Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis
Abstract
Simple Summary
Abstract
1. Introduction
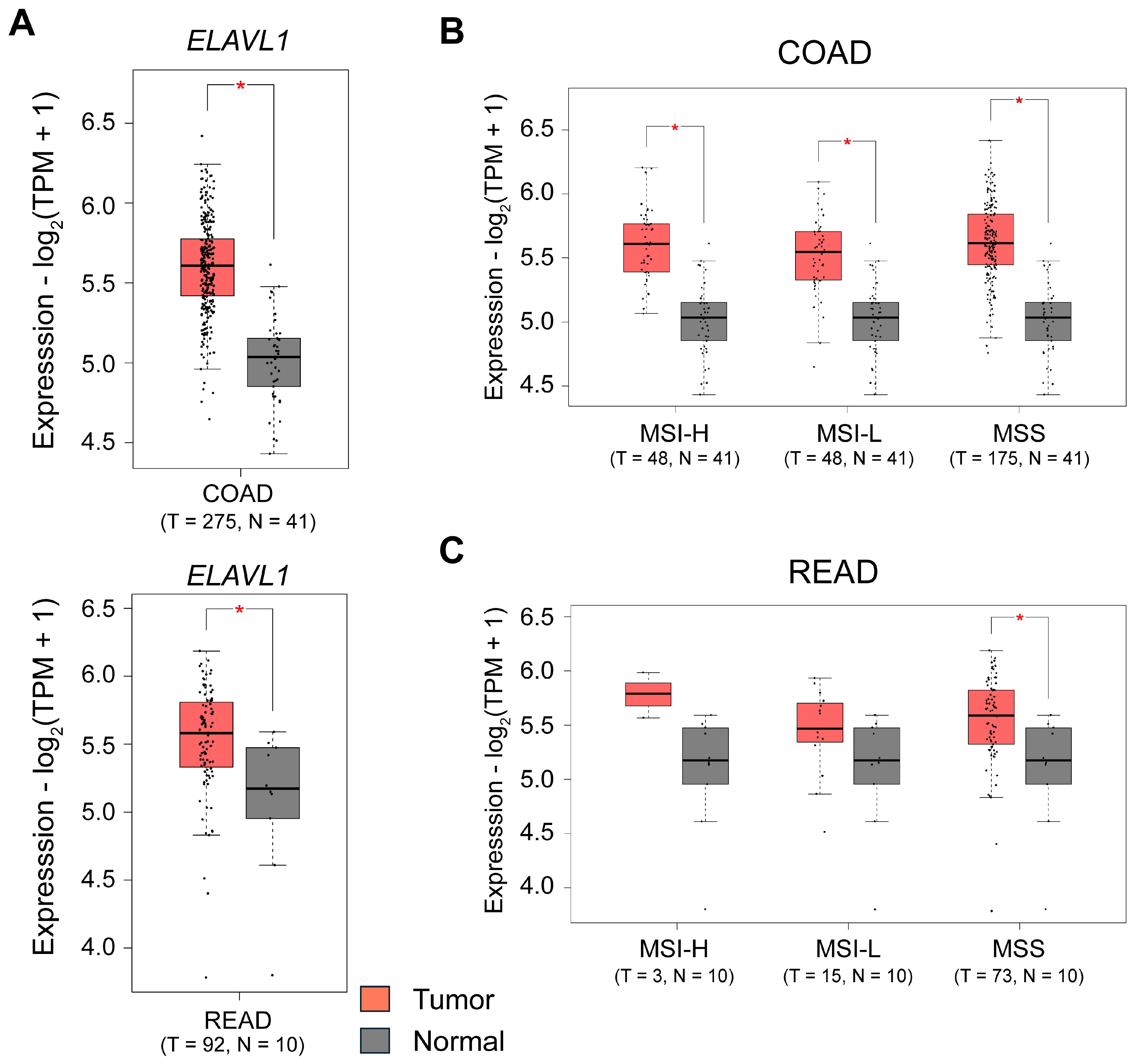
2. The RBP HuR and Its Regulatory Role in CRC
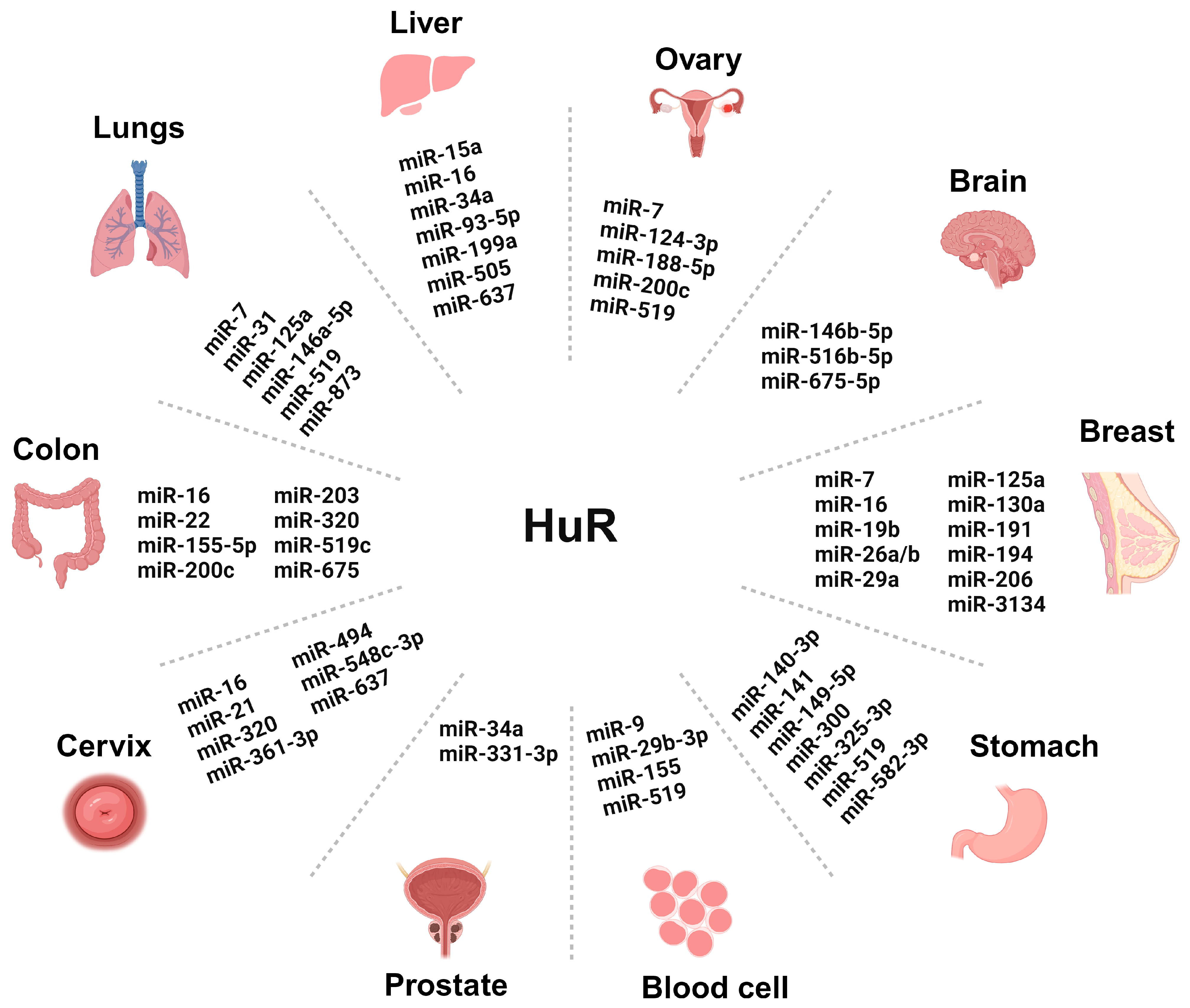
3. Intriguing Role of miRNAs-HuR Axis in CRC
3.1. MiRNAs and Their Role in Cancer Pathogenesis
3.1.1. Role of oncomiRs in HuR Regulation
| No. | microRNA | Function | Reference |
|---|---|---|---|
| 1. | miR-21 | OncomiRs | [74] |
| 2. | miR-92a | OncomiRs | [78] |
| 3. | miR-96 | OncomiRs | [82] |
| 4. | miR-135 | OncomiRs | [86] |
| 5. | miR-16 | Tumor suppressor miR | [87] |
| 6. | miR-22 | Tumor suppressor miR | [88] |
| 7. | miR-34 | Tumor suppressor miR | [89,90] |
| 8. | miR-194 | Tumor suppressor miR | [91,92,93,94] |
| 9. | miR-143 | Tumor suppressor miR | [95,96] |
| 10. | miR-145 | Tumor suppressor miR | [96] |
| 11. | miR-155-5p | Tumor suppressor miR | [97] |
| 12. | miR-519c | Tumor suppressor miR | [98,99] |
| 13. | miR-324-5p | Tumor suppressor miR | [100,101] |
3.1.2. Role of Tumor Suppressor miRs in HuR Regulation
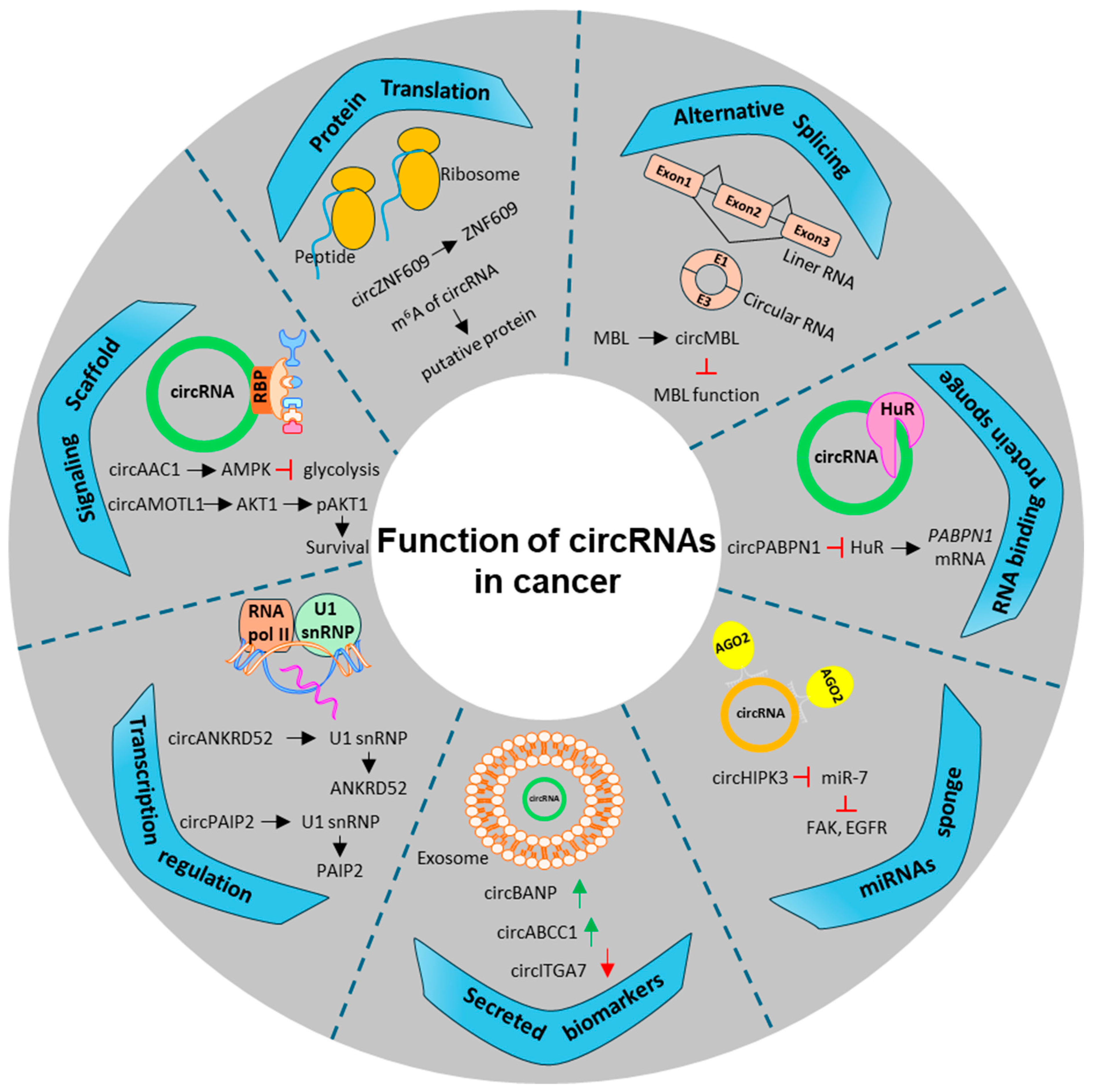
4. CircRNA Mediated Modulation of miRNAs-HuR Axis in CRC
4.1. CircRNA and Their Role in Gene Regulation
4.2. Regulatory Role of circRNA in the Translational and Post-Translational Modification of Their Host Genes
4.3. CircRNA as miRNA Sponges, Decoys, or Scaffolds
4.4. Role of circRNA in the Regulation of miRNA-HuR Axis in CRC
| No. | microRNA (Expression) | Regulation by Other Non-Coding RNA | Reference |
|---|---|---|---|
| 1. | miR-155-5p | circPPFIA1-L, circPPFIA1-S (circRNA) | [151] |
| 2. | miR-373-5p | circ0104103 (circRNA) | [141] |
| 3. | miR-143-3p miR-224-5p | circAGO2 (circRNA) | [142] |
| 4. | miR-212-5p | circNOLC1 (circRNA) | [152] |
| 5. | miR-34b-5p | OIP5-AS1 (lncRNA) | [153] |
| 6. | miR-3121-3p | TNFRSF10A-AS1 (lncRNA) | [154] |
5. Intriguing Role of lncRNA in the Regulation of miRNA-HuR Axis in CRC
6. Conclusions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Siegel, R.L.; Giaquinto, A.N.; Jemal, A. Cancer statistics, 2024. CA Cancer J. Clin. 2024, 74, 12–49. [Google Scholar] [CrossRef] [PubMed]
