Immuno-Transcriptomic Profiling of Blood and Tumor Tissue Identifies Gene Signatures Associated with Immunotherapy Response in Metastatic Bladder Cancer
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Mouse Model of Bladder Cancer
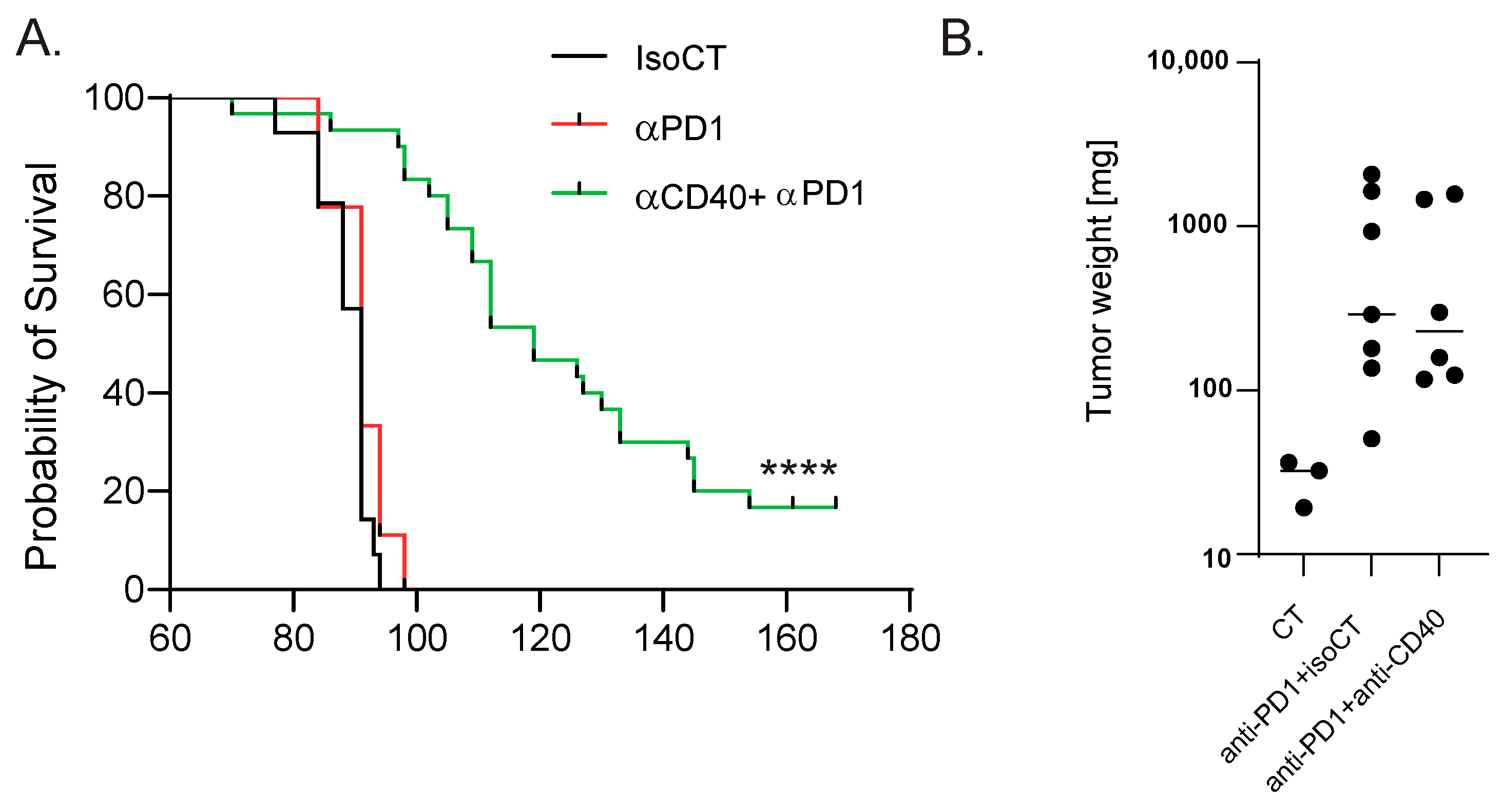
2.2. Therapeutic Schedule and Treatment Groups
2.3. RNA Sequencing and Bioinformatic Analysis
2.4. Data Processing and Quality Check
2.5. Data Transformation and Exploratory Analysis
2.6. Differential Expression Analysis (DEA)
2.7. Modeling
2.8. Single Cell Preparation, FACS Staining and Analysis
3. Results
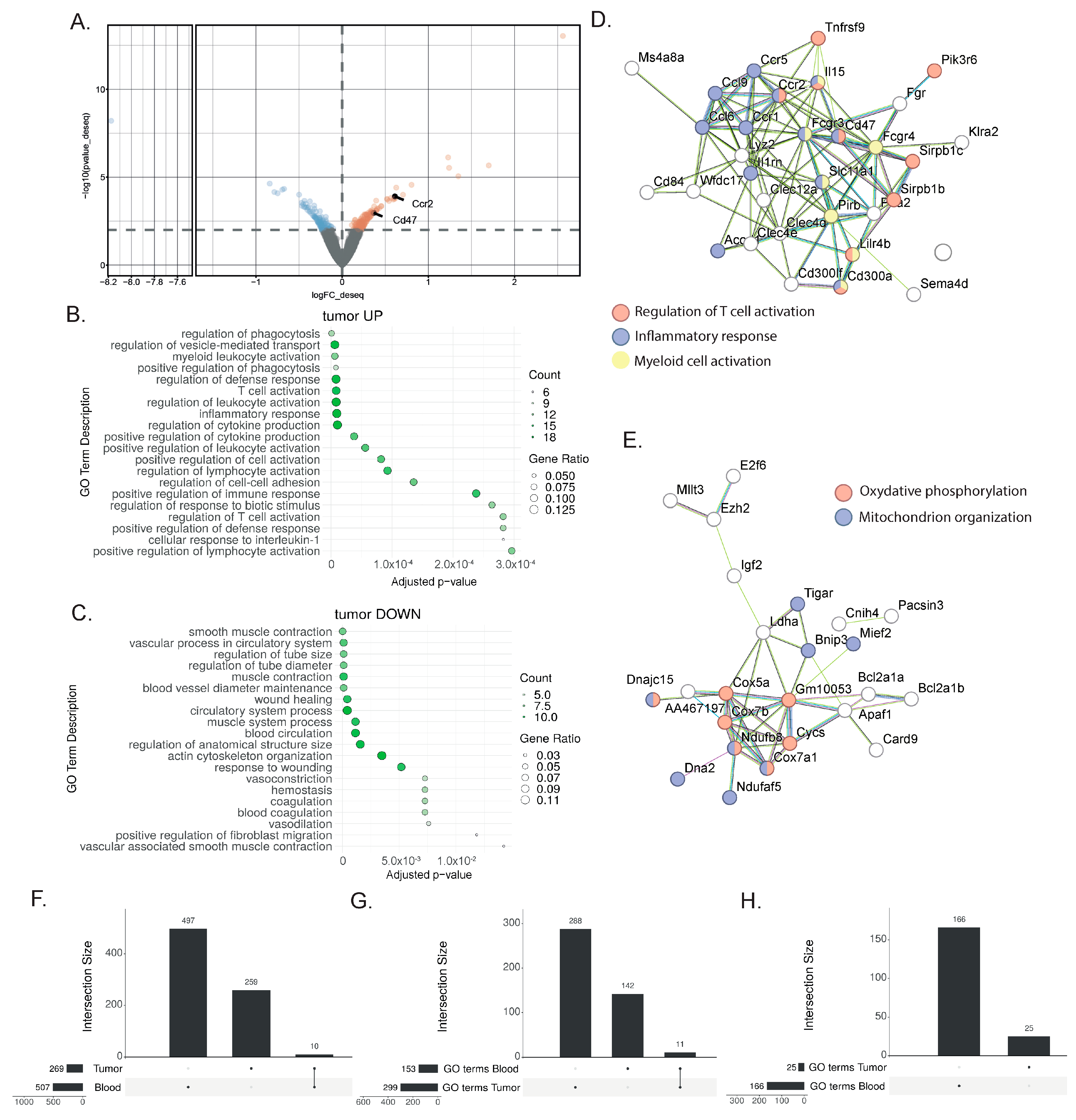
3.1. Identification of Biomarkers Correlating with Immunotherapy Response in Bladder Cancer
3.2. Tumor Analysis Outlined the Complexity of the TME Response to Immunotherapy
3.3. Shared Transcriptional Response to ICIs between Blood and TME
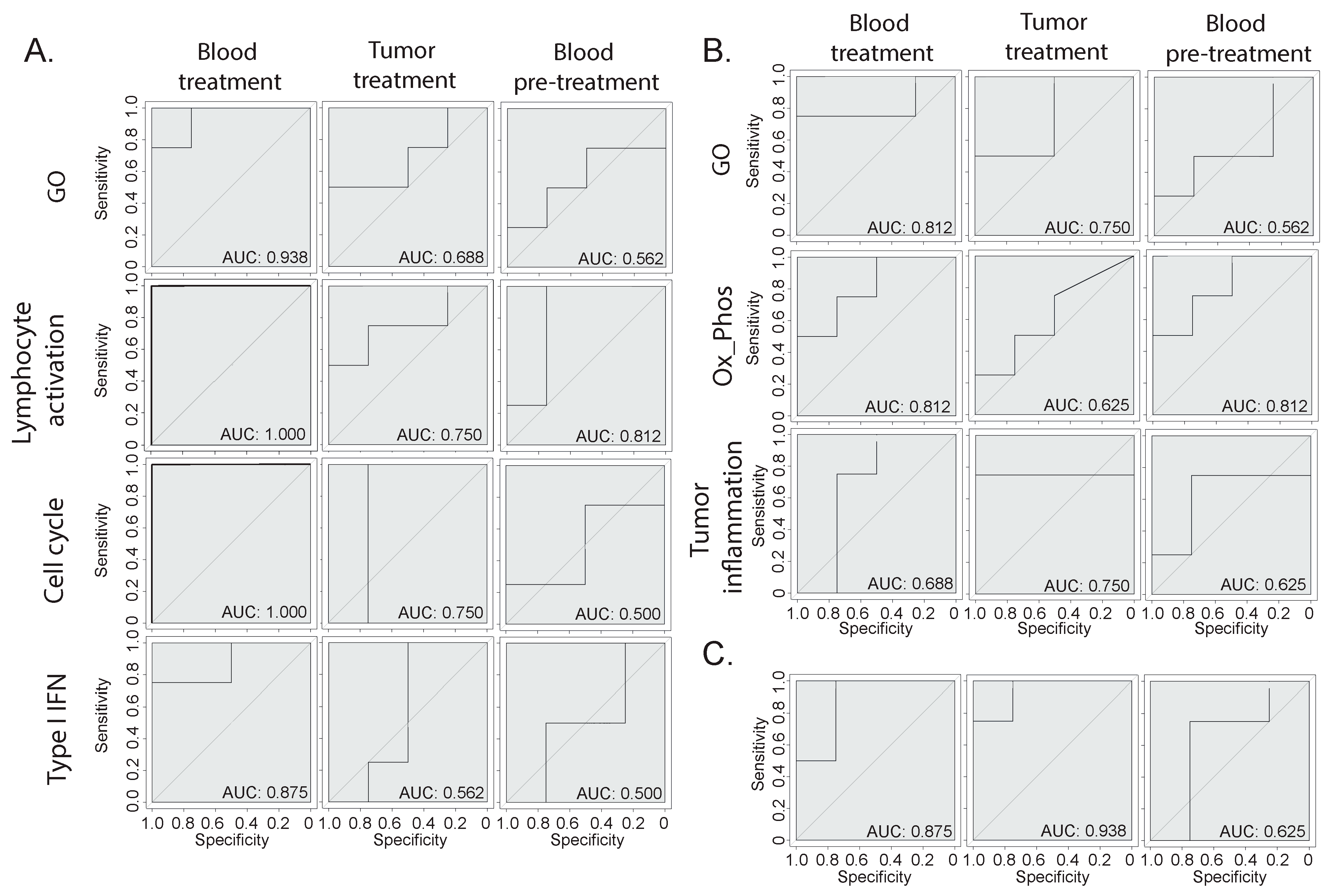
3.4. Prediction of Immunotherapy Response Using Blood or Bladder Signature
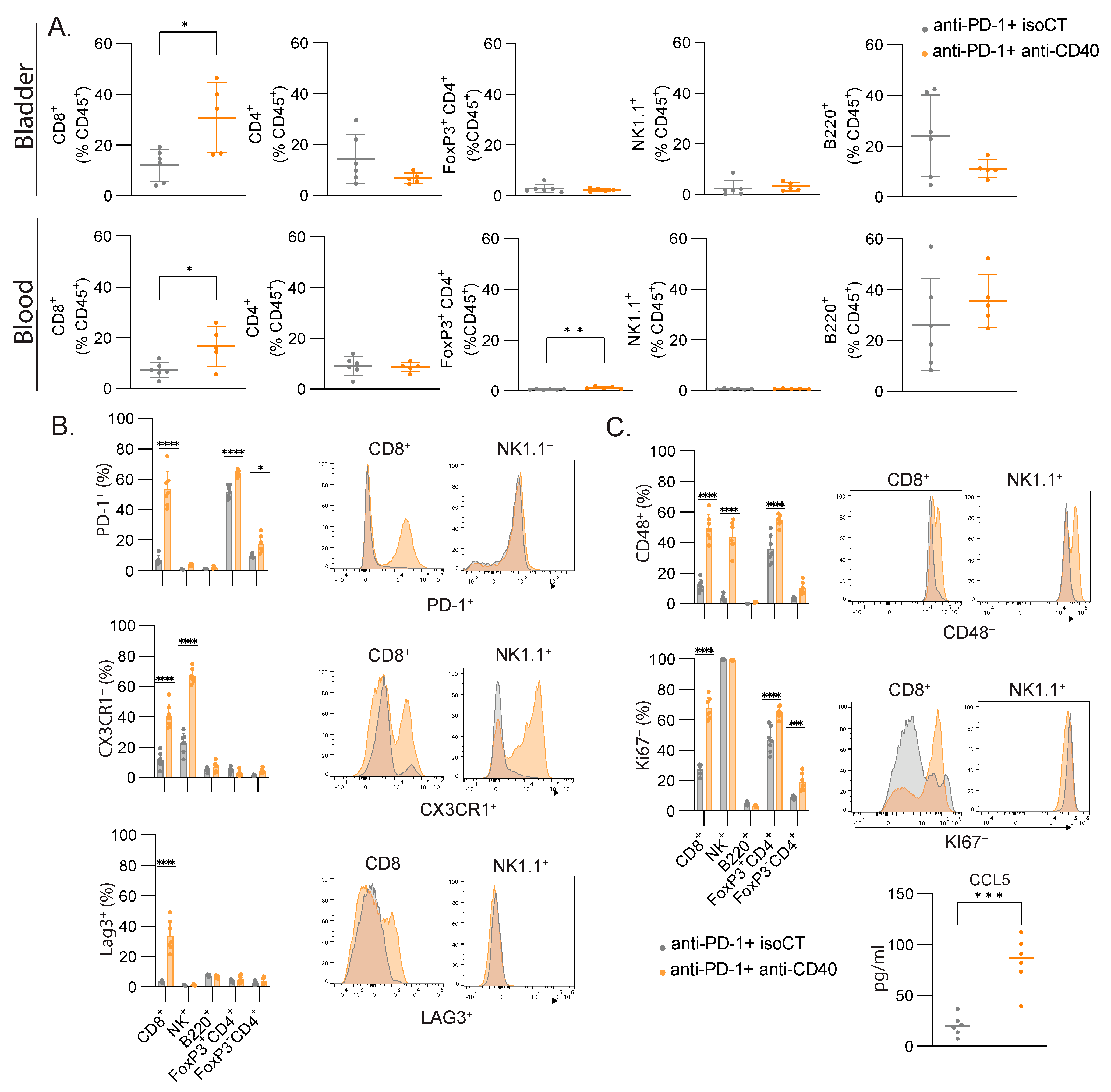
3.5. CD8 T Cells Are the Main Cell Type Showing Increased Proliferation and Expression of Lymphocyte Activation Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ribas, A.; Wolchok, J.D. Cancer immunotherapy using checkpoint blockade. Science 2018, 359, 1350–1355. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Zhang, S.; Deng, Y.; Wang, P.; Hou, Q.; Xu, H. Prognostic Factors for Checkpoint Inhibitor Based Immunotherapy: An Update with New Evidences. Front. Pharmacol. 2018, 9, 1050. [Google Scholar] [CrossRef] [PubMed]
- Hirsch, L.; Zitvogel, L.; Eggermont, A.; Marabelle, A. PD-Loma: A cancer entity with a shared sensitivity to the PD-1/PD-L1 pathway blockade. Br. J. Cancer 2019, 120, 3–5. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; Chen, D.S. Immune escape to PD-L1/PD-1 blockade: Seven steps to success (or failure). Ann. Oncol. 2016, 27, 1492–1504. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Beltran, A.; Cimadamore, A.; Blanca, A.; Massari, F.; Vau, N.; Scarpelli, M.; Cheng, L.; Montironi, R. Immune Checkpoint Inhibitors for the Treatment of Bladder Cancer. Cancers 2021, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Leblond, M.M.; Zdimerova, H.; Desponds, E.; Verdeil, G. Tumor-Associated Macrophages in Bladder Cancer: Biological Role, Impact on Therapeutic Response and Perspectives for Immunotherapy. Cancers 2021, 13, 4712. [Google Scholar] [CrossRef] [PubMed]
