Artificial Intelligence Applied to Non-Invasive Imaging Modalities in Identification of Nonmelanoma Skin Cancer: A Systematic Review
Abstract
Simple Summary
Abstract
1. Background
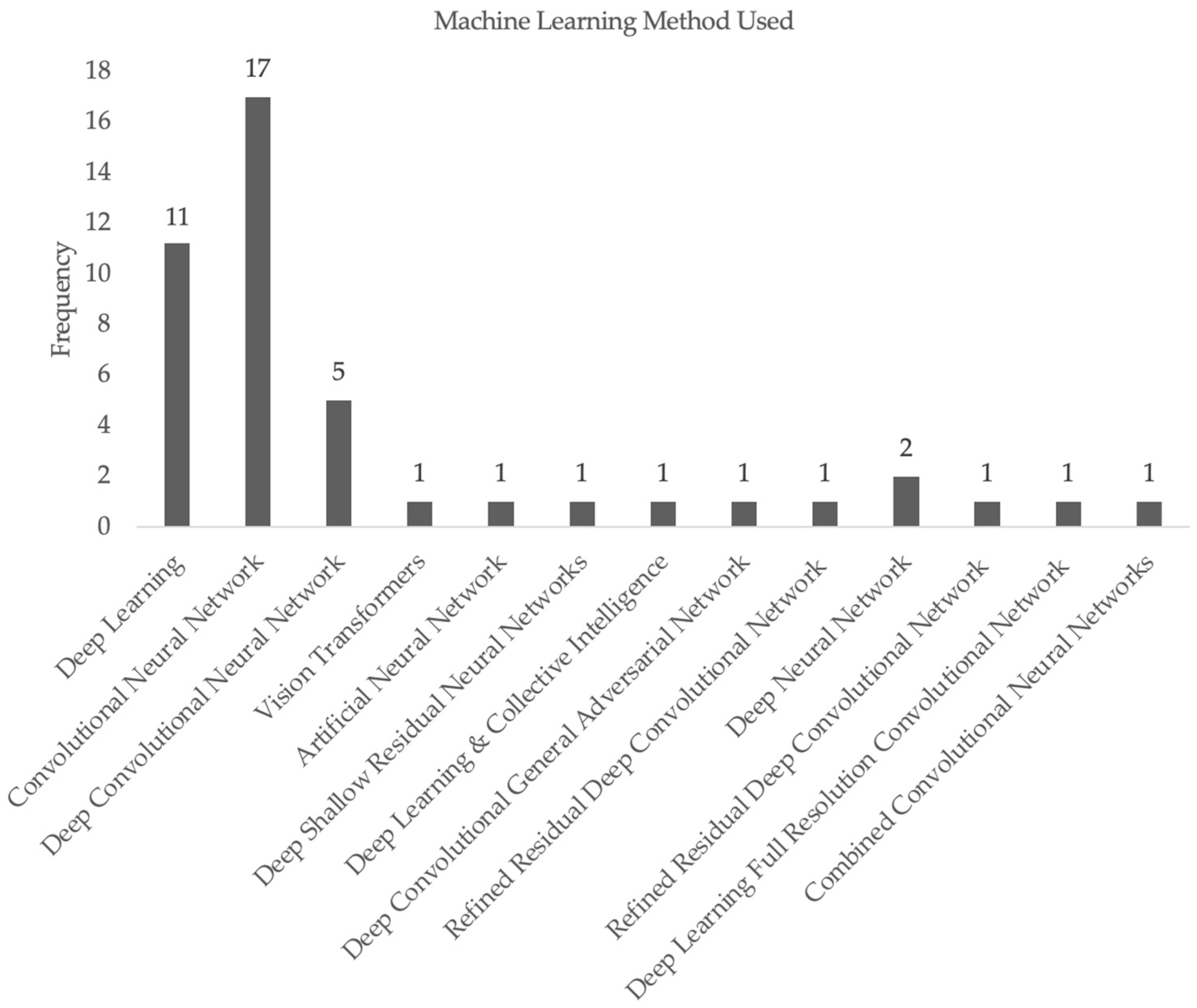
1.1. Common Machine Learning Methods
1.2. AI Applications in Non-Invasive Imaging Modalities
2. Materials and Methods
2.1. Search Strategy
2.2. Study Selection
2.3. Study Analysis
3. Results
| Authors | Image Dataset | Accuracy | Precision | Sensitivity | Specificity | PPV | NPV | AUC | F1 Score |
|---|---|---|---|---|---|---|---|---|---|
| Hosny et al. (2020) [7] | Internal dataset | 98.7% | 95.1% | 95.6% | 99.3% | ||||
| Xin et al. (2022) [13] | HAM1000 | 94.3% | 94.1% | ||||||
| Xin et al. (2022) [13] | Internal dataset | 94.1% | 94.2% | ||||||
| Tang et al. (2022) [14] | Seven Point Checklist | 74.9% | |||||||
| Skreekala et al. (2022) [15] | HAM1000 | 97% | |||||||
| Sangers et al. (2022) [16] | HAM1000 | 86.9% | 70.4% | ||||||
| Samsudin et al. (2022) [17] | HAM1000 | 87.7% | |||||||
| S M et al. (2022) [18] | ISIC 2019 & 2020 | 96.8% * | |||||||
| Reis et al. (2022) [19] | ISIC 2018 | 94.6% | |||||||
| Reis et al. (2022) [19] | ISIC 2019 | 91.9% | |||||||
| Reis et al. (2022) [19] | ISIC 2020 | 90.5% | |||||||
| Razzak et al. (2022) [20] | ISIC 2018 | 98.1% | |||||||
| Qian et al. (2022) [21] | HAM1000 | 91.6% | 73.5% | 96.4% | 97.1% | ||||
| Popescu et al. (2022) [22] | ISIC 2018 | 86.7% | |||||||
| Nguyen et al. (2022) [23] | HAM1000 | 90% | 81% | 99% | 81% | ||||
| Naeem et al. (2022) [24] | ISIC 2019 | 96.9% | |||||||
| Li et al. (2022) [25] | HAM1000 | 95.8% | 96.1% | 95.7% | |||||
| Lee et al. (2022) [26] | ISIC 2018 | 84.4% | 92.8% | 78.5% | 91.2% | ||||
| Laverde-Saad et al. (2022) [27] | HAM1000 | 77.1% | 80% | 86% | 86% | 80% | |||
| La Salvia et al. (2022) [28] | HAM1000 | >80% | >80% | >80% | |||||
| Hosny et al. (2022) [29] | Several datasets | 94.1% * | 91.4% * | 91.2% * | 94.7% * | ||||
| Dascalu et al. (2022) [30] | Internal dataset | 88% | 95.3% | 91.1% | |||||
| Combalia et al. (2019) [31] | HAM1000 | 58.8% | |||||||
| Benyahia et al. (2022) [32] | ISIC 2019 | 92.3% | |||||||
| Bechelli et al. (2022) [33] | HAM1000 | 88% | |||||||
| Bechelli et al. (2022) [33] | HAM1000 | 72% | |||||||
| Afza et al. (2022) [34] | Ph2 | 95.4% | |||||||
| Afza et al. (2022) [34] | ISBI2016 | 91.1% | |||||||
| Afza et al. (2022) [34] | HAM1000 | 85.5% | |||||||
| Afza et al. (2022) [35] | HAM1000 | 93.4% | |||||||
| Afza et al. (2022) [35] | ISIC2018. | 94.4% | |||||||
| Winkler et al. (2021) [36] | HAM1000 | 70% | 70.6% | 69.2% | |||||
| Pacheco et al. (2021) [37] | HAM1000 | 77.1% | |||||||
| Minagawa et al. (2021) [38] | HAM1000 | 85.3% | |||||||