- Siegel, R.L.; Wagle, N.S.; Cercek, A.; Smith, R.A.; Jemal, A. Colorectal cancer statistics, 2023. CA Cancer J. Clin. 2023, 73, 233–254. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Laversanne, M.; Sung, H.; Ferlay, J.; Siegel, R.L.; Soerjomataram, I.; Jemal, A. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2024, 74, 229–263. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Dubois, R.N. Eicosanoids and cancer. Nat. Rev. Cancer 2010, 10, 181–193. [Google Scholar] [CrossRef] [PubMed]
- Ekbom, A.; Helmick, C.; Zack, M.; Adami, H.O. Ulcerative colitis and colorectal cancer. A population-based study. N. Engl. J. Med. 1990, 323, 1228–1233. [Google Scholar] [CrossRef] [PubMed]
- Nadeem, M.S.; Kumar, V.; Al-Abbasi, F.A.; Kamal, M.A.; Anwar, F. Risk of colorectal cancer in inflammatory bowel diseases. Semin. Cancer Biol. 2020, 64, 51–60. [Google Scholar] [CrossRef] [PubMed]
- Shah, S.C.; Itzkowitz, S.H. Colorectal Cancer in Inflammatory Bowel Disease: Mechanisms and Management. Gastroenterology 2022, 162, 715–730.e713. [Google Scholar] [CrossRef] [PubMed]
- Savari, S.; Vinnakota, K.; Zhang, Y.; Sjolander, A. Cysteinyl leukotrienes and their receptors: Bridging inflammation and colorectal cancer. World J. Gastroenterol. 2014, 20, 968–977. [Google Scholar] [CrossRef] [PubMed]
- Satapathy, S.R.; Sjolander, A. Cysteinyl leukotriene receptor 1 promotes 5-fluorouracil resistance and resistance-derived stemness in colon cancer cells. Cancer Lett. 2020, 488, 50–62. [Google Scholar] [CrossRef] [PubMed]
- Kuipers, E.J.; Grady, W.M.; Lieberman, D.; Seufferlein, T.; Sung, J.J.; Boelens, P.G.; van de Velde, C.J.; Watanabe, T. Colorectal cancer. Nat. Rev. Dis. Primers 2015, 1, 15065. [Google Scholar] [CrossRef]
- Cunningham, D.; Atkin, W.; Lenz, H.J.; Lynch, H.T.; Minsky, B.; Nordlinger, B.; Starling, N. Colorectal cancer. Lancet 2010, 375, 1030–1047. [Google Scholar] [CrossRef] [PubMed]
