Microdissection of Distinct Morphological Regions Within Uveal Melanomas Identifies Novel Drug Targets
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Cohort Selection Criteria
2.2. Patient and Tumour Characteristics
2.3. Enucleation Grossing
2.4. Histology and Immunohistochemistry
2.5. Comparative Genomic Hybridisation (CGH)/Single Nucleotide Polymorphism (SNP) Chromosome Microarrays
2.6. Reporting Criteria for CGH/SNP Chromosome Microarray Abnormalities
2.7. Extraction of DNA and RNA from FFPE Tissue
2.8. Targeted Sequencing of DNA and RNA
2.9. Variant Calling, Annotation, and Curation
3. Results
3.1. Tissue Heterogeneity in Large UMs
3.2. Molecular Features of High Metastatic Risk in All Tumours Examined
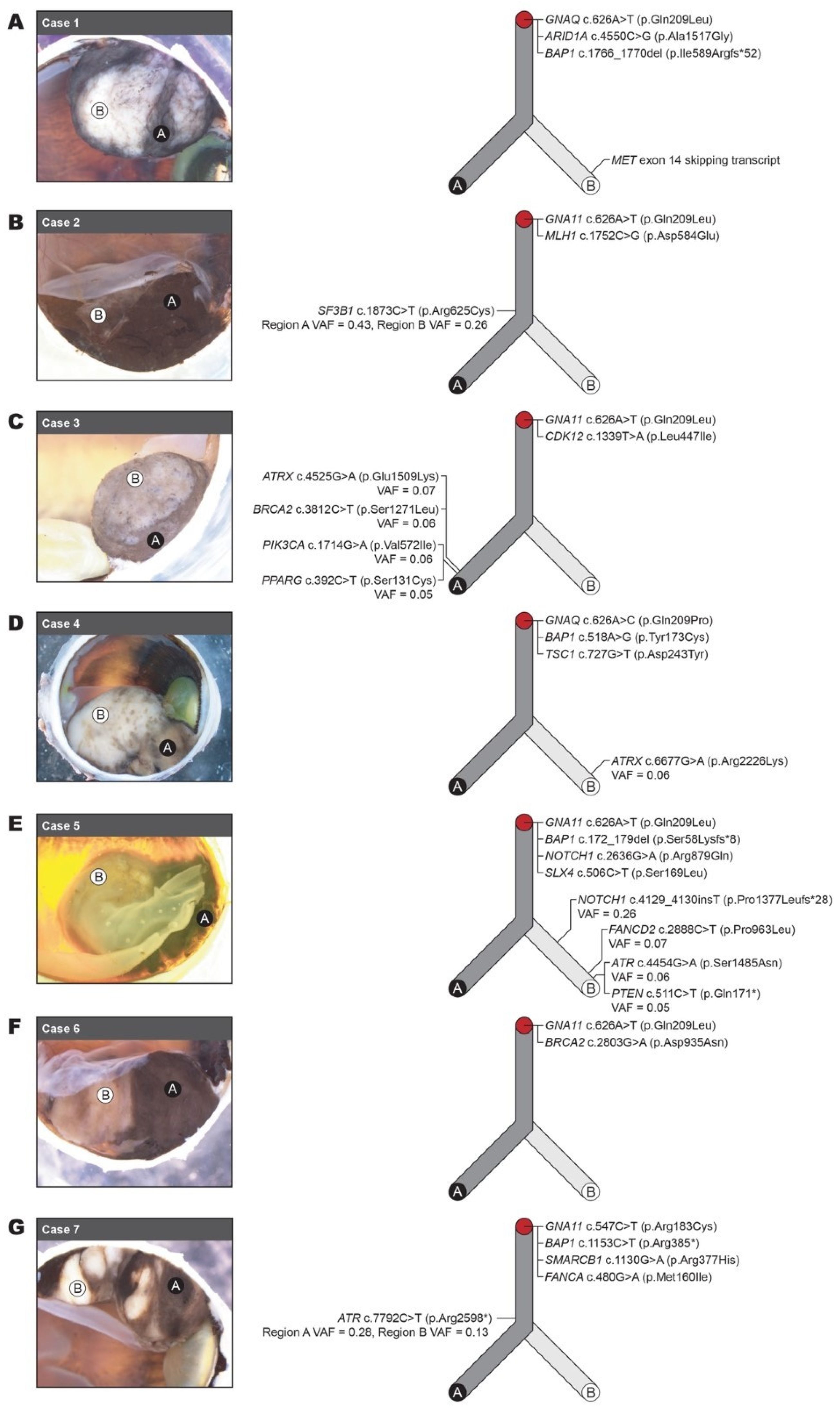
3.3. Molecular Heterogeneity Revealed by Microdissection and Targeted Sequencing
3.4. Recurrently Mutated Genes in UM
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Pandiani, C.; Béranger, G.E.; Leclerc, J.; Ballotti, R.; Bertolotto, C. Focus on cutaneous and uveal melanoma specificities. Genes Dev. 2017, 31, 724–743. [Google Scholar] [CrossRef] [PubMed]
- Royer-Bertrand, B.; Torsello, M.; Rimoldi, D.; El Zaoui, I.; Cisarova, K.; Pescini-Gobert, R.; Raynaud, F.; Zografos, L.; Schalenbourg, A.; Speiser, D.; et al. Comprehensive genetic landscape of uveal melanoma by whole-genome sequencing. Am. J. Hum. Genet. 2016, 99, 1190–1198. [Google Scholar] [CrossRef] [PubMed]
- Robertson, A.G.; Shih, J.; Yau, C.; Gibb, E.A.; Oba, J.; Mungall, K.L.; Hess, J.M.; Uzunangelov, V.; Walter, V.; Danilova, L.; et al. Integrative analysis identifies four molecular and clinical subsets in uveal melanoma. Cancer Cell 2017, 32, 204–220.e15. [Google Scholar] [CrossRef] [PubMed]
- Schvartsman, G.; Taranto, P.; Glitza, I.C.; Agarwala, S.S.; Atkins, M.B.; Buzaid, A.C. Management of metastatic cutaneous melanoma: Updates in clinical practice. Ther. Adv. Med. Oncol. 2019, 11, 1758835919851663. [Google Scholar] [CrossRef] [PubMed]
- Lane, A.M.; Kim, I.K.; Gragoudas, E.S. Survival rates in patients after treatment for metastasis from uveal melanoma. JAMA Ophthalmol. 2018, 136, 981–986. [Google Scholar] [CrossRef]
- Nathan, P.; Hassel, J.C.; Rutkowski, P.; Baurain, J.-F.; Butler, M.O.; Schlaak, M.; Sullivan, R.J.; Ochsenreither, S.; Dummer, R.; Kirkwood, J.M.; et al. Overall Survival Benefit with Tebentafusp in Metastatic Uveal Melanoma. N. Engl. J. Med. 2021, 385, 1196–1206. [Google Scholar] [CrossRef]
- Kujala, E.; Makitie, T.; Kivela, T. Very long-term prognosis of patients with malignant uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 4651–4659. [Google Scholar] [CrossRef]
- Singh, A.D.; Turell, M.E.; Topham, A.K. Uveal melanoma: Trends in incidence, treatment, and survival. Ophthalmology 2011, 118, 1881–1885. [Google Scholar] [CrossRef]