| Iqbal et al. (2021) [39] | HAM1000 | 99.1% | |||||||
| Huang et al. (2021) [40] | HAM1000 | 84.8% | |||||||
| Zhang et al. (2020) [41] | DermIS & Dermquest | 91% | 95% | 92% | 84% | 95% | |||
| Wang et al. (2020) [42] | Several datasets | 80% | 100% | ||||||
| Qin et al. (2020) [43] | HAM1000 | 95.2% | 83.2% | 74.3% | |||||
| Mahbod et al. (2020) [44] | ISIC2019 | 86.2% | |||||||
| Li et al. (2020) [45] | HAM1000 | 78% | 95% | 91% | 87% | ||||
| Gessert et al. (2020) [46] | HAM1000 | 70% | |||||||
| Gessert et al. (2020) [47] | Internal dataset | 53% * | 97.5% * | 94% * | |||||
| Al-masni et al. (2020) [48] | HAM1000 | 89.3% | |||||||
| Ameri et al. (2020) [49] | HAM1000 | 84% | 81% | ||||||
| Tschandl et al. (2019) [50] | Internal dataset | 37.6% | 80.5% | ||||||
| Dascalu et al. (2019) [51] | HAM1000 | 91.7% | 41.8% | 59.9% | 81.4% |
| Authors | Imaging Modality | Accuracy | Sensitivity | Specificity | AUC |
|---|---|---|---|---|---|
| Wodzinski et al. (2019) [8] | RCM | 87% | |||
| Chen et al. (2022) [9] | RCM | 100% (when combined with RS) | 92.4% (when combined with RS) | ||
| Campanella et al. (2022) [10] | RCM | 86.1% | |||
| La Salvia et al. (2022) [11] | HEI | 87% | 88% | 90% |
| Database | Image Type | Total Images | Description of Dataset |
|---|---|---|---|
| HAM1000 | Dermoscopy | 10,015 | Melanoma (MM)—1113 images Vascular—142 images Benign nevus (MN)—6705 images Dermatofibroma (DF)—115 images Seborrheic keratosis (SK)—1099 Basal-cell carcinoma (BCC)—514 images Actinic keratosis (AK)—327 images |
| Xin et al. [13] Internal | Dermoscopy | 1016 | BCC—630 images Squamous-cell carcinoma (SCC)—192 images MM—194 images |
| SPC | Dermoscopy | >2000 | MM, BCC, SK, DF, solar lentigo (SL), vascular, SK Note: Distribution of number of images per lesion type varies in the literature. |
| ISIC 2016 | Dermoscopy | 1279 | Distribution of number of images per lesion type not readily available |
| ISIC 2017 | Dermoscopy | 2000 | MM—374 images SK—254 images Other/unknown—1372 images |
| ISIC 2018 | Dermoscopy | 10,015 | MM—1113 images MN—6705 images BCC—514 images AK—327 images SK—1099 images DF—115 images Vascular—142 images |
| ISIC 2019 | Dermoscopy | 25,331 | MM—4522 images MN—12,875 images BCC—3323 images AK—867 images DF—239 images SK—2624 images SCC—628 images Vascular—253 images |
| ISIC 2020 | Dermoscopy | 33,126 | MM—584 images AMN—1 image Café-au-lait macule—1 image SL—44 Lichenoid keratosis—37 images Other/unknown—27124 images |
| PH2 | Dermoscopy | 200 | Not available |
| Categories | Risk of Bias | Applicability Concerns | |||||
|---|---|---|---|---|---|---|---|
