Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling
Simple Summary
Abstract
1. Introduction
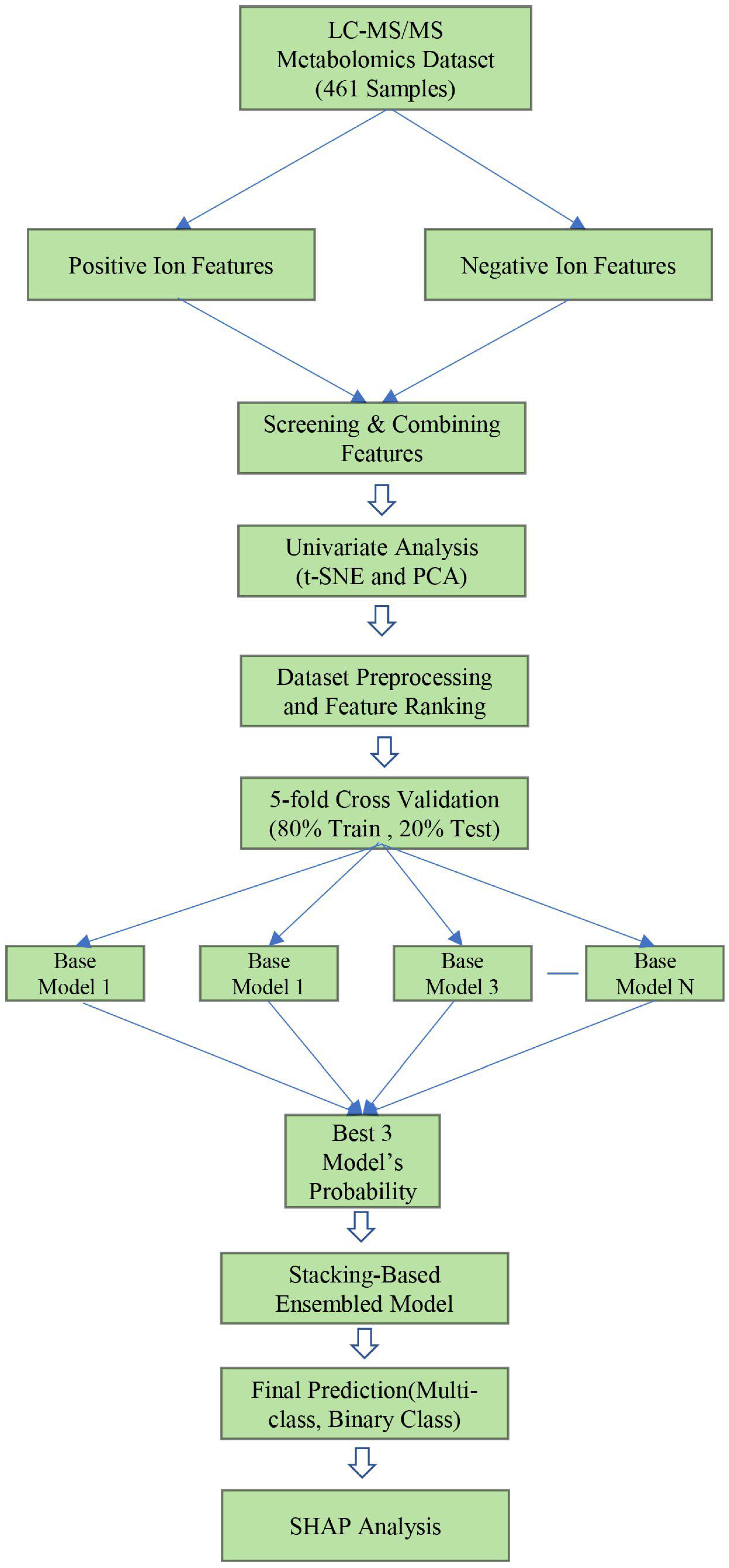
2. Methods and Materials
2.1. Study Cohort, Sample Collection, and Metabolomics Dataset
2.2. Dataset Preprocessing
2.3. Evaluation Metrics
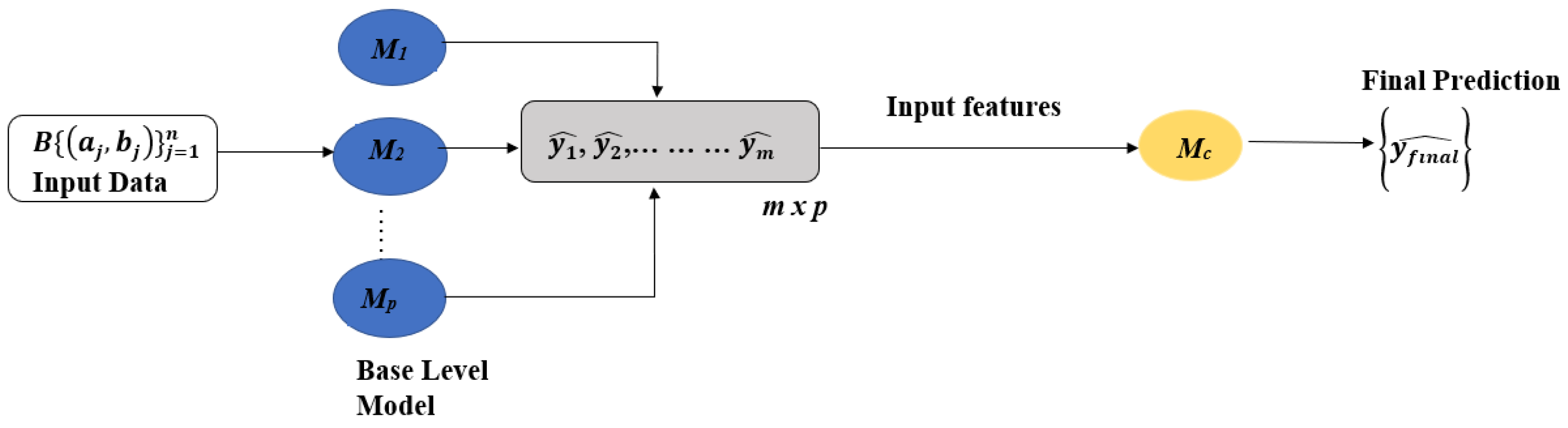
2.4. Development of Machine Learning and Stacking Ensemble Models
3. Results
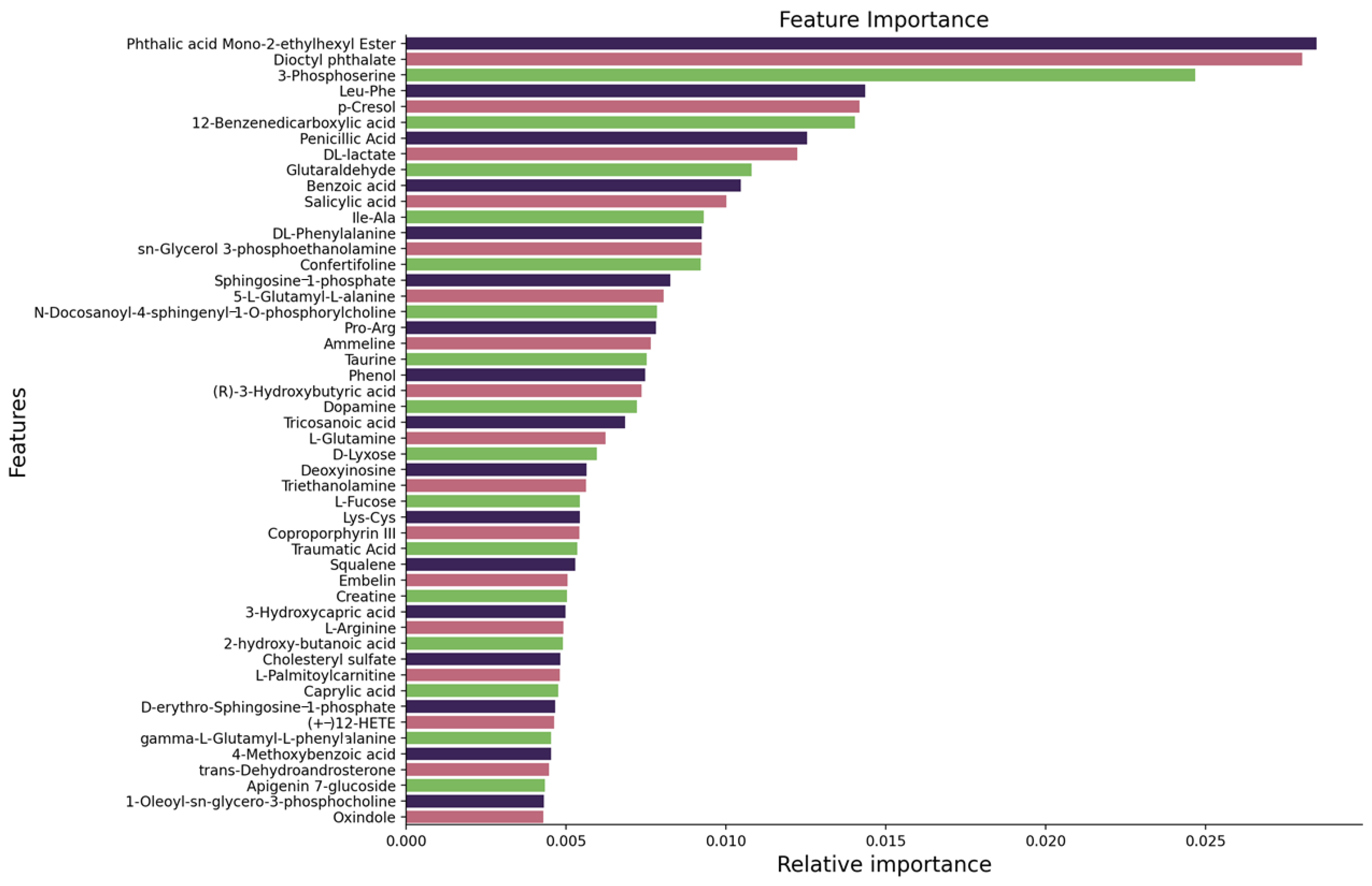
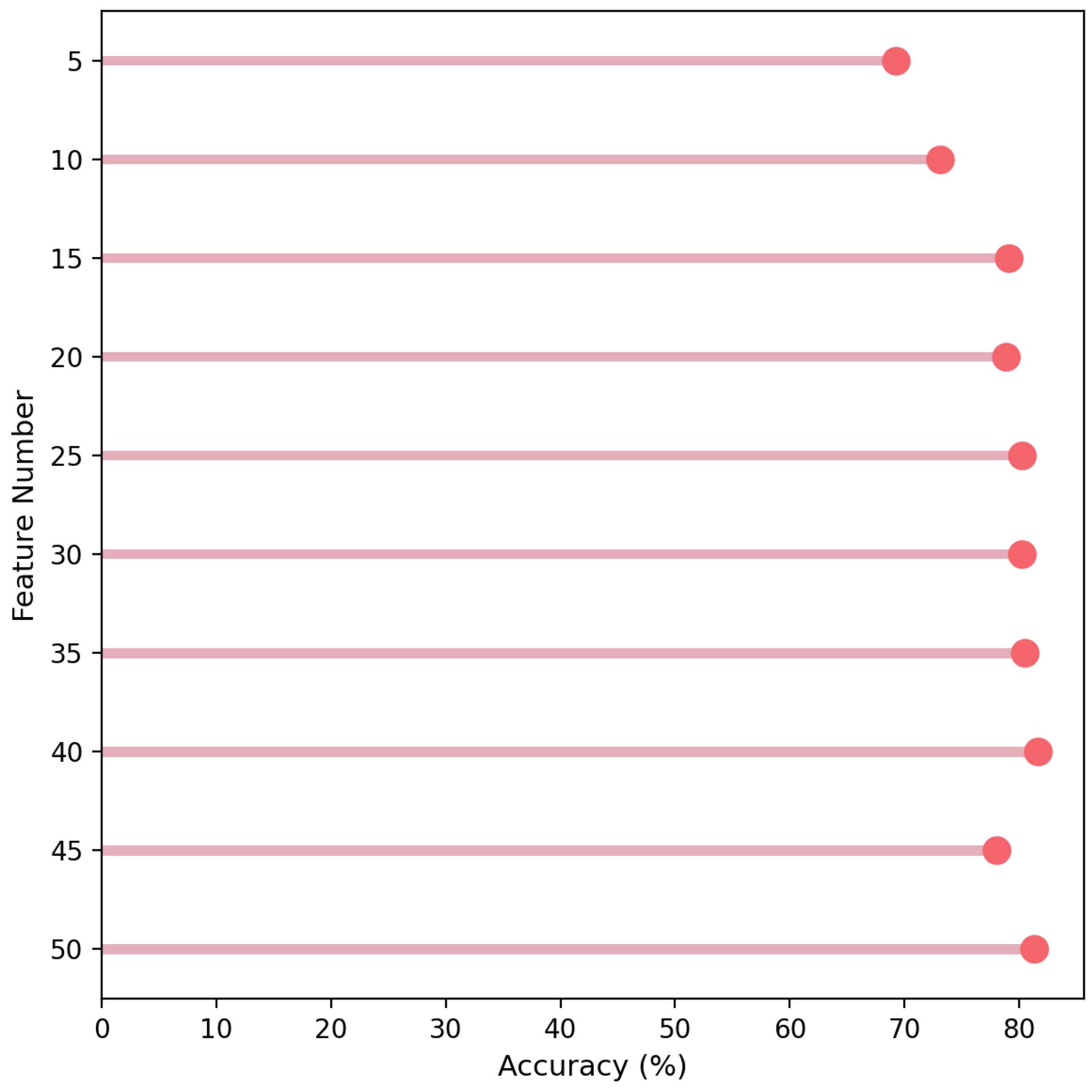
3.1. Feature Ranking
3.2. Multi-Class Classification
3.3. Binary Classification
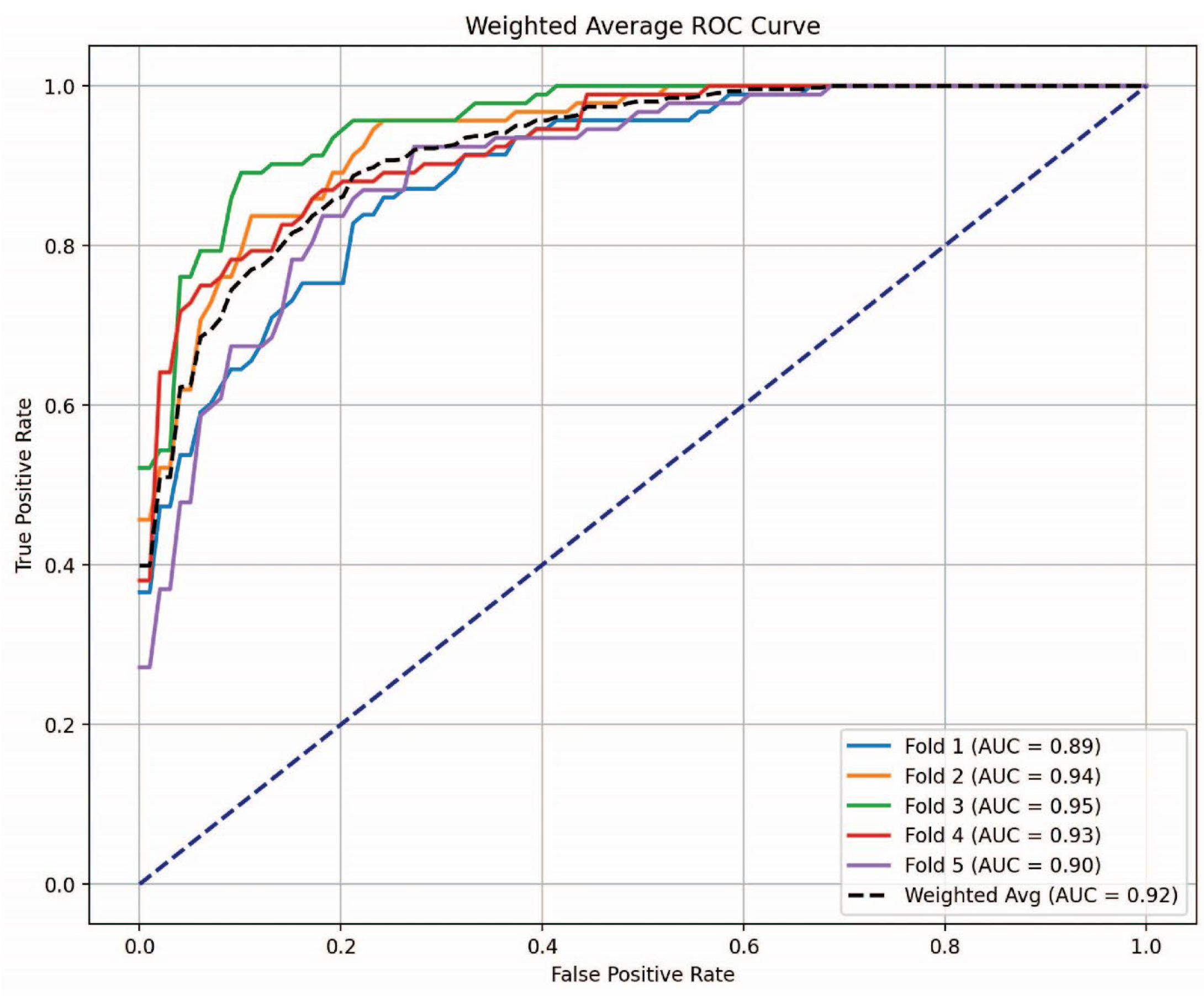
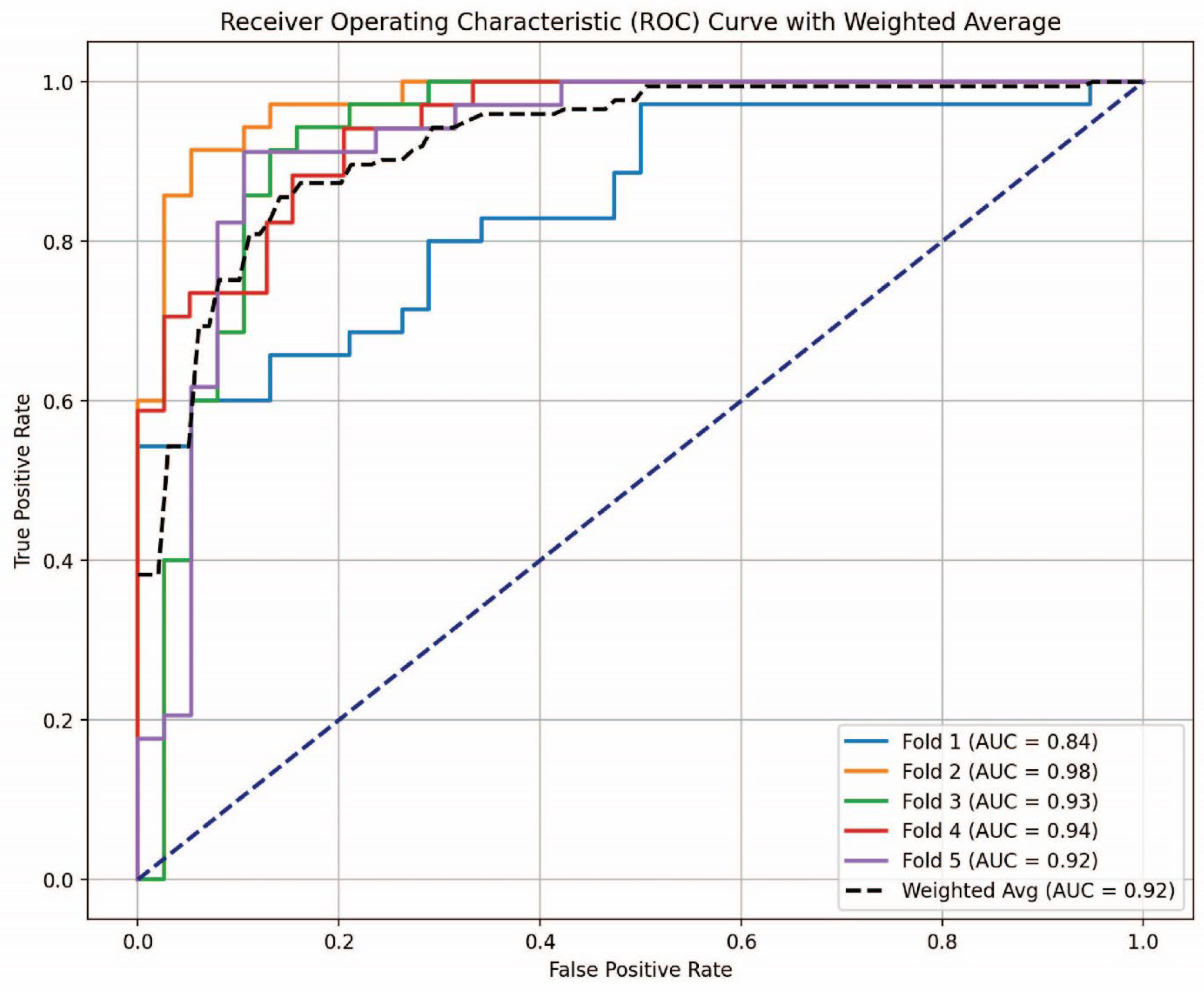
3.4. AUC-ROC Analysis
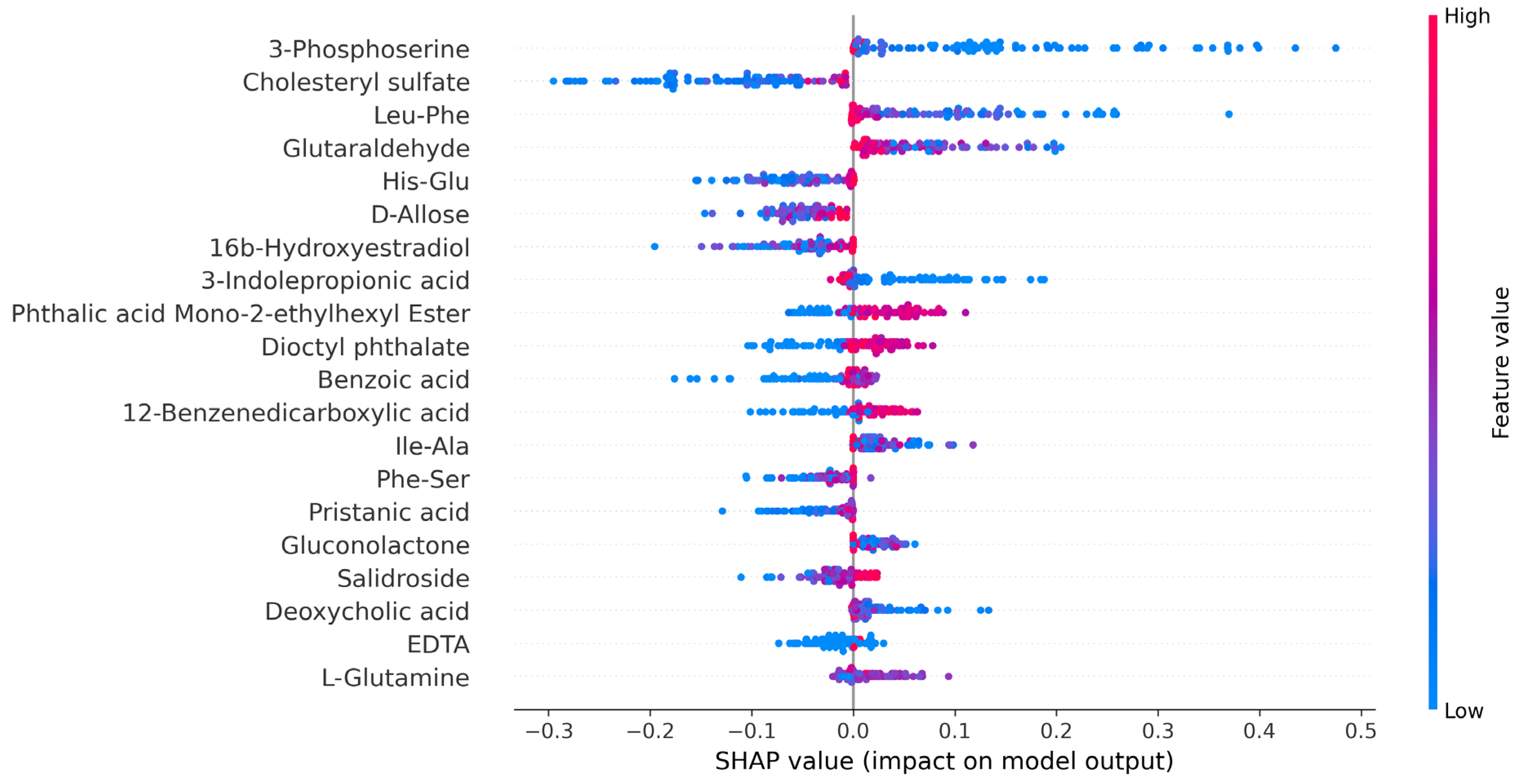
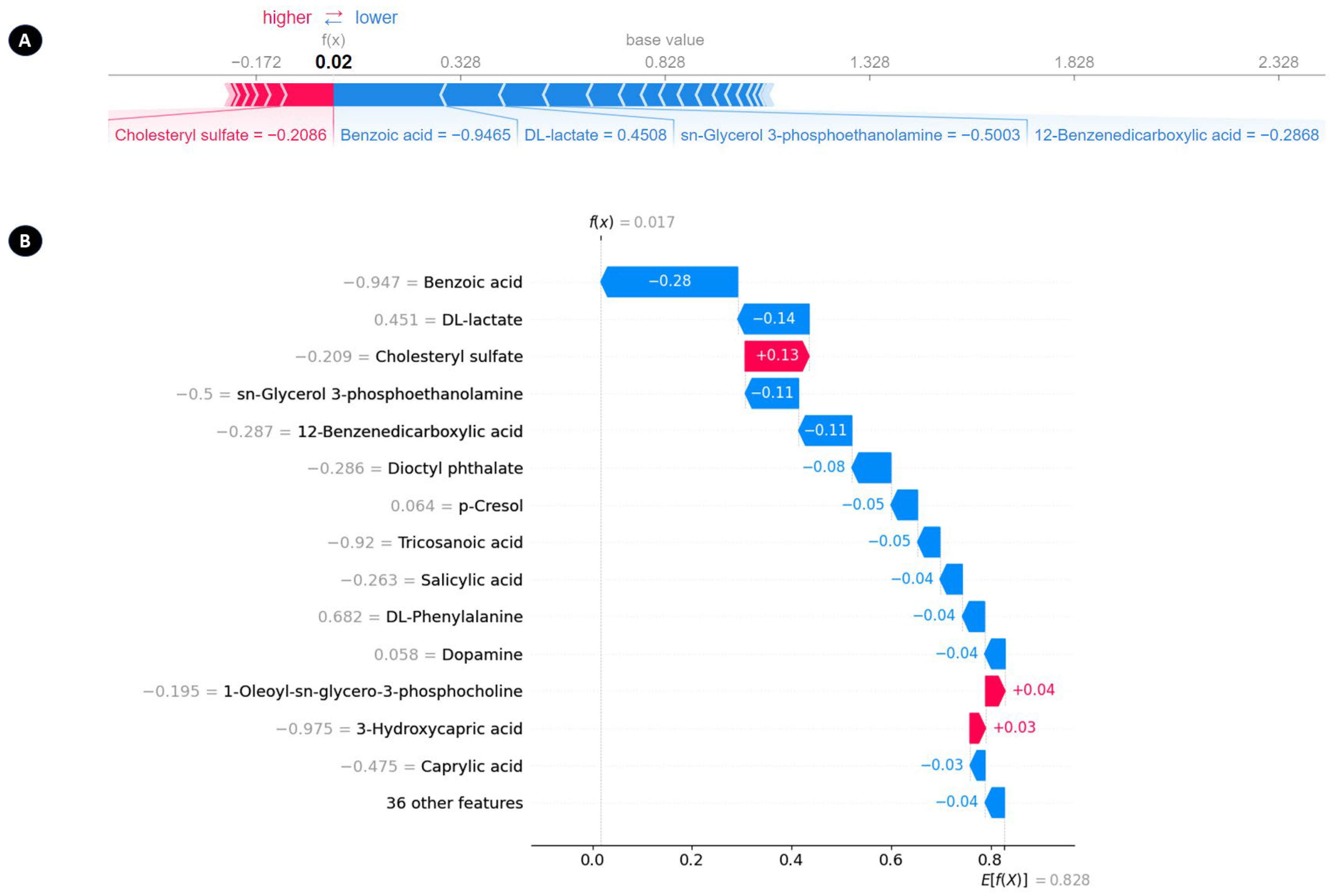
3.5. SHAP Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Lei, S.; Ding, L.; Xu, Y.; Wu, X.; Wang, H.; Zhang, Z.; Gao, T.; Zhang, Y.; Li, L. Global burden and trends of lung cancer incidence and mortality. Chin. Med. J. 2023, 136, 1583–1590. [Google Scholar] [CrossRef]
- Barta, J.A.; Powell, C.A.; Wisnivesky, J.P. Global epidemiology of lung cancer. Ann. Glob. Health 2019, 85, 8. [Google Scholar] [CrossRef] [PubMed]
- Shang, X.; Zhang, C.; Kong, R.; Zhao, C.; Wang, H. Construction of a Diagnostic Model for Small Cell Lung Cancer Combining Metabolomics and Integrated Machine Learning. Oncologist 2024, 29, e392–e401. [Google Scholar] [CrossRef]
- Ayoub, M.; AbuHaweeleh, M.N.; Mahmood, N.; Clelland, C.; Ayoub, M.M.; Saman, H. Small cell lung cancer associated small bowel obstruction, a diagnostic conundrum: A case report. Clin. Case Rep. 2024, 12, e9262. [Google Scholar] [CrossRef]
- Elshoeibi, A.M.; Elsayed, B.; Kaleem, M.Z.; Elhadary, M.R.; Abu-Haweeleh, M.N.; Haithm, Y.; Krzyslak, H.; Vranic, S.; Pedersen, S. Proteomic Profiling of Small-Cell Lung Cancer: A Systematic Review—PubMed. Cancers 2023, 15, 5005. [Google Scholar] [CrossRef]
- Lee, G.; Lee, H.Y.; Park, H.; Schiebler, M.L.; van Beek, E.J.; Ohno, Y.; Seo, J.B.; Leung, A. Radiomics and its emerging role in lung cancer research, imaging biomarkers and clinical management: State of the art. Eur. J. Radiol. 2017, 86, 297–307. [Google Scholar] [CrossRef]
