Simple Summary
Within the rapidly evolving landscape of Machine Learning in the medical field, this paper focuses on the forefront advancements in neuro-oncological radiology. More specifically, it aims to provide the reader with an in-depth exploration of the latest advancements in employing Deep Learning methodologies for the classification of brain tumor radiological images. This review meticulously scrutinizes papers published from 2018 to 2023, unveiling ongoing topics of research while underscoring the main remaining challenges and potential avenues for future research identified by those studies. Beyond the review itself, the paper also underscores the importance of placing the image data modelling provided by Deep Learning techniques within the framework of analytical pipeline research. This means that data quality control and pre-processing should be correctly coupled with modelling itself, in a way that emphasizes the importance of responsible data utilization, as well as the critical need for transparency in data disclosure to ensure trustworthiness and reproducibility of findings.
Abstract
Machine Learning is entering a phase of maturity, but its medical applications still lag behind in terms of practical use. The field of oncological radiology (and neuro-oncology in particular) is at the forefront of these developments, now boosted by the success of Deep-Learning methods for the analysis of medical images. This paper reviews in detail some of the most recent advances in the use of Deep Learning in this field, from the broader topic of the development of Machine-Learning-based analytical pipelines to specific instantiations of the use of Deep Learning in neuro-oncology; the latter including its use in the groundbreaking field of ultra-low field magnetic resonance imaging.
1. Introduction
Although Machine Learning (ML) is entering a phase of maturity, its applications in the medical domain at the point of care are still few and tentative at best. This paradoxical contradiction has been explained according to several different factors. One of them is the lack of experimental reproducibility, a requirement in which ML models in health have been reported to fare badly in comparison to other application areas [1]. One main reason to explain this is the mismatch between a data-centered (and often data-hungry) approach and the scarcity of publicly available and properly curated medical databases, combined with a nascent but insufficient data culture at the clinical level [2]. Another factor has to do with regulatory issues of ML (and Artificial Intelligence in general) in terms of both lack of maturity and geographical heterogeneity [3]. Further elements hampering ML-based tools adoption include data leakage, dataset shift, required model recalibrations, analytical pipeline maintenance failures, or changing medical practice patterns, to name a few [4].
The field of oncological radiology (and neuro-oncology in particular) is arguably at the forefront of the practical use of ML in medicine [5], now boosted by the success of Deep-Learning (DL) methods for the analysis of medical images [6,7]. Unfortunately, though, imaging does not escape the challenges and limitations summarized in the previous paragraph. Central to them, what has been called the “long-tail effect” [8]: pathologies for which only small and scattered datasets exist due to the scarcity of clinical data management strategies (technically complex and expensive) at levels beyond the local (regional, national, international). Associated with this, we must account for the difficulty of achieving standardized labeling (annotation) of imaging databases. An example of how to deal effectively with these problems is Federated Learning, which was used in [9] to gather data from 71 sites from 6 continents, analyzed using ML to address a problem of tumor boundary detection for glioblastoma brain tumors. Please note that the resulting database includes 6314 cases, which is impressive for this medical domain but still modest from an ML perspective. The success of ML in oncological radiology, as summarily stated in [10], will depend on its ability to create value in the delivery of medical care in terms of “increased diagnostic certainty, decreased time on task for radiologists, faster availability of results, and reduced costs of care with better outcomes for patients”.
This paper surveys some of the most recent advances in the use of ML for the analysis of magnetic resonance imaging (MRI) data in neuro-oncology without trying to make an all-encompassing review out of it. Instead, we focus on the most rapidly developing area, which involves the use of methods from the DL family. The variety of approaches sprouting from this family of methods has shaken the standards of data pre-processing or feature engineering before modeling as such. For this reason, we proceed to address the review hierarchically, starting in Section 3 with the broader topic of the development of ML-based analytical pipelines, which addresses the data analysis process beyond specific models and in which we will provide examples from two promising feature engineering approaches, namely source extraction in the form of independent component analysis (ICA) and nonnegative matrix factorization (NMF), and radiomics. The review of DL methods for image data analysis as such is delivered in Section 4. As an addition to this section, we will discuss the potential uses of DL in the groundbreaking field of ultra-low field (ULF) MRI [11]. Before all this, the following section will provide some contextual basic definitions of neuro-oncology concepts and a description of the main challenges and open issues concerning the use of ML in this domain.
2. Open Problems in AI Applied to MRI Analysis
The open problems for the use of ML-based analytical processes in the field of MRI in neuro-oncology can be seen from different perspectives. The first one is the analytical problem itself, according to which the main division is into categorization and segmentation problems. The latter is commented on later in this section.
Categorization can, in turn, be split into diagnosis and prognosis. In diagnosis, the correlation between neuroimaging classifications and histopathological diagnoses was assessed in [12] based on the 2000 version of the WHO classification of brain tumors and in [13] based on the 2007 version. In both studies, the main finding was that the sensitivity was variable among classes, whereas specificity was in the range of 0.85–1. The most difficult categories to diagnose were the glioma subtypes. The study based on the 2000 classification [12] reported a sensitivity of 0.14 for low-grade astrocytoma and 0.15 for low-grade oligodendroglioma. In the study based on the 2007 classification [13], increased sensitivity for low-grade astrocytoma (0.56) was found, but sensitivity was still low for other low-grade gliomas (LGG) such as oligodendroglioma (0.26), or for anaplastic gliomas (astrocytoma, 0.17 or ependymoma, 0.00), and other classes in the long-tail such as meningiomas of grade II and III in aggregate (0.17), or subependymomas and choroid plexus papilloma (0.33 for both). The recently released 2021 WHO classification [14], which incorporates the genetic alterations, opens the door to the reevaluation of these baseline results to accurately estimate the added value of any clinical decision support system (CDSS) based on ML or radiogenomics, over the limits of radiological interpretation of imaging findings. It is reasonable to foresee that the problematic tumor categories will remain so, or even more challenging, given the enhanced stratification of the glial category (e.g., different mutations of IDH1/2, ATRX, TP53, BRAF, H3F3A, CDKN2A/B, TERT and MGMT promoters, EGFR amplification, GFAP, 1p/19q codeletion, etc.).
On the other hand, regarding follow-up, there is no standard of care for recurrent high-grade gliomas, and the currently accepted criteria to assess response are those established by the Response Assessment in Neuro-Oncology Working Group (RANO) [15], which deals with the pseudoprogression phenomenon, defined as the appearance of contrast-enhancing lesions during the first 12 weeks after the end of the concomitant treatment or when the lesion developed within the first 3–6 months after radiation therapy, if it is in the radiation field (inside the 80% isodose line), and especially if it presents as a pattern of enhancement related to radiation-induced necrosis enhancement [16]. Also, with pseudoresponse in those patients treated with antiangiogenics in countries where these are approved [17,18]. Antiangiogenic agents, like bevacizumab, are designed to block the VEGF effect. The mechanism of action may be related to decreased blood supply to the tumor and normalization of tumor vessels, which display increased permeability. These agents are associated with high radiologic responses if we evaluate only the contrast enhancement. The recently published RANO 2.0 criteria [19] refine the former RANO, distinguishing between high-grade and low-grade gliomas. RANO 2.0 also takes into account the IDH status to decide whether the surrounding non-enhancing region should be taken into account or not. In this sense, ML-based pipelines should ideally be designed to allow the evaluation of their added value with respect to medical guidelines for clinical decision-making.
Another viewpoint to approach the open problems in the field has to do with the fact that ML-based analysis is strongly dependent on data pre-processing and the post-processing of results.
A fundamental prerequisite for the successful application of DL models in brain tumor classification is the pre-processing of the MRI data. The key pre-processing steps for the harmonization of MRI data are as follows.
- Resampling: MRI scans can exhibit variations in resolution and voxel sizes depending on the acquisition system. Resampling standardizes the resolution across the MRI images to ensure uniform dimensions.
- Co-registration: entails the alignment of MRI scans with a standardized anatomical template with the purpose of situating different scans within the same anatomical coordinate system.
- Skull-stripping: The main objective of the skull-stripping step is to efficiently isolate the cerebral region of interest from non-cerebral tissues, which enables DL models to focus exclusively on those brain tissues.
- Bias Field Correction: aims to rectify intensity inhomogeneities that are pervasive in MRI scans to guarantee uniformity in intensity values. The technique of choice for bias field correction is N4ITK (N4 Bias Field Correction) [20], which is an improved variant of the N3 (non-parametric nonuniform normalization) retrospective bias correction algorithm [21].
- Normalization: a technique adopted to rescale intensity values of MRI scans to a numeric range, rendering them consistent across the dataset. This process mitigates scale-related disparities. Two prominent approaches commonly applied to MRI data as input for DL models are min-max normalization and z-score normalization. Min-max achieves its goal by rescaling intensity values within MRI scans, spanning their range between 0 and 1. In contrast, z-score, often referred to as standardization, transforms the distribution of intensity values by centering it around a zero mean and standard deviation of value 1.
- Tumor identification: A critical and optional pre-processing step before the classification task involves identifying the tumor region of interest (ROI) through segmentation or by defining a bounding box that encompasses the tumor. Popular DL architectures, such as UNet [22], Faster-RCNN [23], and Mask-RCNN [24] are often employed to perform such segmentation or detection tasks.
The post-processing of results must often address the fact that the DL family of methods is, by their nature, an extreme case of black-box approach, a characteristic that may strongly hamper their medical applicability [25]. This limitation can be addressed using explainability and interpretability strategies; for further details on these, the reader is referred to [25].
3. Ml-Based Analytical Pipelines and Their Use in Neuro-Oncology
Ultimately, the whole point of using ML methods for data-based problems in the area of neuro-oncology is to provide radiologists with evidence-based medical tools at the point of care that can assist them in decision-making processes, especially with ambiguous or borderline cases. This is why it makes sense to embed these methods in Clinical Decision Support Systems (CDSS). A thorough and systematic review of intelligent systems-based CDSS for brain tumor analysis based on magnetic resonance data (spectra or images) is presented in this same Special Issue of Cancers [26]. It reports their increasing use over the last decade, addressing problems that include general ones such as tumor detection, type classification, and grading, but also more specific ones such as physicians’ alerting of treatment change plans.
At the core of ML-based CDSS, we need not just ML methods, models, and techniques but, more formally, ML pipelines. An ML pipeline goes beyond the use of a collection of methods to encompass all stages of the data mining process, including data pre-processing (data cleaning, data transformations potentially including feature selection and extraction, but also other aspects of data curation such as data extraction and standardization, missing data imputation and data clinical validation [27]) and models’ post-processing, potentially including evaluation, implementation and the definition of interpretability and explainability processes [25]. Pipelines can also accommodate specific needs, such as those related to the analysis of “big data”, with their corresponding challenges of standardization and scalability. As described in [28], in a clinical oncology setting, this may require a research infrastructure for federated ML based on the findable, accessible, interoperable, and reusable (FAIR) principles. Alternatively, we can aspire to automate the ML pipeline definition using Automated ML (AutoML) principles, as in [29], where Su and co-workers used a Tree-based Pipeline Optimization Tool (TPOT) in the process of selecting radiomics features predictive of mutations associated with midline gliomas.
An example of an ML pipeline for the specific problem of differentiation of glioblastomas from single brain metastases based on MR spectroscopy (MRS) data can be found in [30]. In this same issue of Cancers, Pitarch and co-workers [31] describe an ML pipeline for glioma grading from MRI data with a focus on the trustworthiness of the predictions generated by the ML models. This entails robustly quantifying the uncertainty of the models regarding their predictions, as well as implementing procedures to avoid data leakage, thus avoiding the risk of unreliable conclusions. All of these can be seen as part of a quest to avoid the pitfalls of implementation of ML-based CDSS that result in the problems of limited reproducibility of analytical results in clinical practice that have been reported in recent studies [1].
As previously explained, the first stages of an ML pipeline, prior to the data modeling itself, involve data pre-processing, and this task may, in turn, involve many sub-problems. As an example of the potential diversity and complexity of this landscape, we comment here on a few recently selected contributions to the problem of feature engineering and extraction following just two particular and completely different approaches: statistical image feature engineering using radiomics and source extraction using ICA- and NMF-based methods.
Radiomics is an image transformation approach that aims to extract either hard-coded statistical or textural features based on expert domain knowledge or feature representations learned from data, often using DL methods. The former may include first-order statistics, size and shape-based features, image intensity histogram descriptors, image textural information, etc. The use of this method for the pre-processing of brain tumor images prior to the use of ML has been recently and exhaustively reviewed in [32]. From that review, it is clear that the predominant problem under analysis is diagnosis, with only a limited number of studies addressing prognosis, survival, and progression. The types of brain tumors under investigation are dominated by the most frequent classes. In particular, glioblastoma, either on its own or combined with metastasis as a super-class of aggressive tumors, is the subject of many studies, with some others also including other frequent super-classes such as low-grade glioma or meningioma, while minority tumor types and grades are only considered in a limited number of studies. Importantly, and related to our previous comments concerning scarce data availability, most of the studies reported in [32] work with very small sample sizes, often not reaching the barrier of 100 cases. The challenge posed by data scarcity is compounded by the fact that most of the studies extract Radiomic features in the hundreds if not the thousands. This means that the ratio of cases-to-features is extremely low, making the use of conventional ML classifiers very difficult. To alleviate this problem, most of the reviewed papers resort to different strategies for qualitative and quantitative feature selection. Image modalities under analysis are dominated by T1, T2, and FLAIR, with few exceptions (PET, or Diffusion- and Perfusion-Weighted Imaging). Most studies are shown to resort to the Area Under the ROC Curve (AUC) as a performance metric, which is a safe choice, as it is far more robust than plain accuracy for small and class-imbalanced datasets.
The use of radiomics as a data transformation strategy in pre-processing is facilitated by the existence of off-the-shelf software such as the open-source PyRadiomics package [33].
Source extraction methods have a very different analytical rationale for data dimensionality reduction as a pre-processing step. They do not achieve it through plain feature transformation, as in radiomics. Instead, they aim to find the underlying and unobserved sources of observed radiological data. In doing so, they achieve dimensionality reduction as a byproduct of a process that may provide insight into the generation of the images themselves.
The ICA technique [34] has a long history in medical applications, most notoriously for the analysis of electroencephalographic signals. Source extraction is natural in this context as a tool for spatially locating sources of the EEG from electric potentials measured in the skull surface. In ICA, we assume that the observed data can be expressed as a linear combination of sources that are estimated to be statistically independent or as independent as possible. This technique has mostly been applied to brain tumor segmentation, but some alternative recent studies have extended its possibilities to dynamic settings, such as that in [35], where dynamic contrast-enhanced MRI is analyzed using temporal ICA (tICA), and in [36], where probabilistic ICA is used for the analysis of dynamic susceptibility contrast (DSC) perfusion MRI.
The NMF technique [37], on the other hand, was originally devised for the extraction of sources from images and assumes data non-negativity but does not assume statistical independence. Data are still approximated by linear combinations of factors. Although NMF and variants of this family of methods have extensively been used for the pre-processing and analysis of MRS and MRS imaging (MRSI) signal [38,39], they have only scarcely been used for the pre-processing of MRI. Some outstanding exceptions include the work in [40] with hierarchical NMF for multi-parametric MRI and the recent proposal of a whole new architecture based on NMF called Factorizer [41], constructed by replacing the self-attention layer of a Vision Transformer (ViT, [42]) block with NMF-based modules.
The technical details of ICA and NMF and their manifold variants are beyond the scope of this review and can be found elsewhere in the literature.
4. Deep Learning in Neuro-Oncology Data Analysis: A Review
In this section, we review existing recent literature to gather evidence about the advantages, challenges, and potential future directions in the use of DL techniques for supervised problems in neuro-oncology. Furthermore, we aim to provide insights into the current state-of-the-art methodologies, address their limitations, and identify areas for further research. Ultimately, our objective is to facilitate the development of robust, responsible, and applicable DL solutions that can effectively contribute to the field of neuro-oncology.
4.1. Overview of the Main DL Methods of Interest
Recent advances in the DL field have brought about new possibilities in medical imaging analysis and diagnosis. One of its arguably most successful models is Convolutional Neural Networks (CNNs), a widely used type of DL algorithm, well known for its ability to capture spatial correlations within image pixel data hierarchically. They have shown promise in medical imaging tasks [43,44,45], enabling improved tumor detection, classification, and prognosis assessment. The input data of a CNN is represented as a tensor with dimensions in the format of (channels, depth, height, width). Notably, the “depth” dimension is specific to 3D images and not applicable to 2D data, and “height” and “width” correspond to the image’s spatial dimensions. In practical terms, the number of channels for color images is translated into three, representing Red, Green, and Blue (RGB) components, while gray-scale images consist of a single channel. The most characteristic operation in a CNN is called convolution, which gives the name to the convolutional layers. These layers capture spatial correlations by applying a set of filters or kernels across all areas of the input image data and compute the weighted sum, resulting in the generation of a feature map as an output. This feature map contains essential characteristics extracted by the actual layer and serves as the input for subsequent layers of processing. Another useful layer used in CNNs is the pooling layer. The pooling operation consists of downsampling the feature maps obtained from the convolution operation. The idea is to reduce the dimensionality without losing significant information. There are mainly two kinds of pooling: max-pooling and average-pooling. The outputs of convolutional layers are often passed through activation functions to introduce non-linearity. The most popular activation functions are ReLU, which inactivates negative values in the output through the formula ; Sigmoid, which maps output values between 0 and 1 using the equation ; and SoftMax, which is the extension of Sigmoid for multi-class problems.
CNNs often consist of multiple layers that work together to learn hierarchical high-level image features. These layers progressively extract more abstract and complex information from the input image data. In the final step, the last feature map is passed through a fully connected layer, resulting in a one-dimensional vector. To obtain the class probabilities, Sigmoid or SoftMax are applied to this vector.
Several networks have made significant contributions to the world of DL. AlexNet [46], GoogLeNet [47], InceptionNet, VGGNet [48], ResNet [49], DenseNet [50], and EfficientNet [51] are among the most widely used CNNs to extract patterns from medical imaging.
DL models are considered data-hungry since they require substantial amounts of data for effective training. In the realm of medical data analysis, a primary challenge, as previously mentioned, is the inherent data scarcity and class imbalance. Common solutions to address this challenge include the application of data-augmentation (DA) methods and transfer-learning (TL) techniques.
Data Augmentation techniques are a crucial strategy to mitigate the challenge of limited annotated data in medical image analysis. These methods encompass a range of transformations applied to existing images, effectively expanding the dataset in terms of both size and diversity. Former approaches involve a wide range of geometric modifications such as rotation, scaling, flipping, cropping, zooming, or color changes. Beyond traditional augmentations, advanced methods like Generative Adversarial Networks (GANs) [52] are used to generate new synthetic and realistic examples.
The idea behind TL is to leverage pre-trained models, typically trained in large and diverse datasets, and adapt them for the specific task at hand, for which we might not have such a representative sample. Widely used pre-trained CNNs, such as ImageNet [53] or MS-COCO [54], have been originally developed from 2D large-scale datasets. However, a notable challenge when dealing with medical image data is the limited availability of large and diverse 3D datasets for universal pre-training [55]. Transferring the knowledge acquired from the 2D to the 3D domain proves to be a non-trivial task, primarily due to the fundamental differences in data structure and representation between these two contexts. To tackle this challenge and address the limitation of limited data, a broadly used strategy is to decompose 3D volumes into individual 2D slices within a determined anatomical plane. However, the decomposition of 3D volumes into individual 2D slices introduces a potential data leakage concern. This issue arises when 2D slices from the same individual inadvertently end up in both the training and testing datasets in an analytical pipeline. Such data leakage can lead to overestimations of model performance and affect the validity of experimental results. In addition, it is important to note that this approach comes with the trade-off of losing the 3D context present in the original data.
Recent efforts have aimed at overcoming these challenges. Banerjee et al. [56] classified low-grade glioma (LGG)and high-grade glioma (HGG) multi-sequence brain MRIs from TCGA and BraTS2017 data using multiple slice-based approaches. In their work, they provided a comparison of the performance obtained with CNNs trained from scratch on 2D image patches (PatchNet), entire 2D slices (SliceNet), and multi-planar slices through a final ensemble method that averages the classification obtained from each anatomical view (VolumeNet). The classification obtained with these models is also compared with pre-trained VGGNet and ResNet on ImageNet. The multi-planar method outperformed the rest of the approaches with an accuracy of 94.74%, and the lowest accuracy (68.07%) was obtained with pre-trained VGGNet. Unfortunately, TCGA and BraTS data share some patient data, which could involve an overlap between training and testing samples and hence be prone to data leakage. Ding et al. [57] combined radiomics and DL features using 2D pre-trained CNNs using single-plane images and performing a subsequent multi-planar fusion. VGG16, in combination with radiomics and RF, achieved the highest accuracy of 80% when combining the information obtained from the three views. Even though the multi-planar approach processes the information gathered from the axial, coronal, and sagittal views, it is still essentially a 2.5D approach, weak at fully capturing 3D contexts. Zhuge et al. [58] presented a properly native 3D CNN for tumor segmentation and subsequent binary glioma grade classification and compared it with a pre-trained 2D ResNet50 on ImageNet with previous tumor detection, employing a Mask R-CNN. The results of the 3D approach were slightly higher than the 2D ones, reporting 97.10% and 96.30% accuracy, respectively. In their study, Chatterjee et al. [59] explored the role of (2+1)D, mixed 2D–3D, and native 3D convolutions based on ResNet. This study highlights the effectiveness of mixed 2D–3D convolutions, achieving an accuracy of 96.98%, surpassing both the (2+1)D and the pure 3D approaches. Furthermore, the use of pre-trained networks demonstrated enhanced performance in the spatial models, yet, intriguingly, the pure 3D model performed better when trained from scratch. A study conducted by Yang et al. [55] introduced ACS convolutions, a novel approach that facilitates TL from models pre-trained on large publicly accessible 2D datasets. In this method, 2D convolutions are divided by channel into three parts and applied separately to the three anatomical views (axial, coronal, and sagittal). The effectiveness of this approach was demonstrated using a publicly available nodule dataset. Subsequently, Baheti et al. [60] further advanced the application of ACS convolutions by showcasing their enhanced performance on 3D MRI brain tumor data. Their study provides evidence of notable improvements in both segmentation and radiogenomic classification tasks.
4.2. Publicly Available Datasets
Access to large and high-quality datasets plays a crucial role in the development and evaluation of robust DL classification algorithms. This section aims to provide a comprehensive review of several publicly accessible datasets that have been widely used in brain tumor classification tasks and DL research. These datasets encompass diverse tumor types, imaging modalities, and annotated labels, facilitating the advancement of computational methods for accurate tumor classification.
Table 1 provides a detailed overview of the most frequently used datasets in the literature.

Table 1.
An overview of publicly available MRI datasets for brain tumor classification benchmarking.
The Brain Tumor Segmentation Challenge (BraTS) and The Computational Precision Medicine: Radiology-Pathology Challenge on Brain Tumor Classification (CPM-RadPath) datasets were created for two popular challenges held at the MICCAI (Medical Image Computing and Computer Assisted Intervention) Conference.
The BraTS Challenge [61] was initially developed in 2012 to benchmark tumor segmentation methods distinguishing glioblastoma from “lower grades”. Notably, this challenge provides not only MRI data but also clinical labels, including a binary classification of glioma grades. Even though their definition does not fully align with WHO’s terminology, they include grades 2 and 3 when referring to “lower grades”.
Throughout the years, the BraTS Challenge has continually evolved, expanding to include additional tasks and diverse datasets. In 2017, the dataset was enriched by integrating data from the TCIA repository, specifically including samples from the TCGA-LGG [71] and TCGA-GBM [70] datasets. It is worth noting that TCGA-LGG data provides labels to differentiate between gliomas of grades 2 and 3. Although the primary focus of the BraTS Challenge has traditionally centered on automated brain tumor segmentation, it has grown to become a widely adopted resource for brain tumor grade classification. Recent challenges have included tasks such as survival prediction and genetic classification, and the 2023 challenge even included image synthesis tasks.
CPM-RadPath [62] from 2019 was designed to evaluate brain tumor classification algorithms in three classes, taking into account the WHO classification of 2016: A (astrocytomas grades II and III, IDH-mutant), O (oligodendroglioma grades II and III, IDH-mutant, 1p/19q codeleted) and G (Glioblastoma and diffuse astrocytic glioma with molecular features of glioblastoma, IDH-wildtype (Grade IV)), interestingly grouping the anaplasic with the low grades in the A and O classes.
This challenge provides participants with paired radiology scans and digitized histopathology images. It is worth noting that the data provided by these challenges are distributed after pre-processing, involving co-registered to the same anatomical template, interpolated to a consistent resolution of 1 mm3, and skull-stripped.
The datasets under consideration encompass a variety of MRI modalities. Specifically, BraTS, CPM-RadPath, REMBRANDT, and TCGA comprise images from four key modalities: T1, T1 post-contrast weighted (T1c), T2-weighted, and Fluid Attenuated Inversion Recovery (FLAIR). The IXI dataset provides not only T1 and T2 but also Proton Density (PD) and Diffusion-weighted (DW) images. Notably, images on Figshare are limited to the T1c modality, while datasets from Kaggle and Radiopaedia lack this information.
The images in the BraTS, CPM-RadPath, IXI, REMBRANDT, and TCGA datasets are stored in 3D structures using widely used medical image formats, specifically NIfTI or DICOM. In contrast, datasets sourced from Kaggle consist of 2D images in PNG format. Notably, Figshare contains 2D images in MATLAB data format. In the Figshare data repository, images are provided alongside a 5-fold CV split at the patient level to mitigate the risk of data leakage. The use of this split ensures that no patient is inadvertently present in both training and testing sets, thus preventing leakage. Moreover, this dataset comprises multiple 2D slices from the same patient in the three distinct anatomical perspectives. Conversely, the datasets sourced from the Kaggle repository lack patient identifier information, making it challenging to ascertain if images are from unique patients or to trace the origin of the data.