- Yang, J.; Manson, D.K.; Marr, B.P.; Carvajal, R.D. Treatment of uveal melanoma: Where are we now? Ther. Adv. Med. Oncol. 2018, 10, 1758834018757175. [Google Scholar] [CrossRef]
- Edition, S.; Edge, S.; Byrd, D. AJCC Cancer Staging Manual, 8th ed.; Springer International Publishing: Cham, Switzerland, 2017. [Google Scholar]
- Collaborative Ocular Melanoma Study Group. Assessment of metastatic disease status at death in 435 patients with large choroidal melanoma in the Collaborative Ocular Melanoma Study (COMS): COMS report no. 15. Arch. Ophthalmol. 2001, 119, 670–676. [Google Scholar] [CrossRef]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Fulco, E.; Alarcon, C.; Shields, J.A. American Joint Committee on Cancer Classification of Posterior Uveal Melanoma (Tumor Size Category) Predicts Prognosis in 7731 Patients. Ophthalmology 2013, 120, 2066–2071. [Google Scholar] [CrossRef] [PubMed]
- Shields, C.L.; Kaliki, S.; Furuta, M.; Mashayekhi, A.; Shields, J.A. Clinical spectrum and prognosis of uveal melanoma based on age at presentation in 8033 cases. Retina 2012, 32, 1363–1372. [Google Scholar] [CrossRef] [PubMed]
- Damato, B.; Eleuteri, A.; Taktak, A.F.; Coupland, S.E. Estimating prognosis for survival after treatment of choroidal melanoma. Prog. Retin. Eye Res. 2011, 30, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Cunha Rola, A.; Taktak, A.; Eleuteri, A.; Kalirai, H.; Heimann, H.; Hussain, R.; Bonnett, L.J.; Hill, C.J.; Traynor, M.; Jager, M.J.; et al. Multicenter External Validation of the Liverpool Uveal Melanoma Prognosticator Online: An OOG Collaborative Study. Cancers 2020, 12, 477. [Google Scholar] [CrossRef] [PubMed]
- Vichitvejpaisal, P.; Dalvin, L.A.; Mazloumi, M.; Ewens, K.G.; Ganguly, A.; Shields, C.L. Genetic Analysis of Uveal Melanoma in 658 Patients Using the Cancer Genome Atlas Classification of Uveal Melanoma as A, B, C, and D. Ophthalmology 2019, 126, 1445–1453. [Google Scholar] [CrossRef]
- Mazloumi, M.; Vichitvejpaisal, P.; Dalvin, L.A.; Yaghy, A.; Ewens, K.G.; Ganguly, A.; Shields, C.L. Accuracy of The Cancer Genome Atlas Classification vs American Joint Committee on Cancer Classification for Prediction of Metastasis in Patients with Uveal Melanoma. JAMA Ophthalmol. 2020, 138, 260–267. [Google Scholar] [CrossRef]
- Sacco, J.J.; Nathan, P.D.; Danson, S.; Lorigan, P.; Nicholson, S.; Ottensmeier, C.; Corrie, P.; Steven, N.; Goodman, A.; Larkin, J.M.G.; et al. Sunitinib versus dacarbazine as first-line treatment in patients with metastatic uveal melanoma. J. Clin. Oncol. 2013, 31, 9031. [Google Scholar] [CrossRef]
- Carvajal, R.D.; Sosman, J.A.; Quevedo, J.F.; Milhem, M.M.; Joshua, A.M.; Kudchadkar, R.R.; Linette, G.P.; Gajewski, T.F.; Lutzky, J.; Lawson, D.H.; et al. Effect of selumetinib vs chemotherapy on progression-free survival in uveal melanoma: A randomized clinical trial. JAMA 2014, 311, 2397–2405. [Google Scholar] [CrossRef]