- Testa, U.; Pelosi, E.; Castelli, G. Colorectal cancer: Genetic abnormalities, tumor progression, tumor heterogeneity, clonal evolution and tumor-initiating cells. Med. Sci. 2018, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Wei, C.; Ensor, J.E.; Smolenski, D.J.; Amos, C.I.; Levin, B.; Berry, D.A. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control. 2013, 24, 1207–1222. [Google Scholar] [CrossRef] [PubMed]
- Vuik, F.E.; Nieuwenburg, S.A.; Bardou, M.; Lansdorp-Vogelaar, I.; Dinis-Ribeiro, M.; Bento, M.J.; Zadnik, V.; Pellisé, M.; Esteban, L.; Kaminski, M.F.; et al. Increasing incidence of colorectal cancer in young adults in Europe over the last 25 years. Gut 2019, 68, 1820–1826. [Google Scholar] [CrossRef]
- Burt, R.W.; DiSario, J.A.; Cannon-Albright, L. Genetics of colon cancer: Impact of inheritance on colon cancer risk. Annu. Rev. Med. 1995, 46, 371–379. [Google Scholar] [CrossRef]
- Tuohy, T.M.; Rowe, K.G.; Mineau, G.P.; Pimentel, R.; Burt, R.W.; Samadder, N.J. Risk of colorectal cancer and adenomas in the families of patients with adenomas: A population-based study in Utah. Cancer 2014, 120, 35–42. [Google Scholar] [CrossRef]
- Baraibar, I.; Ros, J.; Saoudi, N.; Salvà, F.; García, A.; Castells, M.R.; Tabernero, J.; Élez, E. Sex and gender perspectives in colorectal cancer. ESMO Open 2023, 8, 101204. [Google Scholar] [CrossRef]
- Jacobsson, M.; Wagner, V.; Kanneganti, S. Screening for Colorectal Cancer. Surg. Clin. N. Am. 2024, 104, 595–607. [Google Scholar] [CrossRef] [PubMed]
- Robertson, D.J.; Rex, D.K.; Ciani, O.; Drummond, M.F. Colonoscopy vs the Fecal Immunochemical Test: Which is Best? Gastroenterology 2024, 166, 758–771. [Google Scholar] [CrossRef] [PubMed]
- Loupakis, F.; Cremolini, C.; Masi, G.; Lonardi, S.; Zagonel, V.; Salvatore, L.; Cortesi, E.; Tomasello, G.; Ronzoni, M.; Spadi, R.; et al. Initial therapy with FOLFOXIRI and bevacizumab for metastatic colorectal cancer. N. Engl. J. Med. 2014, 371, 1609–1618. [Google Scholar] [CrossRef] [PubMed]
- Jonker, D.J.; O’Callaghan, C.J.; Karapetis, C.S.; Zalcberg, J.R.; Tu, D.; Au, H.J.; Berry, S.R.; Krahn, M.; Price, T.; Simes, R.J.; et al. Cetuximab for the treatment of colorectal cancer. N. Engl. J. Med. 2007, 357, 2040–2048. [Google Scholar] [CrossRef] [PubMed]