Figure 1 summarizes the prevalence of dataset usage in the reviewed literature, including public and private datasets. Datasets that appear in two or fewer papers are grouped under the “Others” category.
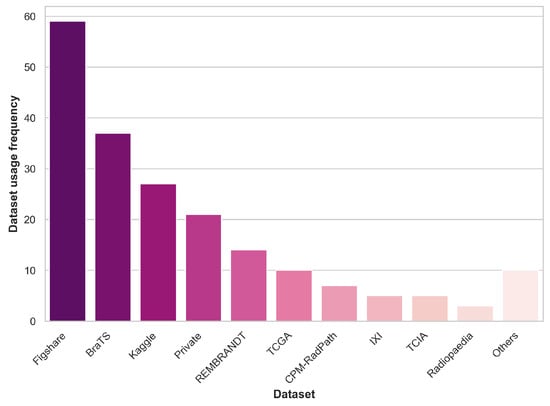
Figure 1.
Dataset usage prevalence across the reviewed literature.
It is worth highlighting that over 85% of the papers reviewed in this analysis make use of public datasets. It is essential to acknowledge that the sample sizes of the datasets, in general, are roughly in the hundreds range. This limited sample size can pose challenges in drawing robust and generalizable conclusions, which is a notable concern within the ML healthcare domain. Addressing the need for larger and more diverse datasets, as previously discussed, is an ongoing challenge in this field.
4.3. Literature Review
Various online repositories of scientific research articles, including PubMed, Google Scholar, and Scopus, were utilized to collect pertinent papers for this review section. The selection was restricted to the years 2018–2023. More specifically, only articles published prior to 30 June 2023 were taken into consideration. The document type was restricted to journal or conference papers. The focal keywords were centered on classifying brain tumors from pre-operative MRI images using DL techniques. While refining our choices, we excluded publications with ambiguous data explanations or lacking methodology details, as the utmost priority was placed on guaranteeing the strength and acuity of our conclusions. An initial identification process yielded a total of 555 papers, with 146 papers remaining after the screening procedure. Figure 2 depicts the distribution of these papers across the years under review, shedding some light on the temporal evolution of research in this domain.
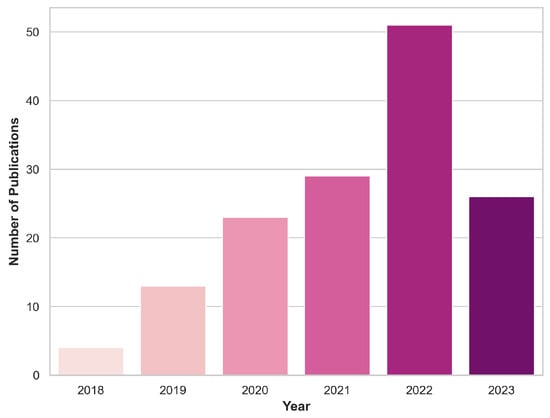
Figure 2.
Yearly inclusion of articles in this review that focus on classifying brain tumors using DL and MRI scans.
In the subsequent analysis, we provide comprehensive insights into the data sources and methodologies employed in the examined papers. Table A1 offers a detailed overview of the datasets, focusing on essential aspects such as the dimensionality of the images, sample size, MRI details, and pre-processing methods used. Table A2 delves into the specifications of the employed DL models, highlighting the brain tumor classification task, data partitioning, architecture, and the reported performance metrics. These tables contribute to a comprehensive understanding of the methodologies employed in the reviewed literature. Table A1 and Table A2 exclusively display the information available from the original authors in the analyzed papers. Any omissions in the table reflect the absence of such details as provided by the original authors in the surveyed papers. Notice that several papers are marked with an asterisk (*), which denotes that not all models have been reported in our tables due to the extensive array of results reported by the authors. Especially in these cases, we recommend readers refer to the original papers for a comprehensive overview of findings.
In this review, we focus on the differentiation of primary brain tumor types, with particular attention to gliomas due to their aggressive nature. Among the 146 examined papers in this section, some address multiple tasks concurrently. Specifically, 77 focused on distinguishing primary brain tumor types, while 27 aimed to identify tumorous images from images of healthy patients. Furthermore, the pursuit of accurate glioma grade classification is assessed in 66 papers, with 41 of them focusing on the binary distinction between low (grades II and III) and high-grade (glioblastoma, grade IV) gliomas. Note that the question asked by these 41 works does not correspond to any of the canonical releases of the WHO classification of brain tumors, as III is, in fact, high-grade. In this sense, such a grouping would facilitate the achievement of good performance results by grouping entities that are more prone not to show contrast enhancement, in contrast to glioblastoma, which always will show contrast enhancement [72]. Additionally, 12 studies delve into the distinction of glioma subtypes.
As previously highlighted, pre-processing techniques are pivotal in medical image processing. Among the 146 papers analyzed in this review, a substantial 80% of them provide insights into the specific pre-processing methodologies that were employed. Within this subset, it was observed that 35% employed registration techniques involving registration to a common anatomical template and co-registration to the same MRI modality. Furthermore, 40% employed segmentation as a critical step to isolate the brain from the surrounding skull structures. Notably, nearly half of the papers embraced normalization techniques to standardize the intensity of the image data before it was fed to the models. Additionally, 30% of the papers undertook the task of brain tumor extraction through methods such as bounding box delineation or tumor segmentation. Moreover, 15% of the papers employed pre-processing integrated image enhancement techniques to improve the contrast and visibility of crucial anatomical structures. In several studies [73,74,75], researchers investigated the advantages of utilizing the tumor area as opposed to the entire image, highlighting the significant benefits of concentrating on the tumor region rather than the entire image.
In the realm of medical research, the size and diversity of the training data sample stand as fundamental factors that substantially influence the performance, generalizability, and robustness of ML models. Several studies have explored the impact of varying the size of the training data sample on model performance [76,77,78,79,80,81,82,83,84]. Their findings highlight the value of ensuring that a substantial volume of data is available for training, as it significantly contributes to the model’s ability to make more accurate and reliable predictions.
Regarding addressing the limitation of data scarcity, approximately 60% of the examined studies employed DA techniques, and 40% incorporated TL in a 2D domain as a viable solution. Several of these investigations [85,86,87,88,89,90,91,92] have demonstrated the advantages of increasing both the quantity and variability of the samples through the inclusion of augmented images. Applying traditional DA techniques, such as geometric variations from original images, was the most widely used strategy, while only a few studies opted for the use of DL generative models [89,93,94,95]. Several studies [73,92,96,97,98,99] have integrated DA as an oversampling technique to address the problem of imbalanced data in the context of brain tumor classification. Furthermore, other works have explored the inclusion of multi-view 2D slices from axial, coronal, and sagittal planes, in addition to employing image flipping and rotations to augment the dataset [100]. Pre-trained models have demonstrated performance enhancements in the classification of glioma grades in several studies [79,101,102]. However, it is noteworthy that not all investigations have reported equivalent advantages when employing pre-trained models to discriminate between healthy and tumorous samples [103] or to differentiate tumor types [104]. These variations in findings underscore the complexity of the observed performance disparities, which may not be solely ascribed to the classification task itself but may also be influenced by intrinsic dataset variations.
The ability of CNNs to automatically extract meaningful features from brain MRI images, as opposed to the conventional need for manual feature engineering in certain ML algorithms like RF, GrB, and SVM, has been emphasized by numerous studies. These studies underscore the potential of CNNs in revolutionizing the landscape of MRI feature extraction for enhanced accuracy and efficiency in brain tumor classification [105,106,107,108,109,110,111]. Most of the reviewed papers (approximately 60%) utilized established state-of-the-art CNN architectures to obtain brain tumor classification. Among these, ResNet and VGGNet backbones were the most prevalent choices, closely followed by AlexNet, GoogLeNet, and Inception. In contrast, the remaining 40% of the papers concentrated on enhancing brain tumor classification by introducing novel model architectures. The inherent black-box nature of CNNs highlights the importance of delving into the comprehension of their predictions, especially in a medical context. Several studies within our review [74,112,113,114,115,116,117] have applied post-processing explainability tools to validate that the network’s decision-making process aligns with the intended diagnostic criteria, therefore enhancing the reliability of CNN-based medical applications.
Additionally, selected studies [57,118,119] explored the synergies of ensemble learning by combining the outputs of radiomics and DL models. Another interesting area of research has considered the opportunity of incorporating ML classifiers as the final layer in CNNs, effectively bypassing the traditional SoftMax layer [76,96,99,103,119,120,121,122,123,124,125,126,127,128].
The integration of information from various data sources has garnered growing interest in the medical field. Brain tumors, due to their distinct features both at the histopathological and radiological level, have motivated numerous studies to explore the synergy between whole slide imaging (WSI) and MRI data [97,129,130,131,132]. These investigations consistently highlight the richer information content in WSI as compared to MRI. However, they also reveal that combining data from both sources leads to improved overall performance in brain tumor characterization. Ensemble learning methods have shown promise in not only integrating information from diverse data types but also in combining predictions from multiple DL models on MRI to improve overall performance [75,82,91,107,108,116,122,133,134,135,136,137,138,139]. As brain tumor diagnosis and prognosis are significantly linked to genetic factors, several studies have undertaken efforts to explore the capabilities of DL models in extracting meaningful MRI features for the classification of these genetic frameworks [56,83,93,100,117,140,141].
Although brain MRIs inherently capture 3D data, a notable observation is that over 80% of the studies conducted their analyses within a 2D domain, focusing on 2D MRI slices. Nonetheless, some investigations have actively explored the significance of incorporating 3D volumetric information into the realm of brain tumor classification [56,58,59,74,97,98,100,112,117,129,130,131,132,135,140,141,142,143,144,145,146,147,148]. Although 3D volumes inherently capture information from the three anatomical planes, 2D slices are restricted to a specific view. Notably, among the studies that adopted a 2D approach, only 44% provided details about the chosen anatomical plane. Among this subset, more than 50% utilized axial, coronal, and sagittal views, while over 40% exclusively employed axial views.
Similarly, close to 70% of the reviewed studies disclosed the MRI modalities utilized for the analysis. Among these, close to 50% exclusively employed the T1c sequence, while 26% used a combination of T1c, T1, T2, and FLAIR sequences, 12% used three sequences, and the rest chose one sequence. Various strategies were employed to integrate information from multiple modalities. The prevalent method involved fusing them as input channels, comparable to the treatment of channels in RGB images. In their study, Ge et al. [100] evaluated the sensitivity of T1c, T2, and FLAIR modalities in glioma grade classification. Their investigation highlighted the T1c sequence as the most informative among these modalities. To further enhance the classification performance, they incorporated information from each source using an aggregation layer within the network architecture. Subsequently, similar ensemble learning approaches were adopted by Gutta et al. [106], Hussain et al. [148], Rui et al. [149]. Notably, Guo et al. [150] directly compared the performance of a modality-fusion approach, where the four MRI modalities were concatenated as a four-channel input, with a decision-fusion approach, where final predictions were derived through a linear weighted sum from the probabilities obtained through four independent pre-trained unimodal models. This study reinforced the notion of the T1c modality’s significance in glioma subtype classification. Moreover, it revealed that any multimodal approach consistently outperformed unimodal models, with the decision-ensemble approach emerging as the most effective strategy.
As previously discussed, decomposing 3D volumes into individual 2D slices may introduce the potential for data leakage. Maintaining the reliability of the analysis is crucial for obtaining robust and trustworthy findings. It is worth noting, however, that only a limited number of studies that use multiple 2D slices [56,76,77,79,93,100,101,106,107,115,118,126,135,149,151,152,153,154,155,156,157], explicitly detailed their approach to data splitting at the patient level, addressing this critical concern. Remarkably, an insightful comparison was carried out in the work of Badža and Barjaktarović [158] between data-splitting strategies at the patient and image levels. The findings elucidate that an image-wise approach yields accuracy results as high as 96% for brain tumor type classification, while a patient-level split demonstrates a higher degree of reliability with an accuracy of 88%. These results underscore the critical importance of utilizing a patient-wise training approach to assess the model’s generalization capacity. Similarly, Ghassemi et al. [85], Ismael et al. [159] also provided evidence of superior performance when using an image-wise split, further reinforcing the importance of thoughtful data splitting. It is also important to note that 3D models operate on complete 3D volumes and are inherently structured at the patient level. This approach substantially reduces the likelihood of data leakage, therefore enhancing the reliability of the analysis and ensuring that the results faithfully represent the model’s performance. This aspect may provide a valuable perspective when interpreting differences in accuracy between 3D and 2D models.
The predominant approach for data partitioning in the examined papers involves the use of hold-out validation with training and validation sets. This was followed by the adoption of K-fold cross-validation, which enhances the robustness of model evaluation. A less frequently employed method was the three-way split, which includes training, validation, and testing sets. In total, only 36% of the studies assessed their final results using an independent test set. Decuyper et al. [140], Gilanie et al. [152], Alanazi et al. [160] took a step further by assessing the generalizability and robustness of their models using external validation sets. Although the authors of the Figshare dataset thoughtfully included a 5-fold CV setup alongside the data to promote comparability and reproducibility, it is still important to remark here that a substantial majority of studies continue to prefer custom data partitioning methods.
5. Machine Learning Applications to Ultra-Low Field Imaging
A completely different area of application of ML to neuroradiology has recently emerged with the availability of ultra-low field magnetic resonance imaging devices for point-of-care applications, typically with <0.1 T permanent magnets [161,162,163]. In the 0.055 T implementation described by Liu et al. [11], DL was used to improve the quality of the acquisition by detecting and canceling external electromagnetic interference (EMI) signals, eliminating the need for radio-frequency shielded rooms. They compared the results of the DL EMI cancelation in 13 patients with brain tumors, both in the 0.055 T and in another 3 T machine, on same-day acquisitions, finding that it was possible to identify the different tumor types. Please note that these processes are, in fact, a completely different use of DL for data pre-processing to those reviewed in Section 3.
Another example is the Hyperfine system, which received FDA clearance in 2020 for brain imaging and in 2021 (K212456) for DL-image reconstruction to enhance the quality of the generated images. In particular, DL is used as part of the image reconstruction pipeline of T1, T2, and FLAIR images. There are two DL steps: the first one is a so-called DL gridding, where the undersampled k-space data are transformed into images not by Fourier transformations but with DL. The transformed images are then combined, and a final post-processing DL step is applied to eliminate noise. However, no details about the specific algorithms are provided. Although the main application seems to be in the neurocritical setting [164], this system is beginning to be compared with the imaging quality at higher fields at different stages, with a particular interest in the early post-operative monitoring after surgical resection (e.g., [165,166]). It is to be expected that Hyperfine brain tumor applications will emerge soon, for example, through the partnership with The Brain Tumor Foundation, to provide the general population with free brain scans.
6. Conclusions
Neuro-oncological radiology relies on non-invasive data acquisition, which makes it the ideal target of data-centered analytics and places it at the forefront of ML-applied developments. In this review paper, we have focused on the most successful instantiation of ML currently, namely DL, and its use for the analysis of imaging data. Emphasis has been put on the fact that DL methods must be seen as only part of analytical pipelines, in which data pre-processing plays a key role.
Promoting the responsible utilization of clinical data is of utmost importance when striving to establish trustworthy conclusions. A fundamental step in this endeavor is the comprehensive disclosure of both the data used and the analytical procedures undertaken. Such transparency not only fosters greater trust in research outcomes but also amplifies the generalizability and reproducibility of the findings. This, in turn, plays a pivotal role in advancing AI-driven solutions in the clinical pathway. Most DL-based analytical solutions depend, to a great extent, not only on data quality but also quantity. For this reason, we argue that the main challenge facing the use of DL in the radiological imaging setting is precisely the creation of sizeable curated image databases for the different problems at hand.
Author Contributions
All authors (C.P., G.U., M.J.-S. and A.V.) contributed to the different phases of article development, including conceptualization, methodology, investigation, and writing. M.J.-S. and A.V. are responsible for supervision and funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by H2020-EU.1.3.—EXCELLENT SCIENCE—Marie Skłodowska-Curie Actions, grant number H2020-MSCA-ITN-2018-813120; Proyectos de investigación en salud 2020, grant number PI20/00064. PID2019-104551RB-I00; Centro de Investigación Biomédica en Red en Bioingeniería, Biomateriales y Nanomedicina (CIBER-BBN (http://www.ciber-bbn.es/en, accessed on 3
November 2023), CB06/01/0010), an initiative of the Instituto de Salud Carlos III (Spain) co-funded by EU Fondo Europeo de Desarrollo Regional (FEDER); Spanish Agencia Española de Investigación (AEI) PID2022-143299OB-I00 grant; XartecSalut 2021-XARDI-00021. Carla Pitarch is a fellow of Eurecat’s “Vicente López” Ph.D. grant program.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data sharing is not applicable.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript; or in the decision to publish the results.
Abbreviations
The following abbreviations are used in this manuscript:
| AUC | Area Under the ROC Curve |
| CDSS | Clinical Decision Support System |
| CNN | Convolutional Neural Network |
| DA | Data Augmentation |
| DL | Deep Learning |
| FAIR | Findable, Accessible, Interoperable, Reusable |
| GAN | Generative Adversarial Network |
| HGG | High-Grade Glioma |
| ICA | Independent Component Analysis |
| LGG | Low-Grade Glioma |
| ML | Machine Learning |
| MRI | Magnetic Resonance Imaging |
| MRS | Magnetic Resonance Spectroscopy |
| MRSI | Magnetic Resonance Spectroscopy Imaging |
| NMF | Nonnegative Matrix Factorization |
| RF | Random Forest |
| RGB | Red, Green, Blue |
| ROI | Region of Interest |
| SVM | Support Vector Machine |
| TCGA | The Cancer Genome Atlas |
| TCIA | The Cancer Imaging Archive |
| tICA | Temporal ICA |
| TL | Transfer Learning |
| TPOT | Tree-based Pipeline Optimization Tool |
| ULF | Ultra-Low Field |
| ViT | Vision Transformer |
| WHO | World Health Organization |
| WSI | Whole Slide Imaging |
Appendix A. Literature Review Summary of Deep-Learning Sources
This appendix comprises Table A1 and Table A2, offering a detailed overview of the datasets and models employed in the papers summarized in Section 4.3. Table A1 provides key details about the datasets used, including image dimensionality, number of patients in the study and images included for the analysis, anatomical plane, MRI modalities, pre-processing procedures, and data-augmentation techniques. On the other hand, Table A2 summarizes analytical aspects, such as the classification task, data-splitting methodology, network architecture, overall performance metrics, and class-specific performance. Hyphens (-) in certain cells indicate that the information was not provided in the original paper.
For papers marked with an asterisk (*), not all outcomes are included in the table due to the extensive range of results reported by the authors. We encourage readers to consult the original papers for a more comprehensive understanding of the findings.

Table A1.
Data Overview: Comprehensive overview of the datasets examined within the DL literature review centered on brain tumor classification tasks and MRI data. Essential information regarding dimensionality, sample size, anatomical plane, MRI modalities, and pre-processing methods are summarized.
Table A1.
Data Overview: Comprehensive overview of the datasets examined within the DL literature review centered on brain tumor classification tasks and MRI data. Essential information regarding dimensionality, sample size, anatomical plane, MRI modalities, and pre-processing methods are summarized.