| Patient Selection | Index Test | Reference Standard | Flow and Timing | Patient Selection | Index Test | Reference Standard | |
| Low Risk | 27/44 | 31/44 | 42/44 | 44/44 | 20/44 | 37/44 | 44/44 |
| High Risk | 0/44 | 0/44 | 0/44 | 0/44 | 10/44 | 0/44 | 0/44 |
| Unclear/Moderate | 17/44 | 13/44 | 2/44 | 0/44 | 14/44 | 7/44 | 0/44 |
4. Discussion
4.1. Limitations
4.2. Future Directions
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Du-Harpur, X.; Watt, F.M.; Luscombe, N.M.; Lynch, M.D. What is AI? Applications of artificial intelligence to dermatology. Br. J. Dermatol. 2020, 183, 423–430. [Google Scholar] [CrossRef]
- Popescu, D.; El-Khatib, M.; El-Khatib, H.; Ichim, L. New Trends in Melanoma Detection Using Neural Networks: A Systematic Review. Sensors 2022, 22, 496. [Google Scholar] [CrossRef]
- Takiddin, A.; Schneider, J.; Yang, Y.; Abd-Alrazaq, A.; Househ, M. Artificial Intelligence for Skin Cancer Detection: Scoping Re-view. J. Med. Internet Res. 2021, 23, e22934. [Google Scholar] [CrossRef] [PubMed]
- Das, K.; Cockerell, C.J.; Patil, A.; Pietkiewicz, P.; Giulini, M. Machine Learning and Its Application in Skin Cancer. Int. J. Environ. Res. Public Health 2021, 18, 13409. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.H.; Foltz, E.A.; Witkowski, A.; Ludzik, J. Analysis of Artificial Intelligence-Based Approaches Applied to Non-Invasive Imaging for Early Detection of Melanoma: A Systematic Review. Cancers 2023, 15, 4694. [Google Scholar] [CrossRef]
- Alzubaidi, L.; Zhang, J.; Humaidi, A.J.; Al-Dujaili, A.; Duan, Y.; Al-Shamma, O.; Santamaría, J.; Fadhel, M.A.; Al-Amidie, M.; Farhan, L. Review of deep learning: Concepts, CNN architectures, challenges, applications, future directions. J. Big Data 2021, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Hosny, K.M.; Kassem, M.A.; Fouad, M.M. Classification of Skin Lesions into Seven Classes Using Transfer Learning with AlexNet. J. Digit. Imaging 2020, 33, 1325–1334. [Google Scholar] [CrossRef] [PubMed]
- Wodzinski, M.; Skalski, A.; Witkowski, A.; Pellacani, G.; Ludzik, J. Convolutional Neural Network Approach to Classify Skin Lesions Using Reflectance Confocal Microscopy. In Proceedings of the 2019 41st Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC), Berlin, Germany, 23–27 July 2019; pp. 4754–4757. [Google Scholar] [CrossRef]
- Chen, M.; Feng, X.; Fox, M.C.; Reichenberg, J.S.; Lopes, F.C.; Sebastian, K.R.; Markey, M.K.; Tunnell, J.W. Deep learning on reflectance confocal microscopy improves Raman spectral diagnosis of basal cell carcinoma. J. Biomed. Opt. 2022, 27, 065004. [Google Scholar] [CrossRef] [PubMed]