- Shestakova, K.M.; Moskaleva, N.E.; Boldin, A.A.; Rezvanov, P.M.; Shestopalov, A.V.; Rumyantsev, S.A.; Zlatnik, E.Y.; Novikova, I.A.; Sagakyants, A.B.; Timofeeva, S.V. Targeted metabolomic profiling as a tool for diagnostics of patients with non-small-cell lung cancer. Sci. Rep. 2023, 13, 11072. [Google Scholar] [CrossRef] [PubMed]
- Amoêdo, N.D.; Valencia, J.P.; Rodrigues, M.F.; Galina, A.; Rumjanek, F.D. How does the metabolism of tumour cells differ from that of normal cells. Biosci. Rep. 2013, 33, e00080. [Google Scholar] [CrossRef] [PubMed]
- Holmes, E.; Wilson, I.D.; Nicholson, J.K. Metabolic phenotyping in health and disease. Cell 2008, 134, 714–717. [Google Scholar] [CrossRef]
- Mariën, H.; Derveaux, E.; Vanhove, K.; Adriaensens, P.; Thomeer, M.; Mesotten, L. Changes in Metabolism as a Diagnostic Tool for Lung Cancer: Systematic Review. Metabolites 2022, 12, 545. [Google Scholar] [CrossRef]
- Noreldeen, H.A.; Liu, X.; Xu, G. Metabolomics of lung cancer: Analytical platforms and their applications. J. Sep. Sci. 2020, 43, 120–133. [Google Scholar] [CrossRef]
- Wei, Z.; Liu, X.; Cheng, C.; Yu, W.; Yi, P. Metabolism of Amino Acids in Cancer. Front. Cell Dev. Biol. 2021, 8, 603837. [Google Scholar] [CrossRef]
- Liberti, M.V.; Locasale, J.W. The Warburg Effect: How Does it Benefit Cancer Cells? Trends Biochem. Sci. 2016, 41, 211–218. [Google Scholar] [CrossRef]
- Valles, I.; Pajares, M.J.; Segura, V.; Guruceaga, E.; Gomez-Roman, J.; Blanco, D.; Tamura, A.; Montuenga, L.M.; Pio, R. Identification of Novel Deregulated RNA Metabolism-Related Genes in Non-Small Cell Lung Cancer. PLoS ONE 2012, 7, e42086. [Google Scholar] [CrossRef] [PubMed]
- Tufail, M.; Jiang, C.; Li, N. Altered metabolism in cancer: Insights into energy pathways and therapeutic targets. Mol. Cancer 2024, 23, 203. [Google Scholar] [CrossRef] [PubMed]
- Albaugh, V.L.; Pinzon-Guzman, C.; Barbul, A. Arginine metabolism and cancer. J. Surg. Oncol. 2017, 115, 273–280. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, J.-M. Altered energy metabolism in cancer: A unique opportunity for therapeutic intervention. Cancer Biol. Ther. 2013, 14, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Mendez, K.M.; Reinke, S.N.; Broadhurst, D.I. A comparative evaluation of the generalised predictive ability of eight machine learning algorithms across ten clinical metabolomics data sets for binary classification. Metabolomics 2019, 15, 150. [Google Scholar] [CrossRef]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Schult, T.A.; Lauer, M.J.; Berker, Y.; Cardoso, M.R.; Vandergrift, L.A.; Habbel, P.; Nowak, J.; Taupitz, M.; Aryee, M.; Mino-Kenudson, M.A. Screening human lung cancer with predictive models of serum magnetic resonance spectroscopy metabolomics. Proc. Natl. Acad. Sci. USA 2021, 118, e2110633118. [Google Scholar] [CrossRef]
