulates the ATPase cycle and stabilizes its closed conformation. Reprinted (adapted) with permission from Ref. [163]. Copyright 2010, Springer Nature.
3.5.2. Targeting the N-Terminal Nucleotide Binding Domain
The nucleotide-binding pocket found in the N-terminal domain (NTD) of HSP90 binds with ATP, and this unique pocket structure is a common feature within the GHKL (gyrase, HSP90, histidine, kinase, MutL) superfamily of proteins. These compounds prevent ATP binding and hydrolysis, leading to the proteasomal degradation of proteins assisted by HSP90. Natural inhibitors, such as radicicol (RD) and geldanamycin (GM), hinder HSP90 by interacting with the N-terminal site, a structural mimicry of ATP [166]. Although these natural HSP90 inhibitors exhibit potential antitumor activities, their clinical research and applications are limited owing to their poor solubility and significant hepatotoxicity [128], which has led to the development of second-generation HSP90 inhibitors using derivatives of natural inhibitors. To improve solubility and reduce toxic side effects, semi-synthetic derivatives of GM, including IPI-504, 17-AAG, and 17-DMAG, have been developed for better clinical features. RD and its derivatives NVP-AUY922, AT-13387, STA-9090 (ganetespib), and KW-2478 have been developed with improved stability and clinical properties. Among the derivatives of GM, IPI-504 was identified as a prodrug of 17-AAG, specifically a hydroquinone hydrochloride formulation, exhibiting enhanced potency and improved toxicity profiles in preclinical investigations compared to its parent compound. Results from Phase 1 clinical trials indicated that KW-2478, used for multiple myeloma either independently or along with bortezomib, exhibited a positive response rate of 39% and a progression-free survival of 26.4 weeks [167]. The availability of X-ray crystallographic structures for HSP90 attached to ATP, geldanamycin, and radicicol has proved pivotal in formulating HSP90 inhibitors, ultimately contributing to the development of synthetic inhibitors. The first purine-based synthetic HSP90 inhibitor, PU3, was identified by Tony Taldone and Gabriela Chiosis [168], along with other inhibitors synthesized later as a common pharmacophore. In addition, other purine analogs that inhibit HSP90 include PU-H71, MPC-3100, Debio 0932/CUDC-305, and CNF2024/BIIB021, which have been synthesized by various research groups.
3.5.3. Targeting the HSP90 C-Terminal Binding Protein
A novel molecular strategy for regulating HSP90 chaperone activity has emerged through the introduction of a fresh ATP-binding site at the C-terminus. This addition induces the modulation of allosteric interactions between the C-terminus and N-terminus of HSP90, presenting a unique approach to controlling its chaperone function [169]. Although most HSP90 inhibitors bind/target the N-terminal ATP-binding site of HSP90, the C-terminus can also be targeted by the well-known natural products of aminocoumarin antibiotics, such as novobiocin, chlorobiocin, and coumermycin A inhibitors of HSP90 [163]. Novobiocin, isolated from Staphylococcus saprophyticus, is a known DNA gyrase inhibitor that inhibits both HSP90 and DNA gyrase B, belonging to the GHKL family [170]. Several synthetic C-terminus HSP90 inhibitors such as Epigallocatechin-3-gallate (EGCG), cisplatin, molybdate, taxol [171,172,173,174], and withaferin A have been identified. The majority of HSP90 inhibitors, both N-terminal HSP90 ATPase inhibitors and C-terminal ATPase inhibitors, are listed in Table 2.

Table 2.
List of commonly used macrocyclic HSP90, N-terminal, and C-terminal ATPase inhibitors.
3.5.4. Targeting Co-Chaperones and Their Interaction with HSP90
The co-chaperone Cdc37 promotes the connection of HSP90 with the kinase protein subset of client proteins to maintain their stability and signaling functions. Interrupting the complex between Cdc37 and HSP90 has surfaced as an alternative focal point for cancer progression. In human colon cancer cells, the silencing of Cdc37 destabilizes various kinase clients (AKT, HER2, CDK4, CDK6, and CRAF) and sensitizes cancer cells to HSP90 inhibitors and apoptosis [179]. Another study reported that the selective inhibitors celastrol and gedunin inactivated the co-chaperone p23 and affected HSP90 function through multiple mechanisms [180,181]. Targeting the interplay between HSP90 and Aha1 offers an alternate approach to modulating HSP90 activity. Aha1, which functions as an activator of HSP90 ATPase, forms associations with the N-terminal and middle domains of HSP90, thereby enhancing the ATPase activity of HSP90. Silencing Aha1 in colon cancer cells through siRNA resulted in a reduction in the activity of Raf-1 kinase and a decrease in the phosphorylation of MEK1/2 kinase and ERK1/2 kinase [182]. In PC3-MM2 prostate cancer cells, HSP90 C-terminal inhibitors, such as KU-135 and novobiocin, disrupted the HSP90α/Aha1 complex, leading to inhibited cell migration, which is a critical factor in tumor metastasis [183].
3.5.5. Targeting HSP90–Client Protein Interactions
Another alternative strategy for inhibiting HSP90 chaperone activity is to target interactions between HSP90 and its client proteins. Androgens drive prostate cancer cell growth through the androgen receptor (AR), which depends on HSP90 for survival and progression. In prostate cancer, HSP90 regulates the activity and stability of the AR by forming the HSP90-AR complex [184]. Nevertheless, disruption of this complex impedes the nuclear translocation of AR, induces cytosolic aggregation, and ultimately leads to AR degradation [185]. Camptothecin, a DNA topoisomerase 1 inhibitor that disrupts the HSP90-AR complex, inhibits AR transcriptional activity and suppresses prostate cancer cell growth [186]. Recent studies have demonstrated that HSP90 interacts with survivin, an inhibitor of apoptosis. However, dissociation or disruption of the HSP90–survivin complex leads to survivin degradation, suppresses cell proliferation, and inhibits mitochondrial apoptosis [158]. Plescia et al. demonstrated that shepherdin, a cell-permeable peptidomimetic compound, is capable of disrupting the HSP90–survivin complex and exhibits the loss of various client proteins, including CDK4, CDK6, Akt, and survival. Conversely, treatment of MCF-7 xenograft models with shepherdin resulted in a nearly complete loss of Akt levels in tumor cells [187].
3.6. Large HSPs and Cancer
The two main large HSPs HSP110 and GRP170 are relatively conserved and distinct sets of stress proteins, whose expression is induced by heat shock and glucose starvation, respectively. Under normal circumstances, the cytoprotective nature of HSP110 prevents stress-induced apoptosis in neuronal cells [188]. The role of HSP110 in apoptosis inhibition by genetically silenced HSP110 in various types of cancer cells that induce apoptosis has been previously reported [189]. HSP110 hinders the release of cytochrome c, a mitochondrial protein, thereby blocking caspase activation and preventing caspase 3- and 9-mediated apoptosis [188,189]. GRP170 is another high-molecular-weight family of HSPs that has been less studied than other conventional HSPs about its role in cancer prognosis and diagnosis. The immune adjuvant properties of GRP170 are gaining importance in cancer therapy owing to its intrinsic ability to induce both innate and adaptive immune responses [190,191].
Large HSPs as a Therapeutic Target
Elevated levels of HSP110 have been detected in numerous cancer types, suggesting that it is a potential therapeutic target for various cancers, including melanoma, colorectal cancer, and non-Hodgkin lymphoma [192,193]. The immunogenic characteristics of large HSPs present an opportunity to develop novel cancer vaccines. In a cervical cancer mouse model, simultaneous administration of the HSP110-E7 epitope complex and T lymphocyte epitope E7 exhibited improved efficacy against tumors. This combined approach resulted in decreased tumor growth and prolonged survival compared with other interventions [194]. Moreover, vaccination with HSP110 yielded favorable outcomes in mouse models of intestinal adenomas and spontaneously developed breast tumors [195]. GRP170 has also been used as a vaccine for the development of potential cancer antigens. Co-administration of specific tumor protein antigens along with the GRP170 complex resulted in a better immune response in the melanoma model [196]. In the first phase of the clinical trial, subjects diagnosed with third- and fourth-stage melanoma were enrolled to assess the efficacy of a vaccine incorporating the chaperone complex comprising human HSP110 and gp100, which had previously demonstrated success in preclinical evaluations (NCT01744171).
3.7. HSF1 as a Potential Cancer Therapeutic Target and Biomarker
HSF1 is a key regulator of the heat shock response (HSR), a mechanism that is retained by addressing proteotoxic stress. This process ensures the timely expression of HSPs, promoting cell viability through enhanced proteostasis [197]. Notably, HSF1 involvement extends to cancer, influencing migration, invasion, and metastasis [198,199]. Its overexpression has been observed in human prostate cancer, particularly in malignant epithelial cells, suggesting that HSF1 is a potential biomarker for various cancer types [200]. Hoang et al. revealed HSF1 upregulation in malignant prostate epithelial cells compared to normal prostate cells in primary cancer specimens [201].
In several malignancies, increased levels of HSF1 are associated with an unfavorable prognosis. Elevated nuclear levels of HSF1 have been linked to increased tumor growth, lymph node invasion, and death in cases of breast cancer, particularly in individuals who test positive for the estrogen receptor (ER) [202]. Similar associations with poor overall and relapse-free survival have been observed in breast cancer patients with high HSF1 mRNA expression. HSF1 also serves as a prognostic biomarker in endometrial cancer, hepatocellular carcinoma (HCC), esophageal carcinomas, and oral squamous cell carcinoma (OSCC) [200,203]. The overexpression of HSF1 has been linked to aggressive disease progression and reduced survival in these cancer types. Additionally, phosphorylated HSF1, specifically phospho-Ser326, has been identified as a potential marker for HSF1 activity and is associated with poor prognosis in ovarian cancer [204]. Collectively, these findings underscore the significance of HSF1 expression as a valuable prognostic indicator in diverse cancers.
Inhibiting transcription factors, such as HSF1, is challenging because of their complex tertiary structures and lack of target sites. HSF1, which is a ligand-less factor with poor druggability, poses difficulties in achieving inhibition specificity. Genetic approaches, such as RNA silencing, have been extensively pursued to overcome this challenge. Currently, both genetic and pharmacological strategies are being explored to inhibit HSF1 [205].
HSF1 regulates cellular stress responses, including HSP expression. The compound KRIBB11 inhibits HSF1 activity by preventing the binding of P-TEFb to the promoter of the HSP70 gene, thus reducing the transcription of stress response genes [206]. In animal models, KRIBB11 demonstrated HSF1 inhibitory effects, lowering HSP70 expression and tumor volume. KRIBB11 successfully prevented lymphatic metastasis in bladder cancer without causing any noticeable toxic effects. When combined with an AURKA inhibitor (danusertib), KRIBB11 exhibited strong anticancer activity against liver cancer by inducing cancer cell apoptosis and triggering an ER stress response [207]. Nonetheless, some studies have reported inconsistent results, indicating that KRIBB11 may not directly inhibit HSF1 in certain contexts, such as ferroptosis-related processes involving PROM2 [208].
Triptolide, a component of Tripterygium wilfordii, acts as an HSF1 inhibitor. In chronic lymphocytic leukemia (CLL), where HSF1 is overexpressed, triptolide causes apoptosis in both primary and cultured CLL B cells. When HSF1 is inhibited by knockdown or triptolide, it reduces HSP90 levels and impairs the cytosolic complex, which consists of deacetylase-histone deacetylase 6 (HDAC6), HSP90, p97, HSF1, and p97. Accordingly, HSF1 inhibition leads to the acetylation of HSP90, which disrupts the function of chaperones and suppresses malignancy in CLL [209].