- Campanella, G.; Navarrete-Dechent, C.; Liopyris, K.; Monnier, J.; Aleissa, S.; Minhas, B.; Scope, A.; Longo, C.; Guitera, P.; Pellacani, G.; et al. Deep Learning for Basal Cell Carcinoma Detection for Reflectance Confocal Microscopy. J. Investig. Dermatol. 2022, 142, 97–103. [Google Scholar] [CrossRef] [PubMed]
- La Salvia, M.; Torti, E.; Leon, R.; Fabelo, H.; Ortega, S.; Balea-Fernandez, F.; Martinez-Vega, B.; Castaño, I.; Almeida, P.; Carretero, G.; et al. Neural Networks-Based On-Site Dermatologic Diagnosis through Hyperspectral Epi-dermal Images. Sensors 2022, 22, 7139. [Google Scholar] [CrossRef]
- Whiting, P.F.; Rutjes, A.W.; Westwood, M.E.; Mallett, S.; Deeks, J.J.; Reitsma, J.B.; Leeflang, M.M.; Sterne, J.A.; Bossuyt, P.M.; QUADAS-2 Group. QUADAS-2: A revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 2011, 155, 529–536. [Google Scholar] [CrossRef]
- Xin, C.; Liu, Z.; Zhao, K.; Miao, L.; Ma, Y.; Zhu, X.; Zhou, Q.; Wang, S.; Li, L.; Yang, F.; et al. An improved transformer network for skin cancer classification. Comput. Biol. Med. 2022, 149, 105939. [Google Scholar] [CrossRef]
- Tang, P.; Yan, X.; Nan, Y.; Xiang, S.; Krammer, S.; Lasser, T. FusionM4Net: A multi-stage multi-modal learning algorithm for multi-label skin lesion classification. Med. Image Anal. 2022, 76, 102307. [Google Scholar] [CrossRef]
- Sreekala, K.; Rajkumar, N.; Sugumar, R.; Sagar, K.V.; Shobarani, R.; Krishnamoorthy, K.P.; Saini, A.K.; Palivela, H.; Yeshitla, A. Skin Diseases Classification Using Hybrid AI Based Localization Approach. Comput. Intell. Neurosci. 2022, 2022, 6138490. [Google Scholar] [CrossRef]
- Sangers, T.; Reeder, S.; van der Vet, S.; Jhingoer, S.; Mooyaart, A.; Siegel, D.M.; Nijsten, T.; Wakkee, M. Validation of a Market-Approved Artificial Intelligence Mobile Health App for Skin Cancer Screening: A Prospective Multicenter Diagnostic Accuracy Study. Dermatology 2022, 238, 649–656. [Google Scholar] [CrossRef]
- Samsudin, S.S.; Arof, H.; Harun, S.W.; Abdul Wahab, A.W.; Idris, M.Y.I. Skin lesion classification using multi-resolution empirical mode decomposition and local binary pattern. PLoS ONE 2022, 17, e0274896. [Google Scholar] [CrossRef]
- S M, J.; P, M.; Aravindan, C.; Appavu, R. Classification of skin cancer from dermoscopic images using deep neural network architectures. Multimed. Tools Appl. 2023, 82, 15763–15778. [Google Scholar] [CrossRef]
- Reis, H.C.; Turk, V.; Khoshelham, K.; Kaya, S. InSiNet: A deep convolutional approach to skin cancer detection and segmentation. Med. Biol. Eng. Comput. 2022, 60, 643–662. [Google Scholar] [CrossRef] [PubMed]
- Razzak, I.; Naz, S. Unit-Vise: Deep Shallow Unit-Vise Residual Neural Networks with Transition Layer For Expert Level Skin Cancer Classification. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 19, 1225–1234. [Google Scholar] [CrossRef] [PubMed]