| No. | Reference | Dim. | Dataset | Sample Size | Plane | MRI Modality | Pre-Processing | Data Augmentation (Augmentation Factor) | |
|---|---|---|---|---|---|---|---|---|---|
| Patients | Images | ||||||||
| 1 | Ge et al. [100] (2018) | 2D | BraTS2017 | 285 (Table 1) | - | Ax, Sag, Cor | T1c, T2, Flair | Tumor mask enhancement | Multi-view images (ax, sag, cor), rotation, flipping |
| 2 | Ge et al. [73] (2018) | 3D | BraTS2017 | 285 (Table 1) | 285 (Table 1) | Ax, Sag, Cor | T1c | None 1 Tumor mask enhancement 2 | (LGG: 2) Flipping |
| 3 | Pereira et al. [74] (2018) | 3D | BraTS2017 | 285 (Table 1) | 285 (Table 1) | Ax, Sag, Cor | T1, T1c, T2, Flair | BFC, z-score normalization (inside brain mask) | Sagittal flipping, rotation, exponential intensity transformation |
| 4 | Yang et al. [101] (2018) | 2D | Private | 113 (LGG: 52, HGG: 61) | 867 (LGG: 368, HGG: 499) | Ax | T1c | Z-score normalization, tumor ROI | (14) HE, random rotation, zooming, adding noise, flipping |
| 5 | Abd-Ellah et al. [167] (2019) | 2D | Brats2017 | - | 1800 (H: 600, LGG: 600, HGG:600) | - | - | - | - |
| 6 | Anaraki et al. [168] (2019) | 2D | IXI REMBRANDT TCGA-GBM TCGA-LGG Private Figshare | 600 130 199 299 60 233 | 16,000 (H: 8000, G.II: 2000, G.III: 2000, G.IV: 4000) 989 | Ax - - - - Ax | T1 T1c T1c T1c T1c T1c | Normalization, resizing | Rotation, translating, scaling, flipping |
| 7 | Deepak and Ameer [76] (2019) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | T1c | Min-max normalization, resizing | Rotation, flipping |
| 8 | Hemanth et al. [169] (2019) | 2D | Private | - | 220 | - | T1, T2, Flair | None | None |
| 9 | Kutlu and Avcı [120] (2019) | 2D | Figshare | 233 (Table 1) | 3064 | Ax, Sag, Cor | T1c | None | None |
| 10 | Lo et al. [102] (2019) | 2D | TCIA | 134 (G.II: 30, G.III: 43, G.IV: 57) | 134 (G.II: 30, G.III: 43, G.IV: 57) | Ax | T1c | Normalization, CE, tumor segmentation | (56) AutoAugment [170] |
| 11 | Muneer et al. [171] (2019) | 2D | Private | 20 | 557 (G.I: 130, G.II: 169, G.III: 103, G.IV: 155) | Ax | T2 | Skull-stripping, tumor segmentation | Resize, reflection, rotation |
| 12 | Rajini [172] (2019) | 2D | IXI REMBRANDT TCGA-GBM TCGA-LGG Figshare | 600 130 “around 200” 299 233 | - - - - - | - - - - - | - - - - T1c | - - - - - | - - - - - |
| 13 | Rahmathunneesa and Muneer [173] (2019) | 2D | Private | - | 760 (G.I: 198, G.II: 205, G.III: 172, G.IV: 185) | Axial | T2 | Skull-stripping, resizing | Resizing, rotation, translation, reflection |
| 14 | Sajjad et al. [174] (2019) | 2D | Radiopaedia Figshare | - 233 | 121 (G.I: 36, G.II: 32, G.III: 25, G.IV: 28) 3064 (Table 1) | - - | - T1c | BFC, Segmentation, Z-score normalization | (30) Rotation, flipping, skewness, shears, gaussian blur, sharpening, edge detection, emboss |
| 15 | Sultan et al. [175] (2019) | 2D | Figshare REMBRANDT | 233 (Table 1) 73 (G.II: 33, G.III: 19, G.IV: 21) | 3064 (Table 1) 516 (G.II: 205, G.III: 129, G.IV: 182) | Ax, Sag, Cor | T1c | Resizing | (5) Rotation, flipping, mirroring, noise |
| 16 | Swati et al. [77] (2019) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | T1c | Min-max normalization, resizing | - |
| 17 | Toğaçar et al. [176] (2019) | 2D | Kaggle-III | - | 253 (Table 1) | - | - | - | Rotation, flipping, brightening, CE, shifting, scaling |
| 18 | Amin et al. [177] (2020) | 2D | BraTS2012 1 BraTS2013 2 BraTS2013 (LB) 3 BraTS2015 4 BraTS2018 5 | 25 (LGG: 5, HGG: 10) 30 (Table 1) 25 (LGG: 4, HGG: 21) 274 (Table 1) 284 (Table 1) | - - - - - | - | T1, T1c, T2, Flair | Noise Removal, tumor enhancement, MRI modality fusion | - |
| 19 | Afshar et al. [178] (2020) | 2D | Figshare | 233 | 3064 | - | - | - | - |
| 20 | Badža and Barjaktarović [158] (2020) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Normalization, Resizing | Rotation, flipping |
| 21 | Banerjee et al. [56] (2020) | 2D | TCGA-GBM TCGA-LGG BraTS2017 | 262 199 285 (Table 1) | 1590 (LGG:750, HGG:840) | Ax 1 Ax, Sag, Cor 2 | T1, T1c, T2, Flair | - | Rotation, shifts, flipping |
| 22 | Bhanothu et al. [179] (2020) | 2D | Figshare | 233 | 2406 (MN: 694, GL: 805, PT: 907) | - | T1c | Min-max normalization | - |
| 23 | Çinar and Yildirim [180] (2020) | 2D | Kaggle-III | - | 253 (Table 1) | Ax, Sag, Cor | - | - | - |
| 24 | Ge et al. [93] (2020) | 2D | BraTS2017 | 285 (Table 1) | - | Ax, Sag, Cor | T1, T1c, T2, Flair | - | GAN |
| 25 | Ghassemi et al. [85] (2020) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Normalization (−1,1) | Rotation, flipping |
| 26 | Ismael et al. [159] (2020) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | - | T1c | Resizing, cropping | Rotation, flipping, shifting, zooming, ZCA whitening, shearing, brightening |
| 27 | Khan et al. [181] (2020) | 2D | Kaggle-III | - | 253 (Table 1) | - | - | Brain cropping | Flipping, rotation, brightness |
| 28 | Ma and Jia [129] (2020) | 3D | CPM-RadPath2019 | 329 (Table 1) | 329 | Ax, Sag, Cor | T1, T1c, T2, Flair | Z-score normalization | Cropping, rotation, zooming, translation, color changes |
| 29 | Mohammed and Al-Ani [182] (2020) | 2D | Radiopaedia | 60 (15 per class) | 1258 (H: 286, MN: 380, E: 311, Med: 281) | Ax, Sag, Cor | - | Resizing, denoising | Rotation, scaling, reflection, translating, cropping |
| 30 | Mzoughi et al. [142] (2020) | 3D | BraTS2018 | 284 (LGG: 75, HGG: 209) | 285 | Ax, Sag, Cor | T1c | Min-max normalization, CE, resizing | flipping |
| 31 | Naser and Deen [183] (2020) | 2D | TCGA-LGG | 108 (G.II: 50, G.III: 58) | 815 (G.II: 400, G.III: 415) | - | T1, T1c, Flair | Cropping, normalization (−1,1), resizing, segmentation | Rotation, zooming, shifting, flipping |
| 32 | Noreen et al. [184] (2020) | 2D | Figshare | 233 | 3064 (Table 1) | T1c | Normalization | - | |
| 33 | Pei et al. [143] (2020) | 3D | CPM-RadPath2020 | 270 (Table 1) | 270 | Ax, Sag, Cor | T1, T1c, T2, Flair | Noise reduction, z-score normalization, tumor segmentation | Rotation, scaling |
| 34 | Rehman et al. [104] (2020) | 2D | Figshare | 233 | 3064 (Table 1) | T1c | CE | Rotation, flipping | |
| 35 | Saxena et al. [185] (2020) | 2D | Kaggle-III | - | 253 (Table 1) | - | - | Brain cropping, resizing | (20) not specified |
| 36 | Sharif et al. [186] (2020) | 2D | BraTS 2013 1 BraTS2015 2 BraTS2017 3 BraTS2018 4 | 30 (Table 1) 274 (Table 1) 285 (Table 1) 284 (Table 1) | - | - | T1, T1c, T2, Flair | CE, tumor segmentation | - |
| 37 | Tandel et al. [105] (2020) | 2D | REMBRANDT | 112 (Table 1) | 2132 (H: 1041, T: 1091) 2156 (H: 1041, LGG: 484, HGG: 631) 2156 (H: 1041, AS: 557, OG: 219, GB: 339) 1115 (AS-II: 356, AS-III: 201, OG-II: 128, OG-III: 91, GB: 339) 2156 (H: 1041, AS-II: 356, AS-III: 201, OG-II: 128, OG-III: 91, GB: 339) | Ax, Sag, Cor | T2 | Skull-stripping | Rotation, scaling |
| 38 | Toğaçar et al. [96] (2020) | 2D | Kaggle-III | - | 253 (Table 1) | - | - | - | Oversampling |
| 39 | Vimal Kurup et al. [187] (2020) | 2D | Figshare | 233 | 3064 (Table 1) | - | T1c | Resizing | Rotation, cropping |
| 40 | Zhuge et al. [58] (2020) | 2D | BraTS2018 TCGA-LGG | 284 (Table 1) 30 | 284 (Table 1) 30 | Ax, Sag, Cor | T1c, T2, Flair | Inhomogeneity correction, z-score normalization, min-max normalization, tumor segmentation | (23) - AutoAugment [170] |
| 3D | BraTS2018 TCGA-LGG | 284 (Table 1) 30 | 284 (Table 1) 30 | Ax, Sag, Cor | T1c, T2, Flair | Rotation, scaling, flipping | |||
| 41 | Alaraimi et al. [78] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | - | - | HE, z-score normalization | Rotation, cropping, flipping, scaling, translation |
| 42 | Ayadi et al. [86] (2021) | 2D | Figshare 1 Radiopaedia 2 REMBRANDT 3 | 233 (Table 1) - 112 (AS-II: 30, AS-III: 17, OG-II: 14, OG-III: 7, GB: 44) | 3064 (Table 1) 121 (MN G.I: 36, GL G.II: 32, GL G.III: 25, GB: 28) - | Ax, Sag, Cor - - | T1c - - | - - - | (17) - Rotation, flipping, gaussian blur, sharpen |
| 43 | Bashir-Gonbadi and Khotanlou [188] (2021) | 2D | IXI BraTS2017 Figshare Private | 582 (healthy) 285 - - | - - 3064 230 | - | - | Skull-stripping, resizing | Flipping, mirroring, shifting, scaling, rotation |
| 44 | Chakrabarty et al. [144] (2021) | 3D | BraTS 2018 BraTS2019 LGG-1p19q Private | 43 LGG 335 (Table 1) 145 1234 (MET: 710, MN: 143, AN: 158, PA: 82, H: 141) | 43 335 159 1234 | Ax, Sag, Cor | T1c | Co-registration, resampling, skull-stripping, z-score normalization, resizing | - |
| 45 | Decuyper et al. [140] (2021) | 3D | TCGA TCGA-1p19q BraTS2019 GUH dataset | 285 (LGG: 121, HGG: 164) 141 202 110 | 285 141 202 110 | Ax, Sag, Cor | T1, T1c, T2, Flair | Tumor segmentation | Rotation, Flipping, Intensity scaling, Elastic transform |
| 46 | Díaz-Pernas et al. [151] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Z-score normalization | (2) Elastic transforms |
| 47 | Gab Allah et al. [94] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Normalization (−1,1) | (12) PGGAN 1 (9) Rotation, mirroring, flipping 2 |
| 48 | Gilanie et al. [152] (2021) | 2D | Private | 180 (AS-1: 50, AS-II: 40, AS-III: 40, AS-IV: 50) | 30,240 (AS-1: 8400, AS-II: 6720, AS-III: 6720, AS-IV: 8400) | T1 & Flair: Ax, T2: Ax, Sag | T1, T2, Flair | BFC, normalization, tumor Segmentation | Rotation |
| 49 | Gu et al. [189] (2021) | 2D | REMBRANDT 1 Figshare 2 | 130 - | 110,020 3064 (Table 1) | - - | - T1c | - - | - - |
| 50 | Guan et al. [153] (2021) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | T1c | CE, tumor ROI, min-max normalization | (3) Rotation, flipping |
| 51 | Gull et al. [154] (2021) | 2D | BraTS2018 1 BraTS2019 2 BraTS2020 3 | - - - | 1425 (LGG: 375, HGG: 1050) 1675 (LGG: 380, HGG: 1295) 2470 (LGG: 645, HGG: 1435, unknown: 390) | - - - | T1, T1c, T2, Flair T1, T1c, T2, Flair T1, T1c, T2, Flair | Grayscaling, median filtering, skull-stripping | - |
| 52 | Gutta et al. [106] (2021) | 2D | Private | 237 (G.I: 17, G.II: 59, G.III: 46, G.IV: 115) | 660 (G.I: 27, G.II: 144, G.III: 184, G.IV: 305) | - | T1, T1c, T2, Flair | Resampling, co-registration, skull-stripping, tumor segmentation | - |
| 53 | Hao et al. [79] (2021) | 2D | BraTS2019 | 335 (Table 1) | 6700 (20 random slices per patient) | Ax, Sag, Cor | T1c, T1, T2 | - | - |
| 54 | Irmak [190] (2021) | 2D | RIDER REMBRANDT TCGA-LGG Figshare | 19 (G.IV) 130 199 233 | (total) 2990 (H: 1350, T: 1640) 3950 (H: 850, MN: 700, GL: 950, PT: 700, MT: 750) 4570 (G.II: 1676, G.III: 1218, G.IV: 1676) | - | T1c, Flair T1c, Flair T1c, Flair T1c | - | - |
| 55 | Kader et al. [191] (2021) | 2D | BraTS2012 BraTS2013 BraTS2014 BraTS2015 | - - - - | 1000 1000 800 700 | - - - - | - - - - | Noise removal, tumor segmentation, resizing | - |
| 56 | Kader et al. [192] (2021) | 2D | Private | - | 17,600 | - | T1, T2, Flair | - | Yes, not specified |
| 57 | Kakarla et al. [193] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | - | T1c | Resizing, min-max normalization, CE | - |
| 58 | Kang et al. [133] (2021) | 2D | Kaggle-III 1 Kaggle-II 2 Kaggle-I 3 | - - - | 253 (Table 1) 3264 (Table 1) 3000 (Table 1) | - - - | - - - | Brain cropping, resizing | Rotation, flipping |
| 59 | Khan et al. [87] (2021) | 2D | BraTS2015 | 274 | 169,880 | Ax, Sag, Cor | T1, T1c, T2, Flair | Z-score normalization, tumor segmentation | (20) Rotation, zooming, geometric transforms, sharpening, noise addition, CE |
| 60 | Kumar et al. [88] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | T1c | - | Rotation | |
| 61 | Masood et al. [194] (2021) | 2D | Figshare Kaggle-III | 233 - | 3064 (Table 1) 253 (Table 1) | - - | T1c - | BFC, CE, tumor ROI | - |
| 62 | Noreen et al. [134] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | - | - | Min-max normalization | - |
| 63 | Özcan et al. [155] (2021) | 2D | Private | 104 (G.II: 50, G.IV: 54) | 518 | Ax, Sag, Cor | Flair | Multiple-cropping, z-score normalization | (20) Rotation, zooming, shearing, flipping, elastic gaussian transforms |
| 64 | Pei et al. [97] (2021) | 3D | CPM-RadPath2020 | 256 (Table 1) | 256 (Table 1) | Ax, Sag, Cor | T1, T1c, T2, Flair | BFC, z-score normalization | (oversampling) |
| 65 | Sadad et al. [195] (2021) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | - | CE, tumor detection | Rotation, flipping |
| 66 | Tandel et al. [107] (2021) | 2D | REMBRANDT | 130 (H: 18, T: 112) | 2156 (H: 1041, T: 1091) 557 (AS-II: 356, AS-III: 201) 219 (OG-II: 218, OG-III: 91) 1115 (LGG: 484, HGG: 631) | Ax, Sag, Cor | T2 | - | Rotation, scaling |
| 67 | Toğaçar et al. [80] (2021) | 2D | Figshare | 233 | 3064 (Table 1) | - | T1c | - | Rotation, scrolling, brightening |
| 68 | Yamashiro et al. [145] (2021) | 3D | BraTS2018 | 284 (Table 1) | 285 (Table 1) | Ax, Sag, Cor | T1c | Tumor segmentation | Flipping, scaling, shifting |
| 69 | Yin et al. [130] (2021) | 3D | CPM-RadPath2020 | 256 (Table 1) | 256 (Table 1) | Ax, Sag, Cor | T1, T1c, T2, Flair | Tumor segmentation, resizing, z-score normalization | Brightness, CE, saturation, hue, flipping, rotation |
| 70 | Aamir et al. [156] (2022) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | T1c | CE, min-max normalization, tumor ROI | (2) Rotation, flipping |
| 71 | Ahmad et al. [89] (2022) | 2D | Figshare | 233 | 3064 (Table 1) | - | T1c | Resizing, normalization | CDA: Rotation, scaling GDA: VAE, GAN |
| 72 | Alanazi et al. [160] (2022) | 2D | Kaggle-I Kaggle-II 1 Figshare 2 | - - 233 | 3000 (H: 1500, T: 1500) 3264 (Table 1) 3064 (Table 1) | - | - | Noise removal, cropping, z-score normalization, resizing | - |
| 73 | Almalki et al. [121] (2022) | 2D | Kaggle-II 1 Figshare 2 | - 233 | 2870 (H: 395, MN: 822, GL: 826, PT: 827) 3064 (Table 1) | - - | - - | Brain cropping, denoising, resizing | - - |
| 74 | Amou et al. [81] (2022) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Min-max normalization, resizing | None |
| 75 | Aurna et al. [82] (2022) | 2D | Figshare Kaggle-II Kaggle [196] | 233 - - | 3064 (Table 1) 3264 (Table 1) 4292 (H: 681, MN: 1318, GL: 1038, PT: 1255) | - - - | - - - | Resizing | Rotation, flipping, zooming, shifting, scaling |
| 76 | Chatterjee et al. [59] (2022) | 2D-3D | BraTS2019 IXI | 332 (LGG: 73, HGG: 259) 259 | 332 259 | 2D: Ax, 3D: Ax, Sag, Cor | T1c | Skull-stripping, normalization (0.5,99.5), resampling | Affine, flipping |
| 77 | Chitnis et al. [197] (2022) | 2D | Kaggle-II | - | 3264 (Table 1) | - | - | Resizing | Autoaugment |
| 78 | Coupet et al. [135] (2022) | 2D-3D | BraTS2018 BraTS2020 | 284 (Table 1) 369 (Table 1) | 50,812 | Ax | T1, T1c, T2, Flair | Histogram & min-max normalization | Rotation, deformations, shearing, zooming, flipping |
| 79 | Dang et al. [98] (2022) | 3D | BraTS2019 | 335 (Table 1) | 335 (Table 1) | Ax, Sag, Cor | T1, T1c, T2, Flair | Segmentation, gamma correction, window setting optimization | (oversampling) Rotation |
| 80 | Danilov et al. [146] (2022) | 3D 2D | Private | 707 (G.I: 189, G.II: 133, G.III: 127, G.IV: 258) | 707 17,730 | Ax, Sag, Cor Ax, Sag, Cor | T1c T1c | Z-score normalization, resampling ImageNet standardization | - Rotation, scaling, mirroring |
| 81 | Ding et al. [57] (2022) | 2D-3D | Private TCIA + Private | 101 (LGG: 58, HGG: 43) 50 (LGG: 25, HGG: 25) | 3 slices as channels | Ax, Sag, Cor | T1c | Tumor ROI, normalization, resizing | - |
| 82 | Ekong et al. [198] (2022) | 2D | BraTS2015 IXI Figshare | - - - | (total) 4000 (H: 1000, MN: 10,000, GL: 1000, PT: 1000) | - - - | - T1, T2 T1c | Resizing, normalization, denoising, BFC, registration, tumor segmentation | Shifting, Rotation, Brightening, Image enlargement, Flipping |
| 83 | Gao et al. [112] (2022) | 3D | Private | 39,210 | 39,210 | Ax, Sag, Cor | T1, T2, T1c | Z-score normalization, resampling | - |
| 84 | Gaur et al. [199] (2022) | 2D | Kaggle-II | - | 2870 | - | - | Resizing | Gaussian Noise |
| 85 | Guo et al. [150] (2022) | 3D | CPM-RadPath2020 | 221 (Table 1) | 221 | Ax, Sag, Cor | T1, T1c, T2, Flair | BFC, skull-stripping, co-registration, tumor segmentation | Rotation, resizing, scaling, gaussian noise, CE |
| 86 | Gupta et al. [95] (2022) | 2D | Kaggle-II | - | 3264 (Table 1) | - | - | CE | CycleGAN |
| 87 | Gurunathan and Krishnan [200] (2022) | 2D | BraTS | - | 260 (LGG: 156, HGG: 104) | Ax, Sag, Cor | T1, T2 | Resizing, tumor segmentation | Rotation, shifts, reflection, flipping, scaling, shearing |
| 88 | Haq et al. [90] (2022) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | - | T1c | Resizing | (2) Zooming |
| 89 | Hsu et al. [131] (2022) | 3D | BraTS2020 CPM-RadPath2020 | 369 (Table 1) 270 (Table 1) | 369 270 | Ax, Sag, Cor | T1, T1c, T2, Flair | Sampling patches, z-score normalization, tumor segmentation | Rotation, flipping, affine translation |
| 90 | Isunuri and Kakarla [201] (2022) | 2D | Figshare | - | 3064 (Table 1) | - | T1c | Resizing, Normalization | - |
| 91 | Jeong et al. [113] (2022) | 2D | BraTS2017 | 285 (Table 1) | 1445 (largest slice ± 8) | Ax, Sag, Cor | T1, T1c, T2, Flair | Resizing, z-score normalization | Rotation, flipping |
| 92 | Kazemi et al. [108] (2022) | 2D | Figshare 1 TCIA 2 | 233 20 | 1500 (MN: 1000, GL: 800, PT: 600) 8798 | - - | T1c T1c | Resizing | - |
| 93 | Khazaee et al. [202] (2022) | 2D | BraTS2019 | - | 26,904 (LGG: 13,671, HGG: 13,233) | - | T1c, T2, Flair | - | Rotation, flipping |
| 94 | Kibriya et al. [122] (2022) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | - | - | Min-max normalization, resizing | (5) Rotation, flipping, mirroring, adding noise |
| 95 | Koli et al. [203] (2022) | 2D | Kaggle-III Figshare | - - | 253 (Table 1) 3064 (Table 1) | - - | - - | - | Rotation |
| 96 | Lakshmi and Rao [204] (2022) | 2D | Figshare | - | 3064 | - | T1c | - | - |
| 97 | Maqsood et al. [114] (2022) | 2D - | Figshare BraTS2018 | 233 284 (Table 1) | 3064 (Table 1) - | - - | T1c - | CE, tumor segmentation, z-score normalization | - |
| 98 | Murthy et al. [205] (2022) | 2D | Kaggle-III | - | 253 (Table 1) | - | - | Median filtering, CE, tumor segmentation | - |
| 99 | Nayak et al. [206] (2022) | 2D | Figshare | - | 3260 (196 H, 3064 Table 1) | Ax, Sag, Cor | T1c | Noise removal, gaussian blurring, min-max normalization | (21) Rotation, Shifting, Zooming |
| 100 | Rajinikanth et al. [124] (2022) | 2D | TCIA | - | 2000 (GL = 1000, GB = 1000) | Ax | - | - | - |
| 101 | Rasool et al. [125] (2022) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | - | Yes, not specified |
| 102 | Raza et al. [207] (2022) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | T1c | Resizing | - |
| 103 | Rizwan et al. [208] (2022) | 2D | Figshare REMBRANDT | 230 (MN: 81, GL: 90, PT: 59) 70 (G.II: 32, G.III: 18, G.IV: 20) | 3061 (MN: 707, GL: 1425, PT: 929) 513 (G.II: 204, G.III: 128, G.IV: 181) | Ax, Sag, Cor - | T1c T1c | Noise, cropping, resizing | (5) Salt-noise, grayscaling |
| 104 | Samee et al. [209] (2022) | 2D | Figshare | 236 (MN: 83, GL: 90, PT: 63) | 3075 (MN: 708, GL: 1427, PT: 940) | Ax, Sag, Cor | T1c | Grayscaling | (16) Rotation, zooming, brightening |
| 105 | Samee et al. [147] (2022) | 3D | BraTS2015 | 65 (LGG: 14, HGG: 51) | 1056 (LGG: 176, HGG: 880) | - | T1, T1c, T2, Flair | Resizing, denoising, CE, tumor segmentation | - |
| 106 | Sangeetha et al. [210] (2022) | 3D | Private | 45 | 45 | Ax, Sag, Cor | T2 | Min-max normalization | (14) Rotation, translation |
| 107 | Saravanan et al. [109] (2022) | 2D | BRATS REMBRANDT | 274 135 | 1200 - | - - | - - | Resizing | - |
| 108 | Sekhar et al. [126] (2022) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | Ax, Sag, Cor | T1c | Min-max normalization, resizing | Yes but not specified |
| 109 | Senan et al. [99] (2022) | 2D | Kaggle-II | - | 3060 (H: 396, MN: 937, GL: 826, PT: 901) | Ax, Sag, Cor | - | Denoising, min-max normalization, resizing, CE | (H: 11, MN: 5, GL:6, PT: 5) Rotation, cutting, zooming, patching, padding, brightening |
| 110 | Srinivas et al. [211] (2022) | 2D | Kaggle | - | 256 (Benign: 158, Malignant: 98) | - | - | Brain cropping, z-score normalization, resizing | Scaling, cropping, resizing, flipping, rotation, geometric transforms |
| 111 | Tandel et al. [75] (2022) | 2D | Rembrandt | 112 (LGG = 44, HGG = 68) | - | Ax | T1, T2, Flair | None 1, Skull-stripping 2, tumor ROI 3 | Scaling, rotation |
| 112 | Tripathi and Bag [83] (2022) | 2D | TCIA | 322 (LGG:159, HGG: 163) | 7392 (LGG: 5088, HGG: 2304) | - | T2 | Skull-stripping, segmentation | Rotation, flipping, scaling, cropping, translation |
| 113 | Tripathi and Bag [141] (2022) | 3D | BraTS2019 TCGA-GBM TCGA-LGG LGG-1p19qdeletion [212] | 202 158 119 138 | (total) 617 (LGG: 331, HGG: 286) | Ax, Sag, Cor | T1c, T2, Flair | Co-registration, skul-stripping, resampling, tumor segmentation | Flipping, shifting, rotation, cropping |
| 114 | Tummala et al. [136] (2022) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | - | - |
| 115 | Vankdothu et al. [213] (2022) | 2D | Kaggle-II | - | 3264 | - | - | Grayscaling, rotation, denoising, tumor ROI | - |
| 116 | Wang et al. [132] (2022) | 3D | CPM-RadPath2020 | 270 | 270 | Ax, Sag, Cor | T1, T1c, T2, Flair | Resizing, brain cropping | Rotation, flipping, scaling, jittering |
| 117 | Xiong et al. [115] (2022) | 2D | Private | 211 (AS: 54, OG: 67, GB: 90) | 633 | Ax, Sag, Cor | ADC, T1c, Flair | Resampling, skull-stripping, z-score normalization, min-max normalization | - |
| 118 | Xu et al. [118] (2022) | 2D | BraTS2020 | 369 (Table 1) | 369 | Ax | T1c, T1, T2 | BFC, skull-stripping, registration, z-score normalization 1 tumor ROI 2 | - |
| 119 | Yazdan et al. [214] (2022) | 2D | Kaggle-II | - | 3264 (Table 1) | - | T1, T2, Flair | Denoising | None |
| 120 | Zahoor et al. [103] (2022) | 2D | Kaggle 1 Figshare 2 | - | 1994 (H) 3064 | - Ax, Sag, Cor | - - | Resizing | Rotation, sharing, scaling, reflection |
| 121 | AlTahhan et al. [127] (2023) | 2D | Figshare Kaggle-II Kaggle-I | - - - | 2880 (H: 396, MN: 825, GL: 829, PT: 830) | - - - | T1c | - | - |
| 122 | Al-Zoghby et al. [137] (2023) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Resizing | - |
| 123 | Anagun [215] (2023) | 2D | Figshare | - | 3064 (Table 1) | Ax, Sag, Cor | T1c | Brain cropping, HE, denoising | (9) Flipping, rotation, shifting, zooming |
| 124 | Anand et al. [91] (2023) | 2D | TCGA-LGG | 110 | 3929 | - | Flair | - | Flipping |
| 125 | Apostolopoulos et al. [216] (2023) | 2D | Kaggle [217] Kaggle [218] | - | 26,249 (H: 2000, MN: 7866, GL: 8208, PT: 8175) | - | - | - | - |
| 126 | Asif et al. [138] (2023) | 2D | Figshare | 233 (Table 1) | 3064 (Table 1) | - | - | Resizing, denoising | - |
| 127 | Athisayamani et al. [110] (2023) | 2D | Figshare | - | - | - | - | Denoising, skull-stripping, brain segmentation | Rotation, flipping |
| 128 | Bairagi et al. [111] (2023) | 2D | BraTS2013 BraTS2015 OPEN-I NLM | - | 65 327 229 | - | T1, T2, Flair | Resizing | (40) Resizing, cropping, rotation, reflection, shear, translation |
| 129 | Deepa et al. [84] (2023) | 2D | BraTS2018 1 Figshare 2 | - - | - 3064 (Table 1) | - | - T1c | Min-max normalization, tumor segmentation | Flipping, translation, rotation, brightening, CE, gaussian noise |
| 130 | El-Wahab et al. [219] (2023) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | - | - |
| 131 | Hossain et al. [116] (2023) | 2D | Kaggle-II | - | 3264 | - | - | - | (4) Rescaling, shearing, zooming, flipping |
| 132 | Hussain et al. [148] (2023) | 3D | BraTS2020 | 369 (Table 1) | 369 (Table 1) | Ax, Sag, Cor | T1, T1c, T2, Flair, Segmentation | Denosing, tumor segmentation | - |
| 133 | Kibriya et al. [119] (2023) | 2D | Kaggle-III 1 Kaggle-I 2 | - - | 253 (Table 1) 3000 (Table 1) | - - | - - | - - | - - |
| 134 | Krishnapriya and Karuna [92] (2023) | 2D | Kaggle-III | - | 253 (Table 1) | - | - | Brain cropping | (Oversampling) Rotation, shifting, rescaling, mirroring |
| 135 | Kumar et al. [128] (2023) | 2D | ACRIN-DSC-MR-BRAIN | - | 1731 | - | T1 | Resizing, grayscaling, CE, tumor segmentation | - |
| 136 | Mahmud et al. [220] (2023) | 2D | Kaggle-II CPTAC-GB ACRIN-FMISO-BRAIN | - 189 45 | 3264 (Table 1) - - | - | - | Normalization, smoothing | Mirroring, rotation, shifting, zooming |
| 137 | Muezzinoglu et al. [221] (2023) | 2D | Kaggle-II | - | 3264 (Table 1) | - | - | Resizing, patch division | - |
| 138 | Özkaraca et al. [222] (2023) | 2D | Kaggle [223] (combines Figshare, Kaggle-I, Kaggle-II) | - | total: 7021 (H: 2002, MN: 1627, GL: 1623, PT: 1769) | - | - | - | - |
| 139 | Özkaya and Şağıroğlu [224] (2023) | 2D | BraTS2020 | 369 | (undersampling slices HGG) | Ax | T1c, T2, Flair | Tumor segmentation, min-max normalization | - |
| 140 | Rasheed et al. [225] (2023) | 2D | Figshare | 233 | 3064 (Table 1) | Ax, Sag, Cor | T1c | Resizing, normalization | None |
| 141 | Rui et al. [149] (2023) | 2D | Private | 42 (G.II: 18, G.III: 10, G.IV: 14) | 1176 (G.II: 504, G.III: 280, G.IV: 392) | Ax | T1c, T2, Flair | Brain cropping, normalization | - |
| 142 | Shirehjini et al. [123] (2023) | 2D | Private | 58 (G.I: 8, G.II: 16, G.III: 10, G.IV: 22) | 1061 (T1c: 229, T1: 251, T2: 299, Flair: 282) | Ax, Sag, Cor | T1, T1c, T2, Flair | Resizing, min-max normalization | - |
| 143 | Srinivasan et al. [226] (2023) | 2D | REMBRANDT | - | 3100 | - | - | Denoising, tumor segmentation | - |
| 144 | Tandel et al. [139] (2023) | 2D | REMBRANDT | 112 (LGG: 44, HGG: 68) | - | Ax, Sag, Cor | T1, T2, Flair | Resizing | Rotation, scaling |
| 145 | van der Voort et al. [117] (2023) | 3D | Erasmus MC [227] Haaglanden Medical Center BraTS REMBRANDT CPTAC-GBM Ivy GAP Amsterdam UMC Brain-tumor-progression University Medical Center Utrecht TCGA-LGG TCGA-GBM | 816 279 168 109 51 39 20 20 6 107 133 | (total) 1412 (G.II: 277, G.III:173, G.IV: 962) | Ax, Sag, Cor | T1, T1c, T2, Flair | Registration, resampling, BFC, skull-stripping, brain cropping, z-score normalization | (2) Cropping, rotation, brightening, CE |
| 146 | Wu et al. [157] (2023) | 2D | BraTS2019 | 326 (LGG:76, HGG: 250) | slices with tumor | - | T1, T1c, T2, Flair | Z-score normalization, center-cropping | Rotation, translation, clipping |
AS: Astrocytoma, Ax: Axial, BFC: Bias Field Correction, CE: Contrast Enhancement, Cor: Coronal, DA: Data Augmentation, E: Ependymoma, Flair: Fluid Attenuated Inversion Recovery, GL: Glioma, GAN: Generative Adversarial Network, GB: Glioblastoma, GDA: Generative Data Augmentation, H: Healthy, HE: Histogram Equalization, HGG: High-Grade Glioma, LGG: Low-Grade Glioma, Med: Medulloblastoma, MN: Meningioma, MT: Metastasis, OG: Oligodendroglioma, PT: Pituitary, ROI: Region of Interest, Sag: Sagittal, T: Tumor, T1c: T1 post-contrast weighted, VAE: Variational Auto-Encoder. Numerical superscripts link datasets with models in Table A2 when different data sources yield individual results.