DTHIB (SISU-102), a synthetic compound identified by screening for interactions with the HSF1 DNA-binding domain, selectively targets and inhibits HSF1 by increasing the degradation of HSF1 trimers (active nuclear). This potent inhibition reduces the excess amount of the multichaperone complex. In prostate cancer, DTHIB surpasses enzalutamide in reducing cell viability, inhibiting androgen receptor signaling, and suppressing PSA expression. DTHIB also hinders cell proliferation and diminishes cancer growth without noticeable side effects in mouse models [210]. Notably, in an acute myeloid leukemia model, DTHIB selectively suppresses the self-renewal of leukemia stem cells while leaving normal progenitor/hematopoietic stem cells. This effect is achieved by inhibiting the targets of HSF1, including HSP90, as well as suppressing mitochondrial oxidative phosphorylation by downregulating SDHC, a significant enzyme complex in the TCA cycle [211].
4. HSP-Based Therapies for Cancer
4.1. HSP-Based Vaccines and Cancer Immunotherapy
HSPs function as chaperones within cells and bind to antigens (peptides) associated with tumors. Moreover, APCs can recognize complexes formed by HSPs and peptides, leading to specific antitumor responses [39]. HSPs play a role in enhancing immune responses during the presentation of tumor antigens. Consequently, there is evidence suggesting that immunization with HSPs obtained from tumor cells can elicit an immune response against the tumor [212]. Various members of the HSP chaperone families, connected with diverse cancer-peptide-based antigens, may be isolated from tumor cells. By purifying the heat-shock-protein–peptide groups/complexes from a patient’s isolated cancer, this complex can serve as a personalized tumor vaccine that delivers antigens derived from tumor cells to the immune system, thereby fostering anticancer immunity [213].
Scientific evidence on the link between HSPs and cancer vaccination has been supported by various studies. Chen et al. recently developed a vaccine composed of fusion proteins such as HSP65 and a peptide (octa) epitope from the six transmembrane epithelial antigens of the prostate 1 (STEAP1) [214]. STEAP1 is highly expressed in multiple types of cancers. The fusion protein HSP65/STEAP1 demonstrated the ability to inhibit or arrest B16F10 melanoma growth (xenograft) and mouse RM-1 prostate cancer [215]. Furthermore, His-HSP65 (HHSP65), a fusion protein, stimulates the expression of TNF-α and facilitates STEAP1 to enhance TNF-α secretion, effectively inhibiting cancer cell proliferation [216]. When combined with IFN-γ, TNF-α triggers the upregulation of MHC-II expression, initiating cellular immune responses and amplifying the cytotoxic activity of various immune cells. Moreover, a vaccine targeting the CD133 epitope coupled with an adjuvant such as gp96 facilitated the transfer of epitope-specific cytotoxic T lymphocytes (CTLs), effectively suppressing leukemia growth in a murine xenograft model [217]. Immunization with bone-marrow-derived dendritic cells derived from bone marrow (BMDCs) stimulated by placental gp96 resulted in reduced tumor growth and enhanced survival in mice. This approach elicits a robust tumor-specific T-cell response, establishing its efficacy as an immunotherapeutic strategy [218].
Recent clinical assessments have broadened the investigation of cancer immunotherapy utilizing HSP-based vaccines. A newly developed self-assembled nanochaperone inspired by HSPs has the potential to significantly augment cancer immunotherapy. This nano-vaccine employs HSP-like microdomains and mannose surface decoration to capture antigens, facilitating their movement into dendritic cells [219]. This process promotes the escape of antigens and improves their cross-presentation in the cytoplasm. This approach has demonstrated its capacity to stimulate responses from the immune system, including both CD8+ T cells (cytotoxic cells) and CD4+ T cells (helper cells), for the prevention of melanoma [219]. The combination of chaperone protein immunotherapy with checkpoint inhibitors of the immune system, such as anti-PD-1/PD-L1, is particularly emphasized in the treatment of melanoma [220].
Furthermore, clinical trials recorded on ClinicalTrials.gov have assessed the effectiveness of a combined intramuscular (IM) vaccination regimen involving pNGVL4a-Sig/E7(detox)/HSP70 DNA and singular intramuscular immunization with TA-CIN to promote clearance of human papillomavirus 16 (ClinicalTrials.gov Identifier: NCT03911076) [221]. Another set of trials investigated the GP96 HSP-peptide complex vaccine as a potential treatment for patients with liver cancer (ClinicalTrials.gov Identifier: NCT04206254) [222].
Glioblastoma, a primary brain cancer with poor or substantial prognosis, is under investigation for its effectiveness [223]. Standard therapy combined with HSPPC96-based vaccination has shown safety and efficacy in patients diagnosed with glioblastoma. TSIR, known as the tumor-specific immune response, predicts the vaccine’s efficacy, with high TSIR correlating with a median overall survival exceeding 40.5 months [224]. Further studies involve the sequencing of T-cell receptors to analyze the T-cell receptor repertoire in tumor-infiltrating lymphocytes (TILs) and identify potential biomarkers for predicting responses to HSPPC96 vaccination [225]. Additionally, a phase II trial in glioblastoma patients (adults) undergoing surgical resection following standard therapy and autologous HSP peptide vaccine (Prophage) administration revealed MGMT promoter methylation as a prognostic factor, with significantly extended overall median survival for methylated tumors compared to unmethylated tumors [226].
4.2. Photothermal and Modulated Electro-Hyperthermia Therapy
Emerging treatments for cancer include photothermal therapy (PTT) and modulated electro-hyperthermia therapy (mEHT). Both approaches to selectively target and eliminate malignant cells involve intentionally raising the temperature, which is referred to as hyperthermia. PTT mostly uses light-absorbing substances, usually nanoparticles. These substances absorb light energy at a certain wavelength and turn it into heat, which raises the temperature in the area, causing localized hyperthermia and damaging or killing cancer cells. Modulated electro-hyperthermia therapy is another form of hyperthermia therapy that involves the application of modulated electric fields to precisely elevate the temperature of cancer cells. Owing to their higher conductivity, cancer cells can consume a larger quantity of energy and experience higher temperatures than normal cells. However, the major problem associated with hyperthermia is the rapid expression of HSPs. However, the main problem with hyperthermia is that it causes rapid production and release of HSPs, which help proteins float inside cells and increase their tolerance [227]. The therapeutic efficacy of PTT is directly influenced by intracellular HSP expression levels, which have both protective and detrimental effects on cancer cells. Thus, the efficacy of hyperthermia therapy can be increased by inhibiting HSPs, which can reduce the thermal resistance of cancer cells and potentiate the cell-killing effect of hyperthermia [228]. Currently, HSP inhibitors are being used in combination with hyperthermia. For instance, Kuo et al. reported that mEHT increased efficiency by combining with the HSP inhibitors nano-curcumin and resveratrol in an in vivo CT26/BALB/c animal tumor model caused by decreased HSP70 expression and the infiltration of immune cells (CD3+ T-cells and F4/80+ macrophages) into tumors receiving this treatment [229].
4.3. Role of Chaperone-Mediated Autophagy in Cancer Diseases
Chaperone-mediated autophagy (CMA) primarily focuses on the degradation of misfolded proteins by translocating these polypeptides across the lysosomal membrane, thereby helping to maintain cellular homeostasis. There are reports on the dual nature of CMA in cancer because of its protumorigenic and antitumorigenic potential [230]. In a pro-tumorigenic role, CMA helps to survive and grow cancer cells through the modulation of the cell cycle. Hypoxia is an important characteristic observed in the tumor microenvironment, which helps activate CMA gene transcription. This CMA assists cancer survival and progression in the hypoxic microenvironment of tumors by hypoxia-inducible factor-1 alpha (HIF-1α) degradation (2). Similarly, the anti-oncogenic role or tumor-suppressive potential of CMA has been reported in the tumor microenvironment, where CMA is involved in protooncogene proteins such as Murine Double Minute 2 (MDM2) and tumor-associated translationally controlled tumor protein (TCTP) upon acetylation [231,232]. Although research in the field of CMA is still being conducted to examine the connection between faulty CMA and cancer, the translation of these results into preventative or therapeutic measures has been hampered by several issues. In the initial stage, every component and modulator of CMA, such as signaling elements and chaperones, participates in a wide range of crucial cellular processes. To date, no research has been conducted to identify a particular component of the CMA pathway that may be used as a target for external manipulation. This significant barrier must be overcome to discover selective chemical modulators of CMA. For instance, LAMP2A knockdown by direct injection of shRNA results in tumor regression and reduced metastasis in human lung cancer xenograft mice [233].
5. Conclusions
HSPs are a conserved family of proteins that are highly expressed in several types of cancer. Cytoprotective functions are crucial for cancer cell survival. HSPs are often associated with cancer progression and response to treatment, such as drug resistance, as well as the overall poor prognosis of patients. The increased prevalence of HSPs in cancer cells has led to over a decade of research, which has made them a potential target for oncologists to develop targeted inhibitors for small HSPs. These small-HSP inhibitors can alter the cellular expression of HSPs and subsequently their activity, thereby inhibiting the proliferation of malignant cells in cancer therapy. They have also been used as chemosensitizing compounds with different chemotherapeutic agents to overcome acquired drug resistance or can yield synergistic cytotoxic effects in cancer therapy. Although there have been a number of reports on their synthesis and application as inhibitors of cancer cell proliferation, sufficient investigation of animal models and the side effects of these inhibitors have not been well reported. Moreover, the molecular mechanisms of action of various HSPs, especially those associated with cancer progression, are yet to be clearly understood, hindering the development of suitable biocompatible HSP inhibitors in cancer therapy.