- Chen, R.; Li, Z.; Yuan, Y.; Zhu, Z.; Zhang, J.; Tian, X.; Zhang, X. A comprehensive analysis of metabolomics and transcriptomics in non-small cell lung cancer. PLoS ONE 2020, 15, e0232272. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Hanash, S.; DeFelice, B.; Miyamoto, S.; Barnett, M.; Zhao, Y.; Goodman, G.; Feng, Z.; Gandara, D.; Fiehn, O. Diacetylspermine is a novel prediagnostic serum biomarker for non–small-cell lung cancer and has additive performance with pro-surfactant protein B. J. Clin. Oncol. 2015, 33, 3880–3886. [Google Scholar] [CrossRef]
- Du, J.; Li, Y.; Wang, L.; Zhou, Y.; Shen, Y.; Xu, F.; Chen, Y. Selective application of neuroendocrine markers in the diagnosis and treatment of small cell lung cancer. Clin. Chim. Acta 2020, 509, 295–303. [Google Scholar] [CrossRef] [PubMed]
- Song, B.; Shi, P.; Xiao, J.; Song, Y.; Zeng, M.; Cao, Y.; Zhu, X. Utility of red cell distribution width as a diagnostic and prognostic marker in non-small cell lung cancer. Sci. Rep. 2020, 10, 15717. [Google Scholar] [CrossRef] [PubMed]
- Schneider, J.; Philipp, M.; Velcovsky, H.-G.; Morr, H.; Katz, N. Pro-gastrin-releasing peptide (ProGRP), neuron specific enolase (NSE), carcinoembryonic antigen (CEA) and cytokeratin 19-fragments (CYFRA 21-1) in patients with lung cancer in comparison to other lung diseases. Anticancer Res. 2003, 23, 885–893. [Google Scholar] [PubMed]
- Yu, Z.; Lu, H.; Si, H.; Liu, S.; Li, X.; Gao, C.; Cui, L.; Li, C.; Yang, X.; Yao, X. A highly efficient gene expression programming (GEP) model for auxiliary diagnosis of small cell lung cancer. PLoS ONE 2015, 10, e0125517. [Google Scholar] [CrossRef] [PubMed]
- Barchiesi, V.; Simeon, V.; Sandomenico, C.; Cantile, M.; Cerasuolo, D.; Chiodini, P.; Morabito, A.; Cavalcanti, E. Circulating progastrin-releasing peptide in the diagnosis of Small Cell Lung Cancer (SCLC) and in therapeutic monitoring. J. Circ. Biomark. 2021, 10, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Jia, G.; Jie, H. Diagnostic value of ProGRP and NSE for small cell lung cancer: A meta-analysis. Zhongguo Fei Ai Za Zhi 2010, 13, 1094–1100. [Google Scholar]