- Piperno-Neumann, S.; Kapiteijn, E.; Larkin, J.M.G.; Carvajal, R.D.; Luke, J.J.; Seifert, H.; Roozen, I.; Zoubir, M.; Yang, L.; Choudhury, S.; et al. Phase I dose-escalation study of the protein kinase C (PKC) inhibitor AEB071 in patients with metastatic uveal melanoma. J. Clin. Oncol. 2014, 32, 9030. [Google Scholar] [CrossRef]
- Shoushtari, A.N.; Kudchadkar, R.R.; Panageas, K.; Murthy, R.K.; Jung, M.; Shah, R.; O’Donnell, B.; Khawaja, T.T.; Shames, Y.; Prempeh-Keteku, N.A.; et al. A randomized phase 2 study of trametinib with or without GSK2141795 in patients with advanced uveal melanoma. J. Clin. Oncol. 2016, 34, 9511. [Google Scholar] [CrossRef]
- Algazi, A.P.; Tsai, K.K.; Shoushtari, A.N.; Munhoz, R.R.; Eroglu, Z.; Piulats, J.M.; Ott, P.A.; Johnson, D.B.; Hwang, J.; Daud, A.I.; et al. Clinical outcomes in metastatic uveal melanoma treated with PD-1 and PD-L1 antibodies. Cancer 2016, 122, 3344–3353. [Google Scholar] [CrossRef] [PubMed]
- Carvajal, R.D.; Schwartz, G.K.; Tezel, T.; Marr, B.; Francis, J.H.; Nathan, P.D. Metastatic disease from uveal melanoma: Treatment options and future prospects. Br. J. Ophthalmol. 2017, 101, 38–44. [Google Scholar] [CrossRef]
- Sandinha, T.; Farquharson, M.; McKay, I.; Roberts, F. Correlation of heterogeneity for chromosome 3 copy number with cell type in choroidal melanoma of mixed-cell type. Investig. Ophthalmol. Vis. Sci. 2006, 47, 5177–5180. [Google Scholar] [CrossRef] [PubMed]
- Dopierala, J.; Damato, B.E.; Lake, S.L.; Taktak, A.F.; Coupland, S.E. Genetic heterogeneity in uveal melanoma assessed by multiplex ligation-dependent probe amplification. Investig. Ophthalmol. Vis. Sci. 2010, 51, 4898–4905. [Google Scholar] [CrossRef] [PubMed]
- Chang, M.Y.; Rao, N.P.; Burgess, B.L.; Johnson, L.; McCannel, T.A. Heterogeneity of monosomy 3 in fine needle aspiration biopsy of choroidal melanoma. Mol. Vis. 2013, 19, 1892. [Google Scholar]
- Augsburger, J.J.; Correa, Z.M.; Augsburger, B.D. Frequency and implications of discordant gene expression profile class in posterior uveal melanomas sampled by fine needle aspiration biopsy. Am. J. Ophthalmol. 2015, 159, 248–256. [Google Scholar] [CrossRef]
- Miller, A.K.; Benage, M.J.; Wilson, D.J.; Skalet, A.H. Uveal melanoma with histopathologic intratumoral heterogeneity associated with gene expression profile discordance. Ocul. Oncol. Pathol. 2017, 3, 156–160. [Google Scholar] [CrossRef]
- Herwig-Carl, M.C.; Sharma, A.; Höller, T.; Holz, F.G.; Schlitter, A.M.; Loeffler, K.U. Spatial intratumor heterogeneity in uveal melanoma: Tumor cell subtypes with a presumed invasive potential exhibit a particular epigenetic staining reaction. Exp. Eye Res. 2019, 182, 175–181. [Google Scholar] [CrossRef]