- Hurwitz, H.; Fehrenbacher, L.; Novotny, W.; Cartwright, T.; Hainsworth, J.; Heim, W.; Berlin, J.; Baron, A.; Griffing, S.; Holmgren, E.; et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004, 350, 2335–2342. [Google Scholar] [CrossRef] [PubMed]
- Le, D.T.; Uram, J.N.; Wang, H.; Bartlett, B.R.; Kemberling, H.; Eyring, A.D.; Skora, A.D.; Luber, B.S.; Azad, N.S.; Laheru, D.; et al. PD-1 Blockade in Tumors with Mismatch-Repair Deficiency. N. Engl. J. Med. 2015, 372, 2509–2520. [Google Scholar] [CrossRef] [PubMed]
- Mohammadi, A.; Mansoori, B.; Baradaran, B. The role of microRNAs in colorectal cancer. Biomed. Pharmacother. 2016, 84, 705–713. [Google Scholar] [CrossRef] [PubMed]
- Balacescu, O.; Sur, D.; Cainap, C.; Visan, S.; Cruceriu, D.; Manzat-Saplacan, R.; Muresan, M.-S.; Balacescu, L.; Lisencu, C.; Irimie, A. The Impact of miRNA in Colorectal Cancer Progression and Its Liver Metastases. Int. J. Mol. Sci. 2018, 19, 3711. [Google Scholar] [CrossRef]
- Luo, X.; Burwinkel, B.; Tao, S.; Brenner, H. MicroRNA Signatures: Novel Biomarker for Colorectal Cancer? Cancer Epidemiol. Biomark. Prev. 2011, 20, 1272–1286. [Google Scholar] [CrossRef]
- Gerstberger, S.; Hafner, M.; Tuschl, T. A census of human RNA-binding proteins. Nat. Rev. Genet. 2014, 15, 829–845. [Google Scholar] [CrossRef]
- Tao, Y.; Zhang, Q.; Wang, H.; Yang, X.; Mu, H. Alternative splicing and related RNA binding proteins in human health and disease. Signal Transduct. Target. Ther. 2024, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Oliveira, C.; Faoro, H.; Alves, L.R.; Goldenberg, S. RNA-binding proteins and their role in the regulation of gene expression in Trypanosoma cruzi and Saccharomyces cerevisiae. Genet. Mol. Biol. 2017, 40, 22–30. [Google Scholar] [CrossRef]
- Pereira, B.; Billaud, M.; Almeida, R. RNA-Binding Proteins in Cancer: Old Players and New Actors. Trends Cancer 2017, 3, 506–528. [Google Scholar] [CrossRef]
- Kang, D.; Lee, Y.; Lee, J.S. RNA-Binding Proteins in Cancer: Functional and Therapeutic Perspectives. Cancers 2020, 12, 2699. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Ni, H.; Liu, Y.; Yuan, Y.; Xi, T.; Li, X.; Zheng, L. RNA-binding proteins in tumor progression. J. Hematol. Oncol. 2020, 13, 90. [Google Scholar] [CrossRef] [PubMed]
