Proteomic Profiling Identifies Predictive Signatures for Progression Risk in Patients with Advanced-Stage Follicular Lymphoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Patients and Methods
2.1. Identification of Differentially Expressed Proteins
2.2. Statistical Analysis
3. Results
3.1. Patient Characteristics
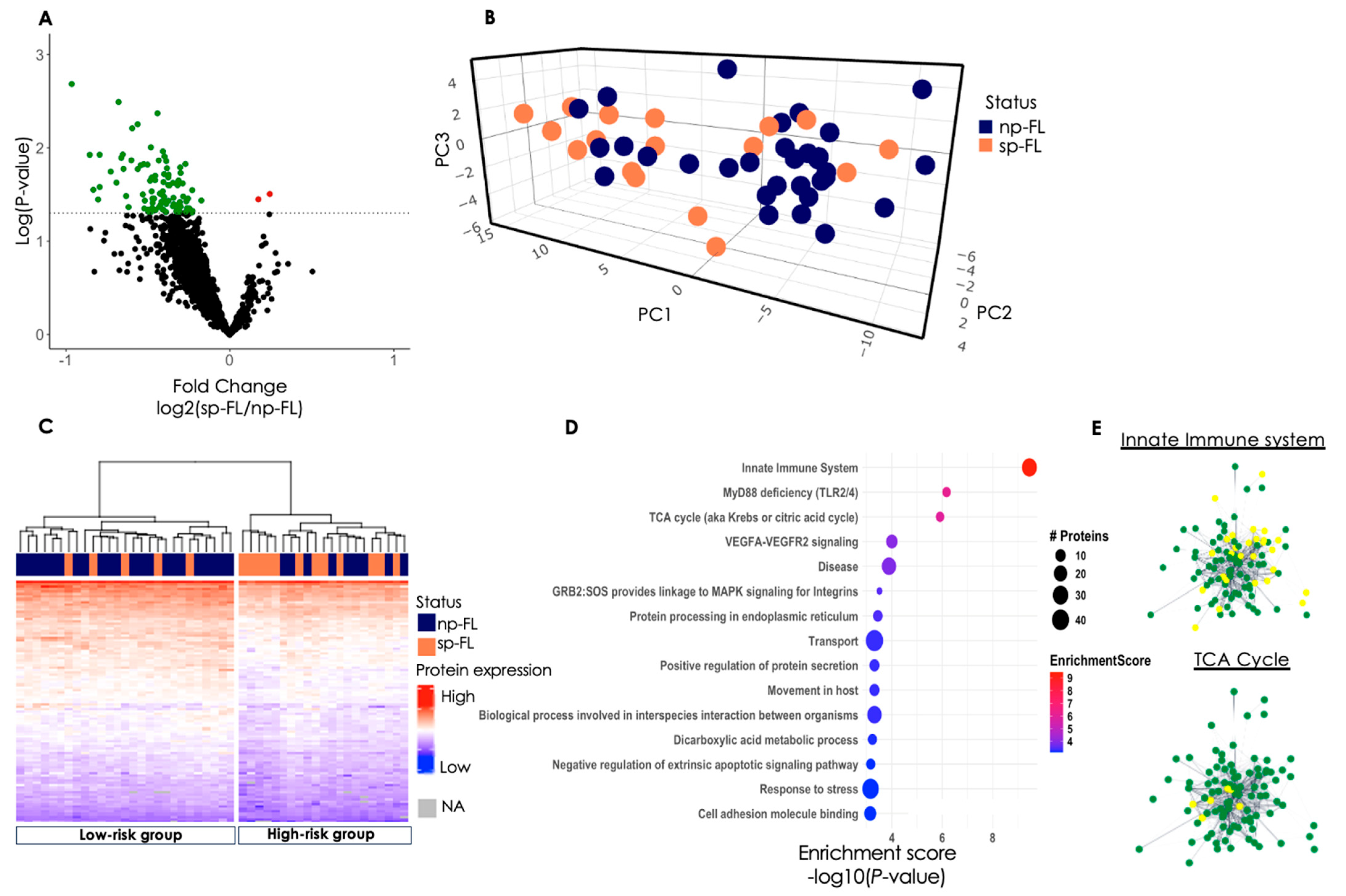
3.2. Proteomic Profiling Reveals Differentially Expressed Proteins
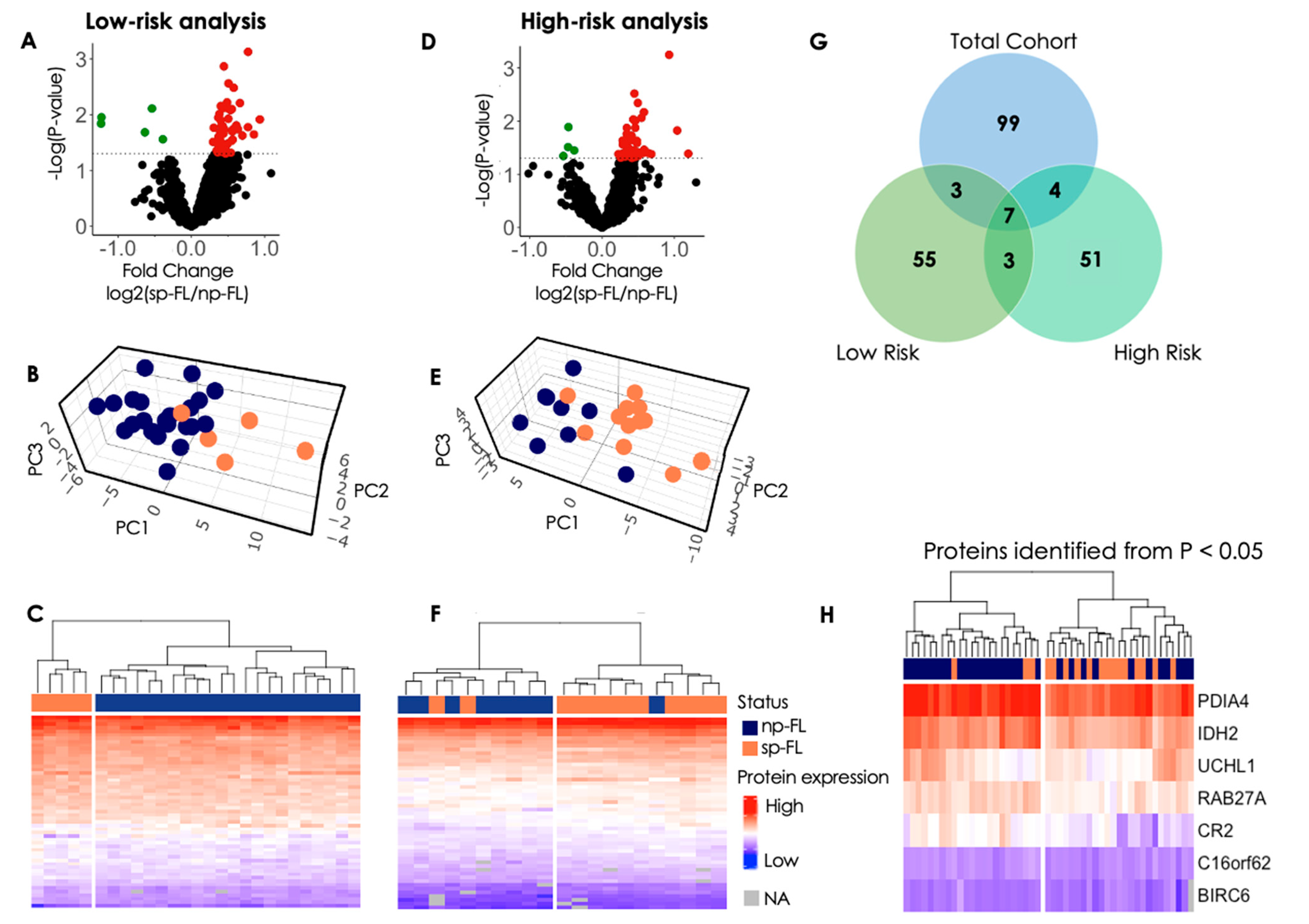
3.2.1. Low-Risk Group Analysis
3.2.2. High-Risk Group Analysis
3.2.3. Insights from Protein Profiles in High- and Low-Risk Groups
3.2.4. Disturbed Cellular Pathways at FL Diagnosis
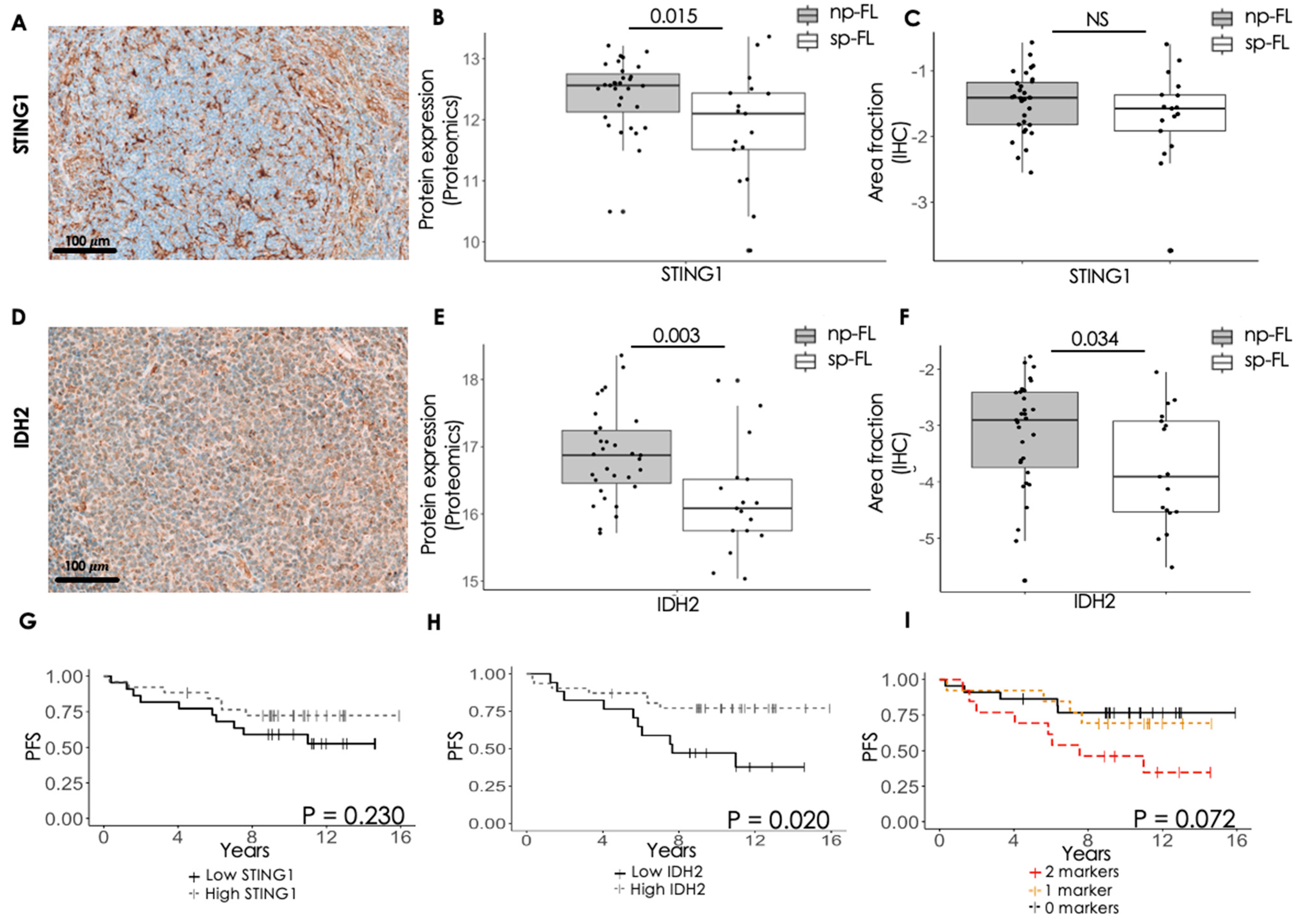
3.3. Immunohistochemical Evaluation of Selected Proteins Identifies Markers Capable of Predicting Progression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Freedman, A.; Jacobsen, E. Follicular lymphoma: 2020 update on diagnosis and management. Am. J. Hematol. 2020, 95, 316–327. [Google Scholar] [CrossRef] [PubMed]
- Swerdlow, S.H.; Campo, E.; Pileri, S.A.; Harris, N.L.; Stein, H.; Siebert, R.; Advani, R.; Ghielmini, M.; Salles, G.A.; Zelenetz, A.D.; et al. The 2016 revision of the World Health Organization classification of lymphoid neoplasms. Blood 2016, 127, 2375–2390. [Google Scholar] [CrossRef] [PubMed]
- Townsend, W.; Buske, C.; Cartron, G.; Cunningham, D.; Dyer, M.J.S.; Gribben, J.G.; Zhang, Z.; Rufibach, K.; Nielsen, T.; Herold, M.; et al. Comparison of efficacy and safety with obinutuzumab plus chemotherapy versus rituximab plus chemotherapy in patients with previously untreated follicular lymphoma: Updated results from the phase III Gallium Study. J. Clin. Oncol. 2020, 38, 15. [Google Scholar] [CrossRef]
- Liu, Q.; Silva, A.; Kridel, R. Predicting early progression in follicular lymphoma. Ann. Lymphoma 2021, 5, 11. [Google Scholar] [CrossRef]
- Casulo, C.; Dixon, J.G.; Le-Rademacher, J.; Hoster, E.; Hochster, H.S.; Hiddemann, W.; Marcus, R.; Kimby, E.; Herold, M.; Sebban, C.; et al. Validation of POD24 as a robust early clinical end point of poor survival in FL from 5225 patients on 13 clinical trials. Blood 2022, 139, 1684–1693. [Google Scholar] [CrossRef]
- Lipof, J.J.; Barr, P.M. Early Progression of Follicular Lymphoma: Biology and Treatment. Hematol. Oncol. Clin. 2020, 34, 757–769. [Google Scholar] [CrossRef]
