Selective Occupation by E2F and RB of Loci Expressed by RNA Polymerase III
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. ChIP-Seq Data
2.1.1. Downloading ChIP-Seq Datasets
2.1.2. Quantification of ChIP-Seq Signals Using Easeq
2.1.3. Data Analysis
Heat Maps
Line Tracks
Binding Overlap Analysis
| Source | Cell Type | ChIP-Seq File Type | Accession |
|---|---|---|---|
| Sanidas et al., 2022 [101] | BJ cells | FLAG-RB-WT | SRR14713166 |
| FLAG-RB-Δcdk | SRR14713167 | ||
| RPE cells | FLAG-RB-WT Input | SRR14713168 | |
| FLAG-RB-Δcdk Input | SRR14713169 | ||
| FLAG-RB-WT | SRR14713063 | ||
| FLAG-RB-Δcdk | SRR14713064 | ||
| FLAG-RB-WT Input | SRR14713079 | ||
| FLAG-RB-Δcdk Input | SRR14713080 | ||
| E2F1 | SRR14713081 | ||
| Michael Snyder, Stanford. 2017 | K562 cells | RB1 | ENCFF305NFS |
| Richard Myers, HAIB. 2023 | E2F1 | ENCFF183WQN | |
| ENCFF193ODF | |||
| E2F4 | ENCFF706ZTX | ||
| ENCFF749XCO | |||
| E2F5 | ENCFF027ECD | ||
| ENCFF915WXK | |||
| Chicas et al., 2010 [102] | Growing IMR90 cells | RB | SRR034478 |
| SRR034479 | |||
| Mock | SRR034492 | ||
| Quiescent IMR90 cells | RB | SRR034480 | |
| SRR034482 | |||
| Mock | SRR034493 | ||
| Senescent IMR90 cells | RB | SRR034484 | |
| SRR034486 | |||
| Mock | SRR034494 | ||
| Quiescent IMR90 cells | p130 | SRR034483 | |
| Ferrari et al., 2012 [103] | Quiescent IMR90 cells | p107 | SRR350272 |
2.2. ChIP-Atlas Data
3. Results
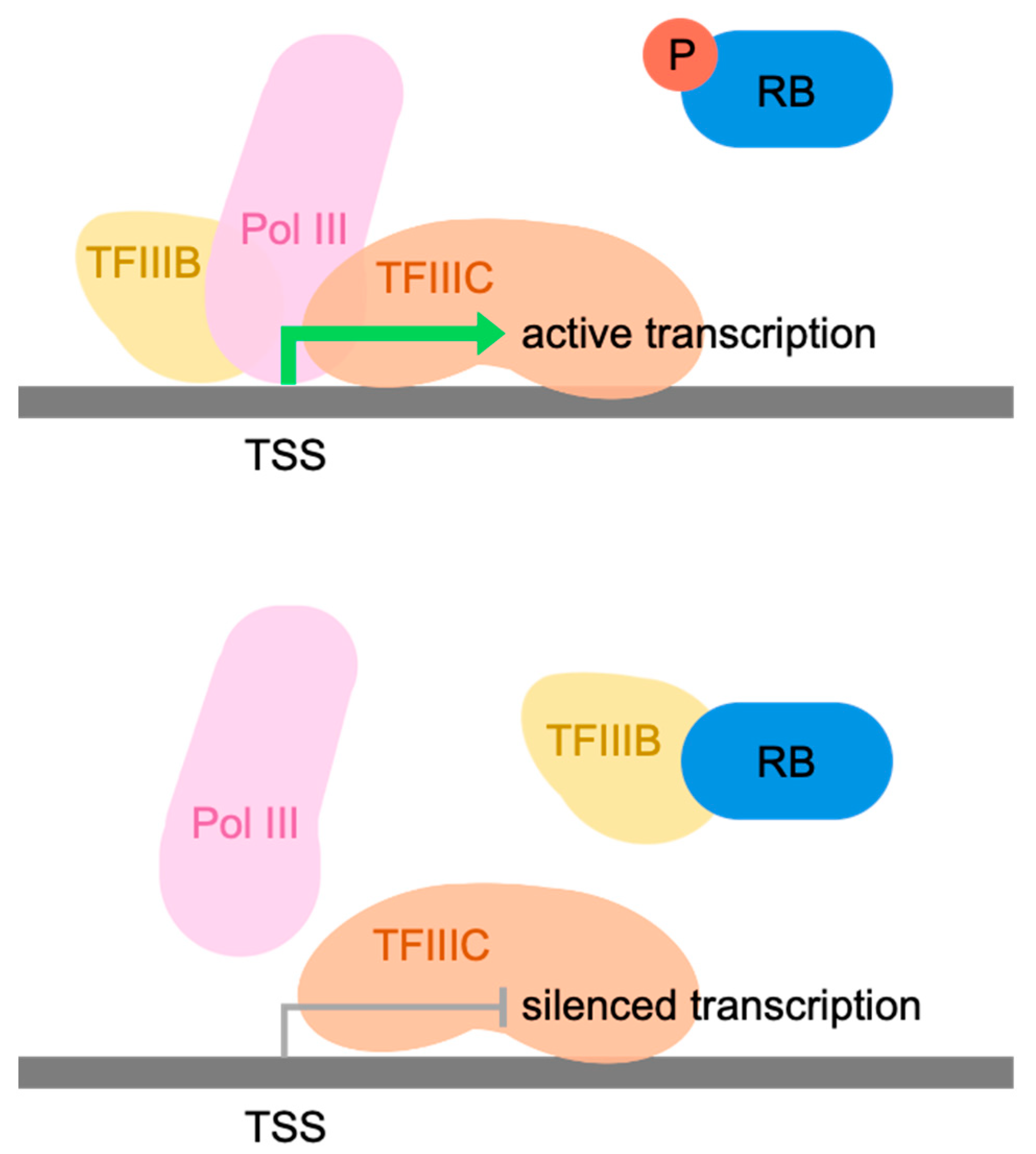
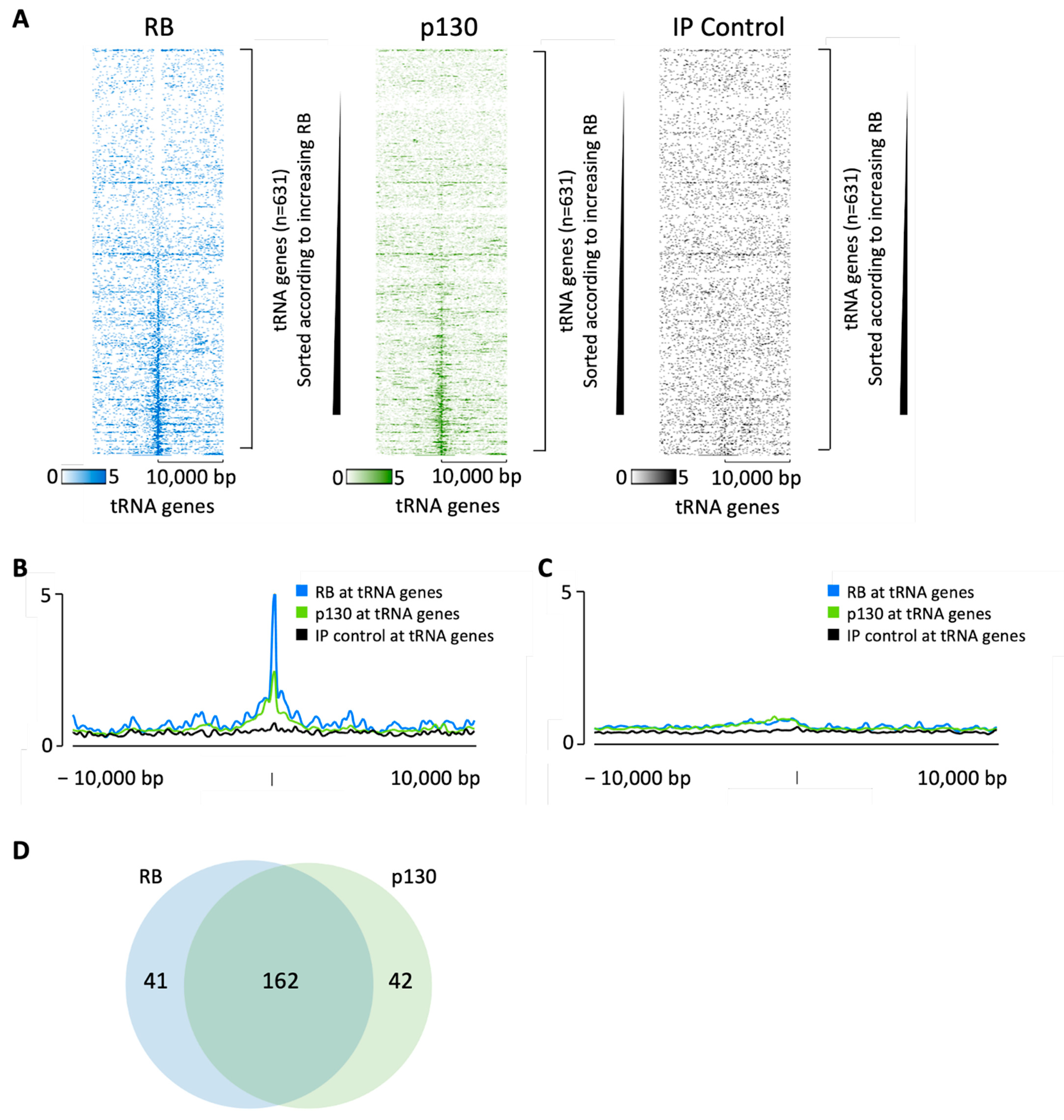
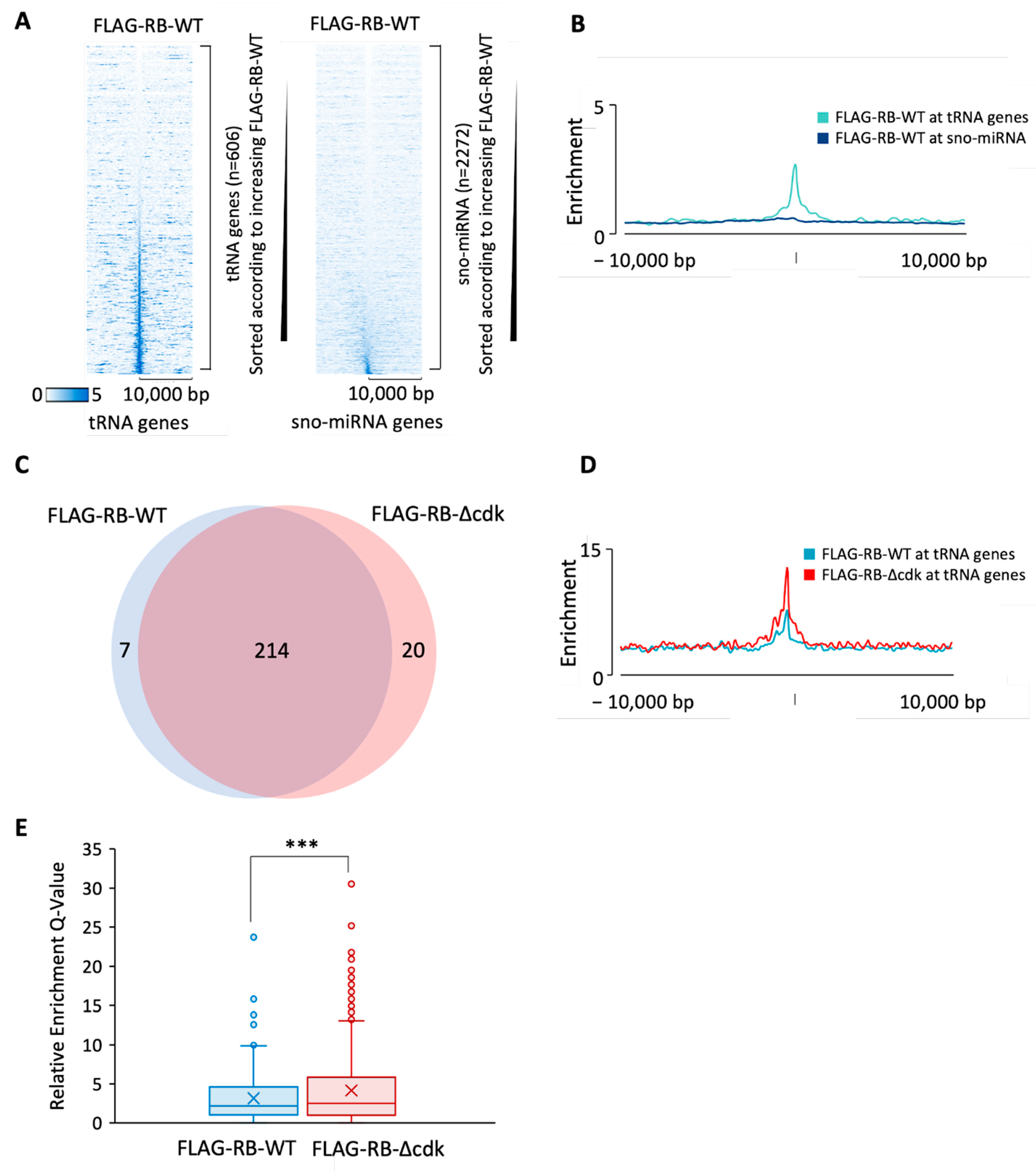
3.1. RB and p130 Associate with Many tRNA Genes in Human Fibroblasts
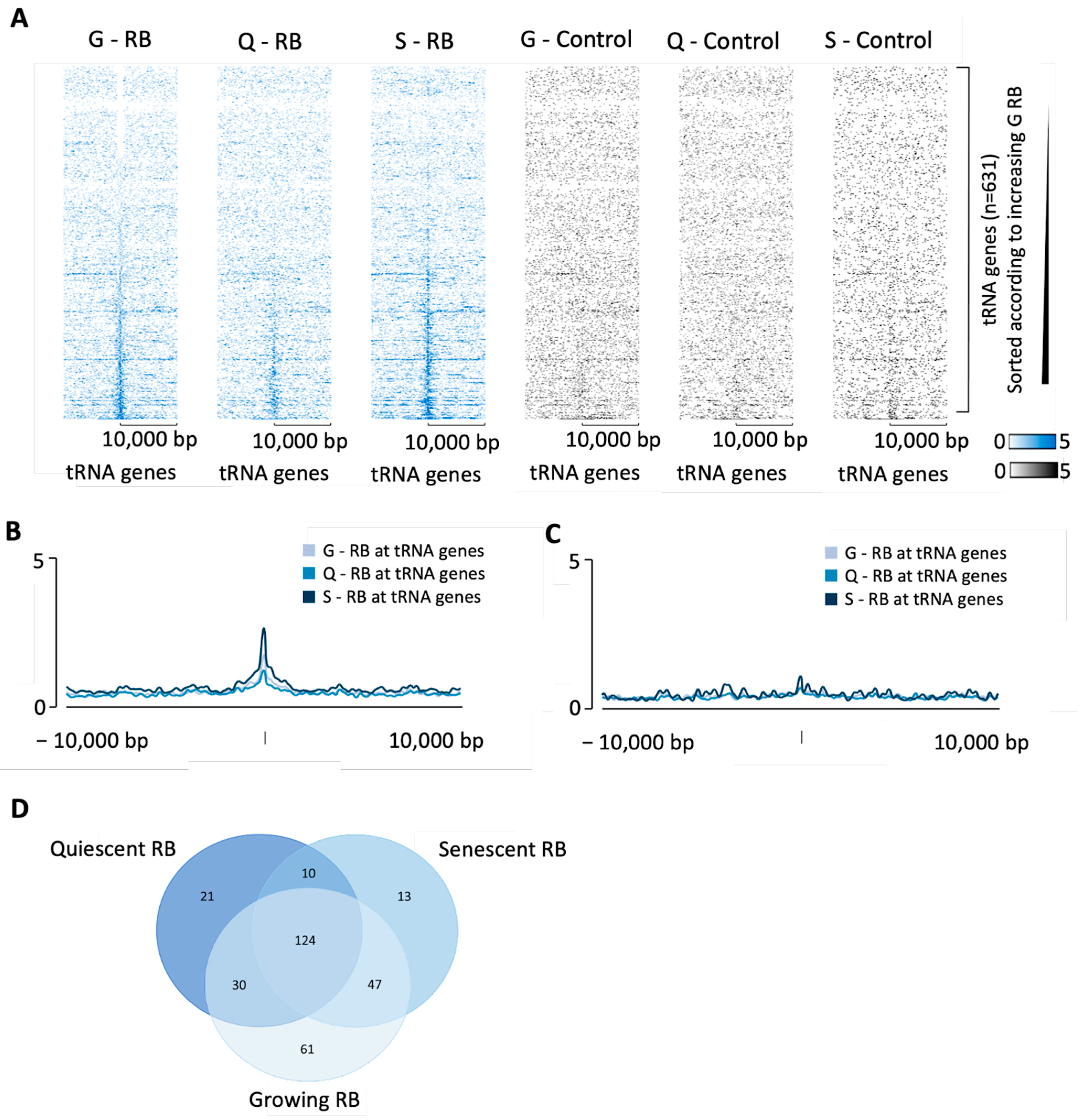
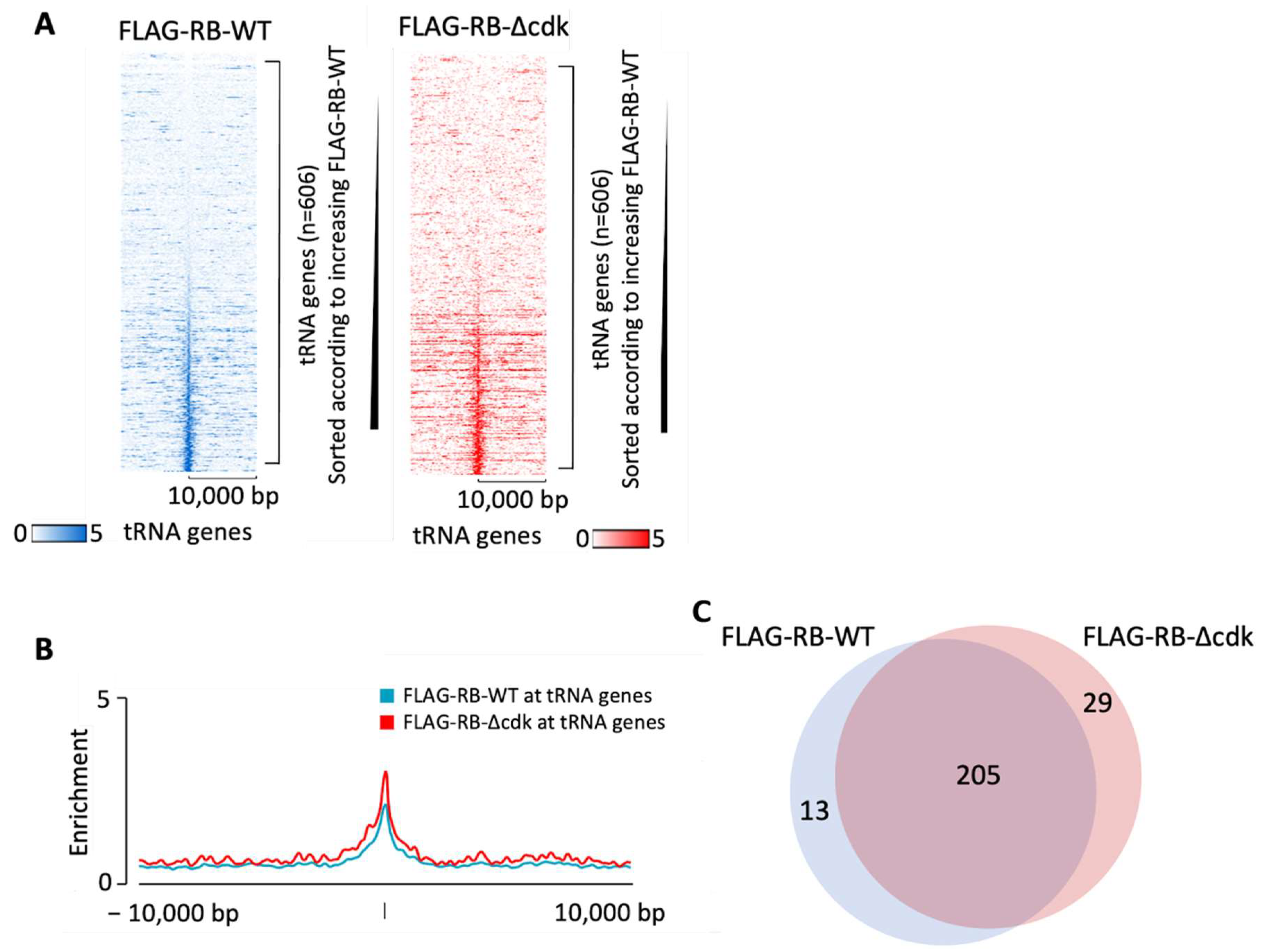
3.2. Phosphoresistant Mutation Stimulates Binding of RB to tRNA Genes
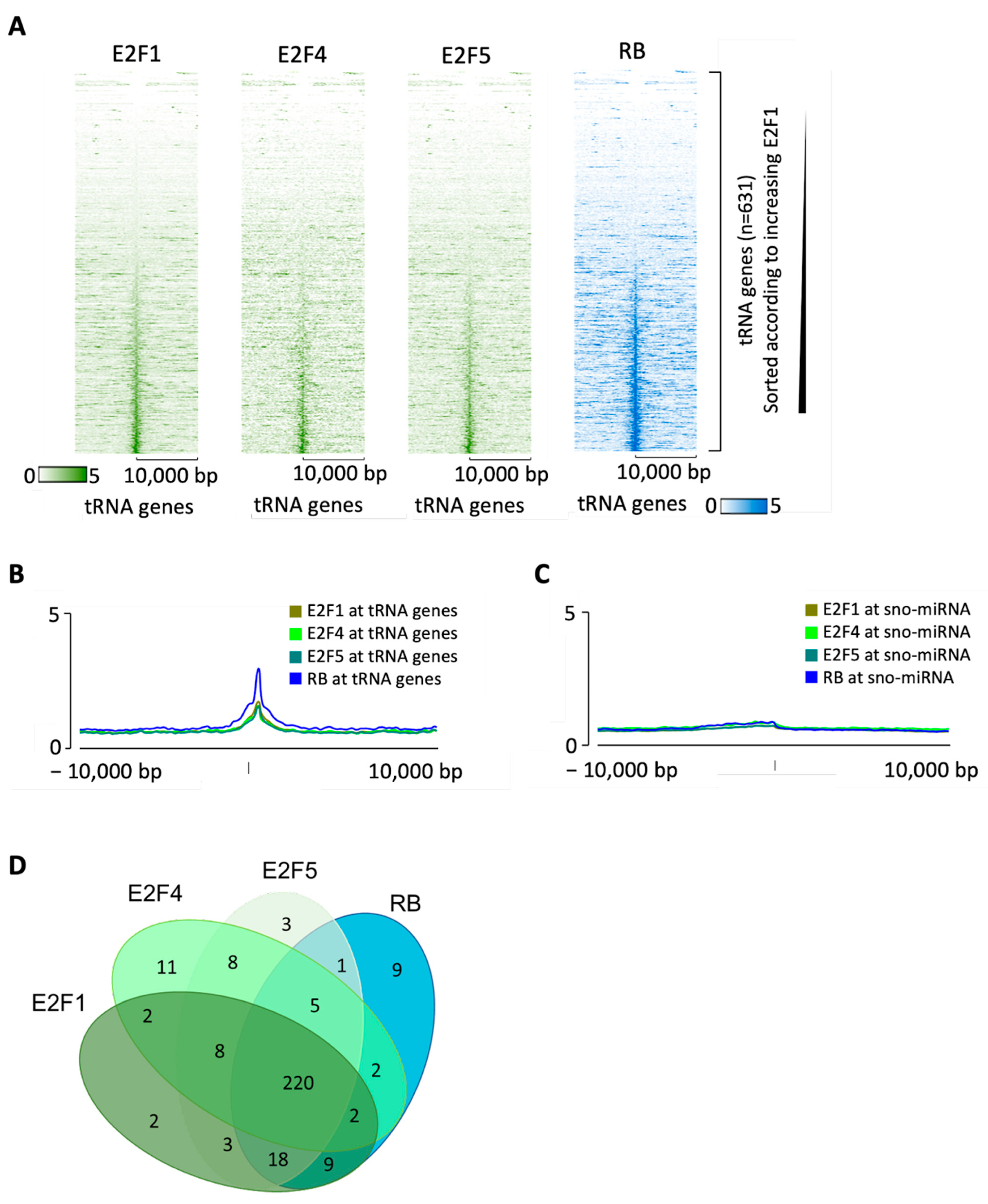
3.3. RB Recruitment to Many tRNA Genes Coincides with Binding Sites for E2F
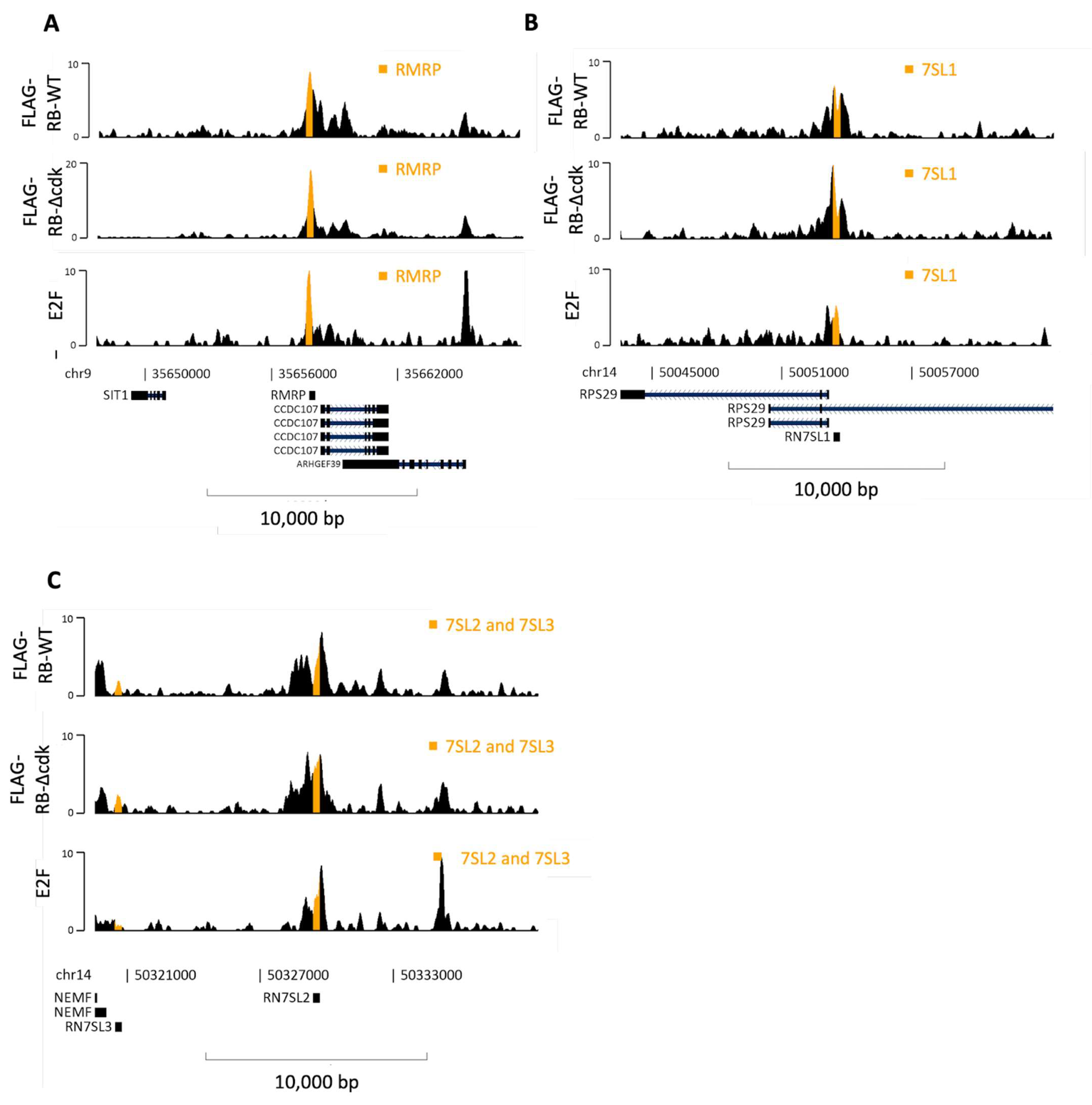
3.4. RB May Be Recruited by E2F to 7SL and RMRP Genes
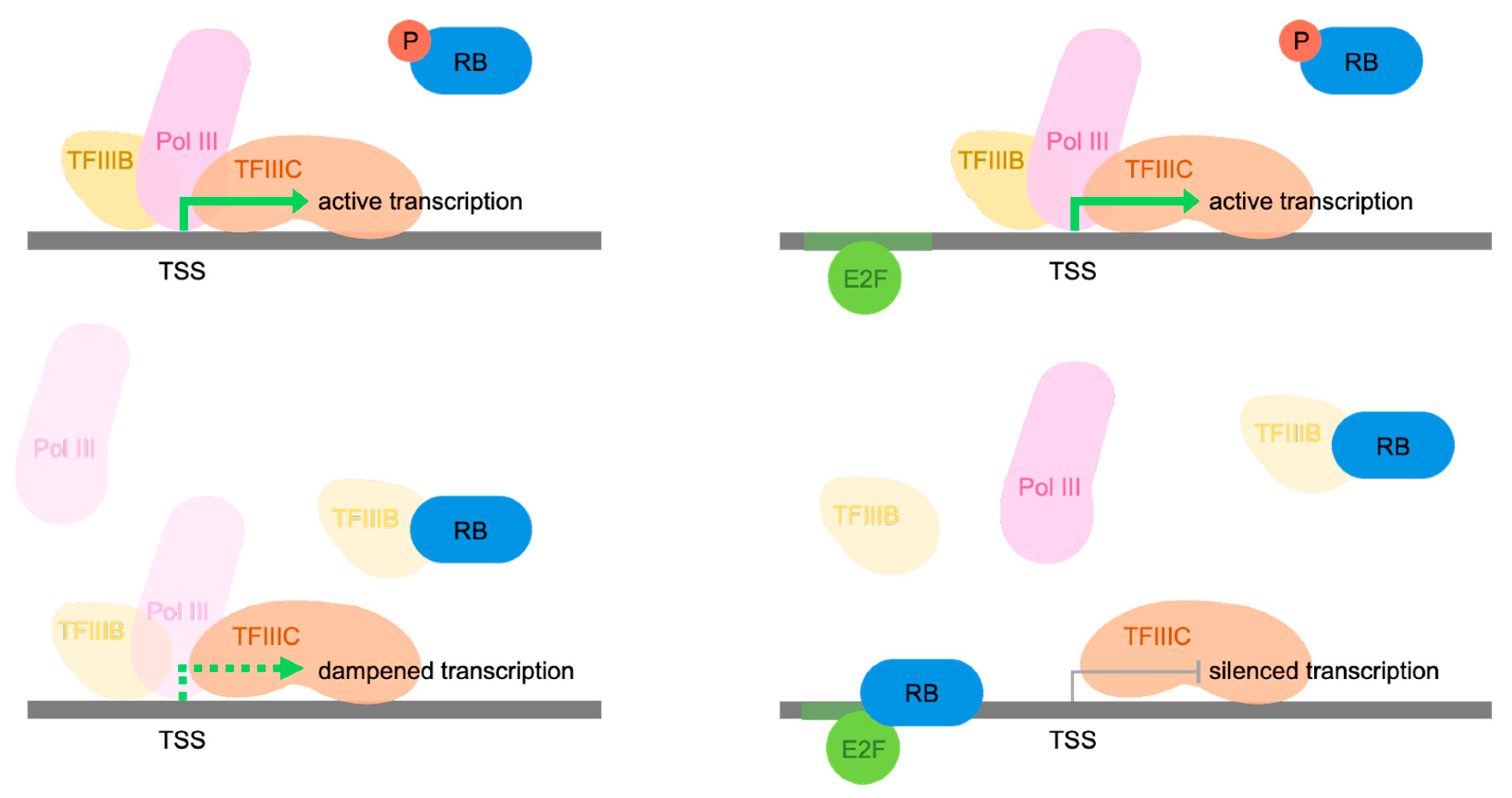
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Friend, S.H.; Bernards, R.; Rogelj, S.; Weinberg, R.A.; Rapaport, J.M.; Alberts, D.M.; Dryja, T.P. A human DNA segment with properties of the gene that predisposes to retinoblastoma and osteosarcoma. Nature 1986, 323, 643–646. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, R.A. The retinoblastoma protein and cell cycle control. Cell 1995, 81, 323–330. [Google Scholar] [CrossRef] [PubMed]
- Whyte, P. The retinoblastoma protein and its relatives. Semin. Cancer Biol. 1995, 6, 83–90. [Google Scholar] [CrossRef] [PubMed]
- Jacks, T.; Fazeli, A.; Schmitt, E.M.; Bronson, R.T.; Goodell, M.A.; Weinberg, R.A. Effects of an Rb mutation in the mouse. Nature 1992, 359, 295–300. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.Y.-H.P.; Chang, C.-Y.; Hu, N.; Wang, Y.-C.J.; Lai, C.-C.; Herrup, K.; Lee, W.-H.; Bradley, A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature 1992, 359, 288–294. [Google Scholar] [CrossRef] [PubMed]
- Hu, N.; Gutsmann, A.; Herbert, D.C.; Bradley, A.; Lee, W.H.; Lee, E.Y. Heterozygous Rb-1D20/+ mice are predisposed to tumours of the pituitary gland with a nearly complete penetrance. Oncogene 1994, 9, 1021–1027. [Google Scholar] [PubMed]
- Maandag, E.; van der Valk, M.; Vlaar, M.; Feltkamp, C.; O’Brien, J.; van Roon, M.; van der Lugt, N.; Berns, A.; Riele, H.T. Developmental rescue of an embryonic-lethal mutation in the retinoblastoma gene in chimeric mice. EMBO J. 1994, 13, 4260–4268. [Google Scholar] [CrossRef]
- Williams, B.; Schmitt, E.; Remington, L.; Bronson, R.; Albert, D.; Weinberg, R.; Jacks, T. Extensive contribution of Rb-deficient cells to adult chimeric mice with limited histopathological consequences. EMBO J. 1994, 13, 4251–4259. [Google Scholar] [CrossRef]