- Kamoun, A.; de Reynies, A.; Allory, Y.; Sjodahl, G.; Robertson, A.G.; Seiler, R.; Hoadley, K.A.; Groeneveld, C.S.; Al-Ahmadie, H.; Choi, W.; et al. A Consensus Molecular Classification of Muscle-invasive Bladder Cancer. Eur. Urol. 2020, 77, 420–433. [Google Scholar] [CrossRef] [PubMed]
- Hiam-Galvez, K.J.; Allen, B.M.; Spitzer, M.H. Systemic immunity in cancer. Nat. Rev. Cancer 2021, 21, 345–359. [Google Scholar] [CrossRef] [PubMed]
- van Wilpe, S.; Wosika, V.; Ciarloni, L.; Hosseinian Ehrensberger, S.; Jeitziner, R.; Angelino, P.; Duiveman-de Boer, T.; Koornstra, R.H.T.; de Vries, I.J.M.; Gerritsen, W.R.; et al. Whole Blood Transcriptome Profiling Identifies DNA Replication and Cell Cycle Regulation as Early Marker of Response to Anti-PD-1 in Patients with Urothelial Cancer. Cancers 2021, 13, 4660. [Google Scholar] [CrossRef] [PubMed]
- Ciarloni, L.; Hosseinian, S.; Monnier-Benoit, S.; Imaizumi, N.; Dorta, G.; Ruegg, C.; Group, D.-C.-S. Discovery of a 29-gene panel in peripheral blood mononuclear cells for the detection of colorectal cancer and adenomas using high throughput real-time PCR. PLoS ONE 2015, 10, e0123904. [Google Scholar] [CrossRef] [PubMed]