Table A2.
Model Overview: Comprehensive summary of the DL architectures employed across the reviewed papers. The table outlines key information, including the brain tumor classification task, data partitioning, architecture, and the reported performance metrics.
Table A2.
Model Overview: Comprehensive summary of the DL architectures employed across the reviewed papers. The table outlines key information, including the brain tumor classification task, data partitioning, architecture, and the reported performance metrics.
| No. | Reference | Classification Task | Data Split | Architecture | Acc% | AUC% | F1% | Class Performance % | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Method | Ratio | Level | ||||||||
| 1 | Ge et al. [100] (2018) | LGG vs. HGG | Three-way | 60:20:20 | Patient | [T1c] CNN [T2] CNN [Flair] CNN [Modality-ensemble] CNN | 83.73 69.74 75.40 90.87 | - - - - | - - - - | LGG = 82.54, HGG = 84.92 LGG = 59.52, HGG = 80.15 LGG = 76.19, HGG = 74.60 LGG = 90.48, HGG = 91.27 |
| 2 | Ge et al. [73] (2018) | LGG vs. HGG | Three-way | 60:20:20 | Patient | Custom CNN 1 [whole image] 2 [tumor ROI] | 84.21 89.47 | - - | - - | - LGG = 90.48, HGG = 86.67 |
| 3 | Pereira et al. [74] (2018) | LGG vs. HGG | Three-way | 60:20:20 | Patient | Custom CNN ROI: brain, Std.: image ROI: brain, Std.: brain ROI: tumor, Std.: image ROI: tumor, Std.: brain | 89.50 89.50 87.70 92.98 | 88.57 89.13 88.41 98.41 | 86.45 86.43 85.08 90.96 | LGG = 80.00, HGG = 92.90 LGG = 80.00, HGG = 92.86 LGG = 86.67, HGG = 88.10 LGG = 86.67, HGG = 95.24 |
| 4 | Yang et al. [101] (2018) | LGG vs. HGG | 5-fold CV, Test | 80:20 | Patient | TL GoogLeNet TL AlexNet GoogLeNet AlexNet | 94.50 92.70 90.90 85.50 | 96.80 96.60 93.90 89.40 | - - - - | - - - - |
| 5 | Abd-Ellah et al. [167] (2019) | H vs. LGG vs. HGG | Three-way | 65:10:25 | - | Parallel CNNs | 97.44 | - | - | (R) 97.00, (S) 98.00 |
| 6 | Anaraki et al. [168] (2019) | H vs. G.II vs. G.III vs. G.IV G.II vs. G.III vs. G.IV MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN + GA | 93.10 90.90 94.20 | - - - | - - - | H = 99.80, G.II = 88.40, G.III = 86.80, G.IV = 97.40 - MN = 87.80, GL = 98.30, PT = 96.5 |
| 7 | Deepak and Ameer [76] (2019) | MN vs. GL vs. PT | 5-fold CV | Patient | TL GoogLeNet -KNN -SVM -SoftMax | 98.00 97.80 92.30 | - - - | - 97.00 - | - MN = 96.00, GL = 97.90, PT = 98.90 - | |
| 8 | Hemanth et al. [169] (2019) | MT vs. MN vs. GL vs. AS | - | - | - | Custom CNN | 96.40 | - | - | MT = 94.00, MN = 93.00, GL = 93.00, AS = 89.00 |
| 9 | Kutlu and Avcı [120] (2019) | Benign vs. Malignant | 5-fold CV | 70:30 | - | TL AlexNet-DWT -LSTM -SVM -KNN | 98.66 92.09 85.91 | 99.00 - - | - - - | B = 99.33, M = 98.66 B = 96.04, M = 92.08 B = 92.95, M = 85.91 |
| 10 | Lo et al. [102] (2019) | G.II vs. G.III vs. G.IV | 10-fold CV | - | TL AlexNet AlexNet | 97.90 61.42 | 99.91 82.22 | - - | G.II = 96.90, G.III = 96.80, G.IV = 99.10 - | |
| 11 | Muneer et al. [171] (2019) | G.I vs. G.II vs. G.III vs. G.IV | Hold-out | 70:30 | - | TL VGG19 Wndchrm | 94.64 92.86 | - - | 93.71 92.32 | - - |
| 12 | Rajini [172] (2019) | H vs. G.II vs G.III vs. G.IV MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN | 96.77 98.16 | 95.65 97.93 | 93.54 97.21 | H = 99.80, G.II = 89.20, G.III = 85.27, G.IV = 98.00 MN = 93.69, GL = 99.15, PT = 99.13 |
| 13 | Rahmathunneesa and Muneer [173] (2019) | G.I vs. G.II vs. G.III vs. G.IV | Hold-out | 70:30 | - | TL AlexNet TL GoogLeNet TL InceptionV3 TL ResNet50 | 92.98 85.96 86.84 96.05 | - - - - | 96.06 91.71 91.62 97.76 | G.I = 96.67, G.II = 93.44, G.III = 92.31, G.IV = 89.09 G.I = 86.67, G.II = 98.36, G.III = 63.46, G.IV = 92.73 G.I = 76.67, G.II = 93.44, G.III = 90.38, G.IV = 87.27 G.I = 93.33, G.II = 91.80, G.III = 100.00, G.IV = 100.00 |
| 14 | Sajjad et al. [174] (2019) | G.I vs. G.II vs. G.III vs. G.IV MN vs. GL vs. PT | Three-way | 50:25:25 | - | TL VGG-19 w/o DA w/ DA w/o DA w/ DA | 87.38 90.67 - 94.58 | - - - - | - - - - | G.I = 90.03, G.II = 89.91, G.III = 84.11, G.IV = 85.50 G.I = 95.54, G.II = 92.66, G.III = 87.77, G.IV = 86.71 MN = 90.22, GL = 93.12, PT = 89.08 MN = 94.05, GL = 96.14, PT = 93.21 |
| 15 | Sultan et al. [175] (2019) | MN vs. GL vs. PT G.II vs. G.III vs. G.IV | Hold-out | 68:32 | - | Custom CNN | 96.13 98.70 | - - | - - | MN = 95.50, GL = 94.40, PT = 93.40 G.II = 100, G.III = 95.00, G.IV = 100.00 |
| 16 | Swati et al. [77] (2019) | MN vs. GL vs. PT | 5-fold CV | Patient | Block-wise TL VGG19 Block-wise TL VGG16 TL AlexNet | 94.82 94.65 89.95 | - - - | 91.73 91.50 86.83 | GL = 95.97, MN = 89.98, PT = 96.81 (R) 93.51, (S) 94.56 (R) 89.10, (S) 89.84 | |
| 17 | Toğaçar et al. [176] (2019) | H vs. T | Hold-out | 70:30 | - | Custom CNN GoogLeNet AlexNet VGG16 | 96.05 89.66 87.93 84.48 | 98.00 - - - | 94.12 90.32 88.52 85.25 | H = 96.00, T = 96.08 H = 84.85, T = 96.00 H = 84.38, T = 92.31 H = 81.25, T = 88.46 |
| 18 | Amin et al. [177] (2020) | H vs. T | Hold-out | 50:50 | - | Custom CNN | 1 97.00 2 98.00 3 100.00 4 96.00 5 97.00 | - - - - - | - - - - - | H = 97.00, T = 97.00 H = 99.00, T = 95.00 H = 100.00, T = 100.00 H = 98.00, T = 92.00 H = 99.00, T = 93.00 |
| 19 | Afshar et al. [178] (2020) | MN vs. GL vs. PT | Hold-out | 80:20 | Custom CNN | 92.45 | 98.00 | - | MN = 75.35, GL = 96.85, PT = 98.90 | |
| 20 | Badža and Barjaktarović [158] (2020) | MN vs. GL vs. PT | 10-fold CV | 60:20:20 | Patient Patient Image Image | Custom CNN w/o DA w/ DA w/o DA w/ DA | 84.45 88.48 95.40 96.56 | - - - - | 81.86 86.97 94.93 96.11 | MN = 62.70, GL = 90.20, PT = 91.30 MN = 71.60, GL = 92.80, PT = 95.00 MN = 89.80, GL = 96.20, PT = 98.40 MN = 90.20, GL = 98.00, PT = 99.20 |
| 21 | Banerjee et al. [56] (2020) | LGG vs. HGG | Hold-out | Patient | 2 VolumeNet 1 SliceNet 1 PatchNet 1 TL ResNet 1 TL VGGNet | 94.74 85.96 82.45 72.30 68.07 | - - - - - | - - - - - | LGG = 94.29, HGG = 96.00 LGG = 80.00, HGG = 88.10 LGG = 74.67, HGG = 85.24 LGG = 72.06, HGG = 71.43 LGG = 69.33, HGG = 67.62 | |
| 22 | Bhanothu et al. [179] (2020) | MN vs. GL vs. PT | Hold-out | 80:20 | - | F-RCNN + VGG16 | - | - | - | (P) GL = 75.18, MN = 68.18, PT = 97.28 |
| 23 | Çinar and Yildirim [180] (2020) | H vs. T | - | - | - | Custom CNN ResNet50 DenseNet201 AlexNet InceptionV3 GoogLeNet | 97.01 92.54 91.04 89.55 88.07 71.64 | - - - - - | 96.90 93.33 92.30 90.05 81.81 66.03 | H = 94.70, T = 100.00 H = 89.74, T = 96.40 H = 85.71, T = 100.00 H = 87.17, T = 92.85 H = 81.81, T = 100.00 H = 66.03, T = 92.85 |
| 24 | Ge et al. [93] (2020) | LGG vs. HGG | Hold-out | 60:20:20 | Patient | Modality-ensemble Semi-supervised CNN w/o DA w/ DA | 89.53 90.70 | - - | - - | LGG = 78.26, HGG = 93.65 LGG = 84.35, HGG = 93.01 |
| 25 | Ghassemi et al. [85] (2020) | MN vs. GL vs. PT | 5-fold CV | Patient Patient Image | Custom CNN w/o pre-training w/ GAN pre-training w/ GAN pre-training | 91.70 93.01 95.60 | - - - | 90.54 92.10 95.10 | MN = 79.86, GL = 94.96, PT = 95.67 MN = 84.82, GL = 94.92, PT = 96.92 MN = 89.98, GL = 96.83, PT = 97.93 | |
| 26 | Ismael et al. [159] (2020) | MN vs. GL vs. PT | Hold-out | 80:20 | Patient Image | ResNet50 | 97.82 99.34 | - - | 97.00 99.00 | MN = 93.00, GL = 99.00, PT = 99.00 MN = 98.00, GL = 99.00, PT = 100.00 |
| 27 | Khan et al. [181] (2020) | H vs. T | Three-way | 70:20:10 | - | Custom CNN VGG16 ResNet50 InceptionV3 | 100.00 96.00 89.00 75.00 | 100.00 96.00 89.00 75.00 | 100.00 97.00 90.00 74.00 | H = 100.00, T = 100.00 H = 92.85, T = 100.00 H = 85.71, T = 92.86 H = 76.92, T = 73.33 |
| 28 | Ma and Jia [129] (2020) | AS vs. OG vs. GB | Three-way | 70:10:20 | Patient | [WSI] 2D ResNet50 [MRI] 3D DenseNet121 [WSI-MRI] Ensemble 2D-3D | 83.33 71.10 88.90 | - - - | 91.40 82.90 94.30 | - - - |
| 29 | Mohammed and Al-Ani [182] (2020) | H vs. EP vs. MN vs. MB | Three-way | 70:10:20 | - | Custom CNN | 96.00 | - | - | - |
| 30 | Mzoughi et al. [142] (2020) | LGG vs. HGG | - | - | Patient | Custom CNN | 96.49 | - | - | - |
| 31 | Naser and Deen [183] (2020) | G.II vs. G.III | 5-fold CV | - | TL VGG16 | 95.00 | 97.00 | - | G.II = 98.00, G.III = 93.00 | |
| 32 | Noreen et al. [184] (2020) | MN vs. GL vs. PT | Hold-out | 80:20 | - | InceptionV3 DenseNet201 | 99.34 99.51 | 99.00 100.00 | - - | MN = 99.00, GL = 100.00, PT = 100.00 MN = 99.00, GL = 100.00, PT = 99.00 |
| 33 | Pei et al. [143] (2020) | AS vs. OG vs. GB | Three-way | 67:11:22 | Patient | Custom CNN | 63.90 | - | - | - |
| 34 | Rehman et al. [104] (2020) | MN vs. GL vs. PT | Three-way | 70:15:15 | - | AlexNet GoogLeNet VGG16 TL AlexNet TL GoogLeNet TL VGG16 | 97.39 98.04 98.69 95.77 95.44 89.79 | - - - - - - | - - - - - - | - - - - - - |
| 35 | Saxena et al. [185] (2020) | H vs. T | Three-way | 70:20:10 | - | TL ResNet50 TL VGG16 TL InceptionV3 | 95.00 90.00 55.00 | 95.00 90.00 55.00 | 95.20 90.90 68.90 | - - - |
| 36 | Sharif et al. [186] (2020) * | LGG vs. HGG | 10-fold CV, Test | 70:30 | - | Ensemble TL InceptionV3-DRLBP | 1 98.30 2 97.80 3 96.90 4 92.50 | - - - - | - - - - | - - - - |
| 37 | Tandel et al. [105] (2020) * | H vs. T H vs. LGG vs. HGG H vs. AS vs. OG vs. GB AS-II vs. AS-III vs. OG-2 vs. OG-3 vs. GB H vs. AS-II vs. AS-III vs. OG-2 vs. OG-3 vs. GB | 2-fold CV, 5-fold CV, 10-fold CV | - | TL AlexNet | 100.00 95.97 96.65 87.14 93.74 | - - - - - | 100.00 94.80 94.78 86.89 91.97 | 100.00 94.85 94.17 84.40 91.51 | |
| 38 | Toğaçar et al. [96] (2020) * | H vs. T | Hold-out | 70:30 | - | Ensemble TL AlexNet-VGG16-RFE-SVM TL AlexNet TL VGG16 | 96.77 90.32 87.10 | - - - | 96.77 89.89 87.23 | (R) 97.83, (S) 95.74 (R) 95.24, (S) 86.27 (R) 87.23, (S) 86.96 |
| 39 | Vimal Kurup et al. [187] (2020) | MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN | 92.60 | 96.33 | 93.33 | GL = 96.00, MN = 94.00, PT = 94.00 |
| 40 | Zhuge et al. [58] (2020) | LGG vs. HGG | 5-fold CV, Test | 60:20:20 | Patient | TL 2D ResNet50 w/o DA w/ DA 3D ConvNet | 89.10 96.30 97.10 | - - - | - - - | (R) 86.40, (S) 91.70 (R) 93.50, (S) 97.20 (R) 94.70, (S) 96.80 |
| 41 | Alaraimi et al. [78] (2021) | MN vs. GL vs. PT | Hold-out | 80:20 | - | TL VGG16 TL GoogLeNet TL AlexNet | 100.00 98.50 94.40 | 98.60 98.10 97.60 | - - - | - - - |
| 42 | Ayadi et al. [86] (2021) | 1 MN vs. GL vs. PT 2 G.I vs. G.II vs. G.III vs. G.IV 3 H vs. T 3 H vs. LGG vs. HGG 3 H vs. AS vs. OG vs. GB 3 AS-II vs. AS-III vs. OG-II vs. OG-III vs. GB 3 H vs. AS-II vs. AS-III vs. OG-II vs. OG-III vs. GB 2 G.I vs. G.II vs. G.III vs. G.IV 3 H vs. T 3 H vs. LGG vs. HGG 3 H vs. AS vs. OG vs. GB 3 AS-II vs. AS-III vs. OG-II vs. OG-III vs. GB 3 H vs. AS-II vs. AS-III vs. OG-II vs. OG-III vs. GB | 5-fold CV | 70:30 | - | Custom CNN [w/o DA] Custom CNN [w/ DA] | 94.74 90.35 100.00 95.00 94.41 86.08 92.09 93.71 100.00 97.22 97.02 88.86 95.72 | - - - - - - - - - - - - - | 94.19 90.38 100.00 91.35 92.89 86.85 89.84 93.88 100.00 95.45 95.75 87.52 91.76 | MN = 89.68, GL = 94.46, PT = 99.03 G.I = 88.23, G.II = 93.33, G.III = 84.00, G.IV = 96.00 H = 100.00, T = 100.00 H = 100.00, LGG = 100.00, HGG = 70.00 H = 99.00, AS = 96.36, OG = 92.00, GB = 80.00 AS-II = 85.71, AS-III = 90.00, OG-II = 86.66, OG-III = 80.00, GB = 85.71 H = 100.00, AS-II = 85.71, AS-III = 90.00, OG-II =86.66, OG-III = 80.00, GB = 82.85 G.I = 90.79, G.II = 95.66, G.III = 90.84, G.IV = 98.22 H = 100.00, T = 100.00 H = 100.00, LGG = 98.40, HGG = 86.00 H = 99.80, AS = 97.09, OG = 90.40, GB = 93.71 AS-II = 88.50, AS-III = 94.00, OG-II = 96.00, OG-III = 62.00, GB = 90.85 H = 100.00, AS-II = 93.14, AS-III = 88.00, OG-II = 98.66, OG-III = 76.00, GB = 94.85 |
| 43 | Bashir-Gonbadi and Khotanlou [188] (2021) | MN vs. GL vs. PT H vs. LGG vs. HGG H vs. AS vs. MN vs. PT vs. LGG vs. HGG | Three-way | - | - | Auto-encoder CNN | 98.50 99.10 99.30 | - - - | 98.6 99.2 99 | MN = 97.90, GL = 99.00, PT = 98.60 H = 98.10, LGG = 99.00, HGG = 97.70 H = 100.00, AS = 100.00, MN = 100.00, PT = 100.00, LGG = 96.60, HGG = 97.80 |
| 44 | Chakrabarty et al. [144] (2021) | LGG vs. HGG vs. MT vs. PT vs. AN vs. H vs. MN | 5-fold CV, Test | 80:20 | Patient | Custom CNN | 91.95 | 96.93 | 93.86 | LGG = 81.50, HGG = 87.00, MT = 98.60, PA = 100.00, AN = 100.00, H = 89.70, MN = 93.30 |
| 45 | Decuyper et al. [140] (2021) | LGG vs. HGG | Three-way | 73:11:16 | Patient | Custom CNN | 90.00 | 93.98 | - | LGG = 89.80, HGG = 90.16 |
| 46 | Díaz-Pernas et al. [151] (2021) | MN vs. GL vs. PT | 5-fold CV | Patient | Custom CNN | 97.30 | - | - | GL = 99.00, MN = 93.00, PT = 98.00 | |
| 47 | Gab Allah et al. [94] (2021) * | MN vs. GL vs. PT | Three-way | 70:15:15 | - | 1 VGG19 2 VGG19 | 98.54 96.59 | - - | - - | GL = 100, MN = 90.20, PT = 96.92 - |
| 48 | Gilanie et al. [152] (2021) | AS-1 vs. AS-II vs. AS-III vs. AS-IV | Hold-out | 50:25:25 | Patient | Custom CNN | 95.56 | - | - | (Acc) G.I = 99.06, G.II = 94.01, G.III = 95.31, G.IV = 97.85 |
| 49 | Gu et al. [189] (2021) | 1 AS vs. OG vs. GB 2 MN vs. GL vs. PT | 5-fold CV | 70:30 | - | Custom CNN | 97.64 96.34 | - - | 94.18 94.69 | AS = 96.86, OG = 91.27, GB = 93.09 MN = 88.75, GL = 94.87, PT = 98.37 |
| 50 | Guan et al. [153] (2021) | MN vs. GL vs. PT | 5-fold CV | 70:30 | Patient | EfficientNet | 98.04 | - | 97.79 | MN = 96.89, GL = 97.82, PT = 99.24 |
| 51 | Gull et al. [154] (2021) | H vs. T | 10-fold CV, Test | 70:10:20 | Patient | GoogLeNet | 1 96.49 2 97.31 3 98.79 | - - - | 97.27 97.92 99.12 | H = 94.17, T = 97.80 H = 95.83, T = 98.14 H = 97.37, T = 99.42 |
| 52 | Gutta et al. [106] (2021) | G.I vs. G.II vs. G.III vs. G.IV | Three-way | 70:15:15 | Patient | Modality-ensemble CNN GrB RF SVM | 87.00 64.00 58.00 56.00 | - - - - | - - - - | G.I = 100.00, G.II = 82.35, G.III = 76.92, G.IV = 92.50 G.I = 0.00, G.II = 23.53, G.III = 42.31, G.IV = 90.74 G.I = 0.00, G.II = 35.23, G.III = 7.69, G.IV = 92.50 G.I = 33.00, G.II = 70.00, G.III = 34.62, G.IV = 72.00 |
| 53 | Hao et al. [79] (2021)* | LGG vs. HGG | Three-way | 60:20:20 | Patient | AlexNet TL AlexNet | - - | 71.93 79.91 | - - | - - |
| 54 | Irmak [190] (2021) | H vs. T H vs. MN vs. GL vs. PT vs. MT G.II vs. G.III vs. G.IV | 5-fold CV, Test | 60:20:20 | - | Custom CNN | 99.33 92.66 98.14 | 99.95 99.81 99.94 | - - - | H = 100, T = 98.80 H = 92.10, MN = 94.20, GL = 94.40, PT = 88.00, MT = 90.00 G.II = 97.91, G.III = 100, G.IV = 97.01 |
| 55 | Kader et al. [191] (2021) | H vs. T | - | - | - | DWAE model | 99.30 | - | 96.55 | H = 96.90, T = 95.60 |
| 56 | Kader et al. [192] (2021) * | H vs. T | 5-fold CV | - | Custom CNN GoogLeNet AlexNet VGG16 | 99.25 89.66 87.66 84.48 | - - - - | 95.23 90.32 88.52 85.25 | (R) 95.89, (S) 93.75 (R) 84.85, (S) 96.00 (R) 84.38, (S) 92.31 (R) 81.25, (S) 8.48 | |
| 57 | Kakarla et al. [193] (2021) | MN vs. GL vs. PT | 5-fold CV, Test | 80:20 | - | Custom CNN | 97.42 | - | - | - |
| 58 | Kang et al. [133] (2021) * | H vs. T H vs. T H vs. MN vs. GL vs. PT | Hold-out | 80:20 | - | Ensemble TL CNNs DenseNet169-InceptionV3-ResNeXt50-AdaBoost DenseNet121-ResNeXt-MnasNet DenseNet169-ShuffleNet-MnasNet | 1 92.16 2 98.83 3 91.58 | - - - | - - - | - - - |
| 59 | Khan et al. [87] (2021) | LGG vs. HGG | - | - | - | VGG19 (w/o DA) VGG19 (w/ DA) | 90.03 94.06 | - - | - - | LGG = 91.05, HGG = 84.03 LGG = 96.05, HGG = 89.09 |
| 60 | Kumar et al. [88] (2021) | MN vs. GL vs. PT | 5-fold CV | - | TL ResNet50 (w/o DA) TL ResNet50 (w/ DA) | 97.48 97.08 | - - | 97.20 97.20 | 97.20 97.20 | |
| 61 | Masood et al. [194] (2021) | MN vs. GL vs. PT H vs. T | Hold-out | 70:30 | - | DenseNet-41-based Mask-RCNN | 98.34 97.90 | - - | - - | (Acc) MN = 97.81, GL = 98.62, PT = 98.60 (Acc) H = 98.06, T = 97.74 |
| 62 | Noreen et al. [134] (2021) * | MN vs. GL vs. PT | 10-fold CV | - | TL InceptionV3 Ensemble InceptionV3-KNN-SVM-RF TL XceptionV3 Ensemble Xception-KNN-SVM-RF | 93.31 94.34 91.63 93.79 | - - - - | 92.67 - 90.00 - | MN = 84.00, GL = 95.00, PT = 98.00 - MN = 78.00, GL = 94.00, PT = 100.00 - | |
| 63 | Özcan et al. [155] (2021) | G.II vs. G.IV | 5-fold CV, Test | 80:20 | Patient | Custom CNN AlexNet GoogLeNet SqueezeNet | 97.10 92.30 93.30 89.40 | 98.90 97.00 98.70 97.50 | 97.00 92.22 93.30 89.30 | G.II = 98.00, G.IV = 96.30 G.II = 94.00, G.IV = 90.70 G.II = 98.00, G.IV = 88.90 G.II = 92.00, G.IV = 87.00 |
| 64 | Pei et al. [97] (2021) | AS vs. OG vs. GB | Hold-out | 85:15 | Patient | [WSI] 2D CNN [MRI] 3D CNN [WSI-MRI] Ensemble 2D-3D CNNs | 77.00 69.80 80.00 | - - - | 88.60 77.10 88.60 | - - - |
| 65 | Sadad et al. [195] (2021) * | MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN | 99.60 | 99.00 | - | - |
| 66 | Tandel et al. [107] (2021) * | H vs. T AS-II vs. AS-III OG-2 vs. OG-3 LGG vs. HGG | 5-fold CV | Patient | Ensemble TL AlexNet, VGG16, ResNet18, GoogleNet, ResNet50 | 96.51 97.70 100.00 98.43 | 96.60 97.04 100.00 98.45 | - - - - | (R) 96.76, (S) 96.43 (R) 94.63, (S) 99.44 (R) 100.00, (S) 100.00 (R) 98.33, (S) 98.57 | |
| 67 | Toğaçar et al. [80] (2021) | MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN | - | - | 96.22 | MN = 94.81, GL = 98.48, PT = 95.38 |
| 68 | Yamashiro et al. [145] (2021) | LGG vs. HGG | Hold-out | 85:15 | Patient | Custom CNN | 91.30 | 92.7 | - | LGG = 69.20, HGG = 100.00 |
| 69 | Yin et al. [130] (2021) | AS vs. OG vs. GB | Hold-out | 86:14 | Patient | [WSI] 2D DenseNet [MRI] 3D DenseNet [WSI-MRI] Ensemble 2D-3D | 88.90 82.00 94.40 | - - - | 94.30 85.70 97.10 | - - - |
| 70 | Aamir et al. [156] (2022) | MN vs. GL vs. PT | 5-fold CV | Patient | Custom CNN | 98.95 | - | 97.98 | MN = 97.31, GL = 99.51, PT = 99.34 | |
| 71 | Ahmad et al. [89] (2022) | MN vs. GL vs. PT | Three-way | 60:20:20 | - | ResNet50 (w/o DA) ResNet50 (w/ CDA) ResNet50 (w/ GDA) ResNet50 (w/ CDA+GDA) | 72.63 77.52 92.30 96.25 | - - - - | 71.07 76.06 91.77 96.97 | MN = 73.94, GL = 76.92, PT = 65.05 MN = 76.76, GL = 82.87, PT = 69.89 MN = 92.25, GL = 96.15, PT = 86.56 MN = 96.47, GL = 96.50, PT = 95.70 |
| 72 | Alanazi et al. [160] (2022) | H vs. T MN vs. GL vs. PT | Three-way | 80:20, 2 Test | - | TL (on Kaggle-I) Custom CNN | 95.75 96.90 | - - | - 99.00 | - MN = 92.00, GL = 98.70, PT = 98.20 |