- Qian, S.; Ren, K.; Zhang, W.; Ning, H. Skin lesion classification using CNNs with grouping of multi-scale attention and class-specific loss weighting. Comput. Methods Programs Biomed. 2022, 226, 107166. [Google Scholar] [CrossRef] [PubMed]
- Popescu, D.; El-Khatib, M.; Ichim, L. Skin Lesion Classification Using Collective Intelligence of Multiple Neural Networks. Sensors 2022, 22, 4399. [Google Scholar] [CrossRef]
- Nguyen, V.D.; Bui, N.D.; Do, H.K. Skin Lesion Classification on Imbalanced Data Using Deep Learning with Soft Attention. Sensors 2022, 22, 7530. [Google Scholar] [CrossRef]
- Naeem, A.; Anees, T.; Fiza, M.; Naqvi, R.A.; Lee, S.W. SCDNet: A Deep Learning-Based Framework for the Multiclassification of Skin Cancer Using Dermoscopy Images. Sensors 2022, 22, 5652. [Google Scholar] [CrossRef]
- Li, H.; Li, W.; Chang, J.; Zhou, L.; Luo, J.; Guo, Y. Dermoscopy lesion classification based on GANs and a fuzzy rank-based ensemble of CNN models. Phys. Med. Biol. 2022, 67, 185005. [Google Scholar] [CrossRef]
- Lee, J.R.H.; Pavlova, M.; Famouri, M.; Wong, A. Cancer-Net SCa: Tailored deep neural network designs for detection of skin cancer from dermoscopy images. BMC Med Imaging 2022, 22, 143. [Google Scholar] [CrossRef]
- Laverde-Saad, A.; Jfri, A.; Garcia, R.; Salgüero, I.; Martínez, C.; Cembrero, H.; Roustán, G.; Alfageme, F. Discriminative deep learning based benignity/malignancy diagnosis of dermatologic ultrasound skin lesions with pretrained artificial intelligence architecture. Skin. Res. Technol. 2022, 28, 35–39. [Google Scholar] [CrossRef]
- La Salvia, M.; Torti, E.; Leon, R.; Fabelo, H.; Ortega, S.; Martinez-Vega, B.; Callico, G.M.; Leporati, F. Deep Convolutional Generative Adversarial Networks to Enhance Artificial Intelligence in Healthcare: A Skin Cancer Application. Sensors 2022, 22, 6145. [Google Scholar] [CrossRef] [PubMed]
- Hosny, K.M.; Kassem, M.A. Refined Residual Deep Convolutional Network for Skin Lesion Classification. J. Digit. Imaging 2022, 35, 258–280. [Google Scholar] [CrossRef] [PubMed]
- Dascalu, A.; Walker, B.N.; Oron, Y.; David, E.O. Non-melanoma skin cancer diagnosis: A comparison between dermoscopic and smartphone images by unified visual and sonification deep learning algorithms. J. Cancer Res. Clin. Oncol. 2022, 148, 2497–2505. [Google Scholar] [CrossRef] [PubMed]
- Combalia, M.; Codella, N.; Rotemberg, V.; Carrera, C.; Dusza, S.; Gutman, D.; Helba, B.; Kittler, H.; Kurtansky, N.R.; Liopyris, K.; et al. Validation of artificial intelligence prediction models for skin cancer diagnosis using dermoscopy images: The 2019 International Skin Imaging Collaboration Grand Challenge. Lancet Digit. Health 2022, 4, e330–e339. [Google Scholar] [CrossRef] [PubMed]