- Shibayama, T.; Ueoka, H.; Nishii, K.; Kiura, K.; Tabata, M.; Miyatake, K.; Kitajima, T.; Harada, M. Complementary roles of pro-gastrin-releasing peptide (ProGRP) and neuron specific enolase (NSE) in diagnosis and prognosis of small-cell lung cancer (SCLC). Lung Cancer 2001, 32, 61–69. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Huang, Y.; Ling, Z.; Chen, J.; Wei, X.; Su, R.; Tang, Z.; Wen, Z.; Deng, Y.; Hu, Z. Lack of efficacy of combined carbohydrate antigen markers for lung cancer diagnosis. Dis. Markers 2020, 2020, 4716793. [Google Scholar] [CrossRef] [PubMed]
- Oremek, G.; Sauer-Eppel, H.; Bruzdziak, T. Value of tumour and inflammatory markers in lung cancer. Anticancer Res. 2007, 27, 1911–1915. [Google Scholar] [PubMed]
- Yang, H.-j.; Gu, Y.; Chen, C.; Xu, C.; Bao, Y.-x. Diagnostic value of pro-gastrin-releasing peptide for small cell lung cancer: A meta-analysis. Clin. Chem. Lab. Med. 2011, 49, 1039–1046. [Google Scholar] [CrossRef]
- Harmsma, M.; Schutte, B.; Ramaekers, F.C. Serum markers in small cell lung cancer: Opportunities for improvement. Biochim. Biophys. Acta (BBA)-Rev. Cancer 2013, 1836, 255–272. [Google Scholar] [CrossRef] [PubMed]
- Sidaway, P. cfDNA monitoring is feasible in SCLC. Nat. Rev. Clin. Oncol. 2020, 17, 7. [Google Scholar] [CrossRef]
- Mondelo-Macía, P.; García-González, J.; León-Mateos, L.; Castillo-García, A.; López-López, R.; Muinelo-Romay, L.; Díaz-Peña, R. Current status and future perspectives of liquid biopsy in small cell lung cancer. Biomedicines 2021, 9, 48. [Google Scholar] [CrossRef]
- Sandfeld-Paulsen, B.; Jakobsen, K.R.; Bæk, R.; Folkersen, B.H.; Rasmussen, T.R.; Meldgaard, P.; Varming, K.; Jørgensen, M.M.; Sorensen, B.S. Exosomal proteins as diagnostic biomarkers in lung cancer. J. Thorac. Oncol. 2016, 11, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Ma, M.; Zhu, H.; Zhang, C.; Sun, X.; Gao, X.; Chen, G. “Liquid biopsy”—ctDNA detection with great potential and challenges. Ann. Transl. Med. 2015, 3, 12–235. [Google Scholar] [CrossRef]
- Mallikharjuna Rao, K.; Saikrishna, G.; Supriya, K. Data preprocessing techniques: Emergence and selection towards machine learning models-a practical review using HPA dataset. Multimed. Tools Appl. 2023, 82, 37177–37196. [Google Scholar] [CrossRef]