- Huang, H.-J.S.; Yee, J.-K.; Shew, J.-Y.; Chen, P.-L.; Bookstein, R.; Friedmann, T.; Lee, E.Y.-H.P.; Lee, W.-H. Suppression of the neoplastic phenotype by replacement of the RB gene in human cancer cells. Science 1988, 242, 1563–1566. [Google Scholar] [CrossRef]
- Bookstein, R.; Shew, J.; Chen, P.; Scully, P.; Lee, W. Suppression of tumorigenicity of human prostate carcinoma cells by replacing a mutated RB gene. Science 1990, 247, 712–715. [Google Scholar] [CrossRef]
- Qin, X.; Chittenden, T.; Livingston, D.M.; Kaelin, W.G. Identification of a growth suppression domain within the retinoblastoma gene product. Genes Dev. 1992, 6, 953–964. [Google Scholar] [CrossRef][Green Version]
- Trimarchi, J.M.; Lees, J.A. Sibling rivalry in the E2F family. Nat. Rev. Mol. Cell Biol. 2001, 3, 11–20. [Google Scholar] [CrossRef]
- Braken, A.P.; Ciro, M.; Cocito, A.; Helin, K. E2F target genes: Unraveling the biology. Trends Biochem. Sci. 2004, 29, 409–417. [Google Scholar] [CrossRef] [PubMed]
- Xie, D.; Pei, Q.; Li, J.; Wan, X.; Ye, T. Emerging Role of E2F Family in Cancer Stem Cells. Front. Oncol. 2021, 11, 723137. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, S.J.; Prater, C.A.; Dean, D.C. Retinoblastoma protein switches the E2F site from positive to negative element. Nature 1992, 358, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Flemington, E.K.; Speck, S.H.; Kaelin, W.G.J. E2F-1-mediated transactivation is inhibited by complex formation with the retinoblastoma susceptibility gene product. Proc. Natl. Acad. Sci. USA 1993, 90, 6914–6918. [Google Scholar] [CrossRef] [PubMed]
- Helin, K.; Harlow, E.; Fattaey, A. Inhibition of E2F-1 transactivation by direct binding of the retinoblastoma protein. Mol. Cell Biol. 1993, 13, 6501–6508. [Google Scholar]
- Weintraub, S.J.; Chow, K.N.B.; Luo, R.X.; Zhang, S.H.; He, S.; Dean, D.C. Mechanism of active transcriptional repression by the retinoblastoma protein. Nature 1995, 375, 812–815. [Google Scholar] [CrossRef]
- Brehm, A.; Miska, E.A.; McCance, D.J.; Reid, J.L.; Bannister, A.J.; Kouzarides, T. Retinoblastoma protein recruits histone deacetylase to repress transcription. Nature 1998, 391, 597–601. [Google Scholar] [CrossRef]
- Luo, R.X.; Postigo, A.A.; Dean, D.C. Rb interacts with histone deacetylase to repress transcription. Cell 1998, 92, 463–473. [Google Scholar] [CrossRef]
- Magnaghi-Jaulin, L.; Groisman, R.; Naguibneva, I.; Robin, P.; Lorain, S.; Le Villain, J.P.; Troalen, F.; Trouche, D.; Harel-Bellan, A. Retinoblastoma protein represses transcription by recruiting a histone deacetylase. Nature 1998, 391, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Brehm, A.; Kouzarides, T. Retinoblastoma protein meets chromatin. Trends Biochem. Sci. 1999, 24, 142–145. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.F.; Liu, X.; Dynlacht, B.D. Mechanism of transcriptional repression of E2F by the retinoblastoma tumour suppressor protein. Mol. Cell 1999, 3, 195–205. [Google Scholar] [CrossRef] [PubMed]
- Robertson, K.D.; Ait-Si-Ali, S.; Yokochi, T.; Wade, P.A.; Jones, P.L.; Wolffe, A.P. DNMT1 forms a complex with Rb, E2F1, and HDAC1 and represses transcription from E2F-responsive promoters. Nat. Genet. 2000, 25, 338–342. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.J.; Schneider, R.; Bauer, U.M.; Bannister, A.J.; Morrison, A.; O’Carroll, D.; Firestein, R.; Cleary, M.; Jenuwein, T.; Herrera, R.E.; et al. Rb targets histone H3 methylation and HP1 to promoters. Nature 2001, 412, 561–565. [Google Scholar] [CrossRef] [PubMed]
- Vandel, L.; Nicolas, E.; Vaute, O.; Ferreira, R.; Ait-Si-Ali, S.; Trouche, D. Transcriptional Repression by the Retinoblastoma Protein through the Recruitment of a Histone Methyltransferase. Mol. Cell Biol. 2001, 21, 6484–6494. [Google Scholar] [CrossRef] [PubMed]