- Benyahia, S.; Meftah, B.; Lezoray, O. Multi-features extraction based on deep learning for skin lesion classification. Tissue Cell 2022, 74, 101701. [Google Scholar] [CrossRef]
- Bechelli, S.; Delhommelle, J. Machine Learning and Deep Learning Algorithms for Skin Cancer Classification from Dermoscopic Images. Bioengineering 2022, 9, 97. [Google Scholar] [CrossRef] [PubMed]
- Afza, F.; Sharif, M.; Mittal, M.; Khan, M.A.; Jude Hemanth, D. A hierarchical three-step superpixels and deep learning framework for skin lesion classification. Methods 2022, 202, 88–102. [Google Scholar] [CrossRef] [PubMed]
- Afza, F.; Sharif, M.; Khan, M.A.; Tariq, U.; Yong, H.S.; Cha, J. Multiclass Skin Lesion Classification Using Hybrid Deep Features Selection and Extreme Learning Machine. Sensors 2022, 22, 799. [Google Scholar] [CrossRef] [PubMed]
- Winkler, J.K.; Sies, K.; Fink, C.; Toberer, F.; Enk, A.; Abassi, M.S.; Fuchs, T.; Blum, A.; Stolz, W.; Coras-Stepanek, B.; et al. Collective human intelligence outperforms artificial intelligence in a skin lesion classification task. J. Dtsch. Dermatol. Ges. 2021, 19, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.G.C.; Krohling, R.A. An Attention-Based Mechanism to Combine Images and Metadata in Deep Learning Models Applied to Skin Cancer Classification. IEEE J. Biomed. Health Inf. 2021, 25, 3554–3563. [Google Scholar] [CrossRef]
- Minagawa, A.; Koga, H.; Sano, T.; Matsunaga, K.; Teshima, Y.; Hamada, A.; Houjou, Y.; Okuyama, R. Dermoscopic diagnostic performance of Japanese dermatologists for skin tumors differs by patient origin: A deep learning convolutional neural network closes the gap. J. Dermatol. 2021, 48, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, I.; Younus, M.; Walayat, K.; Kakar, M.U.; Ma, J. Automated multi-class classification of skin lesions through deep convolutional neural network with dermoscopic images. Comput. Med. Imaging Graph. 2021, 88, 101843. [Google Scholar] [CrossRef] [PubMed]
- Huang, K.; Jiang, Z.; Li, Y.; Wu, Z.; Wu, X.; Zhu, W.; Chen, M.; Zhang, Y.; Zuo, K.; Li, Y.; et al. The Classification of Six Common Skin Diseases Based on Xiangya-Derm: Development of a Chinese Database for Artificial Intelligence. J. Med. Internet Res. 2021, 23, e26025. [Google Scholar] [CrossRef]
- Zhang, L.; Gao, H.J.; Zhang, J.; Badami, B. Optimization of the Convolutional Neural Networks for Automatic Detection of Skin Cancer. Open Med. 2020, 15, 27–37. [Google Scholar] [CrossRef]
- Wang, S.Q.; Zhang, X.Y.; Liu, J.; Tao, C.; Zhu, C.Y.; Shu, C.; Xu, T.; Jin, H.Z. Deep learning-based, computer-aided classifier developed with dermoscopic images shows comparable performance to 164 dermatologists in cutaneous disease diagnosis in the Chinese population. Chin. Med. J. 2020, 133, 2027–2036. [Google Scholar] [CrossRef]