- Durante, M.A.; Rodriguez, D.A.; Kurtenbach, S.; Kuznetsov, J.N.; Sanchez, M.I.; Decatur, C.L.; Snyder, H.; Feun, L.G.; Livingstone, A.S.; Harbour, J.W. Single-cell analysis reveals new evolutionary complexity in uveal melanoma. Nat. Commun. 2020, 11, 496. [Google Scholar] [CrossRef]
- Karlsson, J.; Nilsson, L.M.; Mitra, S.; Alsén, S.; Shelke, G.V.; Sah, V.R.; Forsberg, E.M.V.; Stierner, U.; All-Eriksson, C.; Einarsdottir, B.; et al. Molecular profiling of driver events in metastatic uveal melanoma. Nat. Commun. 2020, 11, 1894. [Google Scholar] [CrossRef]
- Pandiani, C.; Strub, T.; Nottet, N.; Cheli, Y.; Gambi, G.; Bille, K.; Husser, C.; Dalmasso, M.; Béranger, G.; Lassalle, S.; et al. Single-cell RNA sequencing reveals intratumoral heterogeneity in primary uveal melanomas and identifies HES6 as a driver of the metastatic disease. Cell Death Differ. 2021, 28, 1990–2000. [Google Scholar] [CrossRef] [PubMed]
- Rosa, R.; Green, R. Eye. In Surgical Pathology Dissection: An Illustrated Guide; Westra, W., Hubran, R., Phelps, T., Isacson, C., Eds.; Springer: New York, NY, USA, 2003; pp. 194–200. [Google Scholar]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The eighth edition AJCC cancer staging manual: Continuing to build a bridge from a population-based to a more “personalized” approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Conlin, L.K.; Thiel, B.D.; Bonnemann, C.G.; Medne, L.; Ernst, L.M.; Zackai, E.H.; Deardorff, M.A.; Krantz, I.D.; Hakonarson, H.; Spinner, N.B. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum. Mol. Genet. 2010, 19, 1263–1275. [Google Scholar] [CrossRef] [PubMed]
- Li, M.M.; Datto, M.; Duncavage, E.J.; Kulkarni, S.; Lindeman, N.I.; Roy, S.; Tsimberidou, A.M.; Vnencak-Jones, C.L.; Wolff, D.J.; Younes, A.; et al. Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer: A Joint Consensus Recommendation of the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists. J. Mol. Diagn. 2017, 19, 4–23. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Toomey, C.B.; Protopsaltis, N.J.; Phou, S.; Bakhoum, M.F.; Thorson, J.A.; Ediriwickrema, L.S.; Korn, B.S.; Kikkawa, D.O.; Goldbaum, M.H.; Lin, J.H. Prevalence of mismatch repair gene mutations in uveal melanoma. Retina 2020, 40, 2216–2220. [Google Scholar] [CrossRef]
- Johansson, P.; Aoude, L.G.; Wadt, K.; Glasson, W.J.; Warrier, S.K.; Hewitt, A.W.; Kiilgaard, J.F.; Heegaard, S.; Isaacs, T.; Franchina, M.; et al. Deep sequencing of uveal melanoma identifies a recurrent mutation in PLCB4. Oncotarget 2016, 7, 4624–4631. [Google Scholar] [CrossRef]