- Russler-Germain, D.A.; Krysiak, K.; Ramirez, C.; Mosior, M.; Watkins, M.P.; Gomez, F.; Skidmore, Z.L.; Trani, L.; Gao, F.; Geyer, S.; et al. Mutations associated with progression in follicular lymphoma predict inferior outcomes at diagnosis: Alliance A151303. Blood Adv. 2023, 7, 5524–5539. [Google Scholar] [CrossRef]
- Sortais, C.; Lok, A.; Tessoulin, B.; Gastinne, T.; Mahé, B.; Dubruille, V.; Blin, N.; Touzeau, C.; Moreau, A.; Bossard, C.; et al. Progression of disease within 2 years (POD24) is a clinically relevant endpoint to identify high-risk follicular lymphoma patients in real life. Ann. Hematol. 2020, 99, 1595–1604. [Google Scholar] [CrossRef]
- Tobin, J.W.D.; Keane, C.; Gunawardana, J.; Mollee, P.; Birch, S.; Hoang, T.; Lee, J.; Li, L.; Huang, L.; Murigneux, V.; et al. Progression of Disease within 24 Months in Follicular Lymphoma Is Associated With Reduced Intratumoral Immune Infiltration. J. Clin. Oncol. 2019, 37, 3300–3309. [Google Scholar] [CrossRef]
- Wallace, D.; Casulo, C. Early Progressing Follicular Lymphoma. Curr. Oncol. Rep. 2021, 23, 149. [Google Scholar] [CrossRef]
- Jelicic, J.; Stauffer Larsen, T.; Bukumiric, Z.; Andjelic, B. The clinical applicability of current prognostic models in follicular lymphoma: A systematic review. Crit. Rev. Oncol. Hematol. 2021, 164, 103418. [Google Scholar] [CrossRef] [PubMed]
- Bachy, E.; Maurer, M.J.; Habermann, T.M.; Gelas-Dore, B.; Maucort-Boulch, D.; Estell, J.A.; Van den Neste, E.; Bouabdallah, R.; Gyan, E.; Feldman, A.L.; et al. A simplified scoring system in de novo follicular lymphoma treated initially with immunochemotherapy. Blood 2018, 132, 49–58. [Google Scholar] [CrossRef]
- Devan, J.; Janikova, A.; Mraz, M. New concepts in follicular lymphoma biology: From BCL2 to epigenetic regulators and non-coding RNAs. Semin. Oncol. 2018, 45, 291–302. [Google Scholar] [CrossRef]
- Federico, M.; Bellei, M.; Marcheselli, L.; Luminari, S.; Lopez-Guillermo, A.; Vitolo, U.; Pro, B.; Pileri, S.; Pulsoni, A.; Soubeyran, P.; et al. Follicular lymphoma international prognostic index 2: A new prognostic index for follicular lymphoma developed by the international follicular lymphoma prognostic factor project. J. Clin. Oncol. 2009, 27, 4555–4562. [Google Scholar] [CrossRef]
- Pastore, A.; Jurinovic, V.; Kridel, R.; Hoster, E.; Staiger, A.M.; Szczepanowski, M.; Pott, C.; Kopp, N.; Murakami, M.; Horn, H.; et al. Integration of gene mutations in risk prognostication for patients receiving first-line immunochemotherapy for follicular lymphoma: A retrospective analysis of a prospective clinical trial and validation in a population-based registry. Lancet Oncol. 2015, 16, 1111–1122. [Google Scholar] [CrossRef] [PubMed]