- Qin, Z.; Liu, Z.; Zhu, P.; Xue, Y. A GAN-based image synthesis method for skin lesion classification. Comput. Methods Programs Biomed. 2020, 195, 105568. [Google Scholar] [CrossRef]
- Mahbod, A.; Schaefer, G.; Wang, C.; Dorffner, G.; Ecker, R.; Ellinger, I. Transfer learning using a multi-scale and multi-network ensemble for skin lesion classification. Comput. Methods Programs Biomed. 2020, 193, 105475. [Google Scholar] [CrossRef]
- Li, C.X.; Fei, W.M.; Shen, C.B.; Wang, Z.Y.; Jing, Y.; Meng, R.S.; Cui, Y. Diagnostic capacity of skin tumor artificial intelligence-assisted decision-making software in real-world clinical settings. Chin. Med. J. 2020, 133, 2020–2026. [Google Scholar] [CrossRef]
- Gessert, N.; Sentker, T.; Madesta, F.; Schmitz, R.; Kniep, H.; Baltruschat, I.; Werner, R.; Schlaefer, A. Skin Lesion Classification Using CNNs with Patch-Based Attention and Diagno-sis-Guided Loss Weighting. IEEE Trans. Biomed. Eng. 2020, 67, 495–503. [Google Scholar] [CrossRef] [PubMed]
- Gessert, N.; Nielsen, M.; Shaikh, M.; Werner, R.; Schlaefer, A. Skin lesion classification using ensembles of multi-resolution EfficientNets with meta data. MethodsX 2020, 7, 100864. [Google Scholar] [CrossRef] [PubMed]
- Al-Masni, M.A.; Kim, D.H.; Kim, T.S. Multiple skin lesions diagnostics via integrated deep convolutional networks for seg-mentation and classification. Comput. Methods Programs Biomed. 2020, 190, 105351. [Google Scholar] [CrossRef] [PubMed]
- Ameri, A. Deep Learning Approach to Skin Cancer Detection in Dermoscopy Images. J. Biomed. Phys. Eng. 2020, 10, 801–806. [Google Scholar] [CrossRef] [PubMed]
- Tschandl, P.; Rosendahl, C.; Akay, B.N.; Argenziano, G.; Blum, A.; Braun, R.P.; Cabo, H.; Gourhant, J.Y.; Kreusch, J.; Lallas, A.; et al. Expert-Level Diagnosis of Nonpigmented Skin Cancer by Combined Convolutional Neural Networks. JAMA Dermatol. 2019, 155, 58–65. [Google Scholar] [CrossRef] [PubMed]
- Dascalu, A.; David, E.O. Skin cancer detection by deep learning and sound analysis algorithms: A prospective clinical study of an elementary dermoscope. EBioMedicine 2019, 43, 107–113. [Google Scholar] [CrossRef]
- Pruneda, C.; Ramesh, M.; Hope, L.; Hope, R. Nonmelanoma Skin Cancers: Diagnostic Accuracy of Midlevel Providers vs. Dermatologists. Available online: https://www.hmpgloballearningnetwork.com/site/thederm/feature-story/nonmelanoma-skin-cancers-diagnostic-accuracy-midlevel-providers-vs#:~:text=A%20total%20of%2011%2C959%20NMSCs,clinical%20diagnosis%20(Table%201) (accessed on 12 April 2023).