Author Contributions
Conceptualization, P.S. and S.M.; methodology, N.B.; writing—original draft preparation, S.M.; writing—review and editing, N.B.; writing—review and editing, A.K.Y.; visualization, S.P.; supervision, K.-H.B. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (NRF-2021R1F1A1060297).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
This paper contains all relevant data; no additional data required.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Park, C.-J.; Seo, Y.-S. Heat shock proteins: A review of the molecular chaperones for plant immunity. Plant Pathol. J. 2015, 31, 323. [Google Scholar] [CrossRef]
- Lindquist, S.; Craig, E.A. The heat-shock proteins. Annu. Rev. Genet. 1988, 22, 631–677. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Kim, Y.E.; Hipp, M.S.; Bracher, A.; Hayer-Hartl, M.; Ulrich Hartl, F. Molecular chaperone functions in protein folding and proteostasis. Annu. Rev. Biochem. 2013, 82, 323–355. [Google Scholar] [CrossRef] [PubMed]
- Saibil, H. Chaperone machines for protein folding, unfolding and disaggregation. Nat. Rev. Mol. Cell Biol. 2013, 14, 630–642. [Google Scholar] [CrossRef]
- Hartl, F.U.; Bracher, A.; Hayer-Hartl, M. Molecular chaperones in protein folding and proteostasis. Nature 2011, 475, 324–332. [Google Scholar] [CrossRef]
- Hüttner, S.; Strasser, R. Endoplasmic reticulum-associated degradation of glycoproteins in plants. Front. Plant Sci. 2012, 3, 67. [Google Scholar] [CrossRef]
- Shrestha, L.; Bolaender, A.; J Patel, H.; Taldone, T. Heat shock protein (HSP) drug discovery and development: Targeting heat shock proteins in disease. Curr. Top. Med. Chem. 2016, 16, 2753–2764. [Google Scholar] [CrossRef]
- Yahara, I. Stress-inducible cellular responses. Introduction. EXS 1996, 77, XI–XII. [Google Scholar] [PubMed]
- Van Noort, J.M.; Bsibsi, M.; Nacken, P.; Gerritsen, W.H.; Amor, S. The link between small heat shock proteins and the immune system. Int. J. Biochem. Cell Biol. 2012, 44, 1670–1679. [Google Scholar] [CrossRef] [PubMed]
- Kampinga, H.H.; Hageman, J.; Vos, M.J.; Kubota, H.; Tanguay, R.M.; Bruford, E.A.; Cheetham, M.E.; Chen, B.; Hightower, L.E. Guidelines for the nomenclature of the human heat shock proteins. Cell Stress Chaperones 2009, 14, 105–111. [Google Scholar] [CrossRef]
- Haslbeck, M.; Vierling, E. A first line of stress defense: Small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 2015, 427, 1537–1548. [Google Scholar] [CrossRef]
- Bepperling, A.; Alte, F.; Kriehuber, T.; Braun, N.; Weinkauf, S.; Groll, M.; Haslbeck, M.; Buchner, J. Alternative bacterial two-component small heat shock protein systems. Proc. Natl. Acad. Sci. USA 2012, 109, 20407–20412. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Liu, T.; Rios, Z.; Mei, Q.; Lin, X.; Cao, S. Heat shock proteins and cancer. Trends Pharmacol. Sci. 2017, 38, 226–256. [Google Scholar] [CrossRef] [PubMed]
- Shamovsky, I.; Nudler, E. New insights into the mechanism of heat shock response activation. Cell. Mol. Life Sci. 2008, 65, 855–861. [Google Scholar] [CrossRef] [PubMed]
- Garrido, C.; Brunet, M.; Didelot, C.; Zermati, Y.; Schmitt, E.; Kroemer, G. Heat shock proteins 27 and 70: Anti-apoptotic proteins with tumorigenic properties. Cell Cycle 2006, 5, 2592–2601. [Google Scholar] [CrossRef] [PubMed]
- Green, M.; Schuetz, T.J.; Sullivan, E.K.; Kingston, R.E. A heat shock-responsive domain of human HSF1 that regulates transcription activation domain function. Mol. Cell. Biol. 1995, 15, 3354–3362. [Google Scholar] [CrossRef] [PubMed]
- Åkerfelt, M.; Morimoto, R.I.; Sistonen, L. Heat shock factors: Integrators of cell stress, development and lifespan. Nat. Rev. Mol. Cell Biol. 2010, 11, 545–555. [Google Scholar] [CrossRef] [PubMed]
- Fan, G.-C. Role of heat shock proteins in stem cell behavior. Prog. Mol. Biol. Transl. Sci. 2012, 111, 305–322. [Google Scholar] [PubMed]
- De Maio, A. Heat shock proteins: Facts, thoughts, and dreams. Shock 1999, 11, 1–12. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Jolly, C.; Morimoto, R.I. Role of the heat shock response and molecular chaperones in oncogenesis and cell death. J. Natl. Cancer Inst. 2000, 92, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Christensen, J.H.; Nielsen, M.N.; Hansen, J.; Füchtbauer, A.; Füchtbauer, E.-M.; West, M.; Corydon, T.J.; Gregersen, N.; Bross, P. Inactivation of the hereditary spastic paraplegia-associated Hspd1 gene encoding the Hsp60 chaperone results in early embryonic lethality in mice. Cell Stress Chaperones 2010, 15, 851–863. [Google Scholar] [CrossRef]
- Hendrick, J.P.; Hartl, F.-U. Molecular chaperone functions of heat-shock proteins. Annu. Rev. Biochem. 1993, 62, 349–384. [Google Scholar] [CrossRef] [PubMed]
- Mardan-Nik, M.; Pasdar, A.; Jamialahmadi, K.; Biabangard-Zak, A.; Mirhafez, S.R.; Ghalandari, M.; Tajfard, M.; Mohebati, M.; Esmaily, H.; Ferns, G.A. Association of heat shock protein70-2 (HSP70-2) gene polymorphism with coronary artery disease in an Iranian population. Gene 2014, 550, 180–184. [Google Scholar] [CrossRef] [PubMed]
- Sreedhar, A.S.; Kalmár, É.; Csermely, P.; Shen, Y.-F. Hsp90 isoforms: Functions, expression and clinical importance. FEBS Lett. 2004, 562, 11–15. [Google Scholar] [CrossRef]
- Morimoto, R.I. Heat shock: The role of transient inducible responses in cell damage, transformation, and differentiation. Cancer Cells 1991, 3, 295–301. [Google Scholar] [PubMed]
- Oh, H.J.; Chen, X.; Subjeck, J.R. Hsp110 protects heat-denatured proteins and confers cellular thermoresistance. J. Biol. Chem. 1997, 272, 31636–31640. [Google Scholar] [CrossRef]
- Hoang, A.T.; Huang, J.; Rudra-Ganguly, N.; Zheng, J.; Powell, W.C.; Rabindran, S.K.; Wu, C.; Roy-Burman, P. A novel association between the human heat shock transcription factor 1 (HSF1) and prostate adenocarcinoma. Am. J. Pathol. 2000, 156, 857–864. [Google Scholar] [CrossRef]
- Wang, Y.; Theriault, J.R.; He, H.; Gong, J.; Calderwood, S.K. Expression of a dominant negative heat shock factor-1 construct inhibits aneuploidy in prostate carcinoma cells. J. Biol. Chem. 2004, 279, 32651–32659. [Google Scholar] [CrossRef]
- Easton, D.P.; Kaneko, Y.; Subjeck, J.R. The Hsp110 and Grp170 stress proteins: Newly recognized relatives of the Hsp70s. Cell Stress Chaperones 2000, 5, 276. [Google Scholar] [CrossRef]
- Khalil, A.A.; Kabapy, N.F.; Deraz, S.F.; Smith, C. Heat shock proteins in oncology: Diagnostic biomarkers or therapeutic targets? Biochim. Biophys. Acta (BBA)-Rev. Cancer 2011, 1816, 89–104. [Google Scholar] [CrossRef]
- Brown, C.R.; Martin, R.L.; Hansen, W.J.; Beckmann, R.P.; Welch, W.J. The constitutive and stress inducible forms of hsp 70 exhibit functional similarities and interact with one another in an ATP-dependent fashion. J. Cell Biol. 1993, 120, 1101–1112. [Google Scholar] [CrossRef]
- Dean, D.O.; Kent, C.R.; Tytell, M. Constitutive and inducible heat shock protein 70 immunoreactivity in the normal rat eye. Investig. Ophthalmol. Vis. Sci. 1999, 40, 2952–2962. [Google Scholar]
- Bolhassani, A.; Rafati, S. Heat-shock proteins as powerful weapons in vaccine development. Expert Rev. Vaccines 2008, 7, 1185–1199. [Google Scholar] [CrossRef]
- Björk, J.K.; Sistonen, L. Regulation of the members of the mammalian heat shock factor family. FEBS J. 2010, 277, 4126–4139. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.; Rustgi, S.; von Wettstein, D.; Reinbothe, C.; Reinbothe, S. Common functions of the chloroplast and mitochondrial co-chaperones cpDnaJL (CDF1) and mtDnaJ (PAM16) in protein import and ROS scavenging in Arabidopsis thaliana. Commun. Integr. Biol. 2016, 9, e1119343. [Google Scholar] [CrossRef] [PubMed]
- Sin, Y.Y.; Martin, T.P.; Wills, L.; Currie, S.; Baillie, G.S. Small heat shock protein 20 (Hsp20) facilitates nuclear import of protein kinase D 1 (PKD1) during cardiac hypertrophy. Cell Commun. Signal. 2015, 13, 16. [Google Scholar] [CrossRef] [PubMed]
- Calderwood, S.K.; Gong, J.; Murshid, A. Extracellular HSPs: The complicated roles of extracellular HSPs in immunity. Front. Immunol. 2016, 7, 159. [Google Scholar] [CrossRef]
- Tsutsumi, S.; Neckers, L. Extracellular heat shock protein 90: A role for a molecular chaperone in cell motility and cancer metastasis. Cancer Sci. 2007, 98, 1536–1539. [Google Scholar] [CrossRef]
- Patnaik, S.; Nathan, S.; Kar, B.; Gregoric, I.D.; Li, Y.-P. The Role of Extracellular Heat Shock Proteins in Cardiovascular Diseases. Biomedicines 2023, 11, 1557. [Google Scholar] [CrossRef]
- Reddy, V.S.; Madala, S.K.; Trinath, J.; Reddy, G.B. Extracellular small heat shock proteins: Exosomal biogenesis and function. Cell Stress Chaperones 2018, 23, 441–454. [Google Scholar] [CrossRef]
- Schmitt, E.; Gehrmann, M.; Brunet, M.; Multhoff, G.; Garrido, C. Intracellular and extracellular functions of heat shock proteins: Repercussions in cancer therapy. J. Leucoc. Biol. 2007, 81, 15–27. [Google Scholar] [CrossRef]
- Milani, A.; Basirnejad, M.; Bolhassani, A. Heat-shock proteins in diagnosis and treatment: An overview of different biochemical and immunological functions. Immunotherapy 2019, 11, 215–239. [Google Scholar] [CrossRef]
- Kokolakis, G.; Tatari, M.; Zacharopoulou, A.; Mintzas, A.C. The hsp27 gene of the Mediterranean fruit fly, Ceratitis capitata: Structural characterization, regulation and developmental expression. Insect Mol. Biol. 2008, 17, 699–710. [Google Scholar] [CrossRef]