- Press, O.W.; Unger, J.M.; Rimsza, L.M.; Friedberg, J.W.; LeBlanc, M.; Czuczman, M.S.; Kaminski, M.; Braziel, R.M.; Spier, C.; Gopal, A.K.; et al. A comparative analysis of prognostic factor models for follicular lymphoma based on a phase III trial of CHOP-rituximab versus CHOP + 131iodine–tositumomab. Clin. Cancer Res. 2013, 19, 6624–6632. [Google Scholar] [CrossRef]
- Yang, G.; Mills, M.; Kim, Y.; Figura, N.B.; Doyle, C.; Oliver, D.; Grass, G.D.; Robinson, T.; Chavez, J.; Kim, S. Enhancement of the Follicular Lymphoma International Prognostic Index (FLIPI) with lymphopenia (FLIPI-L): A predictor for overall survival and histologic transformation. Blood Cancer J. 2020, 9, 104. [Google Scholar] [CrossRef] [PubMed]
- Mir, F.; Mattiello, F.; Grigg, A.; Herold, M.; Hiddemann, W.; Marcus, R.; Seymour, J.F.; Bolen, C.R.; Knapp, A.; Nielsen, T.; et al. Follicular Lymphoma Evaluation Index (FLEX): A new clinical prognostic model that is superior to existing risk scores for predicting progression-free survival and early treatment failure after frontline immunochemotherapy. Am. J. Hematol. 2020, 95, 1503–1510. [Google Scholar] [CrossRef]
- Enemark, M.B.H.; Wolter, K.; Campbell, A.J.; Andersen, M.D.; Sørensen, E.F.; Hybel, T.E.; Madsen, C.; Lauridsen, K.L.; Plesner, T.L.; Hamilton-Dutoit, S.J.; et al. Proteomics identifies apoptotic markers as predictors of histological transformation in patients with follicular lymphoma. Blood Adv. 2023, 7, 7418–7432. [Google Scholar] [CrossRef]
- Kamper, P.; Ludvigsen, M.; Bendix, K.; Hamilton-Dutoit, S.; Rabinovich, G.A.; Møller, M.B.; Nyengaard, J.R.; Honoré, B.; d’Amore, F. Proteomic analysis identifies galectin-1 as a predictive biomarker for relapsed/refractory disease in classical Hodgkin lymphoma. Blood 2011, 117, 6638–6649. [Google Scholar] [CrossRef]
- Ludvigsen, M.; Campbell, A.J.; Enemark, M.B.; Hybel, T.E.; Karjalainen-Lindsberg, M.-L.; Beiske, K.; Bjerre, M.; Pedersen, L.M.; Holte, H.; Leppä, S.; et al. Proteomics uncovers molecular features for relapse risk stratification in patients with diffuse large B-cell lymphoma. Blood Cancer J. 2023, 13, 161. [Google Scholar] [CrossRef]
- Vase, M.; Ludvigsen, M.; Bendix, K.; Hamilton-Dutoit, S.; Mller, M.B.; Pedersen, C.; Pedersen, G.; Obel, N.; Larsen, C.S.; d’Amore, F.; et al. Proteomic profiling of pretreatment serum from HIV-infected patients identifies candidate markers predictive of lymphoma development. Aids 2016, 30, 1889–1898. [Google Scholar] [CrossRef]