| 73 | Almalki et al. [121] (2022) * | H vs. MN vs. GL vs. PT MN vs. GL vs. PT | Hold-out | 80:20, 2 Test | - | Custom CNN-SVM | 98.00 97.16 | - - | - - | H = 94.70, MN = 97.30, GL = 98.80, PT = 99.40 MN = 99.20, GL = 94.71, PT = 99.40 |
| 74 | Amou et al. [81] (2022) | MN vs. GL vs. PT | Hold-out | 90:10 | - | Custom CNN VGG16 VGG19 DenseNet201 InceptionV3 ResNet50 | 98.70 97.08 96.43 94.81 92.86 89.29 | - - - - - - | 98.60 96.60 95.56 93.60 92.00 89.00 | MN = 97.00, GL = 99.00, PT = 99.00 MN = 97.00, GL = 96.00, PT = 99.00 MN = 93.00, GL = 97.00, PT = 99.00 MN = 85.00, GL = 97.00, PT = 100.00 MN = 82.00, GL = 97.00, PT = 96.00 MN = 57.00, GL = 77.00, PT = 98.00 |
| 75 | Aurna et al. [82] (2022) | H vs. MN vs. GL vs. PT | LOOCV (on dataset) | - | 2-stage Ensemble EfficientNetB0-ResNet50- Custom CNN | 98.96 | 98.90 | 99.00 | H = 100.00, MN = 99.00, GL = 98.00, PT = 99.00 | |
| 76 | Chatterjee et al. [59] (2022) | H vs. LGG vs. HGG | 3-fold CV, Test | 70:30 | Patient | (2+1)D ResNet TL (2+1)D ResNet 2D-3D Mixed ResNet TL 2D-3D Mixed ResNet 3D ResNet18 TL 3D ResNet18 | - - - 96.98 - - | - - - - - - | 90.35 92.37 86.07 93.45 90.95 89.25 | H = 99.04, LGG = 91.43, HGG = 82.29 H = 99.88, LGG = 91.08, HGG = 87.05 H = 97.69, LGG = 88.60, HGG = 75.05 H = 99.51, LGG = 93.19, HGG = 88.37 H = 99.44, LGG = 92.06, HGG = 82.89 H = 99.97, LGG = 85.52, HGG = 83.53 |
| 77 | Chitnis et al. [197] (2022) * | H vs. MN vs. GL vs. PT | Hold-out | 88:12 | - | Custom CNN DenseNet101 VGGNet16 ResNet50 | 90.60 86.80 88.33 85.79 | 95.60 92.84 94.31 94.34 | 91.48 87.84 89.60 86.96 | (R) 91.50, (S) 97.99 (R) 86.14, (S) 96.07 (R) 88.15, (S) 98.61 (R) 85.17, (S) 95.77 |
| 78 | Coupet et al. [135] (2022) * | H vs. T | Three-way | 70:15:15 | Patient | Modality-ensemble TL CNNs TL 3DUNet | 86.38 82.96 | - - | - - | - H = 69.81, T = 96.44 |
| 79 | Dang et al. [98] (2022) | LGG vs. HGG | Three-way | 60:20:20 | - | VGG | 97.44 | - | - | - |
| 80 | Danilov et al. [146] (2022) | LGG vs. HGG G.I vs. G.II vs. G.III vs. G.IV LGG vs. HGG G.I vs. G.II vs. G.III vs. G.IV | Three-way | 80:10:10 | - | (3D) DenseNet (2D) TL ResNet200e | 67.00 83.00 61.00 50.00 | 76.00 95.00 73.00 72.00 | - 80.25 - 35.00 | (R) 58.00, (S) 78.00 G.I = 100.00, G.II = 63.00, G.III = 100.00, G.IV = 85.00 (R) 44.00, (S) 81.00 G.I = 56.00, G.II = 45.00, G.III = 32.00, G.IV = 47.00 |
| 81 | Ding et al. [57] (2022) * | LGG vs. HGG | Hold-out | Patient | Radiomics VGG16 Ensemble Radiomics-VGG16-RF | 74.00 60.00 80.00 | 82.20 71.20 89.80 | - - - | (R) 80.00, (S) 68.00 (R) 68.00, (S) 52.00 (R) 84.00, (S) 76.00 | |
| 82 | Ekong et al. [198] (2022) | H vs. MN vs. GL vs. PT | Three-way | 80:10:10 | - | Bayesian CNN MobileNet AlexNet VGG16 ResNet50 | 94.32 93.42 92.75 89.51 86.58 | - - - - - | 94.00 94.00 93.00 91.00 86.00 | H = 97.50, MN = 92.50, GL = 85.50, PT = 100.00 94.00 93.00 91.00 87.00 |
| 83 | Gao et al. [112] (2022) * | 18 types of tumors * | Three-way | 72:24:4 | Patient | DenseNet | 81.20 | 92.00 | - | (R) 87.60, (S) 84.90 |
| 84 | Gaur et al. [199] (2022) | MN vs. GL vs. PT | Three-way | 80:10:10 | - | Custom CNN | 85.37 | - | - | - |
| 85 | Guo et al. [150] (2022) | AS vs. OG vs. GB | 3-fold CV | - | Radiomics Modality-fusion DenseNet201 Modality-ensemble DenseNet201 | 83.70 84.60 87.80 | 87.00 88.30 90.2 | 83.40 84.60 87.80 | (R) 70.40, (S) 89.90 (R) 73.10, (S) 93.00 (R) 77.20, (S) 93.00 | |
| 86 | Gupta et al. [95] (2022) | H vs. T MN vs. GL vs. PT | Hold-out | 88:12 | - | InceptionResNetV2-RF | 96.66 96.88 | - - | 97.00 96.00 | H = 100.00, T = 93.00 MN = 100.00, GL = 100.00, PT = 85.00 |
| 87 | Gurunathan and Krishnan [200] (2022) | LGG vs. HGG | Hold-out | 75:25 | - | Custom CNN AlexNet VGG19 GoogLeNet | 99.40 98.14 97.97 95.69 | - - - - | 98.10 - - - | (R) 97.20, (S) 98.60 - - - |
| 88 | Haq et al. [90] (2022) * | MN vs. GL vs. PT | Hold-out | 70:30 | - | [w/o DA] TL ResNet50 TL VGG-16 TL InceptionV3 [w/ DA] TL ResNet50 TL VGG-16 TL InceptionV3 | 99.10 98.78 97.78 99.89 98.98 98.50 | 98.78 98.06 97.00 99.56 97.98 98.76 | 99.50 97.49 97.39 99.43 98.79 98.00 | (R) 89.60, (S) 100.00 (R) 84.64, (S) 99.80 (R) 92.23, (S) 96.88 (R) 96.13, (S) 99.08 (R) 97.87, (S) 100.00 (R) 98.56, (S) 100.00 |
| 89 | Hsu et al. [131] (2022) | AS vs. OG vs. GB | Three-way | 67:11:22 | Patient | [WSI] 2D ResNet50 [MRI] 3D ResUNet [WSI-MRI] ResNet50-ResUNet | 77.70 69.80 80.00 | - - - | 88.60 77.10 88.60 | - - - |
| 90 | Isunuri and Kakarla [201] (2022) | MN vs. GL vs. PT | 5-fold CV | - | Custom CNN | 97.52 | - | 97.26 | 97.19 | |
| 91 | Jeong et al. [113] (2022) | LGG vs. HGG | 5-fold CV | - | Custom CNN | 90.91 | 96.34 | - | (R) 92.69, (S) 84.90 | |
| 92 | Kazemi et al. [108] (2022) * | 1 MN vs. GL vs. PT 2 G.II vs. G.III vs. G.IV | Hold-out | 75:25 | - | SVM-KNN AlexNet VGGNet AlexNet-VGGNet SVM-KNN AlexNet VGGNet AlexNet-VGGNet | 80.14 91.88 89.96 98.06 82.44 92.59 90.05 98.99 | 80.93 92.67 90.29 99.14 84.63 92.9 90.51 99.23 | - - - - - - - - | - - - MN = 98.10, GL = 98.88, PT = 98.50 - - - MN = 98.02, GL = 95.90, PT = 98.95 |
| 93 | Khazaee et al. [202] (2022) | LGG vs. HGG | Hold-out | 80:20 | - | TL EfficientNetB0 | 98.87 | - | - | (R) 98.86, (S) 98.79 |
| 94 | Kibriya et al. [122] (2022) | MN vs. GL vs. PT | - | - | - | Ensemble AlexNet-GoogLeNet-ResNet18-SVM | 99.70 | 100.00 | - | MN = 99.80, GL = 98.96, PT = 100.00 |
| 95 | Koli et al. [203] (2022) | H vs. T MN vs. GL vs. PT | Three-way | 70:15:15 | - | TL ResNet50 | 90.00 96.00 | - - | 90.00 95.00 | - MN = 90.00, GL = 98.00, PT = 97.00 |
| 96 | Lakshmi and Rao [204] (2022) | H vs. MN vs. GL vs. PT | Hold-out | 80:20 | - | InceptionV3 | 89.00 | - | - | - |
| 97 | Maqsood et al. [114] (2022) | MN vs. GL vs. PT LGG vs. HGG | 5-fold CV | - | TL MobileNetV2-SVM | 98.92 97.47 | 98.93 - | 97.87 96.71 | MN = 99.03, GL = 98.82, PT = 98.79 (R) 97.22, (S) 97.94 | |
| 98 | Murthy et al. [205] (2022) * | H vs. T | - | - | - | Custom CNN | 95.26 | - | 97.52 | (R) 97.12, (S) 50.00 |
| 99 | Nayak et al. [206] (2022) | MN vs. GL vs. PT | Hold-out | 80:20 | - | TL EfficientNet TL ResNet50 TL MobileNet TL MobileNetV2 | 98.78 96.33 96.94 94.80 | - - - - | 98.75 96.50 97.00 95.00 | H = 98.00, MN = 100.00, GL = 97.00, PT = 100.00 H = 98.00, MN = 98.00, GL = 90.00, PT = 100.00 H = 98.00, MN = 95.00, GL = 94.00, PT = 100.00 H = 96.00, MN = 99.00, GL = 95.00, PT = 90.00 |
| 100 | Rajinikanth et al. [124] (2022) | LGG vs. HGG | 5-fold CV | 90:10 | - | TL VGG16-SoftMax TL VGG16-DT TL VGG16-KNN TL VGG16-SVM | 96.50 96.00 96.50 97.00 | - - - - | 96.55 96.00 96.52 97.00 | (R) 97.03, (S) 95.96 (R) 96.97, (S) 95.05 (R) 97.00, (S) 96.00 (R) 97.00, (S) 97.00 |
| 101 | Rasool et al. [125] (2022) | H vs. MN vs. GL vs. PT | Hold-out | 80:20 | - | TL GoogLeNet GoogLeNet-SVM | 93.10 98.10 | - | - | H = 95.20, MN = 85.10, GL = 97.00, PT = 100.00 H = 98.70, MN = 97.30, GL = 97.80, PT = 98.90 |
| 102 | Raza et al. [207] (2022) | MN vs. GL vs. PT | Hold-out | 70:30 | - | Custom TL GoogLeNet TL AlexNet TL GoogLeNet TL ShuufleNet TL ResNet50 TL MobileNetV2 TL SqueezeNet TL Darknet53 TL ResNet101 TL ExceptionNet | 99.67 97.80 98.26 98.37 98.60 99.00 97.91 99.13 98.91 98.69 | - - - - - - - - - - | 99.66 97.66 98.33 98.33 98.33 99.00 97.66 99.00 98.66 98.00 | (R) 100.00 (R) 97.66 (R) 98.66 (R) 98.66 (R)98.66 (R) 99.00 (R) 98.00 (R) 99.33 (R) 99.00 (R) 98.33 |
| 103 | Rizwan et al. [208] (2022) | MN vs. GL vs. PT G.II vs. G.III vs. G.IV | Train, Val+Test | 65:35 | - | Custom CNN | 99.80 97.14 | - - | - - | (Acc) MN = 98.92, GL = 96.72, PT = 97.81 (Acc) G.II = 99.00, G.III = 96.00, G.IV = 99.00 |
| 104 | Samee et al. [209] (2022) | MN vs. GL vs. PT | Hold-out | 70:30 | - | TL hybrid GoogLeNet-AlexNet TL AlexNet TL VGG16 TL MobileNetV2 TL ResNet TL SqueezeNet | 99.10 96.00 95.00 95.00 94.00 92.00 | 99.00 97.00 95.00 95.00 94.00 92.00 | - - - - - - | MN = 99.00, GL = 99.00, PT = 99.00 MN = 96.00, GL = 96.00, PT = 96.00 MN = 95.00, GL = 95.00, PT = 95.00 MN = 95.00, GL = 95.00, PT = 95.00 MN = 94.00, GL = 94.00, PT = 94.00 MN = 92.00, GL = 92.00, PT = 92.00 |
| 105 | Samee et al. [147] (2022) | LGG vs. HGG | 10-fold CV, Test | 70:15:15 | Patient | Custom CNN | 88.60 | - | - | LGG = 80.00, HGG = 88.60 |
| 106 | Sangeetha et al. [210] (2022) | H vs. T | LOOCV | Patient | TL (in Rembrandt) CNN | 94.00 | - | - | (R) 85.00, (S) 73.00 | |
| 107 | Saravanan et al. [109] (2022) | 1 LGG vs. HGG vs. PIT 2 OLI vs. EP vs. CAM | 10-fold CV | - | SVM-RBF GoogLeNet CDbLNL SVM-RBF GoogLeNet CDbLNL | 85.80 94.60 97.21 84.80 91.60 97.21 | - - - - - - | 85.10 90.90 95.72 84.10 90.10 94.34 | (R) 81.90 (R) 91.50 (R) 95.62 (R) 80.90 (R) 91.50 (R) 93.86 | |
| 108 | Sekhar et al. [126] (2022) | MN vs. GL vs. PT | 5-fold CV | Patient | TL GoogLeNet-SoftMax TL GoogLeNet-SVM TL GoogLeNet-KNN | 94.90 97.60 98.30 | - - - | 94.30 97.35 97.24 | MN = 96.92, GL = 91.13, PT = 97.77 MN = 97.96, GL = 94.59, PT = 100.00 MN = 94.57, GL = 98.02, PT = 99.10 | |
| 109 | Senan et al. [99] (2022) | H vs. MN vs. GL vs. PT | Hold-out | 80:20 | - | AlexNet-SoftMax AlexNet-SVM ResNet18-SoftMax ResNet18-SVM | 93.30 95.10 93.80 91.20 | - - - - | - - - - | H = 91.10, MN = 89.80, GL = 93.30, PT = 97.80 H = 91.10, MN = 89.80, GL = 93.30, PT = 97.80 H = 94.90, MN = 93.60, GL = 93.90, PT = 97.80 H = 87.30, MN = 93.60, GL = 93.30, PT = 97.20 H = 92.40, MN = 86.10, GL = 91.50, PT = 95.60 |
| 110 | Srinivas et al. [211] (2022) | Benign vs. Malignant | Three-way | - | - | TL VGG16 TL InceptionV3 TL ResNet50 | 86.05 64.00 74.00 | - - - | - - - | B = 89.47, M = 87.09 B = 5.55, M = 100.00 B = 89.47, M = 64.52 |
| 111 | Tandel et al. [75] (2022) * | LGG vs. HGG | 5-fold CV | - | TL Ensemble AlexNet, VGGNet, ResNet18, GoogLeNet, ResNet50 [Whole image] [Skull-stripped brain] [Tumor ROI] | 98.43 98.63 99.06 | 98.45 98.63 99.07 | - - - | (R) 98.33, (S) 98.57 (R) 98.63. (S) 98.57 (R) 99.04, (S) 99.10 | |
| 112 | Tripathi and Bag [83] (2022) * | LGG vs. HGG | Hold-out | 70:30 80:20 90:10 Average | - - - - | DST Fusion TL ResNets | 95.64 95.78 96.19 95.87 | - - - - | 92.41 91.91 94.13 92.82 | (R) 92.12, (S) 95.97 (R) 95.12, (S) 95.10 (R) 96.95, (S) 95.77 - |
| 113 | Tripathi and Bag [141] (2022) | LGG vs. HGG | 10-fold CV | Patient | Attention-based CNN | 95.86 | - | 94.84 | (R) 94.82, (S) 96.81 | |
| 114 | Tummala et al. [136] (2022) | MN vs. GL vs. PT | Three-way | 70:10:20 | - | Ensemble ViT | 98.70 | - | - | (R) 97.78, (S) 99.42 |
| 115 | Vankdothu et al. [213] (2022) | H vs. MN vs. GL vs. PIT | Hold-out | 88:12 | - | CNN RNN CNN-LSTM | 89.39 90.02 92.00 | - - - | - - - | (R) 98.30 (R) 98.00 (R) 98.50 |
| 116 | Wang et al. [132] (2022) | AS vs. OG vs. GB | Three-way | 70:10:20 | Patient | [WSI] Ensemble EfficientNet-B2, EfficientNet-B3, SE-ResNext10 [MRI] 3D CNN [WSI-MRI] 2D-3D Ensemble | 82.20 73.30 75.00 | - - - | 88.60 82.90 75.30 | - |
| 117 | Xiong et al. [115] (2022) * | AS vs. OG vs. GB | Three-way | 70:15:15 | Patient | [MRI] TL ResNet34 [MRI-tabular] TL ResNet34 | 67.50 70.00 | - - | - - | AST = 85.70, OLI = 40.00, GBM = 68.80 AST = 85.70, OLI = 30.00, GBM = 81.30 |
| 118 | Xu et al. [118] (2022) * | LGG vs. HGG | Three-way | 60:20:20 | Patient | 1 TL ResNet18 1 TL ResNet18+radiomics 2 TL ResNet18 2 TL ResNet18+radiomics | 83.33 88.10 87.40 94.10 | - - - - | - - - - | (R) 90.8 (R) 90.1 (R) 93.1 (R) 97.1 |
| 119 | Yazdan et al. [214] (2022) * | H vs. MN vs. GL vs. PT | k-fold CV | - | TL AlexNet TL ResNet Multi-scale CNN 1 Multi-scale CNN 2 Multi-scale CNN 3 | 87.89 91.98 89.27 94.19 89.67 | - - - - - | 88.03 91.59 89.41 94.06 89.49 | (R) 87.86, (S) 85.42 (R) 91.44, (S) 89.79 (R) 89.15, (S) 86.91 (R) 93.74, (S) 92.62 (R) 89.24, (S) 88.35 | |
| 120 | Zahoor et al. [103] (2022) * | 1 H vs. T 2 MN vs. GL vs. PT | Hold-out | 60:40 80:20 | - | ResNet18-Softmax TL ResNet18-Softmax TL ResNet18-SVM Custom CNN-SVM Custom CNN-SVM | 97.43 98.91 99.16 99.56 99.20 | - - - 99.90 - | 97.56 98.69 98.94 99.45 99.09 | (R) 98.12 (R) 99.66 (R) 97.99 (R) 98.99 MN = 98.60, GL = 99.30, PT = 99.50 |
| 121 | AlTahhan et al. [127] (2023) | H vs. MN vs. GL vs. PT | Three-way | 70:30:- | - | TL GoogLeNet-SoftMax TL AlexNet-SoftMax TL AlexNet-SVM TL AlexNet-KNN | 88.00 85.00 95.00 97.00 | - - - - | 88.46 86.27 93.62 97.96 | H = 87.50, MN = 88.00, GL = 88.50, PT = 88.00 H = 84.00, MN = 84.60, GL = 88.00, PT = 83.30 H = 92.60, MN = 92.30, GL = 100.00, PT = 96.00 H = 96.20, MN = 96.00, GL = 100.00, PT = 96.00 |
| 122 | Al-Zoghby et al. [137] (2023) | MN vs. GL vs. PT | Hold-out | 80:20 | - | Ensemble TL VGG-16 & Custom CNN | 99.00 | 99.00 | 99.00 | MN = 98.00, GL = 100.00, PT = 99.00 |
| 123 | Anagun [215] (2023) | MN vs. GL vs. PT | Three-way | 80:10:10 | - | TL EfficientNetv2 TL ResNet18 TL ResNet200d TL InceptionV4 | 99.85 99.62 99.83 99.69 | 99.92 99.75 99.84 99.73 | 98.07 96.64 97.72 97.19 | 98.05 96.71 97.66 97.37 |
| 124 | Anand et al. [91] (2023) | H vs. T | Three-way | 76:14:10 | - | TL EfficientNetB0 TL InceptionV3 TL ResNet50 TL VGG19 Custom CNN w/o DA Custom CNN w/ DA Ensemble TL VGG19 & Custom CNN | - - - 95.00 96.00 97.00 98.00 | - - - - - - - | 54.50 91.50 85.00 96.00 96.50 97.00 98.50 | H = 44.00, T = 30.00 H = 90.00, T = 94.00 H = 82.00, T = 81.00 H = 98.00, T = 96.00 H = 95.00, T = 98.00 H = 98.00, T = 96.00 H = 98.50, T = 99.00 |
| 125 | Apostolopoulos et al. [216] (2023) * | H vs. MN vs. GL vs. PT | H10-fold CV | - | Attention VGG19 VGG19 ResNet152 MobileNetV2 InceptionV3 | 93.53 91.08 86.00 86.89 87.13 | 95.3 - - - - | 90.55 - - - - | H = 99.60, MN = 90.62, GL = 96.76, PT = 91.61 - - - - | |
| 126 | Asif et al. [138] (2023) | MN vs. GL vs. PT | Hold-out | 80:20 | - | TL Xception TL VGG16 TL DenseNet201 TL ResNet152V2 TL InceptionResNetV2 Ensemble TL DenseNet201, ResNet152V2, InceptionResNetV2 | 91.83 93.54 97.22 95.58 95.75 98.69 | - - 98.00 98.00 96.00 99.00 | 90.65 93.01 96.81 95.12 94.96 98.39 | MN = 82.98, GL = 92.63, PT = 97.31 MN = 84.40, GL = 96.49, PT = 97.31 MN = 92.91, GL = 98.60, PT = 98.39 MN = 92.91, GL = 94.74, PT = 98.92 MN = 89.36, GL = 97.54, PT = 97.85 MN = 96.45, GL = 99.29, PT = 99.46 |
| 127 | Athisayamani et al. [110] (2023) | MN vs. GL vs. PT | - | - | - | TL ResNet152 CNN SVM | 98.85 97.00 94.00 | 98.00 - - | - - - | MN = 97.00, GL = 98.00, PT = 99.00 (R) 94.00 (R) 94.00 |
| 128 | Bairagi et al. [111] (2023) * | H vs. T | 10-fold CV, Test | 80:20 | - | SVM TL AlexNet TL VGG16 TL GoogLeNet | 89.53 98.67 90.67 91.49 | - - - - | - - - - | - - - - |
| 129 | Deepa et al. [84] (2023) * | H vs. T | Hold-out | 90:10 | - | Custom CJHBA Based DRN | 1 92.10 2 91.84 | - | - - | (R) 93.13, (S) 92.84 (R) 91.55, (S) 91.86 |
| 130 | El-Wahab et al. [219] (2023) | MN vs. GL vs. PT | 5-fold CV, Test | 80:20 | - | TL VGG16 TL VGG19 TL InceptionV3 TL ResNet50 TL MobileNet BTCfCNN TL BTCfCNN (bt folds) TL BTC-fCNN | 92.07 93.05 80.35 74.48 89.16 93.08 98.63 98.86 | - - - - - - - - | - - - - - 92.21 98.46 98.77 | - - - - - (R) 92.01, (S) 96.34 (R) 98.49, (S) 99.31 (R) 98.83, (S) 99.41 |
| 131 | Hossain et al. [116] (2023) | H vs. MN vs. GL vs. PT | three-way | 80:10:10 | - | TL InceptionV3 TL VGG16 TL Xception TL ResNet50 TL VGG19 TL InceptionResNetV2 Ensemble TL VGG16, InceptionV3, Xception | 95.72 95.11 94.50 93.88 94.19 93.58 96.94 | - - - - - - - | 69.00 69.00 69.00 72.00 64.00 70.00 76.00 | H = 100.00, MN = 98.00, GL = 31.00, PT = 70.00 H = 100.00, MN = 99.00, GL = 22.00, PT = 80.00 H = 98.00, MN = 91.00, GL = 39.00, PT = 77.00 H = 100.00, MN = 97.00, GL = 28.00, PT = 72.00 H = 100.00, MN = 97.00, GL = 22.00, PT = 64.00 H = 98.00, MN = 99.00, GL = 33.00, PT = 68.00 H = 100, MN = 93.00, GL = 49.00, PT = 73.00 |
| 132 | Hussain et al. [148] (2023) | LGG vs. HGG | Hold-out | - | Patient | 3D CNN -T1 -T1c -T2 -Flair -Segmentation Ensemble | 94.00 94.00 94.38 93.23 94.38 94.20 | - - - - - - | 95.77 95.77 95.65 95.77 95.77 95.75 | - - - - - - |
| 133 | Kibriya et al. [119] (2023) | H vs. T | Hold-out | 70:30 | - | 1 Radiomics-SVM 1 Radiomics-KNN 1 VGG16-SVM 1 VGG16-KNN 1 Radiomics+VGG16-SVM 1 Radiomics+VGG16-KNN 2 Radiomics-SVM 2 Radiomics-KNN 2 VGG16-SVM 2 VGG16-KNN 2 Radiomics+VGG16-SVM 2 Radiomics+VGG16-KNN | 72.00 84.00 92.10 88.10 93.30 96.00 96.10 96.00 98.00 97.80 99.00 98.70 | - - - - 99.00 99.00 - - - - 100.00 100.00 | - - - - 93.50 94.50 - - - - 99.00 99.00 | - - - - 93.00 95.50 - - - - 99.00 99.00 |
| 134 | Krishnapriya and Karuna [92] (2023) | H vs. T | Hold-out | 70:30 | - | [w/o DA] TL VGG16 TL VGG19 TL ResNet 50 TL InceptionV3 [w/ DA] TL VGG 16 TL VGG19 TL ResNet50 TL InceptionV3 | 90.50 90.70 88.02 66.26 99.00 99.48 97.92 81.25 | - - - - - - - - | - - - - 99.08 99.17 82.24 58.16 | - - - - 98.18 98.76 87.27 63.25 |
| 135 | Kumar et al. [128] (2023) * | Benign vs. Malignant | Hold-out | 90:10 | - | ResNet50-Softmax ResNet50-SVM TL ResNet50 | 86.57 91.24 96.80 | - - - | - - 97.34 | - - Benign = 95.21, Malignant = 97.56 |
| 136 | Mahmud et al. [220] (2023) | H vs. M vs GL vs. PT | Three-way | 80:10:10 | - | Custom CNN ResNet50 VGG16 InceptionV3 | 93.30 81.10 71.60 80.00 | 98.43 94.2 89.6 89.14 | - - - - | 91.13 81.04 70.03 79.81 |
| 137 | Muezzinoglu et al. [221] (2023) | H vs. MN vs. GL vs. PT | 10-fold CV | - | PatchResNet | 98.10 | 98.01 | H = 98.40, MN = 98.51, GL = 95.68, PT = 100.00 | ||
| 138 | Özkaraca et al. [222] (2023) | H vs. MN vs. GL vs. PT | 10-fold CV, Test | 80:20 | - | CNN VGG16 DenseNet Custom CNN | - - - - | - - - - | 92.00 85.75 84.75 96.5 | H = 98.00, MN = 84.00, GL = 90.00, PT = 97.00 H = 96.00, MN = 67.00, GL = 89.00, PT = 94.00 H = 99.00, MN = 83.00, GL = 99.00, PT = 58.00 H = 98.00, MN = 91.00, GL = 97.00, R PT = 99.00 |
| 139 | Özkaya and Şağıroğlu [224] (2023) | LGG vs. HGG | 10-fold CV | - | TL MobileNetV2 TL DenseNet201 TL Xception 99.63 TL InceptionV3 TL EfficientNetV2S 99.24 | 99.85 99.66 99.70 99.63 99.41 | 99.92 99.77 99.64 99.74 99.25 | 99.85 99.67 - 99.64 - | - - - | |