- Singh, D.; Singh, B. Investigating the impact of data normalization on classification performance. Appl. Soft Comput. 2020, 97, 105524. [Google Scholar] [CrossRef]
- Yusuf, M.; Atal, I.; Li, J.; Smith, P.; Ravaud, P.; Fergie, M.; Callaghan, M.; Selfe, J. Reporting quality of studies using machine learning models for medical diagnosis: A systematic review. BMJ Open 2020, 10, e034568. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, P.-H.C.; Krause, J.; Peng, L. How to Read Articles That Use Machine Learning: Users’ Guides to the Medical Literature. JAMA 2019, 322, 1806–1816. [Google Scholar] [CrossRef]
- Sumon, M.S.I.; Hossain, M.S.A.; Al-Sulaiti, H.; Yassine, H.M.; Chowdhury, M.E. Enhancing Influenza Detection through Integrative Machine Learning and Nasopharyngeal Metabolomic Profiling: A Comprehensive Study. Diagnostics 2024, 14, 2214. [Google Scholar] [CrossRef]
- Meyer, D.; Nagler, T.; Hogan, R.J. Copula-based synthetic data augmentation for machine-learning emulators. Geosci. Model Dev. 2021, 14, 5205–5215. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Caesar, L.K.; Kvalheim, O.M.; Cech, N.B. Hierarchical cluster analysis of technical replicates to identify interferents in untargeted mass spectrometry metabolomics. Anal. Chim. Acta 2018, 1021, 69–77. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, F.; Nielsen, F. Hierarchical clustering. In Introduction to HPC with MPI for Data Science; Springer: Berlin/Heidelberg, Germany, 2016; pp. 195–211. [Google Scholar]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Geurts, P.; Ernst, D.; Wehenkel, L. Extremely randomized trees. Mach. Learn. 2006, 63, 3–42. [Google Scholar] [CrossRef]
- Vanderlooy, S.; Hüllermeier, E. A critical analysis of variants of the AUC. Mach. Learn. 2008, 72, 247–262. [Google Scholar] [CrossRef]
- Mangalathu, S.; Hwang, S.-H.; Jeon, J.-S. Failure mode and effects analysis of RC members based on machine-learning-based SHapley Additive exPlanations (SHAP) approach. Eng. Struct. 2020, 219, 110927. [Google Scholar] [CrossRef]
- Horn, L.; Mansfield, A.S.; Szczęsna, A.; Havel, L.; Krzakowski, M.; Hochmair, M.J.; Huemer, F.; Losonczy, G.; Johnson, M.L.; Nishio, M.; et al. First-Line Atezolizumab plus Chemotherapy in Extensive-Stage Small-Cell Lung Cancer. N. Engl. J. Med. 2018, 379, 2220–2229. [Google Scholar] [CrossRef] [PubMed]