- de Lange, M.J.; van Pelt, S.I.; Versluis, M.; Jordanova, E.S.; Kroes, W.G.; Ruivenkamp, C.; van der Burg, S.H.; Luyten, G.P.; van Hall, T.; Jager, M.J.; et al. Heterogeneity revealed by integrated genomic analysis uncovers a molecular switch in malignant uveal melanoma. Oncotarget 2015, 6, 37824. [Google Scholar] [CrossRef]
- Shain, A.H.; Bagger, M.M.; Yu, R.; Chang, D.; Liu, S.; Vemula, S.; Weier, J.F.; Wadt, K.; Heegaard, S.; Bastian, B.C.; et al. The genetic evolution of metastatic uveal melanoma. Nat. Genet. 2019, 51, 1123–1130. [Google Scholar] [CrossRef]
- de Lange, M.J.; Nell, R.J.; van der Velden, P.A. Scientific and clinical implications of genetic and cellular heterogeneity in uveal melanoma. Mol. Biomed. 2021, 2, 25. [Google Scholar] [CrossRef]
- Ou, S.H.; Kwak, E.L.; Siwak-Tapp, C.; Dy, J.; Bergethon, K.; Clark, J.W.; Camidge, D.R.; Solomon, B.J.; Maki, R.G.; Bang, Y.J.; et al. Activity of crizotinib (PF02341066), a dual mesenchymal-epithelial transition (MET) and anaplastic lymphoma kinase (ALK) inhibitor, in a non-small cell lung cancer patient with de novo MET amplification. J. Thorac. Oncol. 2011, 6, 942–946. [Google Scholar] [CrossRef] [PubMed]
- Frampton, G.M.; Ali, S.M.; Rosenzweig, M.; Chmielecki, J.; Lu, X.; Bauer, T.M.; Akimov, M.; Bufill, J.A.; Lee, C.; Jentz, D.; et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015, 5, 850–859. [Google Scholar] [CrossRef] [PubMed]
- Paik, P.K.; Drilon, A.; Fan, P.-D.; Yu, H.; Rekhtman, N.; Ginsberg, M.S.; Borsu, L.; Schultz, N.; Berger, M.F.; Rudin, C.M.; et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015, 5, 842–849. [Google Scholar] [CrossRef]
- Awad, M.M.; Oxnard, G.R.; Jackman, D.M.; Savukoski, D.O.; Hall, D.; Shivdasani, P.; Heng, J.C.; Dahlberg, S.E.; Jänne, P.A.; Verma, S.; et al. MET exon 14 mutations in non-small-cell lung cancer are associated with advanced age and stage-dependent MET genomic amplification and c-Met overexpression. J. Clin. Oncol. 2016, 34, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Pilotto, S.; Gkountakos, A.; Carbognin, L.; Scarpa, A.; Tortora, G.; Bria, E. MET exon 14 juxtamembrane splicing mutations: Clinical and therapeutical perspectives for cancer therapy. Ann. Transl. Med. 2017, 5, 2. [Google Scholar] [CrossRef]
- Johansson, P.A.; Brooks, K.; Newell, F.; Palmer, J.M.; Wilmott, J.S.; Pritchard, A.L.; Broit, N.; Wood, S.; Carlino, M.S.; Leonard, C.; et al. Whole genome landscapes of uveal melanoma show an ultraviolet radiation signature in iris tumours. Nat. Commun. 2020, 11, 2408. [Google Scholar] [CrossRef]
- Hearle, N.; Damato, B.E.; Humphreys, J.; Wixey, J.; Green, H.; Stone, J.; Easton, D.F.; Houlston, R.S. Contribution of germline mutations in BRCA2, P16(INK4A), P14(ARF) and P15 to uveal melanoma. Investig. Ophthalmol. Vis. Sci. 2003, 44, 458–462. [Google Scholar] [CrossRef]