- Pradhan, S.; Kim, G.D. The retinoblastoma gene product interacts with maintenance human DNA (cytosine-5) methyltransferase and modulates its activity. EMBO J. 2002, 21, 779–788. [Google Scholar] [CrossRef]
- Ogawa, H.; Ishiguro, K.; Gaubatz, S.; Livingston, D.M.; Nakatani, Y. A Complex With Chromatin Modifiers That Occupies E2F- and Myc-Responsive Genes in G0 Cells. Science 2002, 296, 1132–1136. [Google Scholar] [CrossRef]
- Selvakumar, T.; Gjidoda, A.; Hovde, S.L.; Henry, R.W. Regulation of human RNA polymerase III transcription by DNMT1 and DNMT3a DNA methyltransferases. J. Biol. Chem. 2012, 287, 7039–7050. [Google Scholar] [CrossRef]
- Adams, P.D.; Kaelin, W.G. Transcriptional control by E2F. Semin. Cancer Biol. 1995, 6, 99–108. [Google Scholar] [CrossRef]
- Herrera, R.E.; Sah, V.P.; Williams, B.O.; Makela, T.P.; Weinberg, R.A.; Jacks, T. Altered cell cycle kinetics, gene expression, and G1 restriction point regulation in Rb-deficient fibroblasts. Mol. Cell Biol. 1996, 16, 2402–2407. [Google Scholar] [CrossRef] [PubMed]
- Hurford, R.K.; Cobrinik, D.; Lee, M.-H.; Dyson, N. pRB and p107/p130 are required for the regulated expression of different sets of E2F responsive genes. Genes Dev. 1997, 11, 1447–1463. [Google Scholar] [CrossRef] [PubMed]
- Pines, J. Cyclins, CDKs and cancer. Semin. Cancer Biol. 1995, 6, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Mittnacht, S. Control of pRB phosphorylation. Curr. Opin. Genet. Dev. 1998, 8, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, G.; Jacks, T. The retinoblastoma gene family: Cousins with overlapping interests. Trends Genet. 1998, 14, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Morris, E.J.; Dyson, N.J. Retinoblastoma protein partners. Adv. Cancer Res. 2001, 82, 1–54. [Google Scholar] [PubMed]
- Sanidas, I.; Morris, R.; Fella, K.A.; Rumde, P.H.; Boukhali, M.; Tai, E.C.; Ting, D.T.; Lawrence, M.S.; Haas, W.; Dyson, N.J. A code of mono-phosphorylation modulates the function of RB. Mol. Cell 2019, 73, 985–1000. [Google Scholar] [CrossRef]
- Schramm, L.; Hernandez, N. Recruitment of RNA polymerase III to its target promoters. Genes Dev. 2002, 16, 2593–2620. [Google Scholar] [CrossRef]
- Larminie, C.G.C.; Cairns, C.A.; Mital, R.; Martin, K.; Kouzarides, T.; Jackson, S.P.; White, R.J. Mechanistic analysis of RNA polymerase III regulation by the retinoblastoma protein. EMBO J. 1997, 16, 2061–2071. [Google Scholar] [CrossRef]
- Chu, W.-M.; Wang, Z.; Roeder, R.G.; Schmid, C.W. RNA polymerase III transcription repressed by Rb through its interactions with TFIIIB and TFIIIC2. J. Biol. Chem. 1997, 272, 14755–14761. [Google Scholar] [CrossRef]
- Larminie, C.G.C.; Sutcliffe, J.E.; Tosh, K.; Winter, A.G.; Felton-Edkins, Z.A.; White, R.J. Activation of RNA polymerase III transcription in cells transformed by simian virus 40. Mol. Cell Biol. 1999, 19, 4927–4934. [Google Scholar] [CrossRef] [PubMed]
- Sutcliffe, J.E.; Cairns, C.A.; McLees, A.; Allison, S.J.; Tosh, K.; White, R.J. RNA polymerase III transcription factor IIIB is a target for repression by pocket proteins p107 and p130. Mol. Cell Biol. 1999, 19, 4255–4261. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Scott, P.H.; Cairns, C.A.; Sutcliffe, J.E.; Alzuherri, H.M.; Mclees, A.; Winter, A.G.; White, R.J. Regulation of RNA polymerase III transcription during cell cycle entry. J. Biol. Chem. 2001, 276, 1005–1014. [Google Scholar] [CrossRef]