| 140 | Rasheed et al. [225] (2023) | MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN VGG16 VGG19 ResNet50 MobileNet InceptionV3 | 98.04 90.70 92.82 94.77 93.47 85.97 | 98.00 93.00 94.00 96.00 95.00 88.00 | 98.00 90.00 93.00 95.00 93.00 85.00 | MN = 95.00, GL = 99.00, PT = 100.00 MN = 79.00, GL = 92.00, PT = 99.00 MN = 85.00, GL = 94.00, PT = 98.00 MN = 89.00, GL = 95.00, PT = 99.00 MN = 90.00, GL = 92.00, PT = 99.00 MN = 66.00, GL = 89.00, PT = 98.00 |
| 141 | Rui et al. [149] (2023) * | LGG vs. HGG | 5-fold CV, Test | - | Patient | Inception CNN [Flair] [T1c] Modality-ensemble | 69.00 74.00 80.00 | - - - | 60.00 70.00 78.00 | (R) 75.00, (S) 60.00 (R) 75.00, (S) 73.00 (R) 76.00, (S) 87.00 |
| 142 | Shirehjini et al. [123] (2023) * | G.I vs. G.II vs. G.III vs. G.IV | Three-way | 70:15:15 | - | TL VGG16-Softmax TL VGG16-LR TL-SVM | 96.93 98.15 99.38 | - - 99.93 | 96.64 98.12 99.09 | (R) 99.29 (R) 97.94 G.I: 96.00, G.II = 100.00, G.III = 100.00, G.IV = 100.00 |
| 143 | Srinivasan et al. [226] (2023) | H vs. MN vs. GL vs. PT | Hold-out | 80:20 | - | Custom CNN UNet ResNet | 98.17 92.61 96.23 | - - - | - - - | (R) 98.79, (S)91.34 (R) 97.56 (S) 81.51 (R) 97.90, (S) 90.23 |
| 144 | Tandel et al. [139] (2023) * | LGG vs. HGG | 5-fold CV | - | Ensemble TL AlexNet, VGG16, ResNet18, GoogLeNet, ResNet50 | [T1] 94.75 [T2] 97.98 [Flair] 98.88 | 94.92 97.99 98.88 | - - - | (R) 94.29, (S) 95.56 (R) 97.60, (S) 98.37 (R) 98.95, (S) 98.80 | |
| 145 | van der Voort et al. [117] (2023) | G.II vs. G.III vs. G.IV LGG vs. HGG | Three-way | 75:15:15 | Patient | UNet | 71.00 84.00 | 81.00 91.00 | - | G.II = 75.00, G.III = 17.00, G.IV = 95.00 (R) 72.00, (S) 93.00 |
| 146 | Wu et al. [157] (2023) | LGG vs. HGG | Three-way | 54:13:33 | Patient | Attention-based custom CNN VGG19 ResNet50 DenseNet201 InceptionV4 | 95.19 - - - - | 98.40 95.80 94.10 95.70 97.00 | 93.34 - - - - | (R) 94.01, (S) 99.53 - - - - |
AS: Astrocytoma, Acc: Accuracy, AUC: Area Under the Receiver Operating Characteristic Curve, CDA: Classic Data Augmentation, CDbLNL: Convolutional Neural Network Database Learning with Neighboring Network Limitation, CJHBA: Chronological Jaya Honey Badger Algorithm, CNN: Convolutional Neural Network, CV: Cross-Validation, DA: Data Augmentation, DRLBP: Dominant Rotated Local Binary Patterns, DRN: Deep Residual Network, DT: Decision Tree, DCGAN: Deep Convolutional Generative Adversarial Network, DWAE: Deep Wavelet Auto-Encoder, DWT: Discrete Wavelet Transform, ELM: Extreme Learning, Machine, EP: Ependymoma, GA: Genetic Algorithm, GAN: Generative Adversarial Network, GB: Glioblastoma, GDA: Generative Data Augmentation, GL: Glioma, GL: Glioma, HGG: High-grade Glioma, KNN: K-Nearest Neighbors, LGG: Low-grade Glioma, LOOCV: Leave-One-Out Cross-Validation, LR: Logistic Regression, LSTM: Long Short-Term Memory, MB: Medulloblastoma, MN: Meningioma, MT: Metastasis, OG: Oligodendroglioma, PT: Pituitary, (P): Precision, (R): Recall, RF: Random Forest, RFE: Recursive Feature Elimination, ROI: Region of Interest, (S): Specificity, Std: Standardization, SVM: Support Vector Machine, TL: Transfer Learning, WSI: Whole Slide Image. Papers highlighted with an asterisk (*) indicate that not all outcomes are reported. For comprehensive details, readers are referred to the original paper. Numerical superscripts link models with datasets in Table A1 when different data sources yield individual results.
References
- Sohn, E. The reproducibility issues that haunt health-care AI. Nature 2023, 613, 402–403. [Google Scholar] [CrossRef] [PubMed]
- McDermott, M.; Wang, S.; Marinsek, N.; Ranganath, R.; Foschini, L.; Ghassemi, M. Reproducibility in machine learning for health research: Still a ways to go. Sci. Transl. Med. 2021, 13, eabb1655. [Google Scholar] [CrossRef] [PubMed]
- Muehlematter, U.; Daniore, P.; Vokinger, K. Approval of artificial intelligence and machine learning-based medical devices in the USA and Europe (2015–20): A comparative analysis. Lancet Digit. Health 2021, 3, e195–e203. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, K.; Moukheiber, L.; Celi, L.; Patel, M.; Mahmood, F.; Gondim, D.; Hogarth, M.; Levenson, R. AI in Pathology: What could possibly go wrong? Semin. Diagn. Pathol. 2023, 40, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Di Nunno, V.; Fordellone, M.; Minniti, G.; Asioli, S.; Conti, A.; Mazzatenta, D.; Balestrini, D.; Chiodini, P.; Agati, R.; Tonon, C.; et al. Machine learning in neuro-oncology: Toward novel development fields. J. Neuro-Oncol. 2022, 159, 333–346. [Google Scholar] [CrossRef] [PubMed]
- Bacciu, D.; Lisboa, P.; Vellido, A. Deep Learning in Biology and Medicine; World Scientific: London, UK, 2022. [Google Scholar]
- Bernal, J.; Kushibar, K.; Clèrigues, A.; Oliver, A.; Lladó, X. Deep learning for medical imaging. In Deep Learning in Biology and Medicine; World Scientific: London, UK, 2022; pp. 11–54. [Google Scholar]
- Xue, H.; Hu, G.; Hong, N.; Dunnick, N.; Jin, Z. How to keep artificial intelligence evolving in the medical imaging world? Challenges and opportunities. Sci. Bull. 2023, 68, 648–652. [Google Scholar] [CrossRef] [PubMed]
- Pati, S.; Baid, U.; Edwards, B.; Sheller, M.; Wang, S.-H.; Reina, G.A.; Foley, P.; Gruzdev, A.; Karkada, D.; Davatzikos, C.; et al. Federated learning enables big data for rare cancer boundary detection. Nat. Commun. 2022, 13, 7346. [Google Scholar] [CrossRef]
- Thrall, J.; Li, X.; Quanzheng, L.; Cruz, C.; Do, S.; Dreyer, K.; Brink, J. Artificial Intelligence and Machine Learning in Radiology: Opportunities, challenges, pitfalls, and criteria for success. J. Am. Coll. Radiol. 2018, 15, 504–508. [Google Scholar] [CrossRef]
- Liu, Y.; Leong, A.; Zhao, Y.; Xiao, L.; Mak, H.; Tsang, A.; Lau, G.; Leung, G.; Wu, E. A low-cost and shielding-free ultra-low-field brain MRI scanner. Nat. Commun. 2021, 12, 7238. [Google Scholar] [CrossRef]
- Julià-Sapé, M.; Acosta, D.; Majós, C.; Moreno-Torres, A.; Wesseling, P.; Acebes, J.; Griffiths, J.R.; Arús, C. Comparison between neuroimaging classifications and histopathological diagnoses using an international multicenter brain tumor magnetic resonance imaging database. J. Neurosurg. 2006, 105, 6–14. [Google Scholar] [CrossRef]
- Arita, K.; Miwa, M.; Bohara, M.; Moinuddin, F.; Kamimura, K.; Yoshimoto, K. Precision of preoperative diagnosis in patients with brain tumor—A prospective study based on “top three list” of differential diagnosis for 1061 patients. Surg. Neurol. Int. 2020, 11, 55. [Google Scholar] [CrossRef] [PubMed]
- Osborn, A.; Louis, D.; Poussaint, T.; Linscott, L.; Salzman, K.L. The 2021 World Health Organization classification of tumors of the central nervous system: What neuroradiologists need to know. Am. J. Neuroradiol. 2022, 43, 928–937. [Google Scholar] [CrossRef]
- Wen, P.Y.; Macdonald, D.R.; Reardon, D.A.; Cloughesy, T.F.; Sorensen, A.G.; Galanis, E.; DeGroot, J.; Wick, W.; Gilbert, M.R.; Lassman, A.B.; et al. Updated response assessment criteria for high-grade gliomas: Response assessment in neuro-oncology working group. J. Clin. Oncol. 2010, 28, 1963–1972. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.; Leeds, N.; Fuller, G.; Van Tassel, P.; Maor, M.; Sawaya, R.; Levin, V. Malignant gliomas: MR imaging spectrum of radiation therapy-and chemotherapy-induced necrosis of the brain after treatment. Radiology 2000, 217, 377–384. [Google Scholar] [CrossRef]
- Segura, P.P.; Quintela, N.V.; García, M.M.; del Barco Berrón, S.; Sarrió, R.G.; Gómez, J.G.; Castaño, A.G.; Martín, L.M.N.; Rubio, O.G.; Losada, E.P. SEOM-GEINO clinical guidelines for high-grade gliomas of adulthood (2022). Clin. Transl. Oncol. 2023, 25, 2634–2646. [Google Scholar] [CrossRef] [PubMed]
- Da Cruz, L.C.H.; Rodriguez, I.; Domingues, R.; Gasparetto, E.; Sorensen, A. Pseudoprogression and Pseudoresponse: Imaging Challenges in the Assessment of Posttreatment Glioma. AJNR Am. J. Neuroradiol. 2011, 32, 1978–1985. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; van den Bent, M.; Youssef, G.; Cloughesy, T.F.; Ellingson, B.M.; Weller, M.; Galanis, E.; Barboriak, D.P.; de Groot, J.; Gilbert, M.R.; et al. RANO 2.0: Update to the response assessment in neuro-oncology criteria for high-and low-grade gliomas in adults. J. Clin. Oncol. 2023, 41, 5187–5199. [Google Scholar] [CrossRef]
- Tustison, N.J.; Avants, B.B.; Cook, P.A.; Zheng, Y.; Egan, A.; Yushkevich, P.A.; Gee, J.C. N4ITK: Improved N3 bias correction. IEEE Trans. Med. Imaging 2010, 29, 1310–1320. [Google Scholar] [CrossRef]
- Sled, J.G.; Zijdenbos, A.P.; Evans, A.C. A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans. Med. Imaging 1998, 17, 87–97. [Google Scholar] [CrossRef]
- Ronneberger, O.; Fischer, P.; Brox, T. U-Net: Convolutional Networks for biomedical image segmentation. In Proceedings of the Medical Image Computing and Computer-Assisted Intervention—MICCAI, Munich, Germany, 5–9 October 2015; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 234–241. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems; Cortes, C., Lawrence, N., Lee, D., Sugiyama, M., Garnett, R., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2015; Volume 28. [Google Scholar]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask R-CNN. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Lisboa, P.; Saralajew, S.; Vellido, A.; Fernández-Domenech, R.; Villmann, T. The Coming of Age of Interpretable and Explainable Machine Learning Models. Neurocomputing 2023, 535, 25–39. [Google Scholar] [CrossRef]
- Mukherjee, T.; Pournik, O.; Lim Choi Keung, S.; Arvanitis, T. Clinical decision support systems for brain tumour diagnosis and prognosis: A systematic review. Cancers 2023, 15, 3523. [Google Scholar] [CrossRef] [PubMed]
- Bertsimas, D.; Wiberg, H. Machine Learning in Oncology: Methods, applications, and challenges. JCO Clin. Cancer Inform. 2020, 4, 885–894. [Google Scholar] [CrossRef]
- Jha, A.; Mithun, S.; Sherkhane, U.B.; Jaiswar, V.; Shi, Z.; Kalendralis, P.; Kulkarni, C.; Dinesh, M.S.; Rajamenakshi, R.; Sunder, G.; et al. Implementation of big imaging data pipeline adhering to FAIR principles for Federated Machine Learning in Oncology. IEEE Trans. Radiat. Plasma Med. Sci. 2022, 6, 207–213. [Google Scholar] [CrossRef]
- Su, X.; Chen, N.; Sun, H.; Liu, Y.; Yang, X.; Wang, W.; Zhang, S.; Tan, Q.; Su, J.; Gong, Q.; et al. Automated Machine Learning based on radiomics features predicts H3 K27M mutation in midline gliomas of the brain. Neuro-Oncology 2020, 22, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Mocioiu, V.; Pedrosa de Barros, N.; Ortega-Martorell, S.; Slotboom, J.; Knecht, U.; Arús, C.; Vellido, A.; Julià-Sapé, M. A Machine Learning pipeline for supporting differentiation of glioblastomas from single brain metastases. In Proceedings of the ESANN 2016, European Symposium on Artificial Neural Networks, Computational Intelligence and Machine Learning (ESANN) Bruges (Belgium), Bruges, Belgium, 5–7 October 2016; pp. 247–252. [Google Scholar]
- Pitarch, C.; Ribas, V.; Vellido, A. AI-Based Glioma Grading for a Trustworthy Diagnosis: An Analytical Pipeline for Improved Reliability. Cancers 2023, 15, 3369. [Google Scholar] [CrossRef]
- Tabassum, M.; Suman, A.; Suero Molina, E.; Pan, E.; Di Ieva, A.; Liu, S. Radiomics and Machine Learning in Brain Tumors and Their Habitat: A Systematic Review. Cancers 2023, 15, 3845. [Google Scholar] [CrossRef] [PubMed]
- Griethuysen, J.; Fedorov, A.; Parmar, C.; Hosny, A.; Aucoin, N.; Narayan, V.; Beets-Tan, R.; Fillon-Robin, J.; Pieper, S.; Aerts, H. Clinical Decision Support Systems for Brain Tumour Diagnosis and Prognosis: A Systematic Review. Cancer Res. 2017, 77, e104–e107. [Google Scholar] [CrossRef]
- Hyvärinen, A.; Oja, E. Independent component analysis: Algorithms and applications. Neural Netw. 2000, 13, 411–430. [Google Scholar] [CrossRef]
- Lee, J.; Zhao, Q.; Kent, M.; Platt, S. Tumor Segmentation using temporal Independent Component Analysis for DCE-MRI. BioRxiv 2022. [Google Scholar] [CrossRef]
- Chakhoyan, A.; Raymond, C.; Chen, J.; Goldman, J.; Yao, J.; Kaprealian, T.; Pouratian, N.; Ellingson, B. Probabilistic independent component analysis of dynamic susceptibility contrast perfusion MRI in metastatic brain tumors. Cancer Imaging 2019, 19, 14. [Google Scholar] [CrossRef]
- Lee, D.; Seung, H. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Martorell, S.; Lisboa, P.; Vellido, A.; Julià-Sapé, M.; Arús, C. Non-negative matrix factorisation methods for the spectral decomposition of MRS data from human brain tumours. BMC Bioinform. 2012, 13, 38. [Google Scholar] [CrossRef]
- Ungan, G.; Arús, C.; Vellido, A.; Julià-Sapé, M. A Comparison of Non-Negative Matrix Underapproximation Methods for the Decomposition of Magnetic Resonance Spectroscopy Data from Human Brain Tumors. NMR Biomed. 2023, 36, e5020. [Google Scholar] [CrossRef]
- Sauwen, N.; Acou, M.; Van Cauter, S.; Sima, D.M.; Veraart, J.; Maes, F.; Himmelreich, U.; Achten, E.; Van Huffel, S. Comparison of unsupervised classification methods for brain tumor segmentation using multi-parametric MRI. Neuroimage Clin. 2016, 12, 753–764. [Google Scholar] [CrossRef] [PubMed]
- Ashtari, P.; Sima, D.; De Lathauwer, L.; Sappey-Marinier, D.; Maes, F.; Van Huffel, S. Factorizer: A scalable interpretable approach to context modeling for medical image segmentation. Med. Image Anal. 2023, 84, 102706. [Google Scholar] [CrossRef]
- Dosovitskiy, A.; Beyer, L.; Kolesnikov, A.; Weissenborn, D.; Zhai, X.; Unterthiner, T.; Dehghani, M.; Minderer, M.; Heigold, G.; Gelly, S.; et al. An image is worth 16 × 16 words: Transformers for image recognition at scale. arXiv 2020, arXiv:2010.11929. [Google Scholar]
- Lundervold, A.S.; Lundervold, A. An overview of deep learning in medical imaging focusing on MRI. Z. Med. Phys. 2019, 29, 102–127. [Google Scholar] [CrossRef] [PubMed]
- Cai, L.; Gao, J.; Zhao, D. A review of the application of deep learning in medical image classification and segmentation. Ann. Transl. Med. 2020, 8, 713. [Google Scholar] [CrossRef]
- Chen, X.; Wang, X.; Zhang, K.; Fung, K.M.; Thai, T.C.; Moore, K.; Mannel, R.S.; Liu, H.; Zheng, B.; Qiu, Y. Recent advances and clinical applications of deep learning in medical image analysis. Med. Image Anal. 2022, 79, 102444. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. ImageNet classification with deep convolutional neural networks. In Proceedings of the Advances in Neural Information Processing Systems; Pereira, F., Burges, C., Bottou, L., Weinberger, K., Eds.; Curran Associates, Inc.: Red Hook, NY, USA, 2012; Volume 25. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going deeper with convolutions. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Nashville, TN, USA, 20–25 June 2014; pp. 1–9. [Google Scholar] [CrossRef]
- Simonyan, K.; Zisserman, A. Very Deep Convolutional Networks for Large-Scale Image Recognition. In Proceedings of the 3rd International Conference on Learning Representations, ICLR 2015—Conference Track Proceedings, San Diego, CA, USA, 7–9 May 2014. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015; pp. 770–778. [Google Scholar] [CrossRef]
- Huang, G.; Liu, Z.; Van Der Maaten, L.; Weinberger, K.Q. Densely connected convolutional networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4700–4708. [Google Scholar]
- Tan, M.; Le, Q. EfficientNet: Rethinking model scaling for convolutional neural networks. In Proceedings of the 36th International Conference on Machine Learning, Long Beach, CA, USA, 9–15 June 2019; Volume 97, pp. 6105–6114. [Google Scholar]
- Goodfellow, I.J.; Pouget-Abadie, J.; Mirza, M.; Xu, B.; Warde-Farley, D.; Ozair, S.; Courville, A.; Bengio, Y. Generative Adversarial Networks. arXiv 2014, arXiv:1406.2661. [Google Scholar] [CrossRef]
- Deng, J.; Dong, W.; Socher, R.; Li, L.J.; Li, K.; Li, F.-F. Imagenet: A large-scale hierarchical image database. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Miami, FL, USA, 20–25 June 2009; pp. 248–255. [Google Scholar]
- Lin, T.Y.; Maire, M.; Belongie, S.; Hays, J.; Perona, P.; Ramanan, D.; Dollár, P.; Zitnick, C.L. Microsoft COCO: Common objects in context. In Proceedings of the Computer Vision—ECCV 2014; Fleet, D., Pajdla, T., Schiele, B., Tuytelaars, T., Eds.; Springer: Cham, Switzerland, 2014; pp. 740–755. [Google Scholar]
- Yang, J.; Huang, X.; He, Y.; Xu, J.; Yang, C.; Xu, G.; Ni, B. Reinventing 2D Convolutions for 3D Images. IEEE J. Biomed. Health Inform. 2021, 25, 3009–3018. [Google Scholar] [CrossRef]
- Banerjee, S.; Mitra, S.; Masulli, F.; Rovetta, S. Glioma classification using deep radiomics. SN Comput. Sci. 2020, 1, 209. [Google Scholar] [CrossRef]
- Ding, J.; Zhao, R.; Qiu, Q.; Chen, J.; Duan, J.; Cao, X.; Yin, Y. Developing and validating a deep learning and radiomic model for glioma grading using multiplanar reconstructed magnetic resonance contrast-enhanced T1-weighted imaging: A robust, multi-institutional study. Quant. Imaging Med. Surg. 2022, 12, 1517. [Google Scholar] [CrossRef] [PubMed]
- Zhuge, Y.; Ning, H.; Mathen, P.; Cheng, J.Y.; Krauze, A.V.; Camphausen, K.; Miller, R.W. Automated glioma grading on conventional MRI images using deep convolutional neural networks. Med. Phys. 2020, 47, 3044–3053. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.; Nizamani, F.A.; Nürnberger, A.; Speck, O. Classification of brain tumours in MR images using deep spatiospatial models. Sci. Rep. 2022, 12, 1505. [Google Scholar] [CrossRef]
- Baheti, B.; Pati, S.; Menze, B.; Bakas, S. Leveraging 2D Deep Learning ImageNet-trained Models for Native 3D Medical Image Analysis. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the BrainLes 2022, Singapore, 18 September 2022; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2023; Volume 13769, pp. 68–79. [Google Scholar] [CrossRef]
- Brain Tumor Segmentation (BraTS) Challenge. Available online: http://www.braintumorsegmentation.org/ (accessed on 10 June 2023).