- Rotow, J.; Bivona, T.G. Understanding and targeting resistance mechanisms in NSCLC—PubMed. Nat. Rev. Cancer 2017, 17, 637–658. [Google Scholar] [CrossRef]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretić, L.; Kong, G.; Leenders, F.; Lu, X.; Fernández-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 7563. [Google Scholar] [CrossRef]
- Butler, L.M.; Perone, Y.; Dehairs, J.; Lupien, L.E.; de Laat, V.; Talebi, A.; Loda, M.; Kinlaw, W.B.; Swinnen, J.V. Lipids and cancer: Emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv. Drug Deliv. Rev. 2020, 159, 245–293. [Google Scholar] [CrossRef]
- Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N.; Munir, R.; Lisec, J.; Swinnen, J.V.; Zaidi, N. Lipid metabolism in cancer cells under metabolic stress. Br. J. Cancer 2019, 120, 12. [Google Scholar] [CrossRef]
- Hayes, J.D.; Dinkova-Kostova, A.T.; Tew, K.D. Oxidative Stress in Cancer—PubMed. Cancer Cell 2020, 38, 167–197. [Google Scholar] [CrossRef]
- Kuo, C.-L.; Ponneri Babuharisankar, A.; Lin, Y.-C.; Lien, H.-W.; Lo, Y.K.; Chou, H.-Y.; Tangeda, V.; Cheng, L.-C.; Cheng, A.N.; Lee, A.Y.-L.; et al. Mitochondrial oxidative stress in the tumor microenvironment and cancer immunoescape: Foe or friend? J. Biomed. Sci. 2022, 29, 74. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. SMOTE: Synthetic minority over-sampling technique. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Mukhamediev, R.I.; Symagulov, A.; Kuchin, Y.; Yakunin, K.; Yelis, M. From classical machine learning to deep neural networks: A simplified scientometric review. Appl. Sci. 2021, 11, 5541. [Google Scholar] [CrossRef]
- Huang, X.; Khetan, A.; Cvitkovic, M.; Karnin, Z. Tabtransformer: Tabular data modeling using contextual embeddings. arXiv 2020, arXiv:2012.06678. [Google Scholar]
- Shwartz-Ziv, R.; Armon, A. Tabular data: Deep learning is not all you need. Inf. Fusion 2022, 81, 84–90. [Google Scholar] [CrossRef]
- Mathé, E.A.; Patterson, A.D.; Haznadar, M.; Manna, S.K.; Krausz, K.W.; Bowman, E.D.; Shields, P.G.; Idle, J.R.; Smith, P.B.; Anami, K.; et al. Noninvasive Urinary Metabolomic Profiling Identifies Diagnostic and Prognostic Markers in Lung Cancer. Cancer Res. 2014, 74, 3259–3270. [Google Scholar] [CrossRef] [PubMed]