- Felton-Edkins, Z.A.; White, R.J. Multiple mechanisms contribute to the activation of RNA polymerase III transcription in cells transformed by papovaviruses. J. Biol. Chem. 2002, 277, 48182–48191. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, H.A.; Jawdekar, G.W.; Lee, K.-A.; Gu, L.; Henry, R.W. Distinct mechanisms for repression of RNA polymerase III transcription by the retinoblastoma tumor suppressor protein. Mol. Cell Biol. 2004, 24, 5989–5999. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Angelis, E.; Garcia, A.; Chan, S.S.; Schenke-Layland, K.; Ren, S.; Goodfellow, S.J.; Jordan, M.C.; Roos, K.P.; White, R.J.; MacLellan, W.R. A cyclin D2-Rb pathway regulates cardiac myocyte size and RNA polymerase III after biomechanical stress in adult myocardium. Circ. Res. 2008, 102, 1222–1229. [Google Scholar] [CrossRef]
- Dieci, G.; Fiorino, G.; Castelnuovo, M.; Teichmann, M.; Pagano, A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007, 23, 614–622. [Google Scholar] [CrossRef] [PubMed]
- White, R.J. RNA polymerases I and III, growth control and cancer. Nat. Rev. Mol. Cell Biol. 2005, 6, 69–78. [Google Scholar] [CrossRef]
- Grewal, S.S. Why should cancer biologists care about tRNAs? tRNA synthesis, mRNA translation and the control of growth. Biochem. Biophys. Acta 2015, 1849, 898–907. [Google Scholar] [CrossRef]
- Huang, S.; Sun, B.; Xiong, Z.; Shu, Y.; Zhou, H.; Zhang, W.; Xiong, J.; Li, Q. The dysregulation of tRNAs and tRNA derivatives in cancer. J. Exp. Clin. Cancer Res. 2018, 37, 101. [Google Scholar] [CrossRef]
- Orellana, E.A.; Siegal, E.; Gregory, R.I. tRNA dysregulation and disease. Nat. Rev. Genet. 2022, 23, 651–664. [Google Scholar] [CrossRef] [PubMed]
- Sangha, A.K.; Kantidakis, T. The aminoacyl-tRNA synthetase and tRNA expression levels are deregulated in cancer and correlate independently with patient survival. Curr. Issues Mol. Biol. 2022, 44, 3001–3017. [Google Scholar] [CrossRef] [PubMed]
- Vincent, C.T.; Schneider, R.J. Selective tRNA charging in breast cancer. Nat. Cell Biol. 2022, 24, 287–289. [Google Scholar] [CrossRef] [PubMed]
- Pinzaru, A.M.; Tavazoie, S.F. Transfer RNAs as dynamic and critical regulators of cancer progression Nat. Rev. Cancer 2023, 23, 746–761. [Google Scholar] [CrossRef]
- Hirsch, H.A.; Gu, L.; Henry, R.W. The retinoblastoma tumor suppressor protein targets distinct general transcription factors to regulate RNA polymerase III gene expression. Mol. Cell Biol. 2000, 20, 9182–9191. [Google Scholar] [CrossRef] [PubMed][Green Version]
- White, R.J.; Trouche, D.; Martin, K.; Jackson, S.P.; Kouzarides, T. Repression of RNA polymerase III transcription by the retinoblastoma protein. Nature 1996, 382, 88–90. [Google Scholar] [CrossRef]
- Brown, T.R.P.; Scott, P.H.; Stein, T.; Winter, A.G.; White, R.J. RNA polymerase III transcription: Its control by tumor suppressors and its deregulation by transforming agents. Gene Expr. 2000, 9, 15–28. [Google Scholar] [CrossRef][Green Version]
- Felton-Edkins, Z.A.; Kenneth, N.S.; Brown, T.R.P.; Daly, N.L.; Gomez-Roman, N.; Grandori, C.; Eisenman, R.N.; White, R.J. Direct regulation of RNA polymerase III transcription by RB, p53 and c-Myc. Cell Cycle 2003, 3, 181–184. [Google Scholar] [CrossRef]
- Grana, X.; Garriga, J.; Mayol, X. Role of the retinoblastoma protein family, pRB, p107 and p130 in the negative control of cell growth. Oncogene 1998, 17, 3365–3383. [Google Scholar] [CrossRef]
- Claudio, P.P.; Howard, C.M.; Baldi, A.; De Luca, A.; Fu, Y.; Condorelli, G.; Sun, Y.; Colburn, N.; Calabretta, B.; Giordano, A. p130/Rb2 has growth suppressive properties similar to yet distinctive from those of retinoblastoma family members pRb and p107. Cancer Res. 1994, 54, 5556–5560. [Google Scholar]
- Woo, M.S.-A.; Sanchez, I.; Dynlacht, B.D. p130 and p107 use a conserved domain to inhibit cellular cyclin-dependent kinase activity. Mol. Cell Biol. 1997, 17, 3566–3579. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; van den Heuvel, S.; Helin, K.; Fattaey, A.; Ewen, M.; Livingston, D.; Dyson, N.; Harlow, E. Inhibition of cell proliferation by p107, a relative of the retinoblastoma protein. Genes Dev. 1993, 7, 1111–1125. [Google Scholar] [CrossRef] [PubMed]