- Meisner, N.-C.; Filipowicz, W. Properties of the regulatory RNA-binding protein HuR and its role in controlling miRNA repression. In Regulation of microRNAs; Springer: New York, NY, USA, 2010; pp. 106–123. [Google Scholar]
- Dalmau, J.; Furneaux, H.M.; Gralla, R.J.; Kris, M.G.; Posner, J.B. Detection of the anti-Hu antibody in the serum of patients with small cell lung cancer—A quantitative western blot analysis. Ann. Neurol. 1990, 27, 544–552. [Google Scholar] [CrossRef] [PubMed]
- Brennan, C.; Steitz*, J. HuR and mRNA stability. Cell. Mol. Life Sci. CMLS 2001, 58, 266–277. [Google Scholar] [CrossRef] [PubMed]
- Finan, J.M.; Sutton, T.L.; Dixon, D.A.; Brody, J.R. Targeting the RNA-Binding Protein HuR in Cancer. Cancer Res. 2023, 83, 3507–3516. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Zhuang, R.; Xiao, L.; Chung, H.K.; Luo, J.; Turner, D.J.; Rao, J.N.; Gorospe, M.; Wang, J.Y. HuR Enhances Early Restitution of the Intestinal Epithelium by Increasing Cdc42 Translation. Mol. Cell Biol. 2017, 37, e00574-16. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Shi, X.; Li, W.; Zhang, F.; Cai, Z. The RNA-Binding Protein HuR in Digestive System Tumors. Biomed. Res. Int. 2020, 2020, 9656051. [Google Scholar] [CrossRef] [PubMed]
- Majumder, M.; Chakraborty, P.; Mohan, S.; Mehrotra, S.; Palanisamy, V. HuR as a molecular target for cancer therapeutics and immune-related disorders. Adv. Drug Deliv. Rev. 2022, 188, 114442. [Google Scholar] [CrossRef]
- Fan, X.C.; Steitz, J.A. Overexpression of HuR, a nuclear–cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998, 17, 3448–3460. [Google Scholar] [CrossRef]
- Dixon, D.A.; Kaplan, C.D.; McIntyre, T.M.; Zimmerman, G.A.; Prescott, S.M. Post-transcriptional control of cyclooxygenase-2 gene expression: The role of the 3′-untranslated region. J. Biol. Chem. 2000, 275, 11750–11757. [Google Scholar] [CrossRef]
- De Silanes, I.L.; Fan, J.; Yang, X.; Zonderman, A.B.; Potapova, O.; Pizer, E.S.; Gorospe, M. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene 2003, 22, 7146–7154. [Google Scholar] [CrossRef]