- Kostenko, S.; Moens, U. Heat shock protein 27 phosphorylation: Kinases, phosphatases, functions and pathology. Cell. Mol. life Sci. 2009, 66, 3289–3307. [Google Scholar] [CrossRef] [PubMed]
- Gobbo, J.; Gaucher-Di-Stasio, C.; Weidmann, S.; Guzzo, J.; Garrido, C. Quantification of HSP27 and HSP70 molecular chaperone activities. In Molecular Chaperones: Methods and Protocols; Springer: Berlin/Heidelberg, Germany, 2011; pp. 137–143. [Google Scholar]
- Stope, M.B.; Klinkmann, G.; Diesing, K.; Koensgen, D.; Burchardt, M.; Mustea, A. Heat shock protein HSP27 secretion by ovarian cancer cells is linked to intracellular expression levels, occurs independently of the endoplasmic reticulum pathway and HSP27′s phosphorylation status, and is mediated by exosome liberation. Dis. Markers 2017, 2017, 1575374. [Google Scholar] [CrossRef] [PubMed]
- Katsogiannou, M.; Andrieu, C.; Rocchi, P. Heat shock protein 27 phosphorylation state is associated with cancer progression. Front. Genet. 2014, 5, 346. [Google Scholar] [CrossRef] [PubMed]
- Ciocca, D.R.; Oesterreich, S.; Chamness, G.C.; MCGuire, W.L.; Fuqua, S.A.W. Biological and clinical implications of heat shock protein 27000 (Hsp27): A review. JNCI J. Natl. Cancer Inst. 1993, 85, 1558–1570. [Google Scholar] [CrossRef]
- Yang, S.; Xiao, H.; Cao, L. Recent advances in heat shock proteins in cancer diagnosis, prognosis, metabolism and treatment. Biomed. Pharmacother. 2021, 142, 112074. [Google Scholar] [CrossRef] [PubMed]
- Assimakopoulou, M. Human meningiomas: Immunohistochemical localization of progesterone receptor and heat shock protein 27 and absence of estrogen receptor and PS2. Cancer Detect. Prev. 2000, 24, 163–168. [Google Scholar]
- Somu, P.; Basavegowda, N.; Gomez, L.A.; Jayaprakash, H.V.; Puneetha, G.K.; Yadav, A.K.; Paul, S.; Baek, K.-H. Crossroad between the Heat Shock Protein and Inflammation Pathway in Acquiring Drug Resistance: A Possible Target for Future Cancer Therapeutics. Biomedicines 2023, 11, 2639. [Google Scholar] [CrossRef]
- Yu, Z.; Zhi, J.; Peng, X.; Zhong, X.; Xu, A. Clinical significance of HSP27 expression in colorectal cancer. Mol. Med. Rep. 2010, 3, 953–958. [Google Scholar]
- Hu, C.; Yang, J.; Qi, Z.; Wu, H.; Wang, B.; Zou, F.; Mei, H.; Liu, J.; Wang, W.; Liu, Q. Heat shock proteins: Biological functions, pathological roles, and therapeutic opportunities. MedComm 2022, 3, e161. [Google Scholar] [CrossRef]
- Daniels, G.A.; Sanchez-Perez, L.; Diaz, R.M.; Kottke, T.; Thompson, J.; Lai, M.; Gough, M.; Karim, M.; Bushell, A.; Chong, H. A simple method to cure established tumors by inflammatory killing of normal cells. Nat. Biotechnol. 2004, 22, 1125–1132. [Google Scholar] [CrossRef] [PubMed]
- Hsu, H.; Lin, J.; Huang, W.; Hsu, T.; Su, K.; Chiou, S.; Tsai, Y.; Hung, S. Chemoresistance of lung cancer stemlike cells depends on activation of Hsp27. Cancer 2011, 117, 1516–1528. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, J.-C.; Tuukkanen, A.; Schroeder, M.; Fahrig, T.; Fahrig, R. RP101 (brivudine) binds to heat shock protein HSP27 (HSPB1) and enhances survival in animals and pancreatic cancer patients. J. Cancer Res. Clin. Oncol. 2011, 137, 1349–1361. [Google Scholar] [CrossRef] [PubMed]
- Gibert, B.; Hadchity, E.; Czekalla, A.; Aloy, M.T.; Colas, P.; Rodriguez-Lafrasse, C.; Arrigo, A.P.; Diaz-Latoud, C. Inhibition of heat shock protein 27 (HspB1) tumorigenic functions by peptide aptamers. Oncogene 2011, 30, 3672–3681. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-K.; Kam, H.; Kim, K.-Y.; Park, S.I.; Lee, Y.-S. Targeting heat shock protein 27 in cancer: A druggable target for cancer treatment? Cancers 2019, 11, 1195. [Google Scholar] [CrossRef] [PubMed]
- McConnell, J.R.; McAlpine, S.R. Heat shock proteins 27, 40, and 70 as combinational and dual therapeutic cancer targets. Bioorg. Med. Chem. Lett. 2013, 23, 1923–1928. [Google Scholar] [CrossRef]
- Asgharzadeh, F.; Moradi-Marjaneh, R.; Marjaneh, M.M. The role of heat shock protein 27 in carcinogenesis and treatment of colorectal cancer. Curr. Pharm. Des. 2022, 28, 2677. [Google Scholar] [CrossRef]
- Heinrich, J.C.; Donakonda, S.; Haupt, V.J.; Lennig, P.; Zhang, Y.; Schroeder, M. New HSP27 inhibitors efficiently suppress drug resistance development in cancer cells. Oncotarget 2016, 7, 68156. [Google Scholar] [CrossRef]
- Hosokawa, N.; Hirayoshi, K.; Kudo, H.; Takechi, H.; Aoike, A.; Ka Wai, K.; Nagata, K. Inhibition of the activation of heat shock factor in vivo and in vitro by flavonoids. Mol. Cell. Biol. 1992, 12, 3490–3498. [Google Scholar]
- Yoshida, M.; Sakai, T.; Hosokawa, N.; Marui, N.; Matsumoto, K.; Fujioka, A.; Nishino, H.; Aoike, A. The effect of quercetin on cell cycle progression and growth of human gastric cancer cells. FEBS Lett. 1990, 260, 10–13. [Google Scholar] [CrossRef]
- Wang, R.E.; Kao, J.L.-F.; Hilliard, C.A.; Pandita, R.K.; Roti Roti, J.L.; Hunt, C.R.; Taylor, J.-S. Inhibition of heat shock induction of heat shock protein 70 and enhancement of heat shock protein 27 phosphorylation by quercetin derivatives. J. Med. Chem. 2009, 52, 1912–1921. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.; Milito, A.; Spagnuolo, C.; Carbone, V.; Rosén, A.; Minasi, P.; Lauria, F.; Russo, G.L. CK2 and PI3K are direct molecular targets of quercetin in chronic lymphocytic leukaemia. Oncotarget 2017, 8, 42571. [Google Scholar] [CrossRef] [PubMed]
- Borgo, C.; Vilardell, J.; Bosello-Travain, V.; Pinna, L.A.; Venerando, A.; Salvi, M. Dependence of HSP27 cellular level on protein kinase CK2 discloses novel therapeutic strategies. Biochim. Biophys. Acta (BBA) Gen. Subj. 2018, 1862, 2902–2910. [Google Scholar] [CrossRef] [PubMed]
- Seigneuric, R.; Gobbo, J.; Colas, P.; Garrido, C. Targeting cancer with peptide aptamers. Oncotarget 2011, 2, 557. [Google Scholar] [CrossRef] [PubMed]
- Kamada, M.; So, A.; Muramaki, M.; Rocchi, P.; Beraldi, E.; Gleave, M. Hsp27 knockdown using nucleotide-based therapies inhibit tumor growth and enhance chemotherapy in human bladder cancer cells. Mol. Cancer Ther. 2007, 6, 299–308. [Google Scholar] [CrossRef] [PubMed]
- Kumano, M.; Furukawa, J.; Shiota, M.; Zardan, A.; Zhang, F.; Beraldi, E.; Wiedmann, R.M.; Fazli, L.; Zoubeidi, A.; Gleave, M.E. Cotargeting stress-activated Hsp27 and autophagy as a combinatorial strategy to amplify endoplasmic reticular stress in prostate cancer. Mol. Cancer Ther. 2012, 11, 1661–1671. [Google Scholar] [CrossRef] [PubMed]
- Hadchity, E.; Aloy, M.-T.; Paulin, C.; Armandy, E.; Watkin, E.; Rousson, R.; Gleave, M.; Chapet, O.; Rodriguez-Lafrasse, C. Heat shock protein 27 as a new therapeutic target for radiation sensitization of head and neck squamous cell carcinoma. Mol. Ther. 2009, 17, 1387–1394. [Google Scholar] [CrossRef]
- Vos, M.J.; Hageman, J.; Carra, S.; Kampinga, H.H. Structural and functional diversities between members of the human HSPB, HSPH, HSPA, and DNAJ chaperone families. Biochemistry 2008, 47, 7001–7011. [Google Scholar] [CrossRef] [PubMed]
- Jang, J.; Lee, S.-H.; Kang, D.-H.; Sim, D.-W.; Ryu, K.-S.; Jo, K.-S.; Lee, J.; Ryu, H.; Kim, E.-H.; Won, H.-S. Structural resemblance of the DNAJA-family protein, Tid1, to the DNAJB-family Hsp40. BMB Rep. 2022, 55, 488. [Google Scholar] [CrossRef] [PubMed]
- Qian, Y.Q.; Patel, D.; Hartl, F.-U.; McColl, D.J. Nuclear magnetic resonance solution structure of the human Hsp40 (HDJ-1) J-domain. J. Mol. Biol. 1996, 260, 224–235. [Google Scholar] [CrossRef]
- Genevaux, P.; Schwager, F.; Georgopoulos, C.; Kelley, W.L. Scanning mutagenesis identifies amino acid residues essential for the in vivo activity of the Escherichia coli DnaJ (Hsp40) J-domain. Genetics 2002, 162, 1045–1053. [Google Scholar] [CrossRef]
- Malinverni, D.; Jost Lopez, A.; De Los Rios, P.; Hummer, G.; Barducci, A. Modeling Hsp70/Hsp40 interaction by multi-scale molecular simulations and coevolutionary sequence analysis. Elife 2017, 6, e23471. [Google Scholar] [CrossRef] [PubMed]
- Qiu, X.-B.; Shao, Y.-M.; Miao, S.; Wang, L. The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones. Cell. Mol. Life Sci. C 2006, 63, 2560–2570. [Google Scholar] [CrossRef]
- Moffatt, N.S.C.; Bruinsma, E.; Uhl, C.; Obermann, W.M.J.; Toft, D. Role of the cochaperone Tpr2 in Hsp90 chaperoning. Biochemistry 2008, 47, 8203–8213. [Google Scholar] [CrossRef]
- Blatch, G.L.; Lässle, M. The tetratricopeptide repeat: A structural motif mediating protein-protein interactions. Bioessays 1999, 21, 932–939. [Google Scholar] [CrossRef]
- Odunuga, O.O.; Hornby, J.A.; Bies, C.; Zimmermann, R.; Pugh, D.J.; Blatch, G.L. Tetratricopeptide repeat motif-mediated Hsc70-mSTI1 interaction: Molecular characterization of the critical contacts for successful binding and specificity. J. Biol. Chem. 2003, 278, 6896–6904. [Google Scholar] [CrossRef]
- Greener, T.; Zhao, X.; Nojima, H.; Eisenberg, E.; Greene, L.E. Role of cyclin G-associated kinase in uncoating clathrin-coated vesicles from non-neuronal cells. J. Biol. Chem. 2000, 275, 1365–1370. [Google Scholar] [CrossRef]
- Sterrenberg, J.N.; Blatch, G.L.; Edkins, A.L. Human DNAJ in cancer and stem cells. Cancer Lett. 2011, 312, 129–142. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Xu, X.; Jiang, Z.; You, Q. Modulation of protein fate decision by small molecules: Targeting molecular chaperone machinery. Acta Pharm. Sin. B 2020, 10, 1904–1925. [Google Scholar] [CrossRef] [PubMed]