- Easton, D.F.; Steele, L.; Fields, P.; Ormiston, W.; Averill, D.; Daly, P.A.; McManus, R.; Neuhausen, S.L.; Ford, D.; Wooster, R.; et al. Cancer risks in two large breast cancer families linked to BRCA2 on chromosome 13q12-13. Am. J. Hum. Genet. 1997, 61, 120–128. [Google Scholar] [CrossRef]
- Houlston, R.S.; Damato, B.E. Genetic predisposition to ocular melanoma. Eye 1999, 13, 43–46. [Google Scholar] [CrossRef]

| Patient and Tumour Characteristics | n/years | |
|---|---|---|
| Age at enucleation (years) | Range | 40–82 |
| Average | 59.3 | |
| Sex | Male | 6 |
| Female | 1 | |
| Patient status | Alive | 5 |
| Deceased | 2 | |
| Affected eye | Left | 4 |
| Right | 3 | |
| Ciliary body involvement | Yes | 6 |
| No | 1 | |
| Tumour thickness | <12 mm | 6 |
| 12–15 mm | 1 | |
| Tumour basal diameter | <15 mm | 1 |
| 15–18 mm | 5 | |
| >18 mm | 1 | |
| Extraocular extension | Absent | 7 |
| Present | 0 | |
| pTNM stage | pT2 | 0 |
| pT3a | 0 | |
| pT3b | 5 | |
| pT4a | 1 | |
| pT4b | 1 | |
| Metastasis | Present | 3 |
| Absent | 4 |
| Case # | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Region | A | B | A | B | A | B | A | B | A | B | A | B | A | B |
| Tumour pigmentation | +++ | − | +++ | ++ | + | − | + | − | +++ | + | +++ | ++ | +++ | + |
| Tumour growth pattern | Solid | Solid | Solid and diffuse | Solid | Solid | Solid | Solid | Solid | Solid | Solid | Solid | Solid | Solid & diffuse | Solid |
| Cell type | Mixed | Mixed | Mixed | Mixed | Mixed | Mixed | Mixed | Mixed | Mixed | Mixed | Mostly spindled | Mostly spindled | Epithelioid | Epithelioid |
| Mitosis per 40 HPF | 21 | ND | 23 | ND | 23 | ND | 23 | ND | 12 | ND | 18 | ND | 19 | ND |
| Microvascular density 1 | Low | Low | High | Mod | Mod | Low | High | Mod | Low | Very low | High | High | Very high | High |
| Extravascular matrix pattern | Loops | LFN | Loops absent | Loops | LFN | LFN | LFN | LFN | LFN | Loops | LFN | LFN | Loops | LFN |
| Vascular lakes | Present | Present | Present | Present | Present | Present | Present | Present | Present | Absent | Present | Present | Absent | Present |
| Infiltrating lymphocytes | Low | Low | Low | Low | High | High | High | High | Low | High | Low | Low | High | Low |
| Infiltrating macrophages | CD68+ High CD163+ Very high | CD68+ Mod CD163+ Mod | CD68+ High CD163+ Low | CD68+ Low CD163+ Low | CD68+ Low CD163+ High | CD68+ Low CD163+ Mod | CD68+ High CD163+ High | CD68+ Very high CD163+ Very high | CD68+ Low CD163+ Low | CD68+ Low CD163+ Mod | CD68+ Mod CD163+ High | CD68+ Mod CD163+ High | CD68+ High CD163+ Very high | CD68+ Low CD163+ High |
| Case # | |||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| BAP1 IHC | Aberrant | Aberrant | Aberrant | Aberrant | Aberrant | Aberrant | Aberrant |
| Chromosome 3 | Monosomy 3 | Loss of 3p (1 copy) * | Monosomy 3 | Monosomy 3 | Monosomy 3 | Monosomy 3 | Loss of 3p (1 copy) * |