- Cobrinik, D.; Lee, M.H.; Hannon, G.; Mulligan, G.; Bronson, R.T.; Dyson, N.; Harlow, E.; Beach, D.; Weinberg, R.A.; Jacks, T. Shared role of the pRB-related p130 and p107 proteins in limb development. Genes Dev. 1996, 10, 1633–1644. [Google Scholar] [CrossRef] [PubMed]
- Clarke, A.R.; Maandag, E.R.; van Roon, M.; van der Lugt, N.M.T.; van der Valk, M.; Hooper, M.L.; Berns, A.; te Riele, H. Requirement for a functional Rb-1 gene in murine development. Nature 1992, 359, 328–330. [Google Scholar] [CrossRef] [PubMed]
- Burkhart, D.L.; Sage, J. Cellular mechanisms of tumour suppression by the retinoblastoma gene. Nat. Rev. Cancer 2008, 8, 671–682. [Google Scholar] [CrossRef] [PubMed]
- Nasmyth, K. Another role rolls in. Nature 1996, 382, 28–29. [Google Scholar] [CrossRef]
- White, R.J. Regulation of RNA polymerases I and III by the retinoblastoma protein: A mechanism for growth control? Trends Biochem. Sci. 1997, 22, 77–80. [Google Scholar] [CrossRef]
- Rideout, E.J.; Marshall, L.; Grewal, S.S. Drosophila RNA polymerase III repressor Maf1 controls body size and developmental timing by modulating tRNAiMet synthesis and systemic insulin signaling. Proc. Natl. Acad. Sci. USA 2012, 109, 1139–1144. [Google Scholar] [CrossRef]
- Pavon-Eternod, M.; Gomes, S.; Rosner, M.R.; Pan, T. Overexpression of initiator methionine tRNA leads to global reprogramming of tRNA expression and increased proliferation in human epithelial cells. RNA 2013, 19, 461–466. [Google Scholar] [CrossRef]
- Goodarzi, H.; Nguyen, H.C.B.; Zhang, S.; Dill, B.D.; Molina, H.; Tavazoie, S.F. Modulated expression of specific tRNAs drives gene expression and cancer progression. Cell 2016, 165, 1416–1427. [Google Scholar] [CrossRef]
- Birch, J.; Clarke, C.J.; Campbell, A.D.; Campbell, K.; Mitchell, L.E.; Liko, D.; Kalna, G.; Strathdee, D.; Sansom, O.J.; Neilson, M.; et al. The initiator methionine tRNA drives cell migration and invasion leading to increased metastatic potential in melanoma. Biol. Open 2016, 5, 1371–1379. [Google Scholar] [CrossRef] [PubMed]
- Clarke, C.J.; Berg, T.J.; Birch, J.; Ennis, D.; Mitchell, L.E.; Cloix, C.; Campbell, A.D.; Sumpton, D.; Nixon, C.; Campbell, K.; et al. The initiator methionine tRNA drives secretion of type II collagen from stromal fibroblasts to promote tumor growth and angiogenesis Curr. Biol. 2016, 26, 755–765. [Google Scholar]
- Macari, F.; El-houfi, Y.; Boldina, G.; Xu, H.; Khoury-Hanna, S.; Ollier, J.; Yazdani, L.; Zheng, G.; Bieche, I.; Legrand, N.; et al. TRIM6/61 connects PKCa with translational control through tRNAiMet stabilization: Impact on tumorigenesis. Oncogene 2016, 35, 1785–1796. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Li, D.; Kovalchuk, I.; Apel, I.J.; Chinnalyan, A.M.; Woycicki, R.K.; Cantor, C.R.; Kovalchuk, O. miR-34a directly targets tRNAiMet precursors and affects cellular proliferation, cell cycle, and apoptosis Proc. Natl. Acad. Sci. USA 2018, 115, 7392–7397. [Google Scholar] [CrossRef]
- Dai, Z.; Liu, H.; Liao, J.; Huang, C.; Ren, X.; Zhu, W.; Zhu, S.; Peng, B.; Li, S.; Lai, J.; et al. N7-Methylguanosine tRNA modification enhances oncogenic mRNA translation and promotes intrahepatic cholangiocarcinoma progression. Mol. Cell 2021, 81, 3339–3355. [Google Scholar] [CrossRef]
- Orellana, E.A.; Liu, Q.; Yankova, E.; Pirouz, M.; De Braekeleer, E.; Zhang, W.; Lim, J.; Aspris, D.; Sendinc, E.; Garyfallos, D.A.; et al. METTL1-mediated m7G modification of Arg-TCT tRNA drives oncogenic transformation. Mol. Cell 2021, 81, 3323–3338. [Google Scholar] [CrossRef]