- Young, L.E.; Sanduja, S.; Bemis–Standoli, K.; Pena, E.A.; Price, R.L.; Dixon, D.A. The mRNA binding proteins HuR and tristetraprolin regulate cyclooxygenase 2 expression during colon carcinogenesis. Gastroenterology 2009, 136, 1669–1679. [Google Scholar] [CrossRef] [PubMed]
- Embade, N.; Fernández-Ramos, D.; Varela-Rey, M.; Beraza, N.; Sini, M.; de Juan, V.G.; Woodhoo, A.; Martínez-López, N.; Rodríguez-Iruretagoyena, B.; Bustamante, F.J. Murine double minute 2 regulates Hu antigen R stability in human liver and colon cancer through NEDDylation. Hepatology 2012, 55, 1237–1248. [Google Scholar] [CrossRef]
- Dixon, D.A.; Tolley, N.D.; King, P.H.; Nabors, L.B.; McIntyre, T.M.; Zimmerman, G.A.; Prescott, S.M. Altered expression of the mRNA stability factor HuR promotes cyclooxygenase-2 expression in colon cancer cells. J. Clin. Investig. 2001, 108, 1657–1665. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Koch, I.; von Keyserlingk, N.; Noske, A.; Niesporek, S.; Dietel, M.; Weichert, W. Expression of the ELAV-like protein HuR in human colon cancer: Association with tumor stage and cyclooxygenase-2. Mod. Pathol. 2006, 19, 1261–1269. [Google Scholar] [CrossRef] [PubMed]
- Sobolewski, C.; Dubuquoy, L.; Legrand, N. MicroRNAs, Tristetraprolin Family Members and HuR: A Complex Interplay Controlling Cancer-Related Processes. Cancers 2022, 14, 3516. [Google Scholar] [CrossRef] [PubMed]
- Tang, Z.; Kang, B.; Li, C.; Chen, T.; Zhang, Z. GEPIA2: An enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res. 2019, 47, W556–W560. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef]
- Iwakawa, H.-o.; Tomari, Y. The functions of microRNAs: mRNA decay and translational repression. Trends Cell Biol. 2015, 25, 651–665. [Google Scholar] [CrossRef]
- Eulalio, A.; Huntzinger, E.; Izaurralde, E. Getting to the root of miRNA-mediated gene silencing. Cell 2008, 132, 9–14. [Google Scholar] [CrossRef]
- O’Brien, J.; Hayder, H.; Zayed, Y.; Peng, C. Overview of microRNA biogenesis, mechanisms of actions, and circulation. Front. Endocrinol. 2018, 9, 388354. [Google Scholar] [CrossRef]