- Gonzalez, M.W.; Kann, M.G. Chapter 4: Protein interactions and disease. PLoS Comput. Biol. 2012, 8, e1002819. [Google Scholar] [CrossRef]
- Arboe, B.; El-Galaly, T.C.; Clausen, M.R.; Munksgaard, P.S.; Stoltenberg, D.; Nygaard, M.K.; Klausen, T.W.; Christensen, J.H.; Gørløv, J.S.; Brown Pde, N. The Danish National Lymphoma Registry: Coverage and Data Quality. PLoS ONE 2016, 11, e0157999. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Dave, S.S.; Wright, G.; Tan, B.; Rosenwald, A.; Gascoyne, R.D.; Chan, W.C.; Fisher, R.I.; Braziel, R.M.; Rimsza, L.M.; Grogan, T.M.; et al. Prediction of survival in follicular lymphoma based on molecular features of tumor-infiltrating immune cells. N. Engl. J. Med. 2004, 351, 2159–2169. [Google Scholar] [CrossRef]
- Zhu, Y.; An, X.; Zhang, X.; Qiao, Y.; Zheng, T.; Li, X. STING: A master regulator in the cancer-immunity cycle. Mol. Cancer 2019, 18, 152. [Google Scholar] [CrossRef]
- Müller, L.; Aigner, P.; Stoiber, D. Type I Interferons and Natural Killer Cell Regulation in Cancer. Front. Immunol. 2017, 8, 304. [Google Scholar] [CrossRef]
- Dahal, L.N.; Dou, L.; Hussain, K.; Liu, R.; Earley, A.; Cox, K.L.; Murinello, S.; Tracy, I.; Forconi, F.; Steele, A.J.; et al. STING Activation Reverses Lymphoma-Mediated Resistance to Antibody Immunotherapy. Cancer Res. 2017, 77, 3619–3631. [Google Scholar] [CrossRef]
- Landmann, R.; Müller, B.; Zimmerli, W. CD14, new aspects of ligand and signal diversity. Microbes Infect. 2000, 2, 295–304. [Google Scholar] [CrossRef]
- Zhang, R.; Kang, R.; Tang, D. STING1 in Different Organelles: Location Dictates Function. Front. Immunol. 2022, 13, 842489. [Google Scholar] [CrossRef]
- Martínez-Reyes, I.; Chandel, N.S. Cancer metabolism: Looking forward. Nat. Rev. Cancer 2021, 21, 669–680. [Google Scholar] [CrossRef]
- Krell, D.; Assoku, M.; Galloway, M.; Mulholland, P.; Tomlinson, I.; Bardella, C. Screen for IDH1, IDH2, IDH3, D2HGDH and L2HGDH mutations in glioblastoma. PLoS ONE 2011, 6, e19868. [Google Scholar] [CrossRef]
- Cardaci, S.; Ciriolo, M.R. TCA Cycle Defects and Cancer: When Metabolism Tunes Redox State. Int. J. Cell Biol. 2012, 2012, 161837. [Google Scholar] [CrossRef]
- Kotredes, K.P.; Razmpour, R.; Lutton, E.; Alfonso-Prieto, M.; Ramirez, S.H.; Gamero, A.M. Characterization of cancer-associated IDH2 mutations that differ in tumorigenicity, chemosensitivity and 2-hydroxyglutarate production. Oncotarget 2019, 10, 2675–2692. [Google Scholar] [CrossRef]