- Trinh, D.L.N.; Elwi, A.N.; Kim, S.-W. Direct interaction between p53 and Tid1 proteins affects p53 mitochondrial localization and apoptosis. Oncotarget 2010, 1, 396. [Google Scholar] [CrossRef] [PubMed]
- Kurzik-Dumke, U.; Hörner, M.; Czaja, J.; Nicotra, M.R.; Simiantonaki, N.; Koslowski, M.; Natali, P.G. Progression of colorectal cancers correlates with overexpression and loss of polarization of expression of the htid-1 tumor suppressor. Int. J. Mol. Med. 2008, 21, 19–31. [Google Scholar] [CrossRef]
- Yu, V.Z.; Wong, V.C.-L.; Dai, W.; Ko, J.M.-Y.; Lam, A.K.-Y.; Chan, K.W.; Samant, R.S.; Lung, H.L.; Shuen, W.H.; Law, S. Nuclear localization of DNAJB6 is associated with survival of patients with esophageal cancer and reduces AKT signaling and proliferation of cancer cells. Gastroenterology 2015, 149, 1825–1836. [Google Scholar] [CrossRef]
- Cui, X.; Choi, H.-K.; Choi, Y.-S.; Park, S.-Y.; Sung, G.-J.; Lee, Y.-H.; Lee, J.; Jun, W.J.; Kim, K.; Choi, K.-C. DNAJB1 destabilizes PDCD5 to suppress p53-mediated apoptosis. Cancer Lett. 2015, 357, 307–315. [Google Scholar] [CrossRef]
- Nishizawa, S.; Hirohashi, Y.; Torigoe, T.; Takahashi, A.; Tamura, Y.; Mori, T.; Kanaseki, T.; Kamiguchi, K.; Asanuma, H.; Morita, R. HSP DNAJB8 controls tumor-initiating ability in renal cancer stem–like cells. Cancer Res. 2012, 72, 2844–2854. [Google Scholar] [CrossRef]
- Yang, T.; Li, X.-N.; Li, X.-G.; Li, M.; Gao, P.-Z. DNAJC6 promotes hepatocellular carcinoma progression through induction of epithelial–mesenchymal transition. Biochem. Biophys. Res. Commun. 2014, 455, 298–304. [Google Scholar] [CrossRef]
- Yamashita, M.; Hirohashi, Y.; Torigoe, T.; Kusumoto, H.; Murai, A.; Imagawa, T.; Sato, N. Dnajb8, a member of the heat shock protein 40 family has a role in the tumor initiation and resistance to docetaxel but is dispensable for stress response. PLoS ONE 2016, 11, e0146501. [Google Scholar] [CrossRef]
- Yun, C.W.; Kim, H.J.; Lim, J.H.; Lee, S.H. Heat shock proteins: Agents of cancer development and therapeutic targets in anti-cancer therapy. Cells 2019, 9, 60. [Google Scholar] [CrossRef]
- Yokota, S.; Kitahara, M.; Nagata, K. Benzylidene lactam compound, KNK437, a novel inhibitor of acquisition of thermotolerance and heat shock protein induction in human colon carcinoma cells. Cancer Res. 2000, 60, 2942–2948. [Google Scholar]
- Loriot, Y.; Mordant, P.; Dorvault, N.; De la Motte Rouge, T.; Bourhis, J.; Soria, J.C.; Deutsch, E. BMS-690514, a VEGFR and EGFR tyrosine kinase inhibitor, shows anti-tumoural activity on non-small-cell lung cancer xenografts and induces sequence-dependent synergistic effect with radiation. Br. J. Cancer 2010, 103, 347–353. [Google Scholar] [CrossRef]
- De La Motte Rouge, T.; Galluzzi, L.; Olaussen, K.A.; Zermati, Y.; Tasdemir, E.; Robert, T.; Ripoche, H.; Lazar, V.; Dessen, P.; Harper, F. A novel epidermal growth factor receptor inhibitor promotes apoptosis in non–small cell lung cancer cells resistant to erlotinib. Cancer Res. 2007, 67, 6253–6262. [Google Scholar] [CrossRef]
- Martin, L.-A.; Head, J.E.; Pancholi, S.; Salter, J.; Quinn, E.; Detre, S.; Kaye, S.; Howes, A.; Dowsett, M.; Johnston, S.R.D. The farnesyltransferase inhibitor R115777 (tipifarnib) in combination with tamoxifen acts synergistically to inhibit MCF-7 breast cancer cell proliferation and cell cycle progression in vitro and in vivo. Mol. Cancer Ther. 2007, 6, 2458–2467. [Google Scholar] [CrossRef]
- Izbicka, E.; Campos, D.; Carrizales, G.; Patnaik, A. Biomarkers of anticancer activity of R115777 (Tipifarnib, ZarnestraTM) in human breast cancer models in vitro. Anticancer Res. 2005, 25, 3215–3223. [Google Scholar]
- Wang, C.-C.; Liao, Y.-P.; Mischel, P.S.; Iwamoto, K.S.; Cacalano, N.A.; McBride, W.H. HDJ-2 as a target for radiosensitization of glioblastoma multiforme cells by the farnesyltransferase inhibitor R115777 and the role of the p53/p21 pathway. Cancer Res. 2006, 66, 6756–6762. [Google Scholar] [CrossRef] [PubMed]
- Cassel, J.A.; Ilyin, S.; McDonnell, M.E.; Reitz, A.B. Novel inhibitors of heat shock protein Hsp70-mediated luciferase refolding that bind to DnaJ. Bioorg. Med. Chem. 2012, 20, 3609–3614. [Google Scholar] [CrossRef] [PubMed]
- Park, S.-Y.; Choi, H.-K.; Seo, J.S.; Yoo, J.-Y.; Jeong, J.-W.; Choi, Y.; Choi, K.-C.; Yoon, H.-G. DNAJB1 negatively regulates MIG6 to promote epidermal growth factor receptor signaling. Biochim. Biophys. Acta (BBA) Mol. Cell Res. 2015, 1853, 2722–2730. [Google Scholar] [CrossRef] [PubMed]
- Van der Weide, K.; de Jonge-Peeters, S.; Huls, G.; Fehrmann, R.S.N.; Schuringa, J.J.; Kuipers, F.; de Vries, E.G.E.; Vellenga, E. Treatment with high-dose simvastatin inhibits geranylgeranylation in AML blast cells in a subset of AML patients. Exp. Hematol. 2012, 40, 177–186. [Google Scholar] [CrossRef] [PubMed]
- Grabocka, E.; Commisso, C.; Bar-Sagi, D. Molecular pathways: Targeting the dependence of mutant RAS cancers on the DNA damage response. Clin. Cancer Res. 2015, 21, 1243–1247. [Google Scholar] [CrossRef] [PubMed]
- Meng, Q.; Li, B.X.; Xiao, X. Toward developing chemical modulators of Hsp60 as potential therapeutics. Front. Mol. Biosci. 2018, 5, 35. [Google Scholar] [CrossRef]
- Jindal, S.; Dudani, A.K.; Singh, B.; Harley, C.B.; Gupta, R.S. Primary structure of a human mitochondrial protein homologous to the bacterial and plant chaperonins and to the 65-kilodalton mycobacterial antigen. Mol. Cell. Biol. 1989, 9, 2279–2283. [Google Scholar] [PubMed]
- Cappello, F.; Marino Gammazza, A.; Palumbo Piccionello, A.; Campanella, C.; Pace, A.; Conway de Macario, E.; Macario, A.J.L. Hsp60 chaperonopathies and chaperonotherapy: Targets and agents. Expert Opin. Ther. Targets 2014, 18, 185–208. [Google Scholar] [CrossRef]
- Ranford, J.C.; Coates, A.R.M.; Henderson, B. Chaperonins are cell-signalling proteins: The unfolding biology of molecular chaperones. Expert Rev. Mol. Med. 2000, 2, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Macario, A.J.L.; de Macario, E.C. Molecular chaperones: Multiple functions, pathologies, and potential applications. Front. Biosci. 2007, 12, 2588–2600. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Dohi, T.; Kang, B.H.; Altieri, D.C. Hsp60 regulation of tumor cell apoptosis. J. Biol. Chem. 2008, 283, 5188–5194. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Zeng, Y.; Graner, M.W.; Katsanis, E. Stressed apoptotic tumor cells stimulate dendritic cells and induce specific cytotoxic T cells. Blood J. Am. Soc. Hematol. 2002, 100, 4108–4115. [Google Scholar] [CrossRef]
- Pockley, A.G.; Muthana, M.; Calderwood, S.K. The dual immunoregulatory roles of stress proteins. Trends Biochem. Sci. 2008, 33, 71–79. [Google Scholar] [CrossRef]
- Tsan, M.-F.; Gao, B. Heat shock protein and innate immunity. Cell. Mol. Immunol. 2004, 1, 274–279. [Google Scholar]
- Osterloh, A.; Meier-Stiegen, F.; Veit, A.; Fleischer, B.; von Bonin, A.; Breloer, M. Lipopolysaccharide-free heat shock protein 60 activates T cells. J. Biol. Chem. 2004, 279, 47906–47911. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Syldath, U.; Bellmann, K.; Burkart, V.; Kolb, H. Human 60-kDa heat-shock protein: A danger signal to the innate immune system. J. Immunol. 1999, 162, 3212–3219. [Google Scholar] [CrossRef] [PubMed]
- Chun, J.N.; Choi, B.; Lee, K.W.; Lee, D.J.; Kang, D.H.; Lee, J.Y.; Song, I.S.; Kim, H.I.; Lee, S.-H.; Kim, H.S. Cytosolic Hsp60 is involved in the NF-κB-dependent survival of cancer cells via IKK regulation. PLoS ONE 2010, 5, e9422. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-H.; Yeh, C.-T. Functional compartmentalization of HSP60-survivin interaction between mitochondria and cytosol in cancer cells. Cells 2019, 9, 23. [Google Scholar] [CrossRef]
- Ghosh, J.C.; Siegelin, M.D.; Dohi, T.; Altieri, D.C. Heat shock protein 60 regulation of the mitochondrial permeability transition pore in tumor cells. Cancer Res. 2010, 70, 8988–8993. [Google Scholar] [CrossRef]
- Tsai, Y.-P.; Yang, M.-H.; Huang, C.-H.; Chang, S.-Y.; Chen, P.-M.; Liu, C.-J.; Teng, S.-C.; Wu, K.-J. Interaction between HSP60 and β-catenin promotes metastasis. Carcinogenesis 2009, 30, 1049–1057. [Google Scholar] [CrossRef]
- Cornford, P.A.; Dodson, A.R.; Parsons, K.F.; Desmond, A.D.; Woolfenden, A.; Fordham, M.; Neoptolemos, J.P.; Ke, Y.; Foster, C.S. Heat shock protein expression independently predicts clinical outcome in prostate cancer. Cancer Res. 2000, 60, 7099–7105. [Google Scholar]
- Pace, A.; Barone, G.; Lauria, A.; Martorana, A.; Palumbo Piccionello, A.; Pierro, P.; Terenzi, A.; Maria Almerico, A.; Buscemi, S.; Campanella, C. Hsp60, a novel target for antitumor therapy: Structure-function features and prospective drugs design. Curr. Pharm. Des. 2013, 19, 2757–2764. [Google Scholar] [CrossRef]
- Wong, C.S.-C.; Wong, V.W.-K.; Chan, C.M.-L.; Ma, B.B.-Y.; Hui, E.P.; Wong, M.C.-K.; Lam, M.Y.-Y.; Au, T.C.-C.; Chan, W.-H.; Cheuk, W. Identification of 5-fluorouracil response proteins in colorectal carcinoma cell line SW480 by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Oncol. Rep. 2008, 20, 89–98. [Google Scholar] [CrossRef]