- Ho, C.J.; Calderon-Delgado, M.; Chan, C.C.; Lin, M.Y.; Tjiu, J.W.; Huang, S.L.; Chen, H.H. Detecting mouse squamous cell carcinoma from submicron full-field optical coherence tomography images by deep learning. J. Biophotonics 2021, 14, e202000271. [Google Scholar] [CrossRef]
- Huynh, T.; Nibali, A.; He, Z. Semi-supervised learning for medical image classification using imbalanced training data. Comput. Methods Programs Biomed. 2022, 216, 106628. [Google Scholar] [CrossRef] [PubMed]
- Rezk, E.; Eltorki, M.; El-Dakhakhni, W. Leveraging Artificial Intelligence to Improve the Diversity of Dermatological Skin Color Pathology: Protocol for an Algorithm Development and Validation Study. JMIR Res. Protoc. 2022, 11, e34896. [Google Scholar] [CrossRef] [PubMed]
- Daneshjou, R.; Vodrahalli, K.; Novoa, R.A.; Jenkins, M.; Liang, W.; Rotemberg, V.; Ko, J.; Swetter, S.M.; Bailey, E.E.; Gevaert, O.; et al. Disparities in dermatology AI performance on a diverse, curated clinical image set. Sci. Adv. 2022, 8, eabq6147. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Foltz, E.A.; Witkowski, A.; Becker, A.L.; Latour, E.; Lim, J.Y.; Hamilton, A.; Ludzik, J. Artificial Intelligence Applied to Non-Invasive Imaging Modalities in Identification of Nonmelanoma Skin Cancer: A Systematic Review. Cancers 2024, 16, 629. https://doi.org/10.3390/cancers16030629
Foltz EA, Witkowski A, Becker AL, Latour E, Lim JY, Hamilton A, Ludzik J. Artificial Intelligence Applied to Non-Invasive Imaging Modalities in Identification of Nonmelanoma Skin Cancer: A Systematic Review. Cancers. 2024; 16(3):629. https://doi.org/10.3390/cancers16030629
Chicago/Turabian StyleFoltz, Emilie A., Alexander Witkowski, Alyssa L. Becker, Emile Latour, Jeong Youn Lim, Andrew Hamilton, and Joanna Ludzik. 2024. "Artificial Intelligence Applied to Non-Invasive Imaging Modalities in Identification of Nonmelanoma Skin Cancer: A Systematic Review" Cancers 16, no. 3: 629. https://doi.org/10.3390/cancers16030629
APA StyleFoltz, E. A., Witkowski, A., Becker, A. L., Latour, E., Lim, J. Y., Hamilton, A., & Ludzik, J. (2024). Artificial Intelligence Applied to Non-Invasive Imaging Modalities in Identification of Nonmelanoma Skin Cancer: A Systematic Review. Cancers, 16(3), 629. https://doi.org/10.3390/cancers16030629