- Lemonnier, F.; Cairns, R.A.; Inoue, S.; Li, W.Y.; Dupuy, A.; Broutin, S.; Martin, N.; Fataccioli, V.; Pelletier, R.; Wakeham, A.; et al. The IDH2 R172K mutation associated with angioimmunoblastic T-cell lymphoma produces 2HG in T cells and impacts lymphoid development. Proc. Natl. Acad. Sci. USA 2016, 113, 15084–15089. [Google Scholar] [CrossRef]
- Anderson, N.M.; Simon, M.C. The tumor microenvironment. Curr. Biol. 2020, 30, R921–R925. [Google Scholar] [CrossRef]
- Newman, J.S.; Francis, I.R.; Kaminski, M.S.; Wahl, R.L. Imaging of lymphoma with PET with 2-[F-18]-fluoro-2-deoxy-D-glucose: Correlation with CT. Radiology 1994, 190, 111–116. [Google Scholar] [CrossRef]
- Dupuy, A.; Lemonnier, F.; Fataccioli, V.; Martin-Garcia, N.; Robe, C.; Pelletier, R.; Poullot, E.; Moktefi, A.; Mokhtari, K.; Rousselet, M.C.; et al. Multiple Ways to Detect IDH2 Mutations in Angioimmunoblastic T-Cell Lymphoma from Immunohistochemistry to Next-Generation Sequencing. J. Mol. Diagn. 2018, 20, 677–685. [Google Scholar] [CrossRef]
- Heavican, T.B.; Bouska, A.; Yu, J.; Lone, W.; Amador, C.; Gong, Q.; Zhang, W.; Li, Y.; Dave, B.J.; Nairismägi, M.L.; et al. Genetic drivers of oncogenic pathways in molecular subgroups of peripheral T-cell lymphoma. Blood 2019, 133, 1664–1676. [Google Scholar] [CrossRef]
- Holst, J.M.; Enemark, M.B.; Pedersen, M.B.; Lauridsen, K.L.; Hybel, T.E.; Clausen, M.R.; Frederiksen, H.; Møller, M.B.; Nørgaard, P.; Plesner, T.L.; et al. Proteomic Profiling Differentiates Lymphoma Patients with and without Concurrent Myeloproliferative Neoplasia. Cancers 2021, 13, 5526. [Google Scholar] [CrossRef] [PubMed]
- Calvert, A.E.; Chalastanis, A.; Wu, Y.; Hurley, L.A.; Kouri, F.M.; Bi, Y.; Kachman, M.; May, J.L.; Bartom, E.; Hua, Y.; et al. Cancer-Associated IDH1 Promotes Growth and Resistance to Targeted Therapies in the Absence of Mutation. Cell Rep. 2017, 19, 1858–1873. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.B.; Dong, D.F.; Wang, M.D.; Gao, K. IDH1 overexpression induced chemotherapy resistance and IDH1 mutation enhanced chemotherapy sensitivity in Glioma cells in vitro and in vivo. Asian Pac. J. Cancer Prev. 2014, 15, 427–432. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; He, Y.; Tan, Z.; Lu, J.; Li, L.; Song, X.; Shi, F.; Xie, L.; You, S.; Luo, X.; et al. Wild-type IDH2 promotes the Warburg effect and tumor growth through HIF1α in lung cancer. Theranostics 2018, 8, 4050–4061. [Google Scholar] [CrossRef]