- Yadav, V.; Jena, M.K.; Parashar, G.; Parashar, N.C.; Joshi, H.; Ramniwas, S.; Tuli, H.S. Emerging role of microRNAs as regulators of protein kinase C substrate MARCKS and MARCKSL1 in cancer. Exp. Cell Res. 2024, 434, 113891. [Google Scholar] [CrossRef] [PubMed]
- Sun, W.; Julie Li, Y.-S.; Huang, H.-D.; Shyy, J.Y.; Chien, S. microRNA: A master regulator of cellular processes for bioengineering systems. Annu. Rev. Biomed. Eng. 2010, 12, 1–27. [Google Scholar] [PubMed]
- Hamilton, M.P.; Rajapakshe, K.; Hartig, S.M.; Reva, B.; McLellan, M.D.; Kandoth, C.; Ding, L.; Zack, T.I.; Gunaratne, P.H.; Wheeler, D.A. Identification of a pan-cancer oncogenic microRNA superfamily anchored by a central core seed motif. Nat. Commun. 2013, 4, 2730. [Google Scholar] [CrossRef]
- Agarwal, V.; Bell, G.W.; Nam, J.-W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Ahn, C.; Han, J.; Choi, H.; Kim, J.; Yim, J.; Lee, J.; Provost, P.; Rådmark, O.; Kim, S. The nuclear RNase III Drosha initiates microRNA processing. Nature 2003, 425, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Denli, A.M.; Tops, B.B.; Plasterk, R.H.; Ketting, R.F.; Hannon, G.J. Processing of primary microRNAs by the Microprocessor complex. Nature 2004, 432, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [PubMed]
- Treiber, T.; Treiber, N.; Meister, G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat. Rev. Mol. Cell Biol. 2019, 20, 5–20. [Google Scholar] [CrossRef]
- Lee, Y.; Kim, M.; Han, J.; Yeom, K.H.; Lee, S.; Baek, S.H.; Kim, V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004, 23, 4051–4060. [Google Scholar] [CrossRef]
- Borchert, G.M.; Lanier, W.; Davidson, B.L. RNA polymerase III transcribes human microRNAs. Nat. Struct. Mol. Biol. 2006, 13, 1097–1101. [Google Scholar] [CrossRef] [PubMed]
- MacFarlane, L.-A.; R Murphy, P. MicroRNA: Biogenesis, function and role in cancer. Curr. Genom. 2010, 11, 537–561. [Google Scholar] [CrossRef]
- Peng, Y.; Croce, C.M. The role of MicroRNAs in human cancer. Signal Transduct. Target. Ther. 2016, 1, 15004. [Google Scholar] [CrossRef] [PubMed]