- Tanabe, M.; Ishida, R.; Izuhara, F.; Komatsuda, A.; Wakui, H.; Sawada, K.; Otaka, M.; Nakamura, N.; Itoh, H. The ATPase Activity of Molecular Chaperone HSP60 Is Inhibited by Immunosuppressant Mizoribine; University of Embu: Embu, Kenya, 2012. [Google Scholar]
- Chang, C.-L.; Hsu, Y.-T.; Wu, C.-C.; Yang, Y.-C.; Wang, C.; Wu, T.-C.; Hung, C.-F. Immune mechanism of the antitumor effects generated by bortezomib. J. Immunol. 2012, 189, 3209–3220. [Google Scholar] [CrossRef]
- Ban, H.S.; Shimizu, K.; Minegishi, H.; Nakamura, H. Identification of HSP60 as a Primary Target of o-Carboranylphenoxyacetanilide, an HIF-1α Inhibitor. J. Am. Chem. Soc. 2010, 132, 11870–11871. [Google Scholar] [CrossRef] [PubMed]
- Nobili, S.; Mini, E.; Landini, I.; Gabbiani, C.; Casini, A.; Messori, L. Gold compounds as anticancer agents: Chemistry, cellular pharmacology, and preclinical studies. Med. Res. Rev. 2010, 30, 550–580. [Google Scholar] [CrossRef] [PubMed]
- Cappello, F.; Conway de Macario, E.; Marasà, L.; Zummo, G.; Macario, A.J.L. Hsp60 expression, new locations, functions, and perspectives for cancer diagnosis and therapy. Cancer Biol. Ther. 2008, 7, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Murphy, M.E. The HSP70 family and cancer. Carcinogenesis 2013, 34, 1181–1188. [Google Scholar] [CrossRef] [PubMed]
- Zuiderweg, E.R.P.; Bertelsen, E.B.; Rousaki, A.; Mayer, M.P.; Gestwicki, J.E.; Ahmad, A. Allostery in the Hsp70 chaperone proteins. In Molecular Chaperones; Springer: Berlin/Heidelberg, Germany, 2013; pp. 99–153. [Google Scholar]
- Jego, G.; Hazoumé, A.; Seigneuric, R.; Garrido, C. Targeting heat shock proteins in cancer. Cancer Lett. 2013, 332, 275–285. [Google Scholar] [CrossRef] [PubMed]
- Lipp, A.-M.; Nimmervoll, B.; Sonnleitner, A.; Hesse, J.; Haselgruebler, T.; Balogi, Z. The complex function of hsp70 in metastatic cancer. Cancers 2014, 6, 42–66. [Google Scholar]
- Stankiewicz, A.R.; Lachapelle, G.; Foo, C.P.Z.; Radicioni, S.M.; Mosser, D.D. Hsp70 inhibits heat-induced apoptosis upstream of mitochondria by preventing Bax translocation. J. Biol. Chem. 2005, 280, 38729–38739. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Sigua, C.; Bali, P.; George, P.; Fiskus, W.; Scuto, A.; Annavarapu, S.; Mouttaki, A.; Sondarva, G.; Wei, S. Mechanistic role of heat shock protein 70 in Bcr-Abl–mediated resistance to apoptosis in human acute leukemia cells. Blood 2005, 105, 1246–1255. [Google Scholar] [CrossRef]
- Yaglom, J.A.; Gabai, V.L.; Sherman, M.Y. High levels of heat shock protein Hsp72 in cancer cells suppress default senescence pathways. Cancer Res. 2007, 67, 2373–2381. [Google Scholar] [CrossRef]
- Colvin, T.A.; Gabai, V.L.; Gong, J.; Calderwood, S.K.; Li, H.; Gummuluru, S.; Matchuk, O.N.; Smirnova, S.G.; Orlova, N.V.; Zamulaeva, I.A. Hsp70–Bag3 interactions regulate cancer-related signaling networks. Cancer Res. 2014, 74, 4731–4740. [Google Scholar] [CrossRef]
- Shevtsov, M.; Multhoff, G.; Mikhaylova, E.; Shibata, A.; Guzhova, I.; Margulis, B. Combination of anti-cancer drugs with molecular chaperone inhibitors. Int. J. Mol. Sci. 2019, 20, 5284. [Google Scholar] [CrossRef]
- Budina-Kolomets, A.; Basu, S.; Belcastro, L.; Murphy, M.E. The Hsp70 family of heat shock proteins in tumorigenesis: From molecular mechanisms to therapeutic opportunities. In Stress Response Pathways in Cancer: From Molecular Targets to Novel Therapeutics; Springer: Berlin/Heidelberg, Germany, 2015; pp. 203–224. [Google Scholar]
- Ravagnan, L.; Gurbuxani, S.; Susin, S.A.; Maisse, C.; Daugas, E.; Zamzami, N.; Mak, T.; Jäättelä, M.; Penninger, J.M.; Garrido, C. Heat-shock protein 70 antagonizes apoptosis-inducing factor. Nat. Cell Biol. 2001, 3, 839–843. [Google Scholar] [CrossRef]
- Schmitt, E.; Maingret, L.; Puig, P.-E.; Rerole, A.-L.; Ghiringhelli, F.; Hammann, A.; Solary, E.; Kroemer, G.; Garrido, C. Heat shock protein 70 neutralization exerts potent antitumor effects in animal models of colon cancer and melanoma. Cancer Res. 2006, 66, 4191–4197. [Google Scholar] [CrossRef] [PubMed]
- Leu, J.-J.; Pimkina, J.; Frank, A.; Murphy, M.E.; George, D.L. A small molecule inhibitor of inducible heat shock protein 70. Mol. Cell 2009, 36, 15–27. [Google Scholar] [CrossRef] [PubMed]
- Massey, A.J.; Williamson, D.S.; Browne, H.; Murray, J.B.; Dokurno, P.; Shaw, T.; Macias, A.T.; Daniels, Z.; Geoffroy, S.; Dopson, M. A novel, small molecule inhibitor of Hsc70/Hsp70 potentiates Hsp90 inhibitor induced apoptosis in HCT116 colon carcinoma cells. Cancer Chemother. Pharmacol. 2010, 66, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Rousaki, A.; Miyata, Y.; Jinwal, U.K.; Dickey, C.A.; Gestwicki, J.E.; Zuiderweg, E.R.P. Allosteric drugs: The interaction of antitumor compound MKT-077 with human Hsp70 chaperones. J. Mol. Biol. 2011, 411, 614–632. [Google Scholar] [CrossRef] [PubMed]
- Propper, D.J.; Braybrooke, J.P.; Taylor, D.J.; Lodi, R.; Styles, P.; Cramer, J.A.; Collins, W.C.J.; Levitt, N.C.; Talbot, D.C.; Ganesan, T.S. Phase I trial of the selective mitochondrial toxin MKT 077 in chemo-resistant solid tumours. Ann. Oncol. 1999, 10, 923–927. [Google Scholar] [CrossRef] [PubMed]
- Koren, J., III; Miyata, Y.; Kiray, J.; O’Leary, J.C., III; Nguyen, L.; Guo, J.; Blair, L.J.; Li, X.; Jinwal, U.K.; Cheng, J.Q. Rhodacyanine derivative selectively targets cancer cells and overcomes tamoxifen resistance. PLoS ONE 2012, 7, e35566. [Google Scholar]
- Nadler, S.G.; Eversole, A.C.B.; Tepper, M.A.; Cleaveland, J.S. Elucidating the mechanism of action of the immunosuppressant 15-deoxyspergualin. Ther. Drug Monit. 1995, 17, 700–703. [Google Scholar] [CrossRef] [PubMed]
- Fewell, S.W.; Smith, C.M.; Lyon, M.A.; Dumitrescu, T.P.; Wipf, P.; Day, B.W.; Brodsky, J.L. Small molecule modulators of endogenous and co-chaperone-stimulated Hsp70 ATPase activity. J. Biol. Chem. 2004, 279, 51131–51140. [Google Scholar] [CrossRef]
- Rodina, A.; Patel, P.D.; Kang, Y.; Patel, Y.; Baaklini, I.; Wong, M.J.H.; Taldone, T.; Yan, P.; Yang, C.; Maharaj, R. Identification of an allosteric pocket on human hsp70 reveals a mode of inhibition of this therapeutically important protein. Chem. Biol. 2013, 20, 1469–1480. [Google Scholar] [CrossRef] [PubMed]
- Rérole, A.-L.; Gobbo, J.; De Thonel, A.; Schmitt, E.; Pais de Barros, J.P.; Hammann, A.; Lanneau, D.; Fourmaux, E.; Deminov, O.; Micheau, O. Peptides and aptamers targeting HSP70: A novel approach for anticancer chemotherapy. Cancer Res. 2011, 71, 484–495. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Botzler, C.; Wiesnet, M.; Müller, E.; Meier, T.; Wilmanns, W.; Issels, R.D. A stress-inducible 72-kDa heat-shock protein (HSP72) is expressed on the surface of human tumor cells, but not on normal cells. Int. J. cancer 1995, 61, 272–279. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Hightower, L.E. Cell surface expression of heat shock proteins and the immune response. Cell Stress Chaperones 1996, 1, 167. [Google Scholar] [CrossRef] [PubMed]
- Multhoff, G.; Hightower, L.E. Distinguishing integral and receptor-bound heat shock protein 70 (Hsp70) on the cell surface by Hsp70-specific antibodies. Cell Stress Chaperones 2011, 16, 251–255. [Google Scholar] [CrossRef] [PubMed]
- Stangl, S.; Gehrmann, M.; Dressel, R.; Alves, F.; Dullin, C.; Themelis, G.; Ntziachristos, V.; Staeblein, E.; Walch, A.; Winkelmann, I. In vivo imaging of CT26 mouse tumours by using cmHsp70. 1 monoclonal antibody. J. Cell. Mol. Med. 2011, 15, 874–887. [Google Scholar] [CrossRef]
- Yi, F.; Regan, L. A novel class of small molecule inhibitors of Hsp90. ACS Chem. Biol. 2008, 3, 645–654. [Google Scholar] [CrossRef]
- Roodveldt, C.; Bertoncini, C.W.; Andersson, A.; Van Der Goot, A.T.; Hsu, S.; Fernández-Montesinos, R.; de Jong, J.; van Ham, T.J.; Nollen, E.A.; Pozo, D. Chaperone proteostasis in Parkinson’s disease: Stabilization of the Hsp70/α-synuclein complex by Hip. EMBO J. 2009, 28, 3758–3770. [Google Scholar] [CrossRef]
- Kamal, A.; Thao, L.; Sensintaffar, J.; Zhang, L.; Boehm, M.F.; Fritz, L.C.; Burrows, F.J. A high-affinity conformation of Hsp90 confers tumour selectivity on Hsp90 inhibitors. Nature 2003, 425, 407–410. [Google Scholar] [CrossRef]
- Chiosis, G.; Neckers, L. Tumor selectivity of Hsp90 inhibitors: The explanation remains elusive. ACS Chem. Biol. 2006, 1, 279–284. [Google Scholar] [CrossRef] [PubMed]
- Whitesell, L.; Lindquist, S.L. HSP90 and the chaperoning of cancer. Nat. Rev. Cancer 2005, 5, 761–772. [Google Scholar] [CrossRef]
- Mollapour, M.; Bourboulia, D.; Beebe, K.; Woodford, M.R.; Polier, S.; Hoang, A.; Chelluri, R.; Li, Y.; Guo, A.; Lee, M.-J. Asymmetric Hsp90 N domain SUMOylation recruits Aha1 and ATP-competitive inhibitors. Mol. Cell 2014, 53, 317–329. [Google Scholar] [CrossRef]
- Solit, D.B.; Chiosis, G. Development and application of Hsp90 inhibitors. Drug Discov. Today 2008, 13, 38–43. [Google Scholar] [CrossRef]
- Fortugno, P.; Beltrami, E.; Plescia, J.; Fontana, J.; Pradhan, D.; Marchisio, P.C.; Sessa, W.C.; Altieri, D.C. Regulation of survivin function by Hsp90. Proc. Natl. Acad. Sci. USA 2003, 100, 13791–13796. [Google Scholar] [CrossRef]
- Meli, M.; Pennati, M.; Curto, M.; Daidone, M.G.; Plescia, J.; Toba, S.; Altieri, D.C.; Zaffaroni, N.; Colombo, G. Small-molecule targeting of heat shock protein 90 chaperone function: Rational identification of a new anticancer lead. J. Med. Chem. 2006, 49, 7721–7730. [Google Scholar] [CrossRef]