- Nawrocki, S.T.; Carew, J.S.; Maclean, K.H.; Courage, J.F.; Huang, P.; Houghton, J.A.; Cleveland, J.L.; Giles, F.J.; McConkey, D.J. Myc regulates aggresome formation, the induction of Noxa, and apoptosis in response to the combination of bortezomib and SAHA. Blood 2008, 112, 2917–2926. [Google Scholar] [CrossRef]
- Gulmann, C.; Espina, V.; Petricoin, E., 3rd; Longo, D.L.; Santi, M.; Knutsen, T.; Raffeld, M.; Jaffe, E.S.; Liotta, L.A.; Feldman, A.L. Proteomic analysis of apoptotic pathways reveals prognostic factors in follicular lymphoma. Clin. Cancer Res. 2005, 11, 5847–5855. [Google Scholar] [CrossRef]
- Weinkauf, M.; Christopeit, M.; Hiddemann, W.; Dreyling, M. Proteome- and microarray-based expression analysis of lymphoma cell lines identifies a p53-centered cluster of differentially expressed proteins in mantle cell and follicular lymphoma. Electrophoresis 2007, 28, 4416–4426. [Google Scholar] [CrossRef]
- Duś-Szachniewicz, K.; Rymkiewicz, G.; Agrawal, A.K.; Kołodziej, P.; Wiśniewski, J.R. Large-Scale Proteomic Analysis of Follicular Lymphoma Reveals Extensive Remodeling of Cell Adhesion Pathway and Identifies Hub Proteins Related to the Lymphomagenesis. Cancers 2021, 13, 630. [Google Scholar] [CrossRef]
- O’Shea, D.; O’Riain, C.; Taylor, C.; Waters, R.; Carlotti, E.; Macdougall, F.; Gribben, J.; Rosenwald, A.; Ott, G.; Rimsza, L.M.; et al. The presence of TP53 mutation at diagnosis of follicular lymphoma identifies a high-risk group of patients with shortened time to disease progression and poorer overall survival. Blood 2008, 112, 3126–3129. [Google Scholar] [CrossRef]
- Kridel, R.; Sehn, L.H.; Gascoyne, R.D. Can histologic transformation of follicular lymphoma be predicted and prevented? Blood 2017, 130, 258–266. [Google Scholar] [CrossRef]
- Deng, Y.; Ma, J.; Zhao, S.; Yang, M.; Sun, Y.; Zhang, Q. Expression of glucose transporter-1 in follicular lymphoma affected tumor-infiltrating immunocytes and was related to progression of disease within 24 months. Transl. Oncol. 2023, 28, 101614. [Google Scholar] [CrossRef]
- Honoré, B. Proteomic Protocols for Differential Protein Expression Analyses. In Xenotransplantation: Methods and Protocols; Costa, C., Ed.; Springer: New York, NY, USA, 2020; pp. 47–58. [Google Scholar]
- Zougman, A.; Selby, P.J.; Banks, R.E. Suspension trapping (STrap) sample preparation method for bottom-up proteomics analysis. Proteomics 2014, 14, 1006. [Google Scholar] [CrossRef]