- Computational Precision Medicine: Radiology-Pathology Challenge on Brain Tumor Classification 2019 (CPM-RadPath). Available online: https://www.med.upenn.edu/cbica/cpm-rad-path-2019/ (accessed on 30 August 2023).
- Figshare Brain Tumor Dataset. Available online: https://figshare.com/articles/dataset/brain_tumor_dataset/1512427 (accessed on 1 June 2023).
- IXI Dataset. Available online: https://brain-development.org/ixi-dataset/ (accessed on 10 June 2023).
- Hamada, A. Br35H Brain Tumor Detection 2020 Dataset. Available online: https://www.kaggle.com/datasets/ahmedhamada0/brain-tumor-detection (accessed on 1 June 2023).
- Bhuvaji, S.; Kadam, A.; Bhumkar, P.; Dedge, S. Brain Tumor Classification (MRI). Available online: https://www.kaggle.com/datasets/sartajbhuvaji/brain-tumor-classification-mri (accessed on 1 June 2023).
- Chakrabarty, N. Brain MRI Images Dataset for Brain Tumor Detection, Kaggle. 2019. Available online: https://www.kaggle.com/datasets/navoneel/brain-mri-images-for-brain-tumor-detection (accessed on 1 June 2023).
- Radiopaedia. Available online: https://radiopaedia.org/cases/system/central-nervous-system (accessed on 1 June 2023).
- Scarpace, L.; Flanders, A.E.; Jain, R.; Mikkelsen, T.; Andrews, D.W. Data From REMBRANDT [Data set]. The Cancer Imaging Archive. 2019. Available online: https://www.cancerimagingarchive.net/collection/rembrandt/ (accessed on 20 April 2023).
- Scarpace, L.; Mikkelsen, T.; Cha, S.; Rao, S.; Tekchandani, S.; Gutman, D.; Saltz, J.H.; Erickson, B.J.; Pedano, N.; Flanders, A.E.; et al. The Cancer Genome Atlas Glioblastoma Multiforme Collection (TCGA-GBM) (Version 4) [Data set]. The Cancer Imaging Archive. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-gbm/ (accessed on 4 March 2023).
- Pedano, N.; Flanders, A.E.; Scarpace, L.; Mikkelsen, T.; Eschbacher, J.M.; Hermes, B.; Sisneros, V.; Barnholtz-Sloan, J.; Ostrom, Q. The Cancer Genome Atlas Low Grade Glioma Collection (TCGA-LGG) (Version 3) [Data set]. The Cancer Imaging Archive. 2016. Available online: https://www.cancerimagingarchive.net/collection/tcga-lgg/ (accessed on 5 March 2023).
- Upadhyay, N.; Waldman, A.D. Conventional MRI evaluation of gliomas. Br. J. Radiol. 2011, 84, S107. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Qu, Q.; Gu, I.Y.H.; Store Jakola, A. 3D Multi-scale convolutional networks for glioma grading using MR images. In Proceedings of the 2018 25th IEEE International Conference on Image Processing (ICIP), Athens, Greece, 7–10 October 2018; pp. 141–145. [Google Scholar] [CrossRef]
- Pereira, S.; Meier, R.; Alves, V.; Reyes, M.; Silva, C.A. Automatic brain tumor grading from MRI data using convolutional neural networks and quality assessment. In Understanding and Interpreting Machine Learning in Medical Image Computing Applications, Proceedings of the MLCN 2018, DLF 2018, and iMIMIC 2018, Granada, Spain, 16–20 September 2018; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2018; Volume 11038, pp. 106–114. [Google Scholar] [CrossRef]
- Tandel, G.S.; Tiwari, A.; Kakde, O. Performance enhancement of MRI-based brain tumor classification using suitable segmentation method and deep learning-based ensemble algorithm. Biomed. Signal Process. Control. 2022, 78, 104018. [Google Scholar] [CrossRef]
- Deepak, S.; Ameer, P.M. Brain tumor classification using deep CNN features via transfer learning. Comput. Biol. Med. 2019, 111, 103345. [Google Scholar] [CrossRef]
- Swati, Z.N.K.; Zhao, Q.; Kabir, M.; Ali, F.; Ali, Z.; Ahmed, S.; Lu, J. Brain tumor classification for MR images using transfer learning and fine-tuning. Comput. Med. Imaging Graph. 2019, 75, 34–46. [Google Scholar] [CrossRef]
- Alaraimi, S.; Okedu, K.E.; Tianfield, H.; Holden, R.; Uthmani, O. Transfer learning networks with skip connections for classification of brain tumors. Int. J. Imaging Syst. Technol. 2021, 31, 1564–1582. [Google Scholar] [CrossRef]
- Hao, R.; Namdar, K.; Liu, L.; Khalvati, F. A Transfer Learning—Based Active Learning Framework for Brain Tumor Classification. Front. Artif. Intell. 2021, 4, 61. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. Tumor type detection in brain MR images of the deep model developed using hypercolumn technique, attention modules, and residual blocks. Med. Biol. Eng. Comput. 2021, 59, 57–70. [Google Scholar] [CrossRef]
- Amou, M.A.; Xia, K.; Kamhi, S.; Mouhafid, M. A Novel MRI Diagnosis Method for Brain Tumor Classification Based on CNN and Bayesian Optimization. Healthcare 2022, 10, 494. [Google Scholar] [CrossRef] [PubMed]
- Aurna, N.F.; Yousuf, M.A.; Taher, K.A.; Azad, A.K.M.; Moni, M.A. A classification of MRI brain tumor based on two stage feature level ensemble of deep CNN models. Comput. Biol. Med. 2022, 146, 105539. [Google Scholar] [CrossRef] [PubMed]
- Tripathi, P.C.; Bag, S. A computer-aided grading of glioma tumor using deep residual networks fusion. Comput. Methods Programs Biomed. 2022, 215, 106597. [Google Scholar] [CrossRef] [PubMed]
- Deepa, S.; Janet, J.; Sumathi, S.; Ananth, J.P. Hybrid Optimization Algorithm Enabled Deep Learning Approach Brain Tumor Segmentation and Classification Using MRI. J. Digit. Imaging 2023, 36, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Ghassemi, N.; Shoeibi, A.; Rouhani, M. Deep neural network with generative adversarial networks pre-training for brain tumor classification based on MR images. Biomed. Signal Process. Control. 2020, 57, 101678. [Google Scholar] [CrossRef]
- Ayadi, W.; Elhamzi, W.; Charfi, I.; Atri, M. Deep CNN for brain tumor classification. Neural Process. Lett. 2021, 53, 671–700. [Google Scholar] [CrossRef]
- Khan, A.R.; Khan, S.; Harouni, M.; Abbasi, R.; Iqbal, S.; Mehmood, Z. Brain tumor segmentation using K-means clustering and deep learning with synthetic data augmentation for classification. Microsc. Res. Tech. 2021, 84, 1389–1399. [Google Scholar] [CrossRef]
- Kumar, R.L.; Kakarla, J.; Isunuri, B.V.; Singh, M. Multi-class brain tumor classification using residual network and global average pooling. Multimed. Tools Appl. 2021, 80, 13429–13438. [Google Scholar] [CrossRef]
- Ahmad, B.; Sun, J.; You, Q.; Palade, V.; Mao, Z. Brain Tumor Classification Using a Combination of Variational Autoencoders and Generative Adversarial Networks. Biomedicines 2022, 10, 223. [Google Scholar] [CrossRef] [PubMed]
- Haq, A.U.; Li, J.P.; Kumar, R.; Ali, Z.; Khan, I.; Uddin, M.I.; Agbley, B.L.Y. MCNN: A multi-level CNN model for the classification of brain tumors in IoT-healthcare system. J. Ambient. Intell. Humaniz. Comput. 2022, 14, 4695–4706. [Google Scholar] [CrossRef]
- Anand, V.; Gupta, S.; Gupta, D.; Gulzar, Y.; Xin, Q.; Juneja, S.; Shah, A.; Shaikh, A. Weighted Average Ensemble Deep Learning Model for Stratification of Brain Tumor in MRI Images. Diagnostics 2023, 13, 1320. [Google Scholar] [CrossRef] [PubMed]
- Krishnapriya, S.; Karuna, Y. Pre-trained deep learning models for brain MRI image classification. Front. Hum. Neurosci. 2023, 17, 1150120. [Google Scholar] [CrossRef] [PubMed]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Deep semi-supervised learning for brain tumor classification. BMC Med. Imaging 2020, 20, 1–11. [Google Scholar] [CrossRef]
- Gab Allah, A.M.; Sarhan, A.M.; Elshennawy, N.M. Classification of brain MRI tumor images based on deep learning PGGAN augmentation. Diagnostics 2021, 11, 2343. [Google Scholar] [CrossRef] [PubMed]
- Gupta, R.K.; Bharti, S.; Kunhare, N.; Sahu, Y.; Pathik, N. Brain Tumor Detection and Classification Using Cycle Generative Adversarial Networks. Interdiscip. Sci. Comput. Life Sci. 2022, 14, 485–502. [Google Scholar] [CrossRef]
- Toğaçar, M.; Cömert, Z.; Ergen, B. Classification of brain MRI using hyper column technique with convolutional neural network and feature selection method. Expert Syst. Appl. 2020, 149, 113274. [Google Scholar] [CrossRef]
- Pei, L.; Hsu, W.W.; Chiang, L.A.; Guo, J.M.; Iftekharuddin, K.M.; Colen, R. A Hybrid Convolutional Neural Network Based-Method for Brain Tumor Classification Using mMRI and WSI. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the BrainLes 2020, Lima, Peru, 4 October 2020; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2021; Volume 12659, pp. 487–496. [Google Scholar]
- Dang, K.; Vo, T.; Ngo, L.; Ha, H. A deep learning framework integrating MRI image preprocessing methods for brain tumor segmentation and classification. IBRO Neurosci. Rep. 2022, 13, 523–532. [Google Scholar] [CrossRef]
- Senan, E.M.; Jadhav, M.E.; Rassem, T.H.; Aljaloud, A.S.; Mohammed, B.A.; Al-Mekhlafi, Z.G. Early Diagnosis of Brain Tumour MRI Images Using Hybrid Techniques between Deep and Machine Learning. Comput. Math. Methods Med. 2022, 2022, 8330833. [Google Scholar] [CrossRef]
- Ge, C.; Gu, I.Y.H.; Jakola, A.S.; Yang, J. Deep learning and multi-sensor fusion for glioma classification using multistream 2D convolutional networks. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, EMBS, Sarawak, Malaysia, 18–21 July 2018; pp. 5894–5897. [Google Scholar] [CrossRef]
- Yang, Y.; Yan, L.F.; Zhang, X.; Han, Y.; Nan, H.Y.; Hu, Y.C.; Hu, B.; Yan, S.L.; Zhang, J.; Cheng, D.L.; et al. Glioma grading on conventional MR images: A deep learning study with transfer learning. Front. Neurosci. 2018, 12, 804. [Google Scholar] [CrossRef] [PubMed]
- Lo, C.M.; Chen, Y.C.; Weng, R.C.; Hsieh, K.L.C. Intelligent Glioma Grading Based on Deep Transfer Learning of MRI Radiomic Features. Appl. Sci. 2019, 9, 4926. [Google Scholar] [CrossRef]
- Zahoor, M.M.; Qureshi, S.A.; Bibi, S.; Khan, S.H.; Khan, A.; Ghafoor, U.; Bhutta, M.R. A New Deep Hybrid Boosted and Ensemble Learning-Based Brain Tumor Analysis Using MRI. Sensors 2022, 22, 2726. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Naz, S.; Razzak, M.I.; Akram, F.; Imran, M. A Deep Learning-Based Framework for Automatic Brain Tumors Classification Using Transfer Learning. Circuits Syst. Signal Process. 2020, 39, 757–775. [Google Scholar] [CrossRef]
- Tandel, G.S.; Balestrieri, A.; Jujaray, T.; Khanna, N.N.; Saba, L.; Suri, J.S. Multiclass magnetic resonance imaging brain tumor classification using artificial intelligence paradigm. Comput. Biol. Med. 2020, 122, 103804. [Google Scholar] [CrossRef]
- Gutta, S.; Acharya, J.; Shiroishi, M.S.; Hwang, D.; Nayak, K.S. Improved Glioma Grading Using Deep Convolutional Neural Networks. Am. J. Neuroradiol. 2021, 42, 233–239. [Google Scholar] [CrossRef]
- Tandel, G.S.; Tiwari, A.; Kakde, O.G. Performance optimisation of deep learning models using majority voting algorithm for brain tumour classification. Comput. Biol. Med. 2021, 135, 104564. [Google Scholar] [CrossRef]
- Kazemi, A.; Shiri, M.E.; Sheikhahmadi, A.; Khodamoradi, M. Classifying tumor brain images using parallel deep learning algorithms. Comput. Biol. Med. 2022, 148, 105775. [Google Scholar] [CrossRef]
- Saravanan, S.; Kumar, V.V.; Sarveshwaran, V.; Indirajithu, A.; Elangovan, D.; Allayear, S.M. Computational and Mathematical Methods in Medicine Glioma Brain Tumor Detection and Classification Using Convolutional Neural Network. Comput. Math. Methods Med. 2022, 2022, 4380901. [Google Scholar] [CrossRef]
- Athisayamani, S.; Antonyswamy, R.S.; Sarveshwaran, V.; Almeshari, M.; Alzamil, Y.; Ravi, V. Feature Extraction Using a Residual Deep Convolutional Neural Network (ResNet-152) and Optimized Feature Dimension Reduction for MRI Brain Tumor Classification. Diagnostics 2023, 13, 668. [Google Scholar] [CrossRef]
- Bairagi, V.K.; Gumaste, P.P.; Rajput, S.H.; Chethan, K.S. Automatic brain tumor detection using CNN transfer learning approach. Med. Biol. Eng. Comput. 2023, 61, 1821–1836. [Google Scholar] [CrossRef]
- Gao, P.; Shan, W.; Guo, Y.; Wang, Y.; Sun, R.; Cai, J.; Li, H.; Chan, W.S.; Liu, P.; Yi, L.; et al. Development and Validation of a Deep Learning Model for Brain Tumor Diagnosis and Classification Using Magnetic Resonance Imaging. JAMA Netw. Open 2022, 5, e2225608. [Google Scholar] [CrossRef] [PubMed]
- Jeong, S.W.; Cho, H.H.; Lee, S.; Park, H. Robust multimodal fusion network using adversarial learning for brain tumor grading. Comput. Methods Programs Biomed. 2022, 226, 107165. [Google Scholar] [CrossRef]
- Maqsood, S.; Damaševičius, R.; Maskeliūnas, R. Multi-Modal Brain Tumor Detection Using Deep Neural Network and Multiclass SVM. Medicina 2022, 58, 1090. [Google Scholar] [CrossRef] [PubMed]
- Xiong, D.; Ren, X.; Huang, W.; Wang, R.; Ma, L.; Gan, T.; Ai, K.; Wen, T.; Li, Y.; Wang, P.; et al. Noninvasive Classification of Glioma Subtypes Using Multiparametric MRI to Improve Deep Learning. Diagnostics 2022, 12, 3063. [Google Scholar] [CrossRef] [PubMed]
- Hossain, S.; Chakrabarty, A.; Gadekallu, T.R.; Alazab, M.; Piran, M.J. Vision Transformers, Ensemble Model, and Transfer Learning Leveraging Explainable AI for Brain Tumor Detection and Classification. IEEE J. Biomed. Health Inform. 2023. [Google Scholar] [CrossRef] [PubMed]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Nandoe Tewarie, R.; Lycklama, G.J.; De Witt Hamer, P.C.; Eijgelaar, R.S.; et al. Combined molecular subtyping, grading, and segmentation of glioma using multi-task deep learning. Neuro-Oncology 2023, 25, 279–289. [Google Scholar] [CrossRef] [PubMed]
- Xu, C.; Peng, Y.; Zhu, W.; Chen, Z.; Li, J.; Tan, W.; Zhang, Z.; Chen, X. An automated approach for predicting glioma grade and survival of LGG patients using CNN and radiomics. Front. Oncol. 2022, 12, 969907. [Google Scholar] [CrossRef]
- Kibriya, H.; Amin, R.; Kim, J.; Nawaz, M.; Gantassi, R. A Novel Approach for Brain Tumor Classification Using an Ensemble of Deep and Hand-Crafted Features. Sensors 2023, 23, 4693. [Google Scholar] [CrossRef]
- Kutlu, H.; Avcı, E. A Novel Method for Classifying Liver and Brain Tumors Using Convolutional Neural Networks, Discrete Wavelet Transform and Long Short-Term Memory Networks. Sensors 2019, 19, 1992. [Google Scholar] [CrossRef]
- Almalki, Y.E.; Ali, M.U.; Kallu, K.D.; Masud, M.; Zafar, A.; Alduraibi, S.K.; Irfan, M.; Basha, M.A.A.; Alshamrani, H.A.; Alduraibi, A.K.; et al. Isolated Convolutional-Neural-Network-Based Deep-Feature Extraction for Brain Tumor Classification Using Shallow Classifier. Diagnostics 2022, 12, 1793. [Google Scholar] [CrossRef] [PubMed]
- Kibriya, H.; Amin, R.; Alshehri, A.H.; Masood, M.; Alshamrani, S.S.; Alshehri, A. A Novel and Effective Brain Tumor Classification Model Using Deep Feature Fusion and Famous Machine Learning Classifiers. Comput. Intell. Neurosci. 2022, 2022, 7897669. [Google Scholar] [CrossRef] [PubMed]
- Shirehjini, O.F.; Mofrad, F.B.; Shahmohammadi, M.; Karami, F. Grading of gliomas using transfer learning on MRI images. Magn. Reson. Mater. Phys. Biol. Med. 2023, 36, 43–53. [Google Scholar] [CrossRef] [PubMed]
- Rajinikanth, V.; Kadry, S.; Damaševičius, R.; Sujitha, R.A.; Balaji, G.; Mohammed, M.A. Glioma/glioblastoma detection in brain MRI using pre-trained deep-learning scheme. In Proceedings of the 2022 Third International Conference on Intelligent Computing Instrumentation and Control Technologies (ICICICT), Guangzhou, China, 12–14 August 2022; pp. 987–990. [Google Scholar]
- Rasool, M.; Ismail, N.; Boulila, W.; Ammar, A.; Samma, H.; Yafooz, W.S.; Emara, A.H. A Hybrid Deep Learning Model for Brain Tumour Classification. Entropy 2022, 24, 799. [Google Scholar] [CrossRef] [PubMed]
- Sekhar, A.; Biswas, S.; Hazra, R.; Sunaniya, A.K.; Mukherjee, A.; Yang, L. Brain Tumor Classification Using Fine-Tuned GoogLeNet Features and Machine Learning Algorithms: IoMT Enabled CAD System. IEEE J. Biomed. Health Inform. 2022, 26, 983–991. [Google Scholar] [CrossRef]
- AlTahhan, F.E.; Khouqeer, G.A.; Saadi, S.; Elgarayhi, A.; Sallah, M. Refined Automatic Brain Tumor Classification Using Hybrid Convolutional Neural Networks for MRI Scans. Diagnostics 2023, 13, 864. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Choudhary, S.; Jain, A.; Singh, K.; Ahmadian, A.; Bajuri, M.Y. Brain Tumor Classification Using Deep Neural Network and Transfer Learning. Brain Topogr. 2023, 36, 305–318. [Google Scholar] [CrossRef]
- Ma, X.; Jia, F. Brain tumor classification with multimodal MR and pathology images. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Proceedings of the BrainLes 2019, Shenzhen, China, 17 October 2019; Lecture Notes in Computer Science; Springer: Cham, Switzerland, 2020; Volume 11993, pp. 343–352. [Google Scholar] [CrossRef]
- Yin, B.; Cheng, H.; Wang, F.; Wang, Z. Brain tumor classification based on MRI images and noise reduced pathology images. In Proceedings of the Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries: 6th International Workshop, BrainLes 2020, Lima, Perú, 4–8 October 2020; Springer: Berlin/Heidelberg, Germany, 2021; pp. 465–474. [Google Scholar]
- Hsu, W.W.; Guo, J.M.; Pei, L.; Chiang, L.A.; Li, Y.F.; Hsiao, J.C.; Colen, R.; Liu, P. A weakly supervised deep learning-based method for glioma subtype classification using WSI and mpMRIs. Sci. Rep. 2022, 12, 6111. [Google Scholar] [CrossRef]
- Wang, X.; Wang, R.; Yang, S.; Zhang, J.; Wang, M.; Zhong, D.; Zhang, J.; Han, X. Combining Radiology and Pathology for Automatic Glioma Classification. Front. Bioeng. Biotechnol. 2022, 10, 841958. [Google Scholar] [CrossRef]
- Kang, J.; Ullah, Z.; Gwak, J. MRI-Based Brain Tumor Classification Using Ensemble of Deep Features and Machine Learning Classifiers. Sensors 2021, 21, 2222. [Google Scholar] [CrossRef]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Alassafi, M.O. Brain Tumor Classification Based on Fine-Tuned Models and the Ensemble Method. Comput. Mater. Contin. 2021, 67, 3967–3982. [Google Scholar] [CrossRef]
- Coupet, M.; Urruty, T.; Leelanupab, T.; Naudin, M.; Bourdon, P.; Maloigne, C.F.; Guillevin, R. A multi-sequences MRI deep framework study applied to glioma classfication. Multimed. Tools Appl. 2022, 81, 13563–13591. [Google Scholar] [CrossRef] [PubMed]
- Tummala, S.; Kadry, S.; Bukhari, S.A.C.; Rauf, H.T. Classification of Brain Tumor from Magnetic Resonance Imaging Using Vision Transformers Ensembling. Curr. Oncol. 2022, 29, 7498–7511. [Google Scholar] [CrossRef] [PubMed]
- Al-Zoghby, A.M.; Al-Awadly, E.M.K.; Moawad, A.; Yehia, N.; Ebada, A.I. Dual Deep CNN for Tumor Brain Classification. Diagnostics 2023, 13, 2050. [Google Scholar] [CrossRef] [PubMed]
- Asif, S.; Zhao, M.; Chen, X.; Zhu, Y. BMRI-NET: A Deep Stacked Ensemble Model for Multi-class Brain Tumor Classification from MRI Images. Interdiscip. Sci. Comput. Life Sci. 2023, 15, 499–514. [Google Scholar] [CrossRef]
- Tandel, G.S.; Tiwari, A.; Kakde, O.G.; Gupta, N.; Saba, L.; Suri, J.S. Role of Ensemble Deep Learning for Brain Tumor Classification in Multiple Magnetic Resonance Imaging Sequence Data. Diagnostics 2023, 13, 481. [Google Scholar] [CrossRef]
- Decuyper, M.; Bonte, S.; Deblaere, K.; Holen, R.V. Automated MRI based pipeline for segmentation and prediction of grade, IDH mutation and 1p19q co-deletion in glioma. Comput. Med. Imaging Graph. 2021, 88, 101831. [Google Scholar] [CrossRef]
- Tripathi, P.C.; Bag, S. An attention-guided CNN framework for segmentation and grading of glioma using 3D MRI scans. IEEE/ACM Trans. Comput. Biol. Bioinform. 2022, 3, 1890–1904. [Google Scholar] [CrossRef]
- Mzoughi, H.; Njeh, I.; Wali, A.; Slima, M.B.; BenHamida, A.; Mhiri, C.; Mahfoudhe, K.B. Deep Multi-Scale 3D Convolutional Neural Network (CNN) for MRI Gliomas Brain Tumor Classification. J. Digit. Imaging 2020, 33, 903–915. [Google Scholar] [CrossRef]
- Pei, L.; Vidyaratne, L.; Rahman, M.M.; Iftekharuddin, K.M. Context aware deep learning for brain tumor segmentation, subtype classification, and survival prediction using radiology images. Sci. Rep. 2020, 10, 19726. [Google Scholar] [CrossRef]
- Chakrabarty, S.; Sotiras, A.; Milchenko, M.; Lamontagne, P.; Hileman, M.; Marcus, D. MRI-based identification and classification of major intracranial tumor types by using a 3D convolutional neural network: A retrospective multi-institutional analysis. Radiol. Artif. Intell. 2021, 3, e200301. [Google Scholar] [CrossRef] [PubMed]
- Yamashiro, H.; Teramoto, A.; Saito, K.; Fujita, H. Development of a Fully Automated Glioma-Grading Pipeline Using Post-Contrast T1-Weighted Images Combined with Cloud-Based 3D Convolutional Neural Network. Appl. Sci. 2021, 11, 5118. [Google Scholar] [CrossRef]
- Danilov, G.; Korolev, V.; Shifrin, M.; Ilyushin, E.; Maloyan, N.; Saada, D.; Ishankulov, T.; Afandiev, R.; Shevchenko, A.; Konakova, T.; et al. Noninvasive Glioma Grading with Deep Learning: A Pilot Study. Stud. Health Technol. Inform. 2022, 290, 675–678. [Google Scholar] [CrossRef] [PubMed]
- Samee, N.A.; Ahmad, T.; Mahmoud, N.F.; Atteia, G.; Abdallah, H.A.; Rizwan, A. Clinical Decision Support Framework for Segmentation and Classification of Brain Tumor MRIs Using a U-Net and DCNN Cascaded Learning Algorithm. Healthcare 2022, 10, 2340. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.; Haider, S.; Maqsood, S.; Damaševičius, R.; Maskeliūnas, R.; Khan, M. ETISTP: An Enhanced Model for Brain Tumor Identification and Survival Time Prediction. Diagnostics 2023, 13, 1456. [Google Scholar] [CrossRef]
- Rui, W.; Zhang, S.; Shi, H.; Sheng, Y.; Zhu, F.; Yao, Y.; Chen, X.; Cheng, H.; Zhang, Y.; Aili, A.; et al. Deep Learning-Assisted Quantitative Susceptibility Mapping as a Tool for Grading and Molecular Subtyping of Gliomas. Phenomics 2023, 3, 243–254. [Google Scholar] [CrossRef]
- Guo, S.; Wang, L.; Chen, Q.; Wang, L.; Zhang, J.; Zhu, Y. Multimodal MRI Image Decision Fusion-Based Network for Glioma Classification. Front. Oncol. 2022, 12, 819673. [Google Scholar] [CrossRef]
- Díaz-Pernas, F.J.; Martínez-Zarzuela, M.; Antón-Rodríguez, M.; González-Ortega, D. A deep learning approach for brain tumor classification and segmentation using a multiscale convolutional neural network. Healthcare 2021, 9, 153. [Google Scholar] [CrossRef]
- Gilanie, G.; Bajwa, U.I.; Waraich, M.M.; Anwar, M.W. Risk-free WHO grading of astrocytoma using convolutional neural networks from MRI images. Multimed. Tools Appl. 2021, 80, 4295–4306. [Google Scholar] [CrossRef]
- Guan, Y.; Aamir, M.; Rahman, Z.; Ali, A.; Abro, W.A.; Dayo, Z.A.; Bhutta, M.S.; Hu, Z.; Guan, Y.; Aamir, M.; et al. A framework for efficient brain tumor classification using MRI images. Math. Biosci. Eng. 2021, 18, 5790–5815. [Google Scholar] [CrossRef]
- Gull, S.; Akbar, S.; Khan, H.U. Automated Detection of Brain Tumor through Magnetic Resonance Images Using Convolutional Neural Network. BioMed Res. Int. 2021, 2021, 3365043. [Google Scholar] [CrossRef]
- Özcan, H.; Emiroğlu, B.G.; Sabuncuoğlu, H.; Özdoğan, S.; Soyer, A.; Saygı, T. A comparative study for glioma classification using deep convolutional neural networks. Math. Biosci. Eng. 2021, 18, 1550–1572. [Google Scholar] [CrossRef] [PubMed]
- Aamir, M.; Rahman, Z.; Dayo, Z.A.; Abro, W.A.; Uddin, M.I.; Khan, I.; Imran, A.S.; Ali, Z.; Ishfaq, M.; Guan, Y.; et al. A deep learning approach for brain tumor classification using MRI images. Comput. Electr. Eng. 2022, 101, 108105. [Google Scholar] [CrossRef]
- Wu, P.; Wang, Z.; Zheng, B.; Li, H.; Alsaadi, F.E.; Zeng, N. AGGN: Attention-based glioma grading network with multi-scale feature extraction and multi-modal information fusion. Comput. Biol. Med. 2023, 152, 106457. [Google Scholar] [CrossRef]
- Badža, M.M.; Barjaktarović, M.Č. Classification of brain tumors from MRI images using a convolutional neural network. Appl. Sci. 2020, 10, 1999. [Google Scholar] [CrossRef]
- Ismael, S.A.A.; Mohammed, A.; Hefny, H. An enhanced deep learning approach for brain cancer MRI images classification using residual networks. Artif. Intell. Med. 2020, 102, 101779. [Google Scholar] [CrossRef]
- Alanazi, M.F.; Ali, M.U.; Hussain, S.J.; Zafar, A.; Mohatram, M.; Irfan, M.; AlRuwaili, R.; Alruwaili, M.; Ali, N.H.; Albarrak, A.M. Brain tumor/mass classification framework using magnetic-resonance-imaging-based isolated and developed transfer deep-learning model. Sensors 2022, 22, 372. [Google Scholar] [CrossRef]
- O’Reilly, T.; Teeuwisse, W.M.; de Gans, D.; Koolstra, K.; Webb, A.G. In vivo 3D brain and extremity MRI at 50 mT using a permanent magnet Halbach array. Magn. Reson. Med. 2021, 85, 495–505. [Google Scholar] [CrossRef]
- Cooley, C.Z.; McDaniel, P.C.; Stockmann, J.P.; Srinivas, S.A.; Cauley, S.F.; Śliwiak, M.; Sappo, C.R.; Vaughn, C.F.; Guerin, B.; Rosen, M.S.; et al. A portable scanner for magnetic resonance imaging of the brain. Nat. Biomed. Eng. 2020, 5, 229–239. [Google Scholar] [CrossRef]
- Man, C.; Lau, V.; Su, S.; Zhao, Y.; Xiao, L.; Ding, Y.; Leung, G.K.; Leong, A.T.; Wu, E.X. Deep learning enabled fast 3D brain MRI at 0.055 tesla. Sci. Adv. 2023, 9, eadi9327. [Google Scholar] [CrossRef]
- Swoop Portable MR System. Available online: https://hyperfine.io.assets/pdfs/Swoop (accessed on 6 November 2023).