- Campbell, J.D.; Alexandrov, A.; Kim, J.; Wala, J.; Berger, A.H.; Pedamallu, C.S.; Shukla, S.A.; Guo, G.; Brooks, A.N.; Murray, B.A.; et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas—PubMed. Nat. Genet. 2016, 48, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Gao, M.; Zhao, L.; Zhang, Z.; Wang, J.; Wang, C. Using a stacked ensemble learning framework to predict modulators of protein-protein interactions. Comput. Biol. Med. 2023, 161, 107032. [Google Scholar] [CrossRef] [PubMed]
- Liang, M.; Chang, T.; An, B.; Duan, X.; Du, L.; Wang, X.; Miao, J.; Xu, L.; Gao, X.; Zhang, L.; et al. A Stacking Ensemble Learning Framework for Genomic Prediction. Front. Genet. 2021, 12, 600040. [Google Scholar] [CrossRef] [PubMed]



| Initial Results | Stacking Ensemble Results | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Models | A | P | R | S | F1 | AUC | Models | A | P | R | S | F1 | AUC |
| CatBoost | 84.81 | 84.86 | 84.81 | 90.08 | 84.82 | 94.82 | SVM | 85.03 | 85.05 | 85.03 | 90.25 | 85.04 | 92.47 |
| RandomForest | 84.59 | 84.55 | 84.59 | 90.04 | 84.57 | 94.64 | MLPClassifier | 84.82 | 84.87 | 84.81 | 90.14 | 84.82 | 94.17 |
| ExtraTrees | 80.69 | 80.71 | 80.69 | 87.44 | 80.64 | 93.45 | LDA | 84.82 | 84.84 | 84.81 | 90.11 | 84.82 | 94.06 |
| GradientBoosting | 80.47 | 80.76 | 80.47 | 87.23 | 80.54 | 92.01 | LogisticRegression | 84.6 | 84.6 | 84.6 | 89.94 | 84.6 | 94.29 |
| XGB | 79.82 | 79.57 | 79.82 | 87.67 | 79.66 | 92.65 | ExtraTrees | 84.6 | 84.6 | 84.6 | 89.94 | 84.6 | 94.25 |
| LGBM | 79.17 | 79.34 | 79.17 | 86.4 | 79.24 | 92.37 | ElasticNet | 84.6 | 84.6 | 84.6 | 89.94 | 84.6 | 94.29 |
| LogisticRegression | 78.09 | 78.22 | 78.09 | 85.68 | 78.1 | 91.58 | XGBClassifier | 84.16 | 84.17 | 84.17 | 89.65 | 84.17 | 93.43 |
| SVM | 77 | 77.35 | 77 | 84.97 | 77.08 | 89.26 | LGBM | 83.51 | 83.51 | 83.51 | 89.17 | 83.5 | 93.8 |
| MLPClassifier | 78.741 | 78.832 | 78.741 | 86.202 | 78.759 | 91.37 | RandomForest | 83.08 | 83.11 | 83.08 | 88.98 | 83.08 | 93.4 |
| ElasticNet | 76.13 | 76.06 | 76.13 | 84.67 | 76.06 | 91.18 | CatBoost | 81.34 | 81.37 | 81.34 | 87.84 | 81.35 | 93.67 |
| Initial Results | Stacking Ensemble Results | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Models | A | P | R | S | F1 | AUC | Models | A | P | R | S | F1 | AUC |
| MLPClassifier | 85.43 | 85.43 | 85.43 | 85.43 | 85.43 | 92.04 | ExtraTreesClassifier | 88.19 | 88.27 | 88.19 | 88.32 | 88.19 | 92.65 |