- Santos, M.; Fidalgo, A.; Varanda, A.S.; Soares, A.R.; Almeida, G.M.; Martins, D.; Mendes, N.; Oliveira, C.; Santos, M.A.S. Upregulation of tRNA-Ser-AGA-2-1 promotes malignant behavior in normal bronchial cells. Front. Mol. Biosci. 2022, 9, 809985. [Google Scholar] [CrossRef]
- Krishnan, P.; Ghosh, S.; Wang, B.; Heyns, M.; Li, D.; Mackey, J.R.; Kovalchuk, O.; Damaraju, S. Genome-wide profiling of transfer RNAs and their role as novel prognostic markers for breast cancer. Sci. Rep. 2016, 6, 32843. [Google Scholar] [CrossRef]
- Zhang, Z.; Ye, Y.; Gong, J.; Ruan, H.; Liu, C.-J.; Xiang, Y.; Cai, C.; Guo, A.-Y.; Ling, J.; Diao, L.; et al. Global analysis of tRNA and translation factor expression reveals a dynamic landscape of translational regulation in human cancers. Comm. Biol. 2018, 1, 234. [Google Scholar] [CrossRef]
- Butterfield, S.P.; Sizer, R.E.; Rand, E.; White, R.J. Selection of tRNA Genes in Human Breast Tumours Varies Substantially between Individuals. Cancers 2023, 15, 3576. [Google Scholar] [CrossRef]
- Nabet, B.Y.; Qiu, Y.; Shabason, J.E.; Wu, T.J.; Yoon, T.; Kim, B.C.; Benci, J.L.; DeMichele, A.M.; Tchou, J.; Marcotrigiano, J.; et al. Exosome RNA unshielding couples stromal activation to pattern recognition receptor signaling in cancer. Cell 2017, 170, 352–366. [Google Scholar] [CrossRef] [PubMed]
- Thiel, C.T.; Horn, D.; Zabel, B.; Ekici, A.B.; Salinas, K.; Gebhart, E.; Ruschendorf, F.; Sticht, H.; Spranger, J.; Muller, D.; et al. Severely incapacitating mutations in patients with extreme short stature identify RNA-processing endoribonuclease RMRP as an essential cell growth regulator. Am. J. Hum. Genet. 2005, 77, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Goldfarb, K.C.; Cech, T.R. Targeted CRISPR disruption reveals a role for RNase MRP RNA in human preribosomal RNA processing. Genes Dev. 2017, 31, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Rheinbay, E.; Parasuraman, P.; Grimsby, J.; Tiao, G.; Engreitz, J.M.; Kim, J. Recurrent and functional regulatory mutations in breast cancer. Nature 2017, 547, 55–60. [Google Scholar] [CrossRef] [PubMed]
- Vakkilainen, S.; Skoog, T.; Einarsdottir, E.; Middleton, A.; Pekkinen, M.; Ohman, T.; Katayama, S.; Krjutskov, K.; Kovanen, P.E.; Varjosalo, M.; et al. The human long non-coding RNA gene RMRP has pleiotropic effects and regulates cell-cycle progression at G2. Sci. Rep. 2019, 9, 13758. [Google Scholar] [CrossRef] [PubMed]
- Rheinbay, E.; Nielsen, M.M.; Abascal, F.; Wala, J.A.; Shapira, O.; Tiao, G.; Hornshøj, H.; Hess, J.M.; Juul, R.I.; Lin, Z.; et al. Analyses of non-coding somatic drivers in 2,658 cancer whole genomes. Nature 2020, 578, 102–111. [Google Scholar] [CrossRef]
- Geiduschek, E.P.; Kassavetis, G.A. The RNA polymerase III transcription apparatus. J. Mol. Biol. 2001, 310, 1–26. [Google Scholar] [CrossRef]
- Arimbasseri, A.; Maraia, R.J. RNA polymerase III advances: Structural and tRNA functional views. Trends Biochem. Sci. 2016, 41, 546–559. [Google Scholar] [CrossRef]
- Ramsay, E.P.; Vannini, A. Structural rearrangements of the RNA polymerase III machinery during tRNA transcription initiation. BBA—Gene Regul. Mech. 2017, 1861, 285–294. [Google Scholar] [CrossRef]
- Kassavetis, G.A.; Braun, B.R.; Nguyen, L.H.; Geiduschek, E.P.S. S. cerevisiae TFIIIB is the transcription initiation factor proper of RNA polymerase III, while TFIIIA and TFIIIC are assembly factors. Cell 1990, 60, 235–245. [Google Scholar] [CrossRef]
- Sutcliffe, J.E.; Brown, T.R.P.; Allison, S.J.; Scott, P.H.; White, R.J. Retinoblastoma protein disrupts interactions required for RNA polymerase III transcription. Mol. Cell Biol. 2000, 20, 9192–9202. [Google Scholar] [CrossRef] [PubMed]
- Gjidoda, A.; Henry, R.W. RNA polymerase III repression by the retinoblastoma tumor suppressor protein. Biochim. Biophys. Acta 2013, 1829, 385–392. [Google Scholar] [CrossRef] [PubMed]