- Altaf, A.; Baqai, M.W.S.; Urooj, F.; Alam, M.S.; Aziz, H.F.; Mubarak, F.; Knopp, E.A.; Siddiqui, K.M.; Enam, S.A. Utilization of an ultra-low-field, portable magnetic resonance imaging for brain tumor assessment in lower middle-income countries. Surg. Neurol. Int. 2023, 14, 260. [Google Scholar] [CrossRef] [PubMed]
- Altaf, A.; Baqai, M.W.S.; Urooj, F.; Alam, M.S.; Aziz, H.F.; Mubarak, F.; Knopp, E.; Siddiqui, K.; Enam, S.A. Intraoperative use of ultra-low-field, portable magnetic resonance imaging—First report. Surg. Neurol. Int. 2023, 14, 212. [Google Scholar] [CrossRef] [PubMed]
- Abd-Ellah, M.K.; Awad, A.I.; Hamed, H.F.; Khalaf, A.A. Parallel deep CNN structure for glioma detection and classification via brain MRI Images. In Proceedings of the 2019 31st International Conference on Microelectronics (ICM), Cairo, Egypt, 15–18 December 2019; pp. 304–307. [Google Scholar]
- Anaraki, A.K.; Ayati, M.; Kazemi, F. Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern. Biomed. Eng. 2019, 39, 63–74. [Google Scholar] [CrossRef]
- Hemanth, D.J.; Anitha, J.; Naaji, A.; Geman, O.; Popescu, D.E.; Son, L.H. A Modified Deep Convolutional Neural Network for Abnormal Brain Image Classification. IEEE Access 2019, 7, 4275–4283. [Google Scholar] [CrossRef]
- Cubuk, E.D.; Zoph, B.; Mane, D.; Vasudevan, V.; Le, Q.V. AutoAugment: Learning Augmentation Policies from Data. arXiv 2018. [Google Scholar] [CrossRef]
- Muneer, K.V.A.; Rajendran, V.R.; Joseph, K.P. Glioma Tumor Grade Identification Using Artificial Intelligent Techniques. J. Med. Syst. 2019, 43, 1–12. [Google Scholar] [CrossRef]
- Rajini, N.H. Brain Tumor Image Classification and Grading Using Convolutional Neural Network and Particle Swarm Optimization Algorithm. Int. J. Eng. Adv. Technol. (IJEAT) 2019, 8, 2249–8958. [Google Scholar]
- Rahmathunneesa, A.P.; Muneer, K.V.A. Performance analysis of pre-trained deep learning networks for brain tumor categorization. In Proceedings of the 2019 9th International Conference on Advances in Computing and Communication (ICACC), Changsha, China, 18–20 October 2019; pp. 253–257. [Google Scholar] [CrossRef]
- Sajjad, M.; Khan, S.; Muhammad, K.; Wu, W.; Ullah, A.; Baik, S.W. Multi-grade brain tumor classification using deep CNN with extensive data augmentation. J. Comput. Sci. 2019, 30, 174–182. [Google Scholar] [CrossRef]
- Sultan, H.H.; Salem, N.M.; Al-Atabany, W. Multi-Classification of Brain Tumor Images Using Deep Neural Network. IEEE Access 2019, 7, 69215–69225. [Google Scholar] [CrossRef]
- Toğaçar, M.; Ergen, B.; Cömert, Z. BrainMRNet: Brain tumor detection using magnetic resonance images with a novel convolutional neural network model. Med. Hypotheses 2020, 134, 109531. [Google Scholar] [CrossRef]
- Amin, J.; Sharif, M.; Gul, N.; Yasmin, M.; Shad, S.A. Brain tumor classification based on DWT fusion of MRI sequences using convolutional neural network. Pattern Recognit. Lett. 2020, 129, 115–122. [Google Scholar] [CrossRef]
- Afshar, P.; Plataniotis, K.N.; Mohammadi, A. BoostCaps: A boosted capsule network for brain tumor classification. In Proceedings of the Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Virtual, 20–24 July 2020; pp. 1075–1079. [Google Scholar] [CrossRef]
- Bhanothu, Y.; Kamalakannan, A.; Rajamanickam, G. Detection and classification of brain tumor in MRI images using deep convolutional network. In Proceedings of the 2020 6th International Conference on Advanced Computing and Communication Systems (ICACCS), Coimbatore, India, 6–7 March 2020; pp. 248–252. [Google Scholar] [CrossRef]
- Çinar, A.; Yildirim, M. Detection of tumors on brain MRI images using the hybrid convolutional neural network architecture. Med. Hypotheses 2020, 139, 109684. [Google Scholar] [CrossRef] [PubMed]
- Khan, H.A.; Jue, W.; Mushtaq, M.; Mushtaq, M.U.; Khan, H.A.; Jue, W.; Mushtaq, M.; Mushtaq, M.U. Brain tumor classification in MRI image using convolutional neural network. Math. Biosci. Eng. 2020, 17, 6203–6216. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, B.A.; Al-Ani, S. An efficient approach to diagnose brain tumors through deep CNN. Math. Biosci. Eng. 2020, 18, 851–867. [Google Scholar] [CrossRef] [PubMed]
- Naser, M.A.; Deen, M.J. Brain tumor segmentation and grading of lower-grade glioma using deep learning in MRI images. Comput. Biol. Med. 2020, 121, 103758. [Google Scholar] [CrossRef] [PubMed]
- Noreen, N.; Palaniappan, S.; Qayyum, A.; Ahmad, I.; Imran, M.; Shoaib, M. A deep learning model based on concatenation approach for the diagnosis of brain tumor. IEEE Access 2020, 8, 55135–55144. [Google Scholar] [CrossRef]
- Saxena, P.; Maheshwari, A.; Maheshwari, S. Predictive Modeling of Brain Tumor: A Deep Learning Approach. Adv. Intell. Syst. Comput. 2020, 1189, 275–285. [Google Scholar] [CrossRef]
- Sharif, M.I.; Li, J.P.; Khan, M.A.; Saleem, M.A. Active deep neural network features selection for segmentation and recognition of brain tumors using MRI images. Pattern Recognit. Lett. 2020, 129, 181–189. [Google Scholar] [CrossRef]
- Vimal Kurup, R.; Sowmya, V.; Soman, K. Effect of data pre-processing on brain tumor classification using capsulenet. In Proceedings of the ICICCT 2019—System Reliability, Quality Control, Safety, Maintenance and Management: Applications to Electrical, Electronics and Computer Science and Engineering, Hyderabad, India, 9–11 January 2019; Springer: Berlin/Heidelberg, Germany, 2020; pp. 110–119. [Google Scholar]
- Bashir-Gonbadi, F.; Khotanlou, H. Brain tumor classification using deep convolutional autoencoder-based neural network: Multi-task approach. Multimed. Tools Appl. 2021, 80, 19909–19929. [Google Scholar] [CrossRef]
- Gu, X.; Shen, Z.; Xue, J.; Fan, Y.; Ni, T. Brain Tumor MR Image Classification Using Convolutional Dictionary Learning with Local Constraint. Front. Neurosci. 2021, 15, 679847. [Google Scholar] [CrossRef]
- Irmak, E. Multi-Classification of Brain Tumor MRI Images Using Deep Convolutional Neural Network with Fully Optimized Framework. Iran. J. Sci. Technol. Trans. Electr. Eng. 2021, 45, 1015–1036. [Google Scholar] [CrossRef]
- Kader, I.A.E.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Ahmad, I.S.; Kamhi, S. Brain Tumor Detection and Classification on MR Images by a Deep Wavelet Auto-Encoder Model. Diagnostics 2021, 11, 1589. [Google Scholar] [CrossRef]
- Kader, I.A.E.; Xu, G.; Shuai, Z.; Saminu, S.; Javaid, I.; Ahmad, I.S. Differential Deep Convolutional Neural Network Model for Brain Tumor Classification. Brain Sci. 2021, 11, 352. [Google Scholar] [CrossRef]
- Kakarla, J.; Isunuri, B.V.; Doppalapudi, K.S.; Bylapudi, K.S.R. Three-class classification of brain magnetic resonance images using average-pooling convolutional neural network. Int. J. Imaging Syst. Technol. 2021, 31, 1731–1740. [Google Scholar] [CrossRef]
- Masood, M.; Nazir, T.; Nawaz, M.; Mehmood, A.; Rashid, J.; Kwon, H.Y.; Mahmood, T.; Hussain, A. A novel deep learning method for recognition and classification of brain tumors from MRI images. Diagnostics 2021, 11, 744. [Google Scholar] [CrossRef] [PubMed]
- Sadad, T.; Rehman, A.; Munir, A.; Saba, T.; Tariq, U.; Ayesha, N.; Abbasi, R. Brain tumor detection and multi-classification using advanced deep learning techniques. Microsc. Res. Tech. 2021, 84, 1296–1308. [Google Scholar] [CrossRef] [PubMed]
- MohamedMetwalySherif. Brain Tumor Dataset. 2020. Available online: https://www.kaggle.com/datasets/mohamedmetwalysherif/braintumordataset (accessed on 10 June 2023).
- Chitnis, S.; Hosseini, R.; Xie, P. Brain tumor classification based on neural architecture search. Sci. Rep. 2022, 12, 19206. [Google Scholar] [CrossRef] [PubMed]
- Ekong, F.; Yu, Y.; Patamia, R.A.; Feng, X.; Tang, Q.; Mazumder, P.; Cai, J. Bayesian Depth-Wise Convolutional Neural Network Design for Brain Tumor MRI Classification. Diagnostics 2022, 12, 1657. [Google Scholar] [CrossRef]
- Gaur, L.; Bhandari, M.; Razdan, T.; Mallik, S.; Zhao, Z. Explanation-Driven Deep Learning Model for Prediction of Brain Tumour Status Using MRI Image Data. Front. Genet. 2022, 13, 448. [Google Scholar] [CrossRef]
- Gurunathan, A.; Krishnan, B. A Hybrid CNN-GLCM Classifier For Detection And Grade Classification Of Brain Tumor. Brain Imaging Behav. 2022, 16, 1410–1427. [Google Scholar] [CrossRef]
- Isunuri, B.V.; Kakarla, J. Three-class brain tumor classification from magnetic resonance images using separable convolution based neural network. Concurr. Comput. Pract. Exp. 2022, 34, e6541. [Google Scholar] [CrossRef]
- Khazaee, Z.; Langarizadeh, M.; Ahmadabadi, M.E.S. Developing an Artificial Intelligence Model for Tumor Grading and Classification, Based on MRI Sequences of Human Brain Gliomas. Int. J. Cancer Manag. 2022, 15, 120638. [Google Scholar] [CrossRef]
- Koli, R.; Lotya, S.; Govekar, P.; Sachdev, K.; Bhatia, G. Detection and classification of brain tumor using MRI images. In Proceedings of the ICT Analysis and Applications, Goa, India, 29–30 July 2022; Springer: Berlin/Heidelberg, Germany, 2022; pp. 393–402. [Google Scholar]
- Lakshmi, M.J.; Rao, S.N. Brain tumor magnetic resonance image classification: A deep learning approach. Soft Comput. 2022, 26, 6245–6253. [Google Scholar] [CrossRef]
- Murthy, M.Y.B.; Koteswararao, A.; Babu, M.S. Adaptive fuzzy deformable fusion and optimized CNN with ensemble classification for automated brain tumor diagnosis. Biomed. Eng. Lett. 2022, 12, 37–58. [Google Scholar] [CrossRef] [PubMed]
- Nayak, D.R.; Padhy, N.; Mallick, P.K.; Zymbler, M.; Kumar, S. Brain Tumor Classification Using Dense Efficient-Net. Axioms 2022, 11, 34. [Google Scholar] [CrossRef]
- Raza, A.; Ayub, H.; Khan, J.A.; Ahmad, I.; S. Salama, A.; Daradkeh, Y.I.; Javeed, D.; Ur Rehman, A.; Hamam, H. A hybrid deep learning-based approach for brain tumor classification. Electronics 2022, 11, 1146. [Google Scholar] [CrossRef]
- Rizwan, M.; Shabbir, A.; Javed, A.R.; Shabbir, M.; Baker, T.; Obe, D.A.J. Brain Tumor and Glioma Grade Classification Using Gaussian Convolutional Neural Network. IEEE Access 2022, 10, 29731–29740. [Google Scholar] [CrossRef]
- Samee, N.A.; Mahmoud, N.F.; Atteia, G.; Abdallah, H.A.; Alabdulhafith, M.; Al-Gaashani, M.S.; Ahmad, S.; Muthanna, M.S.A. Classification Framework for Medical Diagnosis of Brain Tumor with an Effective Hybrid Transfer Learning Model. Diagnostics 2022, 12, 2541. [Google Scholar] [CrossRef]
- Sangeetha, S.K.; Muthukumaran, V.; Deeba, K.; Rajadurai, H.; Maheshwari, V.; Dalu, G.T. Multiconvolutional Transfer Learning for 3D Brain Tumor Magnetic Resonance Images. Comput. Intell. Neurosci. 2022, 2022, 8722476. [Google Scholar] [CrossRef]
- Srinivas, C.; Nandini, N.P.; Zakariah, M.; Alothaibi, Y.A.; Shaukat, K.; Partibane, B.; Awal, H. Deep Transfer Learning Approaches in Performance Analysis of Brain Tumor Classification Using MRI Images. J. Healthc. Eng. 2022, 2022, 3264367. [Google Scholar] [CrossRef]
- Erickson, B.; Akkus, Z.; Sedlar, J.; Korfiatis, P. Data from LGG-1p19qDeletion (Version 2) [Data set]. The Cancer Imaging Archive. 2017. Available online: https://www.cancerimagingarchive.net/collection/lgg-1p19qdeletion/ (accessed on 14 July 2023).
- Vankdothu, R.; Hameed, M.A.; Fatima, H. A brain tumor identification and classification using deep learning based on CNN-LSTM method. Comput. Electr. Eng. 2022, 101, 107960. [Google Scholar] [CrossRef]
- Yazdan, S.A.; Ahmad, R.; Iqbal, N.; Rizwan, A.; Khan, A.N.; Kim, D.H. An Efficient Multi-Scale Convolutional Neural Network Based Multi-Class Brain MRI Classification for SaMD. Tomography 2022, 8, 1905–1927. [Google Scholar] [CrossRef] [PubMed]
- Anagun, Y. Smart brain tumor diagnosis system utilizing deep convolutional neural networks. Multimed. Tools Appl. 2023, 82, 44527–44553. [Google Scholar] [CrossRef] [PubMed]
- Apostolopoulos, I.D.; Aznaouridis, S.; Tzani, M. An Attention-Based Deep Convolutional Neural Network for Brain Tumor and Disorder Classification and Grading in Magnetic Resonance Imaging. Information 2023, 14, 174. [Google Scholar] [CrossRef]
- Komaravolu, A. Brain Tumor MRI Images. Available online: https://www.kaggle.com/datasets/adityakomaravolu/brain-tumor-mri-images (accessed on 10 June 2023).
- Yaseen, R. Brain Tumor Data MRI. Available online: https://www.kaggle.com/datasets/roroyaseen/brain-tumor-data-mri (accessed on 10 June 2023).
- El-Wahab, B.S.A.; Nasr, M.E.; Khamis, S.; Ashour, A.S. BTC-fCNN: Fast Convolution Neural Network for Multi-class Brain Tumor Classification. Health Inf. Sci. Syst. 2023, 11, 3. [Google Scholar] [CrossRef]
- Mahmud, M.I.; Mamun, M.; Abdelgawad, A. A Deep Analysis of Brain Tumor Detection from MR Images Using Deep Learning Networks. Algorithms 2023, 16, 176. [Google Scholar] [CrossRef]
- Muezzinoglu, T.; Baygin, N.; Tuncer, I.; Barua, P.D.; Baygin, M.; Dogan, S.; Tuncer, T.; Palmer, E.E.; Cheong, K.H.; Acharya, U.R. PatchResNet: Multiple Patch Division–Based Deep Feature Fusion Framework for Brain Tumor Classification Using MRI Images. J. Digit. Imaging 2023, 12, 973–987. [Google Scholar] [CrossRef] [PubMed]
- Özkaraca, O.; İhsan Bağrıaçık, O.; Gürüler, H.; Khan, F.; Hussain, J.; Khan, J.; e Laila, U. Multiple Brain Tumor Classification with Dense CNN Architecture Using Brain MRI Images. Life 2023, 13, 349. [Google Scholar] [CrossRef]
- Nickparvar, M. Brain Tumor MRI Dataset. 2021. Available online: https://www.kaggle.com/datasets/masoudnickparvar/brain-tumor-mri-dataset?select=Training (accessed on 4 June 2023).
- Özkaya, C.; Şağıroğlu, C. Glioma Grade Classification Using CNNs and Segmentation with an Adaptive Approach Using Histogram Features in Brain MRIs. IEEE Access 2023, 11, 52275–52287. [Google Scholar] [CrossRef]
- Rasheed, Z.; Ma, Y.K.; Ullah, I.; Shloul, T.A.; Tufail, A.B.; Ghadi, Y.Y.; Khan, M.Z.; Mohamed, H.G. Automated Classification of Brain Tumors from Magnetic Resonance Imaging Using Deep Learning. Brain Sci. 2023, 13, 602. [Google Scholar] [CrossRef]
- Srinivasan, S.; Bai, P.S.M.; Mathivanan, S.K.; Muthukumaran, V.; Babu, J.C.; Vilcekova, L. Grade Classification of Tumors from Brain Magnetic Resonance Images Using a Deep Learning Technique. Diagnostics 2023, 13, 1153. [Google Scholar] [CrossRef] [PubMed]
- van der Voort, S.R.; Incekara, F.; Wijnenga, M.M.; Kapsas, G.; Gahrmann, R.; Schouten, J.W.; Dubbink, H.J.; Vincent, A.J.; van den Bent, M.J.; French, P.J.; et al. The Erasmus Glioma Database (EGD): Structural MRI scans, WHO 2016 subtypes, and segmentations of 774 patients with glioma. Data Brief 2021, 37, 107191. [Google Scholar] [CrossRef] [PubMed]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).