- Ciarloni, L.; Ehrensberger, S.H.; Imaizumi, N.; Monnier-Benoit, S.; Nichita, C.; Myung, S.J.; Kim, J.S.; Song, S.Y.; Kim, T.I.; van der Weg, B.; et al. Development and Clinical Validation of a Blood Test Based on 29-Gene Expression for Early Detection of Colorectal Cancer. Clin. Cancer Res. 2016, 22, 4604–4611. [Google Scholar] [CrossRef] [PubMed]
- Leblond, M.M.; Tille, L.; Nassiri, S.; Gilfillan, C.B.; Imbratta, C.; Schmittnaegel, M.; Ries, C.H.; Speiser, D.E.; Verdeil, G. CD40 Agonist Restores the Antitumor Efficacy of Anti-PD1 Therapy in Muscle-Invasive Bladder Cancer in an IFN I/II-Mediated Manner. Cancer Immunol. Res. 2020, 8, 1180–1192. [Google Scholar] [CrossRef] [PubMed]
- Puzio-Kuter, A.M.; Castillo-Martin, M.; Kinkade, C.W.; Wang, X.; Shen, T.H.; Matos, T.; Shen, M.M.; Cordon-Cardo, C.; Abate-Shen, C. Inactivation of p53 and Pten promotes invasive bladder cancer. Genes. Dev. 2009, 23, 675–680. [Google Scholar] [CrossRef] [PubMed]
- Seitz, R.S.; Hurwitz, M.E.; Nielsen, T.J.; Bailey, D.B.; Varga, M.G.; Ring, B.Z.; Metts, C.F.; Schweitzer, B.L.; McGregor, K.; Ross, D.T. Translation of the 27-gene immuno-oncology test (IO score) to predict outcomes in immune checkpoint inhibitor treated metastatic urothelial cancer patients. J. Transl. Med. 2022, 20, 370. [Google Scholar] [CrossRef] [PubMed]