| Chromosome 8q | Gain (5 copies) | Disomy | Gain (3 copies) | Gain (4–5 copies) | Gain (3 copies) | Disomy | Gain (3 copies) |
| BAP1 sequence variants | Detected c.1766_1770 del | Not detected | Not detected | Detected c.518A>G | Detected c.172_179del | Not detected | Detected c.1153C>T |
| Case # | Gene and Variant (HGVS) | Region A VAF | Region B VAF | gnomAD AF (Allele Count [Homozygotes]) | AMP/ASCO/CAP Tier | Gene Implicated Previously in UM |
|---|---|---|---|---|---|---|
| 1 | NM_002072.4(GNAQ):c.626A>T (p.Gln209Leu) | 0.51 | 0.48 | Not present | Tier 2C | Yes [3,34,37] |
| NM_006015.5(ARID1A):c.4550C>G (p.Ala1517Gly) | 0.90 | 0.94 | Not present | Tier 2C | No | |
| NM_004656.4(BAP1):c.1766_1770del (p.Ile589Argfs*52) | 0.80 | 0.81 | Not present | Tier 2C | Yes [3,34,37] | |
| NM_001127500.2:MET exon 14 skipping transcript variant Δ | ND | Detected | NA | Tier 2C | No | |
| 2 | NM_002067.4(GNA11):c.626A>T (p.Gln209Leu) | 0.44 | 0.33 | Not present | Tier 2C | Yes [3,34,37] |
| NM_012433.3(SF3B1):c.1873C>T (p.Arg625Cys) | 0.43 | 0.26 | Not present | Tier 2C | Yes [3,34,37] | |
| NM_000249.3(MLH1):c.1752C>G (p.Asp584Glu) | 0.87 | 0.72 | Not present | Tier 3 | No | |
| 3 | NM_002067.4(GNA11):c.626A>T (p.Gln209Leu) | 0.48 | 0.40 | Not present | Tier 2C | Yes [3,34,37] |
| NM_016507.3(CDK12):c.1339T>A (p.Leu447Ile) | 0.51 | 0.51 | 0.0001315 (20[0]) | Tier 3 | Yes [3] | |
| NM_000059.3(BRCA2):c.3812C>T (p.Ser1271Leu) | 0.06 | ND | Not present | Tier 2C | Yes, [3] | |
| NM_000489.5(ATRX):c.4525G>A (p.Glu1509Lys) | 0.07 | ND | Not present | Tier 2C | Yes [37] | |
| NM_006218.3(PIK3CA):c.1714G>A (p.Val572Ile) | 0.06 | ND | Not present | Tier 3 | No | |
| NM_138712.3(PPARG):c.392C>T (p.Ser131Cys) | 0.05 | ND | Not present | Tier 3 | No | |
| 4 | NM_002072.4(GNAQ):c.626A>C (p.Gln209Pro) | 0.46 | 0.54 | Not present | Tier 2C | Yes [3,37] |
| NM_004656.4(BAP1):c.518A>G (p.Tyr173Cys) | 0.86 | 0.76 | Not present | Tier 2C | Yes [3,37] | |
| NM_000368.4(TSC1):c.727G>T (p.Asp243Tyr) | 0.48 | 0.47 | Not present | Tier 3 | No | |
| NM_000489.5(ATRX):c.6677G>A (p.Arg2226Lys) | ND | 0.06 | Not present | Tier 2C | Yes [37] | |
| 5 | NM_002067.4(GNA11):c.626A>T (p.Gln209Leu) | 0.33 | 0.39 | Not present | Tier 2C | Yes |
| NM_004656.4(BAP1):c.172_179del (p.Ser58Lysfs*8) | 0.41 | 0.78 | Not present | Tier 2C | Yes | |
| NM_017617.5(NOTCH1):c.2636G>A (p.Arg879Gln) | 0.44 | 0.52 | 0.0001051 (16[0]) | Tier 3 | Yes [37] | |
| NM_032444.3(SLX4):c.506C>T (p.Ser169Leu) | 0.36 | 0.36 | Not present | Tier 2C | Yes [34,37] | |
| NM_017617.5(NOTCH1):c.4129_4130insT (p.Pro1377Leufs*28) | ND | 0.26 | Not present | Tier 3 | Yes [37] | |
| NM_033084.4(FANCD2):c.2888C>T (p.Pro963Leu) | ND | 0.07 | Not present | Tier 3 | No | |