- Schramm, L.; Pendergrast, P.S.; Sun, Y.; Hernandez, N. Different human TFIIIB activities direct RNA polymerase III transcription from TATA-containing and TATA-less promoters. Genes Dev. 2000, 14, 2650–2663. [Google Scholar] [CrossRef] [PubMed]
- Moqtaderi, Z.; Wang, J.; Raha, D.; White, R.J.; Snyder, M.; Weng, Z.; Struhl, K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010, 17, 635–640. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M. The Sequence Read Archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef]
- Press, A.R. Ultrafast and Memory-Efficient Alignment of Short DNA Sequences to the Human Genome; CreateSpace Independent Publishing Platform: Scotts Valley, CA, USA, 2015; p. 38. [Google Scholar]
- Consortium, T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Karolchik, D.; Hinrichs, A.S.; Furey, T.S.; Roskin, K.M.; Sugnet, C.W.; Haussler, D.; Kent, W.J. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004, 32, D493–D496. [Google Scholar] [CrossRef]
- Lerdrup, M.; Johansen, J.V.; Agrawal-Singh, S.; Hansen, K. An interactive environment for agile analysis and visualization of ChIP-sequencing data. Nat. Struct. Mol. Biol. 2016, 23, 349–357. [Google Scholar] [CrossRef]
- Sanidas, I.; Lee, H.; Rumde, P.H.; Boulay, G.; Morris, R.; Golczer, G.; Stanzione, M.; Hajizadeh, S.; Zhong, J.; Ryan, M.B.; et al. Chromatin-bound RB targets promoters, enhancers, and CTCF-bound loci and is redistributed by cell-cycle progression. Mol. Cell 2022, 82, 3333–3349. [Google Scholar] [CrossRef]
- Chicas, A.; Wang, X.; Zhang, C.; McCurrach, M.; Zhao, Z.; Mert, O.; Dickens, R.A.; Narita, M.; Zhang, M.; Lowe, S.W. Dissecting the unique role of the retinoblastoma tumor suppressor during cellular senescence. Cancer Cell 2010, 17, 376–387. [Google Scholar] [CrossRef] [PubMed]
- Ferrari, R.; Su, T.; Li, B.; Bonora, G.; Oberai, A.; Chan, Y.; Sasidharan, R.; Berk, A.J.; Pellegrini, M.; Kurdistani, S.K. Reorganization of the host epigenome by a viral oncogene. Genome Res. 2012, 22, 1212–1221. [Google Scholar] [CrossRef] [PubMed]
- Oki, S.; Ohta, T.; Shioi, G.; Hatanaka, H.; Ogasawara, O.; Okuda, Y.; Kawaji, H.; Nakaki, R.; Sese, J.; Meno, C. ChIP-Atlas: A data-mining suite powered by full integration of public ChIP-seq data. EMBO Rep. 2018, 19, e46255. [Google Scholar] [CrossRef] [PubMed]
- Zou, Z.; Ohta, T.; Miura, F.; Oki, S. ChIP-Atlas 2021 update: A data-mining suite for exploring epigenomic landscapes by fully integrating ChIP-seq, ATAC-seq and Bisulfite-seq data. Nucleic Acids Res. 2022, 50, W175–W182. [Google Scholar] [CrossRef] [PubMed]
- Wells, J.; Boyd, K.E.; Fry, C.J.; Bartley, S.M.; Farnham, P.J. Target gene specificity of E2F and pocket protein family members in living cells. Mol. Cell Biol. 2000, 20, 5797–5807. [Google Scholar] [CrossRef] [PubMed]
- Schmitt, M.E.; Clayton, D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1993, 13, 7935–7941. [Google Scholar] [PubMed]
- Clayton, D.A. A nuclear function for RNase MRP. Proc. Natl. Acad. Sci. USA 1994, 91, 4615–4617. [Google Scholar] [CrossRef]
- Lygerou, Z.; Allmang, C.; Tollervey, D.; Seraphin, B. Accurate processing of a eukaryotic precursor ribosomal RNA by ribonuclease MRP in vitro. Science 1996, 272, 268–270. [Google Scholar] [CrossRef]
- Maida, Y.; Yasukawa, M.; Furuuchi, M.; Lassmann, T.; Possemato, R.; Okamoto, N.; Kasim, V.; Hayashizaki, Y.; Hahn, W.C.; Matsutomi, K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009, 461, 230–235. [Google Scholar] [CrossRef]
- Ridanpaa, M.; van Eenennaam, H.; Pelin, K.; Chadwick, R.; Johnson, C.; Yuan, B.; vanVenrooij, W.; Pruijn, G.; Samela, R.; Rockas, S.; et al. Mutations in the RNA component of RNase MRP cause a pleiotropic human disease, cartilage-hair hypoplasia. Cell 2001, 104, 195–203. [Google Scholar] [CrossRef]