- Vanguri, R.S.; Smithy, J.W.; Li, Y.; Zhuang, M.; Maher, C.A.; Aleynick, N.; Peng, X.; Al-Ahmadie, H.; Funt, S.A.; Rosenberg, J.E.; et al. Integration of peripheral blood- and tissue-based biomarkers of response to immune checkpoint blockade in urothelial carcinoma. J. Pathol. 2023, 261, 349–360. [Google Scholar] [CrossRef] [PubMed]
- Deng, H.; Deng, D.; Qi, T.; Liu, Z.; Wu, L.; Yuan, J. An IFN-gamma-related signature predicts prognosis and immunotherapy response in bladder cancer: Results from real-world cohorts. Front. Genet. 2022, 13, 1100317. [Google Scholar] [CrossRef] [PubMed]
- de Visser, K.E.; Joyce, J.A. The evolving tumor microenvironment: From cancer initiation to metastatic outgrowth. Cancer Cell 2023, 41, 374–403. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Desponds, E.; Croci, D.; Wosika, V.; Hadadi, N.; Fonseca Costa, S.S.; Ciarloni, L.; Ongaro, M.; Zdimerova, H.; Leblond, M.M.; Hosseinian Ehrensberger, S.; et al. Immuno-Transcriptomic Profiling of Blood and Tumor Tissue Identifies Gene Signatures Associated with Immunotherapy Response in Metastatic Bladder Cancer. Cancers 2024, 16, 433. https://doi.org/10.3390/cancers16020433