| NM_001184.3(ATR):c.4454G>A (p.Ser1485Asn) | ND | 0.06 | Not present | Tier 2C | No | |
| NM_000314.7(PTEN):c.511C>T (p.Gln171*) | ND | 0.05 | Not present | Tier 3 | No | |
| 6 | NM_002067.4(GNA11):c.626A>T (p.Gln209Leu) | 0.46 | 0.42 | Not present | Tier 2C | Yes [3,34,37] |
| NM_000059.3(BRCA2):c.2803G>A (p.Asp935Asn) | 0.48 | 0.50 | 0.0006506 (99[0]) | Tier 2C | Yes [3] | |
| 7 | NM_004656.4(BAP1):c.1153C>T (p.Arg385*) | 0.64 | 0.75 | Not present | Tier 2C | Yes [3,34,37] |
| NM_002067.4(GNA11):c.547C>T (p.Arg183Cys) | 0.41 | 0.44 | Not present | Tier 2C | Yes [3,34,37] | |
| NM_003073.4(SMARCB1):c.1130G>A (p.Arg377His) | 0.36 | 0.44 | 0.000006568 (1[0]) | Tier 3 | No | |
| NM_000135.4(FANCA):c.480G>A (p.Met160Ile) | 0.46 | 0.42 | 0.0002891 (44[0]) | Tier 2C | Yes [3,34,37] | |
| NM_001184.3(ATR):c.7792C>T (p.Arg2598*) | 0.28 | 0.13 | Not present | Tier 2C | No |
| Clinical Trials (Phase) | Tumour Type | Gene(s) |
|---|---|---|
| NCT03947385 (Phase I/II) | UM | GNAQ, GNA11 |
| NCT02693535 (Phase II) | Advanced solid tumours | MET exon 14 skipping transcript * |
| NCT03207347 (Phase I/II) | UM | ARID1A, ATR *, BAP1, PTEN *, SLX4 |
| NCT03767075 (Phase II) | Advanced solid tumours | DNA repair genes (ATRX *, ATR *, BAP1, BRCA2 *, SLX4, ARID1A) |
| NCT01971515 (Phase I) | Advanced malignancies | PIK3CA *, PTEN *, TSC1 |
| NCT02961283 (Phase I) | Advanced solid tumours | PIK3CA *, PTEN * |
| NCT04171700 (Phase II) | Advanced solid tumours | BRCA2 *, FANCA |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toumi, E.; Hesson, L.B.; Lin, V.; Wright, D.; Hajdu, E.; Lim, L.-A.S.; Giblin, M.; Zhou, F.; Hoffmeister, A.; Zabih, F.; et al. Microdissection of Distinct Morphological Regions Within Uveal Melanomas Identifies Novel Drug Targets. Cancers 2024, 16, 4152. https://doi.org/10.3390/cancers16244152
Toumi E, Hesson LB, Lin V, Wright D, Hajdu E, Lim L-AS, Giblin M, Zhou F, Hoffmeister A, Zabih F, et al. Microdissection of Distinct Morphological Regions Within Uveal Melanomas Identifies Novel Drug Targets. Cancers. 2024; 16(24):4152. https://doi.org/10.3390/cancers16244152
Chicago/Turabian StyleToumi, Elsa, Luke B. Hesson, Vivian Lin, Dale Wright, Elektra Hajdu, Li-Anne S. Lim, Michael Giblin, Fanfan Zhou, Alexandra Hoffmeister, Farida Zabih, and et al. 2024. "Microdissection of Distinct Morphological Regions Within Uveal Melanomas Identifies Novel Drug Targets" Cancers 16, no. 24: 4152. https://doi.org/10.3390/cancers16244152
APA StyleToumi, E., Hesson, L. B., Lin, V., Wright, D., Hajdu, E., Lim, L.-A. S., Giblin, M., Zhou, F., Hoffmeister, A., Zabih, F., Fung, A. T., Conway, R. M., & Cherepanoff, S. (2024). Microdissection of Distinct Morphological Regions Within Uveal Melanomas Identifies Novel Drug Targets. Cancers, 16(24), 4152. https://doi.org/10.3390/cancers16244152