- Street, T.O.; Lavery, L.A.; Verba, K.A.; Lee, C.-T.; Mayer, M.P.; Agard, D.A. Cross-monomer substrate contacts reposition the Hsp90 N-terminal domain and prime the chaperone activity. J. Mol. Biol. 2012, 415, 3–15. [Google Scholar] [CrossRef][Green Version]
- Roe, S.M.; Ali, M.M.U.; Meyer, P.; Vaughan, C.K.; Panaretou, B.; Piper, P.W.; Prodromou, C.; Pearl, L.H. The mechanism of Hsp90 regulation by the protein kinase-specific cochaperone p50cdc37. Cell 2004, 116, 87–98. [Google Scholar] [CrossRef] [PubMed]
- Wayne, N.; Bolon, D.N. Dimerization of Hsp90 is required for in vivo function. J. Biol. Chem. 2007, 282, 35386–35395. [Google Scholar] [CrossRef] [PubMed]
- Trepel, J.; Mollapour, M.; Giaccone, G.; Neckers, L. Targeting the dynamic HSP90 complex in cancer. Nat. Rev. Cancer 2010, 10, 537–549. [Google Scholar] [CrossRef] [PubMed]
- Scheufler, C.; Brinker, A.; Bourenkov, G.; Pegoraro, S.; Moroder, L.; Bartunik, H.; Hartl, F.U.; Moarefi, I. Structure of TPR domain–peptide complexes: Critical elements in the assembly of the Hsp70–Hsp90 multichaperone machine. Cell 2000, 101, 199–210. [Google Scholar] [CrossRef]
- Gonzalez-Angulo, A.M.; Meric-Bernstam, F.; Chawla, S.; Falchook, G.; Hong, D.; Akcakanat, A.; Chen, H.; Naing, A.; Fu, S.; Wheler, J. Weekly nab-Rapamycin in patients with advanced nonhematologic malignancies: Final results of a phase I trial. Clin. Cancer Res. 2013, 19, 5474–5484. [Google Scholar] [CrossRef] [PubMed]
- Sidera, K.; Patsavoudi, E. HSP90 inhibitors: Current development and potential in cancer therapy. Recent Pat. Anticancer. Drug Discov. 2014, 9, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Nakashima, T.; Ishii, T.; Tagaya, H.; Seike, T.; Nakagawa, H.; Kanda, Y.; Akinaga, S.; Soga, S.; Shiotsu, Y. New molecular and biological mechanism of antitumor activities of KW-2478, a novel nonansamycin heat shock protein 90 inhibitor, in multiple myeloma cells. Clin. Cancer Res. 2010, 16, 2792–2802. [Google Scholar] [CrossRef] [PubMed]
- Taldone, T.; Chiosis, G. Purine-scaffold Hsp90 inhibitors. Curr. Top. Med. Chem. 2009, 9, 1436–1446. [Google Scholar] [CrossRef]
- Donnelly, A.; Blagg, B.S.J. Novobiocin and additional inhibitors of the Hsp90 C-terminal nucleotide-binding pocket. Curr. Med. Chem. 2008, 15, 2702–2717. [Google Scholar] [CrossRef] [PubMed]
- Marcu, M.G.; Chadli, A.; Bouhouche, I.; Catelli, M.; Neckers, L.M. The heat shock protein 90 antagonist novobiocin interacts with a previously unrecognized ATP-binding domain in the carboxyl terminus of the chaperone. J. Biol. Chem. 2000, 275, 37181–37186. [Google Scholar] [CrossRef] [PubMed]
- Palermo, C.M.; Westlake, C.A.; Gasiewicz, T.A. Epigallocatechin gallate inhibits aryl hydrocarbon receptor gene transcription through an indirect mechanism involving binding to a 90 kDa heat shock protein. Biochemistry 2005, 44, 5041–5052. [Google Scholar] [CrossRef]
- Itoh, H.; Ogura, M.; Komatsuda, A.; Wakui, H.; Miura, A.B.; Tashima, Y. A novel chaperone-activity-reducing mechanism of the 90-kDa molecular chaperone HSP90. Biochem. J. 1999, 343, 697–703. [Google Scholar] [CrossRef]
- Hartson, S.D.; Thulasiraman, V.; Huang, W.; Whitesell, L.; Matts, R.L. Molybdate inhibits hsp90, induces structural changes in its C-terminal domain, and alters its interactions with substrates. Biochemistry 1999, 38, 3837–3849. [Google Scholar] [CrossRef]
- Byrd, C.A.; Bornmann, W.; Erdjument-Bromage, H.; Tempst, P.; Pavletich, N.; Rosen, N.; Nathan, C.F.; Ding, A. Heat shock protein 90 mediates macrophage activation by Taxol and bacterial lipopolysaccharide. Proc. Natl. Acad. Sci. USA 1999, 96, 5645–5650. [Google Scholar] [CrossRef]
- Belletti, D.; Pederzoli, F.; Forni, F.; Vandelli, M.A.; Tosi, G.; Ruozi, B. Protein cage nanostructure as drug delivery system: Magnifying glass on apoferritin. Expert Opin. Drug Deliv. 2017, 14, 825–840. [Google Scholar] [CrossRef]
- Darvishi, S.; Tavakoli, S.; Kharaziha, M.; Girault, H.H.; Kaminski, C.F.; Mela, I. Advances in the sensing and treatment of wound biofilms. Angew. Chemie 2022, 134, e202112218. [Google Scholar] [CrossRef]
- Sanchez, J.; Carter, T.R.; Cohen, M.S.; Blagg, B.S.J. Old and new approaches to target the Hsp90 chaperone. Curr. Cancer Drug Targets 2020, 20, 253–270. [Google Scholar] [CrossRef]
- Garg, G.; Khandelwal, A.; Blagg, B.S.J. Anticancer inhibitors of Hsp90 function: Beyond the usual suspects. Adv. Cancer Res. 2016, 129, 51–88. [Google Scholar]
- Smith, J.R.; Clarke, P.A.; de Billy, E.; Workman, P. Silencing the cochaperone CDC37 destabilizes kinase clients and sensitizes cancer cells to HSP90 inhibitors. Oncogene 2009, 28, 157–169. [Google Scholar] [CrossRef] [PubMed]
- Chadli, A.; Felts, S.J.; Wang, Q.; Sullivan, W.P.; Botuyan, M.V.; Fauq, A.; Ramirez-Alvarado, M.; Mer, G. Celastrol inhibits Hsp90 chaperoning of steroid receptors by inducing fibrillization of the Co-chaperone p23. J. Biol. Chem. 2010, 285, 4224–4231. [Google Scholar] [CrossRef] [PubMed]
- Patwardhan, C.A.; Fauq, A.; Peterson, L.B.; Miller, C.; Blagg, B.S.J.; Chadli, A. Gedunin Inactivates the Co-chaperone p23 Protein Causing Cancer Cell Death by Apoptosis. J. Biol. Chem. 2013, 288, 7313–7325. [Google Scholar] [CrossRef] [PubMed]
- Holmes, J.L.; Sharp, S.Y.; Hobbs, S.; Workman, P. Silencing of HSP90 cochaperone AHA1 expression decreases client protein activation and increases cellular sensitivity to the HSP90 inhibitor 17-allylamino-17-demethoxygeldanamycin. Cancer Res. 2008, 68, 1188–1197. [Google Scholar] [CrossRef]
- Ghosh, S.; Shinogle, H.E.; Garg, G.; Vielhauer, G.A.; Holzbeierlein, J.M.; Dobrowsky, R.T.; Blagg, B.S.J. Hsp90 C-terminal inhibitors exhibit antimigratory activity by disrupting the Hsp90α/Aha1 complex in PC3-MM2 cells. ACS Chem. Biol. 2015, 10, 577–590. [Google Scholar] [CrossRef]
- Centenera, M.M.; Fitzpatrick, A.K.; Tilley, W.D.; Butler, L.M. Hsp90: Still a viable target in prostate cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2013, 1835, 211–218. [Google Scholar] [CrossRef] [PubMed]
- Georget, V.; Térouanne, B.; Nicolas, J.-C.; Sultan, C. Mechanism of antiandrogen action: Key role of hsp90 in conformational change and transcriptional activity of the androgen receptor. Biochemistry 2002, 41, 11824–11831. [Google Scholar] [CrossRef]
- Liu, S.; Yuan, Y.; Okumura, Y.; Shinkai, N.; Yamauchi, H. Camptothecin disrupts androgen receptor signaling and suppresses prostate cancer cell growth. Biochem. Biophys. Res. Commun. 2010, 394, 297–302. [Google Scholar] [CrossRef]
- Plescia, J.; Salz, W.; Xia, F.; Pennati, M.; Zaffaroni, N.; Daidone, M.G.; Meli, M.; Dohi, T.; Fortugno, P.; Nefedova, Y. Rational design of shepherdin, a novel anticancer agent. Cancer Cell 2005, 7, 457–468. [Google Scholar] [CrossRef] [PubMed]
- Hatayama, T.; Yamagishi, N.; Minobe, E.; Sakai, K. Role of hsp105 in protection against stress-induced apoptosis in neuronal PC12 cells. Biochem. Biophys. Res. Commun. 2001, 288, 528–534. [Google Scholar] [CrossRef] [PubMed]
- Hosaka, S.; Nakatsura, T.; Tsukamoto, H.; Hatayama, T.; Baba, H.; Nishimura, Y. Synthetic small interfering RNA targeting heat shock protein 105 induces apoptosis of various cancer cells both in vitro and in vivo. Cancer Sci. 2006, 97, 623–632. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Lee, A.S. Glucose regulated proteins in cancer progression, drug resistance and immunotherapy. Cancer Biol. Ther. 2006, 5, 741–744. [Google Scholar] [CrossRef] [PubMed]
- Manjili, M.H.; Park, J.; Facciponte, J.G.; Wang, X.; Subjeck, J.R. Immunoadjuvant chaperone, GRP170, induces ‘danger signals’ upon interaction with dendritic cells. Immunol. Cell Biol. 2006, 84, 203–208. [Google Scholar] [CrossRef] [PubMed]
- Dorard, C.; De Thonel, A.; Collura, A.; Marisa, L.; Svrcek, M.; Lagrange, A.; Jego, G.; Wanherdrick, K.; Joly, A.L.; Buhard, O. Expression of a mutant HSP110 sensitizes colorectal cancer cells to chemotherapy and improves disease prognosis. Nat. Med. 2011, 17, 1283–1289. [Google Scholar] [CrossRef] [PubMed]
- Zappasodi, R.; Bongarzone, I.; Ghedini, G.C.; Castagnoli, L.; Cabras, A.D.; Messina, A.; Tortoreto, M.; Tripodo, C.; Magni, M.; Carlo-Stella, C. Serological identification of HSP105 as a novel non-Hodgkin lymphoma therapeutic target. Blood J. Am. Soc. Hematol. 2011, 118, 4421–4430. [Google Scholar] [CrossRef] [PubMed]
- Ren, F.; Xu, Y.; Mao, L.; Ou, R.; Ding, Z.; Zhang, X.; Tang, J.; Li, B.; Jia, Z.; Tian, Z. Heat shock protein 110 improves the anti-tumor effects of the cytotoxic T lymphocyte epitope E7 in mice. Cancer Biol. Ther. 2010, 9, 134–141. [Google Scholar] [CrossRef]
- Yokomine, K.; Nakatsura, T.; Senju, S.; Nakagata, N.; Minohara, M.; Kira, J.; Motomura, Y.; Kubo, T.; Sasaki, Y.; Nishimura, Y. Regression of intestinal adenomas by vaccination with heat shock protein 105-pulsed bone marrow-derived dendritic cells in ApcMin/+ mice. Cancer Sci. 2007, 98, 1930–1935. [Google Scholar] [CrossRef]
- Wang, X.-Y.; Sun, X.; Chen, X.; Facciponte, J.; Repasky, E.A.; Kane, J.; Subjeck, J.R. Superior antitumor response induced by large stress protein chaperoned protein antigen compared with peptide antigen. J. Immunol. 2010, 184, 6309–6319. [Google Scholar] [CrossRef]