- Ridanpaa, M.; Sistonen, P.; Rockas, S.; Rimoin, D.L.; Makitie, O.; Kaitila, I. Worldwide mutation spectrum in cartilage-hair hypoplasia: Ancient founder origin of the major 70A->G mutation of the untranslated RMRP. Eur. J. Hum. Genet. 2002, 10, 439–447. [Google Scholar] [CrossRef] [PubMed]
- Walter, P.; Blobel, G. Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 1982, 299, 691–698. [Google Scholar] [CrossRef]
- Chen, W.; Bocker, W.; Brosius, J.; Tiedge, H. Expression of neural BC200 RNA in human tumours. J. Pathol. 1997, 183, 345–351. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Panda, A.C.; Kang, M.-J.; Guo, R.; Kim, J.; Grammatikakis, I.; Yoon, J.-H.; Dudekula, D.B.; Noh, J.H.; Yang, X.-L.; et al. 7SL RNA represses p53 translation by competing with HuR. Nucleic Acids Res. 2014, 42, 10099–10111. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Vavrova-Anderson, J.; Oler, A.J.; Cowling, V.H.; Cairns, B.R.; White, R.J. SINE transcription by RNA polymerase III is suppressed by histone methylation but not by DNA methylation. Nat. Commun. 2015, 6, 6569. [Google Scholar] [CrossRef] [PubMed]
- Rapino, F.; Delaunay, S.; Rambow, F.; Zhou, Z.; Tharun, L.; De Tullio, P.; Sin, O.; Shostak, K.; Schmitz, S.; Piepers, J.; et al. Codon-specific translation reprogramming promotes resistance to targeted therapy. Nature 2018, 558, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Thandapani, P.; Kloetgen, A.; Witkowski, M.T.; Glytsou, C.; Lee, A.K.; Wang, E.; Wang, J.; LeBoeuf, S.E.; Avrampou, K.; Papagiannakopoulos, T.; et al. Valine tRNA levels and availability regulate complex I assembly in leukaemia. Nature 2022, 601, 428–433. [Google Scholar] [CrossRef]
- Hanson, G.; Coller, J. Codon optimality, bias and usage in translation and mRNA decay. Nat. Rev. Mol. Cell Biol. 2018, 19, 20–30. [Google Scholar] [CrossRef]
- Gingold, H.; Tehler, D.; Christoffersen, N.R.; Nielsen, M.M.; Asmar, F.; Kooistra, S.M.; Christophersen, N.S.; Christensen, L.L.; Borre, M.; Sorensen, K.D.; et al. A dual program for translation regulation in cellular proliferation and differentiation. Cell 2014, 158, 1281–1292. [Google Scholar] [CrossRef]
- White, R.J. Transcription by RNA polymerase III—More complex than we thought. Nat. Rev. Genet. 2011, 12, 459–463. [Google Scholar] [CrossRef]
- Yang, J.; Smith, D.K.; Ni, H.; Wu, K.Y.; Huang, D.; Pan, S.; Sathe, A.A.; Tang, Y.; Liu, M.-L.; Xing, C.; et al. SOX4-mediated repression of specific tRNAs inhibits proliferation of human glioblastoma cells. Proc. Natl. Acad. Sci. USA 2020, 117, 5782–5790. [Google Scholar] [CrossRef] [PubMed]
- White, R.J.; Gottlieb, T.M.; Downes, C.S.; Jackson, S.P. Cell cycle regulation of RNA polymerase III transcription. Mol. Cell Biol. 1995, 15, 6653–6662. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sizer, R.E.; Butterfield, S.P.; Hancocks, L.A.; Gato De Sousa, L.; White, R.J. Selective Occupation by E2F and RB of Loci Expressed by RNA Polymerase III. Cancers 2024, 16, 481. https://doi.org/10.3390/cancers16030481
Sizer RE, Butterfield SP, Hancocks LA, Gato De Sousa L, White RJ. Selective Occupation by E2F and RB of Loci Expressed by RNA Polymerase III. Cancers. 2024; 16(3):481. https://doi.org/10.3390/cancers16030481
Chicago/Turabian StyleSizer, Rebecca E., Sienna P. Butterfield, Lucy A. Hancocks, Leonor Gato De Sousa, and Robert J. White. 2024. "Selective Occupation by E2F and RB of Loci Expressed by RNA Polymerase III" Cancers 16, no. 3: 481. https://doi.org/10.3390/cancers16030481
APA StyleSizer, R. E., Butterfield, S. P., Hancocks, L. A., Gato De Sousa, L., & White, R. J. (2024). Selective Occupation by E2F and RB of Loci Expressed by RNA Polymerase III. Cancers, 16(3), 481. https://doi.org/10.3390/cancers16030481