- Cehofski, L.J.; Kojima, K.; Terao, N.; Kitazawa, K.; Thineshkumar, S.; Grauslund, J.; Vorum, H.; Honoré, B. Aqueous Fibronectin Correlates with Severity of Macular Edema and Visual Acuity in Patients With Branch Retinal Vein Occlusion: A Proteome Study. Investig. Ophthalmol. Vis. Sci. 2020, 61, 6. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Cox, J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat. Protoc. 2016, 11, 2301–2319. [Google Scholar] [CrossRef]
- Tyanova, S.; Temu, T.; Sinitcyn, P.; Carlson, A.; Hein, M.Y.; Geiger, T.; Mann, M.; Cox, J. The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 2016, 13, 731–740. [Google Scholar] [CrossRef]
- Doncheva, N.T.; Morris, J.H.; Gorodkin, J.; Jensen, L.J. Cytoscape StringApp: Network Analysis and Visualization of Proteomics Data. J. Proteome Res. 2019, 18, 623–632. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, D607–D613. [Google Scholar] [CrossRef]
- Beck Enemark, M.; Monrad, I.; Madsen, C.; Lystlund Lauridsen, K.; Honoré, B.; Plesner, T.L.; Hamilton-Dutoit, S.J.; d’Amore, F.; Ludvigsen, M. PD-1 Expression in Pre-Treatment Follicular Lymphoma Predicts the Risk of Subsequent High-Grade Transformation. OncoTargets Ther. 2021, 14, 481–489. [Google Scholar] [CrossRef]
- Enemark, M.B.; Hybel, T.E.; Madsen, C.; Lauridsen, K.L.; Honoré, B.; Plesner, T.L.; Hamilton-Dutoit, S.; d’Amore, F.; Ludvigsen, M. Tumor-Tissue Expression of the Hyaluronic Acid Receptor RHAMM Predicts Histological Transformation in Follicular Lymphoma Patients. Cancers 2022, 14, 1316. [Google Scholar] [CrossRef]
- Hybel, T.E.; Vase, M.Ø.; Maksten, E.F.; Enemark, M.B.; Lauridsen, K.L.; Hamilton-Dutoit, S.; Andersen, C.; Møller, M.B.; Sørensen, S.S.; Jespersen, B.; et al. Intratumoral expression of CD38 in patients with post-transplant lymphoproliferative disorder. Acta Oncol. 2021, 60, 1637–1642. [Google Scholar] [CrossRef]
| Total, n = 48 n (%) | sp-FL, n = 17 n (%) | np-FL, n = 31 n (%) | p-Value | |
|---|---|---|---|---|
| Sex | NS | |||
| Male | 26 (54) | 10 (59) | 16 (52) | |
| Female | 22 (46) | 7 (41) | 15 (48) | |
| Age at diagnosis, y | NS | |||
| Median | 59 | 56 | 60 | |
| Range | 30–82 | 30–78 | 39–82 | |
| Age ≤ 60 | 0.034 | |||
| No | 26 (54) | 13 (76) | 13 (42) | |
| Yes | 22 (46) | 4 (24) | 18 (58) | |
| Site of Biopsy | NS | |||
| Lymph node | 45 (94) | 17 (100) | 28 (91) | |
| Gl. parotidea | 1 (2) | 0 (0) | 1 (3) | |
| Ileum | 1 (2) | 0 (0) | 1 (3) | |
| Nasal cavity | 1 (2) | 0 (0) | 1 (3) | |
| FL grade | NS | |||
| 1 | 20 (42) | 8 (47) | 12 (39) | |
| 2 | 20 (42) | 8 (47) | 12 (39) | |
| 3A | 7 (15) | 1 (6) | 6 (19) | |