- Prince, T.L.; Lang, B.J.; Guerrero-Gimenez, M.E.; Fernandez-Muñoz, J.M.; Ackerman, A.; Calderwood, S.K. HSF1: Primary factor in molecular chaperone expression and a major contributor to cancer morbidity. Cells 2020, 9, 1046. [Google Scholar] [CrossRef]
- Dai, C.; Sampson, S.B. HSF1: Guardian of proteostasis in cancer. Trends Cell Biol. 2016, 26, 17–28. [Google Scholar] [CrossRef] [PubMed]
- Alasady, M.J.; Mendillo, M.L. The multifaceted role of HSF1 in tumorigenesis. In HSF1 and Molecular Chaperones in Biology and Cancer; Springer: Berlin/Heidelberg, Germany, 2020; pp. 69–85. [Google Scholar]
- Carpenter, R.L.; Gökmen-Polar, Y. HSF1 as a cancer biomarker and therapeutic target. Curr. Cancer Drug Targets 2019, 19, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Jia, G.; Wu, W.; Chen, L.; Yu, Y.; Tang, Q.; Liu, H.; Jiang, Q.; Han, B. HSF1 is a novel prognostic biomarker in high-risk prostate cancer that correlates with ferroptosis. Discov. Oncol. 2023, 14, 107. [Google Scholar] [CrossRef] [PubMed]
- Vydra, N.; Janus, P.; Kus, P.; Stokowy, T.; Mrowiec, K.; Toma-Jonik, A.; Krzywon, A.; Cortez, A.J.; Wojtas, B.; Gielniewski, B. Heat shock factor 1 (HSF1) cooperates with estrogen receptor α (ERα) in the regulation of estrogen action in breast cancer cells. Elife 2021, 10, e69843. [Google Scholar] [CrossRef] [PubMed]
- Ishiwata, J.; Kasamatsu, A.; Sakuma, K.; Iyoda, M.; Yamatoji, M.; Usukura, K.; Ishige, S.; Shimizu, T.; Yamano, Y.; Ogawara, K. State of heat shock factor 1 expression as a putative diagnostic marker for oral squamous cell carcinoma. Int. J. Oncol. 2012, 40, 47–52. [Google Scholar] [PubMed]
- Yasuda, K.; Hirohashi, Y.; Mariya, T.; Murai, A.; Tabuchi, Y.; Kuroda, T.; Kusumoto, H.; Takaya, A.; Yamamoto, E.; Kubo, T. Phosphorylation of HSF1 at serine 326 residue is related to the maintenance of gynecologic cancer stem cells through expression of HSP27. Oncotarget 2017, 8, 31540. [Google Scholar] [CrossRef]
- McConnell, J.R.; Buckton, L.K.; McAlpine, S.R. Regulating the master regulator: Controlling heat shock factor 1 as a chemotherapy approach. Bioorg. Med. Chem. Lett. 2015, 25, 3409–3414. [Google Scholar] [CrossRef] [PubMed]
- Yoon, Y.J.; Kim, J.A.; Shin, K.D.; Shin, D.-S.; Han, Y.M.; Lee, Y.J.; Lee, J.S.; Kwon, B.-M.; Han, D.C. KRIBB11 inhibits HSP70 synthesis through inhibition of heat shock factor 1 function by impairing the recruitment of positive transcription elongation factor b to the hsp70 promoter. J. Biol. Chem. 2011, 286, 1737–1747. [Google Scholar] [CrossRef]
- Shen, Z.; Yin, L.; Zhou, H.; Ji, X.; Jiang, C.; Zhu, X.; He, X. Combined inhibition of AURKA and HSF1 suppresses proliferation and promotes apoptosis in hepatocellular carcinoma by activating endoplasmic reticulum stress. Cell. Oncol. 2021, 44, 1035–1049. [Google Scholar] [CrossRef]
- Brown, C.W.; Chhoy, P.; Mukhopadhyay, D.; Karner, E.R.; Mercurio, A.M. Targeting prominin2 transcription to overcome ferroptosis resistance in cancer. EMBO Mol. Med. 2021, 13, e13792. [Google Scholar] [CrossRef]
- Ganguly, S.; Home, T.; Yacoub, A.; Kambhampati, S.; Shi, H.; Dandawate, P.; Padhye, S.; Saluja, A.K.; McGuirk, J.; Rao, R. Targeting HSF1 disrupts HSP90 chaperone function in chronic lymphocytic leukemia. Oncotarget 2015, 6, 31767. [Google Scholar] [CrossRef]
- Dong, B.; Jaeger, A.M.; Hughes, P.F.; Loiselle, D.R.; Hauck, J.S.; Fu, Y.; Haystead, T.A.; Huang, J.; Thiele, D.J. Targeting therapy-resistant prostate cancer via a direct inhibitor of the human heat shock transcription factor 1. Sci. Transl. Med. 2020, 12, eabb5647. [Google Scholar] [CrossRef] [PubMed]
- Dong, Q.; Xiu, Y.; Wang, Y.; Hodgson, C.; Borcherding, N.; Jordan, C.; Buchanan, J.; Taylor, E.; Wagner, B.; Leidinger, M. HSF1 is a driver of leukemia stem cell self-renewal in acute myeloid leukemia. Nat. Commun. 2022, 13, 6107. [Google Scholar] [CrossRef]
- Binder, R.J. Functions of heat shock proteins in pathways of the innate and adaptive immune system. J. Immunol. 2014, 193, 5765–5771. [Google Scholar] [CrossRef] [PubMed]
- Murshid, A.; Gong, J.; Stevenson, M.A.; Calderwood, S.K. Heat shock proteins and cancer vaccines: Developments in the past decade and chaperoning in the decade to come. Expert Rev. Vaccines 2011, 10, 1553–1568. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, R.; Chen, A.; Wang, Y.; Wang, Y.; Zhou, J.; Cao, R. Inhibition of mouse RM-1 prostate cancer and B16F10 melanoma by the fusion protein of HSP65 & STEAP1 186–193. Biomed. Pharmacother. 2019, 111, 1124–1131. [Google Scholar]
- Guo, L.; Xie, H.; Zhang, Z.; Wang, Z.; Peng, S.; Niu, Y.; Shang, Z. Fusion protein vaccine based on Ag85B and STEAP1 induces a protective immune response against prostate cancer. Vaccines 2021, 9, 786. [Google Scholar] [CrossRef] [PubMed]
- Sakai, T.; Hisaeda, H.; Ishikawa, H.; Maekawa, Y.; Zhang, M.; Nakao, Y.; Takeuchi, T.; Matsumoto, K.; Good, R.A.; Himeno, K. Expression and role of heat-shock protein 65 (HSP65) in macrophages during Trypanosoma cruzi infection: Involvement of HSP65 in prevention of apoptosis of macrophages. Microbes Infect. 1999, 1, 419–427. [Google Scholar] [CrossRef]
- Wang, S.; Fan, H.; Li, Y.; Zheng, H.; Li, X.; Li, C.; Chen, L.; Ju, Y.; Meng, S. CD133 epitope vaccine with gp96 as adjuvant elicits an antitumor T cell response against leukemia. Chin. J. Biotechnol. 2017, 33, 1006–1017. [Google Scholar]
- Zheng, H.; Liu, L.; Zhang, H.; Kan, F.; Wang, S.; Li, Y.; Tian, H.; Meng, S. Dendritic cells pulsed with placental gp96 promote tumor-reactive immune responses. PLoS ONE 2019, 14, e0211490. [Google Scholar] [CrossRef]
- Li, X.; Cai, X.; Zhang, Z.; Ding, Y.; Ma, R.; Huang, F.; Liu, Y.; Liu, J.; Shi, L. Mimetic heat shock protein mediated immune process to enhance cancer immunotherapy. Nano Lett. 2020, 20, 4454–4463. [Google Scholar] [CrossRef]
- Das, J.K.; Xiong, X.; Ren, X.; Yang, J.-M.; Song, J. Heat shock proteins in cancer immunotherapy. J. Oncol. 2019, 2019, 3267207. [Google Scholar] [CrossRef]
- Einstein, M.H.; Roden, R.B.S.; Ferrall, L.; Akin, M.; Blomer, A.; Wu, T.C.; Chang, Y.-N. Safety run-in of intramuscular pNGVL4a-Sig/E7 (detox)/HSP70 DNA and TA-CIN protein vaccination as treatment for HPV16+ ASC-US, ASC-H, or LSIL/CIN1. Cancer Prev. Res. 2023, 16, 219–227. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Du, Y.; Zhang, Y.; Ji, N. Immunotherapy with heat shock protein 96 to treat gliomas. Chin. Neurosurg. J. 2021, 7, 53–57. [Google Scholar] [CrossRef] [PubMed]
- Bloch, O.; Lim, M.; Sughrue, M.E.; Komotar, R.J.; Abrahams, J.M.; O’Rourke, D.M.; D’Ambrosio, A.; Bruce, J.N.; Parsa, A.T. Autologous heat shock protein peptide vaccination for newly diagnosed glioblastoma: Impact of peripheral PD-L1 expression on response to therapy. Clin. Cancer Res. 2017, 23, 3575–3584. [Google Scholar] [CrossRef] [PubMed]
- Ji, N.; Zhang, Y.; Liu, Y.; Xie, J.; Wang, Y.; Hao, S.; Gao, Z. Heat shock protein peptide complex-96 vaccination for newly diagnosed glioblastoma: A phase I, single-arm trial. JCI Insight 2018, 3, e99145. [Google Scholar] [CrossRef] [PubMed]
- Valpione, S.; Mundra, P.A.; Galvani, E.; Campana, L.G.; Lorigan, P.; De Rosa, F.; Gupta, A.; Weightman, J.; Mills, S.; Dhomen, N. The T cell receptor repertoire of tumor infiltrating T cells is predictive and prognostic for cancer survival. Nat. Commun. 2021, 12, 4098. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Mudgal, P.; Wang, L.; Wu, H.; Huang, N.; Alexander, P.B.; Gao, Z.; Ji, N.; Li, Q.-J. T cell receptor repertoire as a prognosis marker for heat shock protein peptide complex-96 vaccine trial against newly diagnosed glioblastoma. Oncoimmunology 2020, 9, 1749476. [Google Scholar] [CrossRef] [PubMed]
- Viana, P.; Hamar, P. Targeting the heat shock response induced by modulated electro-hyperthermia (mEHT) in cancer. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2024, 1879, 189069. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, C.; Messaoudi, S.; Fattal, E.; Vergnaud-Gauduchon, J. Cancer drug resistance: Rationale for drug delivery systems and targeted inhibition of HSP90 family proteins. Cancer Drug Resist. 2019, 2, 381. [Google Scholar] [CrossRef]
- Kuo, I.-M.; Lee, J.-J.; Wang, Y.-S.; Chiang, H.-C.; Huang, C.-C.; Hsieh, P.-J.; Han, W.; Ke, C.-H.; Liao, A.T.C.; Lin, C.-S. Potential enhancement of host immunity and anti-tumor efficacy of nanoscale curcumin and resveratrol in colorectal cancers by modulated electro-hyperthermia. BMC Cancer 2020, 20, 603. [Google Scholar] [CrossRef] [PubMed]
- Molina, M.L.; García-Bernal, D.; Martinez, S.; Valdor, R. Autophagy in the immunosuppressive perivascular microenvironment of glioblastoma. Cancers 2019, 12, 102. [Google Scholar] [CrossRef]
- Lu, T.-L.; Huang, G.-J.; Wang, H.-J.; Chen, J.-L.; Hsu, H.-P.; Lu, T.-J. Hispolon promotes MDM2 downregulation through chaperone-mediated autophagy. Biochem. Biophys. Res. Commun. 2010, 398, 26–31. [Google Scholar] [CrossRef]
- Bonhoure, A.; Vallentin, A.; Martin, M.; Senff-Ribeiro, A.; Amson, R.; Telerman, A.; Vidal, M. Acetylation of translationally controlled tumor protein promotes its degradation through chaperone-mediated autophagy. Eur. J. Cell Biol. 2017, 96, 83–98. [Google Scholar] [CrossRef]
- Kon, M.; Kiffin, R.; Koga, H.; Chapochnick, J.; Macian, F.; Varticovski, L.; Cuervo, A.M. Chaperone-mediated autophagy is required for tumor growth. Sci. Transl. Med. 2011, 3, 109ra117. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).