Desponds E, Croci D, Wosika V, Hadadi N, Fonseca Costa SS, Ciarloni L, Ongaro M, Zdimerova H, Leblond MM, Hosseinian Ehrensberger S, et al. Immuno-Transcriptomic Profiling of Blood and Tumor Tissue Identifies Gene Signatures Associated with Immunotherapy Response in Metastatic Bladder Cancer. Cancers. 2024; 16(2):433. https://doi.org/10.3390/cancers16020433
Chicago/Turabian StyleDesponds, Emma, Davide Croci, Victoria Wosika, Noushin Hadadi, Sara S. Fonseca Costa, Laura Ciarloni, Marco Ongaro, Hana Zdimerova, Marine M. Leblond, Sahar Hosseinian Ehrensberger, and et al. 2024. "Immuno-Transcriptomic Profiling of Blood and Tumor Tissue Identifies Gene Signatures Associated with Immunotherapy Response in Metastatic Bladder Cancer" Cancers 16, no. 2: 433. https://doi.org/10.3390/cancers16020433
APA StyleDesponds, E., Croci, D., Wosika, V., Hadadi, N., Fonseca Costa, S. S., Ciarloni, L., Ongaro, M., Zdimerova, H., Leblond, M. M., Hosseinian Ehrensberger, S., Romero, P., & Verdeil, G. (2024). Immuno-Transcriptomic Profiling of Blood and Tumor Tissue Identifies Gene Signatures Associated with Immunotherapy Response in Metastatic Bladder Cancer. Cancers, 16(2), 433. https://doi.org/10.3390/cancers16020433