| Unknown | 1 (1) | 0 (0) | 1 (3) | |
| Ann Arbor Stage | NS | |||
| III | 24 (50) | 8 (47) | 16 (52) | |
| IV | 24 (50) | 9 (53) | 15 (48) | |
| B-Symptoms | NS | |||
| No | 31 (65) | 11 (65) | 20 (65) | |
| Yes | 17 (35) | 6 (35) | 11 (35) | |
| Bulky disease | NS | |||
| No | 30 (63) | 10 (59) | 20 (65) | |
| Yes | 16 (33) | 6 (35) | 10 (32) | |
| Unknown | 2 (4) | 1 (6) | 1 (3) | |
| LDH-elevation | NS | |||
| No | 32 (67) | 13 (76) | 19 (61) | |
| Yes | 16 (33) | 4 (24) | 12 (39) | |
| FLIPI | NS | |||
| Low/intermediate | 20 (42) | 9 (53) | 11 (35) | |
| High | 28 (58) | 8 (47) | 20 (65) | |
| Anemia | NS | |||
| No | 43 (90) | 15 (88) | 28 (90) | |
| Yes | 5 (10) | 2 (12) | 3 (10) | |
| Nodal sites | NS | |||
| ≥4 | 5 (10) | 0 (0) | 5 (16) | |
| <4 | 43 (90) | 17 (100) | 26 (84) | |
| POD24 | 0.001 | |||
| No | 41 (85) | 10 (59) | 31 (100) | |
| Yes | 7 (15) | 7 (41) | 0 (0) | |
| Transformation | NS | |||
| No | 47 (98) | 16 (94) | 31 (100) | |
| Yes | 1 (2) | 1 (6) | 0 (0) | |
| Death | NS | |||
| No | 47 (98) | 17 (100) | 30 (97) | |
| Yes | 1 (2) | 0 (0) | 1 (3) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hemmingsen, J.K.; Enemark, M.H.; Sørensen, E.F.; Lauridsen, K.L.; Hamilton-Dutoit, S.J.; Kridel, R.; Honoré, B.; Ludvigsen, M. Proteomic Profiling Identifies Predictive Signatures for Progression Risk in Patients with Advanced-Stage Follicular Lymphoma. Cancers 2024, 16, 3278. https://doi.org/10.3390/cancers16193278
Hemmingsen JK, Enemark MH, Sørensen EF, Lauridsen KL, Hamilton-Dutoit SJ, Kridel R, Honoré B, Ludvigsen M. Proteomic Profiling Identifies Predictive Signatures for Progression Risk in Patients with Advanced-Stage Follicular Lymphoma. Cancers. 2024; 16(19):3278. https://doi.org/10.3390/cancers16193278
Chicago/Turabian StyleHemmingsen, Jonas Klejs, Marie Hairing Enemark, Emma Frasez Sørensen, Kristina Lystlund Lauridsen, Stephen Jacques Hamilton-Dutoit, Robert Kridel, Bent Honoré, and Maja Ludvigsen. 2024. "Proteomic Profiling Identifies Predictive Signatures for Progression Risk in Patients with Advanced-Stage Follicular Lymphoma" Cancers 16, no. 19: 3278. https://doi.org/10.3390/cancers16193278
APA StyleHemmingsen, J. K., Enemark, M. H., Sørensen, E. F., Lauridsen, K. L., Hamilton-Dutoit, S. J., Kridel, R., Honoré, B., & Ludvigsen, M. (2024). Proteomic Profiling Identifies Predictive Signatures for Progression Risk in Patients with Advanced-Stage Follicular Lymphoma. Cancers, 16(19), 3278. https://doi.org/10.3390/cancers16193278