| SVM | 84.61 | 84.67 | 84.61 | 84.61 | 84.62 | 93.06 | XGBClassifier | 87.36 | 87.4 | 87.36 | 87.41 | 87.37 | 92.22 |
| ElasticNet | 84.61 | 84.62 | 84.61 | 84.61 | 84.61 | 93.19 | LogisticRegression | 86.81 | 86.82 | 86.81 | 86.81 | 86.82 | 92.65 |
| RandomForest | 83.51 | 83.61 | 83.51 | 83.51 | 83.52 | 89.71 | CatBoost | 86.81 | 86.88 | 86.81 | 86.91 | 86.82 | 92.76 |
| LogisticRegression | 83.51 | 83.52 | 83.51 | 83.51 | 83.52 | 92.49 | ElasticNet | 86.81 | 86.82 | 86.81 | 86.81 | 86.82 | 92.72 |
| LinearDiscriminantAnalysis | 83.24 | 83.36 | 83.24 | 83.24 | 83.25 | 91.47 | LinearDiscriminantAnalysis | 86.54 | 86.58 | 86.53 | 86.61 | 86.54 | 92.3 |
| CatBoost | 82.69 | 82.86 | 82.69 | 82.69 | 82.7 | 90.05 | SVM | 86.54 | 86.56 | 86.54 | 86.56 | 86.54 | 92.27 |
| ExtraTrees | 81.86 | 81.86 | 81.86 | 81.86 | 81.85 | 90.14 | LGBM | 86.54 | 86.63 | 86.54 | 86.17 | 86.5 | 92 |
| AdaBoostClassifier | 79.94 | 80.02 | 79.94 | 79.94 | 79.95 | 85.92 | MLPClassifier | 85.99 | 86.16 | 85.99 | 86.22 | 85.99 | 92.25 |
| LGBM | 76.64 | 76.86 | 76.64 | 76.64 | 76.65 | 82.72 | RandomForest | 85.99 | 86.03 | 85.99 | 86.06 | 85.99 | 91.62 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sumon, M.S.I.; Malluhi, M.; Anan, N.; AbuHaweeleh, M.N.; Krzyslak, H.; Vranic, S.; Chowdhury, M.E.H.; Pedersen, S. Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling. Cancers 2024, 16, 4225. https://doi.org/10.3390/cancers16244225
Sumon MSI, Malluhi M, Anan N, AbuHaweeleh MN, Krzyslak H, Vranic S, Chowdhury MEH, Pedersen S. Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling. Cancers. 2024; 16(24):4225. https://doi.org/10.3390/cancers16244225
Chicago/Turabian StyleSumon, Md. Shaheenur Islam, Marwan Malluhi, Noushin Anan, Mohannad Natheef AbuHaweeleh, Hubert Krzyslak, Semir Vranic, Muhammad E. H. Chowdhury, and Shona Pedersen. 2024. "Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling" Cancers 16, no. 24: 4225. https://doi.org/10.3390/cancers16244225
APA StyleSumon, M. S. I., Malluhi, M., Anan, N., AbuHaweeleh, M. N., Krzyslak, H., Vranic, S., Chowdhury, M. E. H., & Pedersen, S. (2024). Integrative Stacking Machine Learning Model for Small Cell Lung Cancer Prediction Using Metabolomics Profiling. Cancers, 16(24), 4225. https://doi.org/10.3390/cancers16244225