- Majidinia, M.; Darband, S.G.; Kaviani, M.; Nabavi, S.M.; Jahanban-Esfahlan, R.; Yousefi, B. Cross-regulation between Notch signaling pathway and miRNA machinery in cancer. DNA Repair 2018, 66, 30–41. [Google Scholar] [CrossRef] [PubMed]
- Hayes, J.; Peruzzi, P.P.; Lawler, S. MicroRNAs in cancer: Biomarkers, functions and therapy. Trends Mol. Med. 2014, 20, 460–469. [Google Scholar] [CrossRef] [PubMed]
- Calin, G.; Croce, C. MicroRNAs and chromosomal abnormalities in cancer cells. Oncogene 2006, 25, 6202–6210. [Google Scholar] [CrossRef] [PubMed]
- Lujambio, A.; Calin, G.A.; Villanueva, A.; Ropero, S.; Sánchez-Céspedes, M.; Blanco, D.; Montuenga, L.M.; Rossi, S.; Nicoloso, M.S.; Faller, W.J. A microRNA DNA methylation signature for human cancer metastasis. Proc. Natl. Acad. Sci. USA 2008, 105, 13556–13561. [Google Scholar] [CrossRef] [PubMed]
- van Kouwenhove, M.; Kedde, M.; Agami, R. MicroRNA regulation by RNA-binding proteins and its implications for cancer. Nat. Rev. Cancer 2011, 11, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Mehrgou, A.; Ebadollahi, S.; Seidi, K.; Ayoubi-Joshaghani, M.H.; Yazdi, A.A.; Zare, P.; Jaymand, M.; Jahanban-Esfahlan, R. Roles of miRNAs in colorectal cancer: Therapeutic implications and clinical opportunities. Adv. Pharm. Bull. 2021, 11, 233. [Google Scholar] [CrossRef]
- Zhou, H.; Rao, Y.; Sun, Q.; Liu, Y.; Zhou, X.; Chen, Y.; Chen, J. MiR-4458/human antigen R (HuR) modulates PBX3 mRNA stability in melanoma tumorigenesis. Arch. Dermatol. Res. 2020, 312, 665–673. [Google Scholar] [CrossRef]
- Hussen, B.M.; Hidayat, H.J.; Salihi, A.; Sabir, D.K.; Taheri, M.; Ghafouri-Fard, S. MicroRNA: A signature for cancer progression. Biomed. Pharmacother. 2021, 138, 111528. [Google Scholar] [CrossRef]
- Thomas, J.; Ohtsuka, M.; Pichler, M.; Ling, H. MicroRNAs: Clinical relevance in colorectal cancer. Int. J. Mol. Sci. 2015, 16, 28063–28076. [Google Scholar] [CrossRef] [PubMed]
- Asangani, I.A.; Rasheed, S.A.; Nikolova, D.; Leupold, J.; Colburn, N.; Post, S.; Allgayer, H. MicroRNA-21 (miR-21) post-transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene 2008, 27, 2128–2136. [Google Scholar] [CrossRef] [PubMed]
- Xiong, B.; Cheng, Y.; Ma, L.; Zhang, C. MiR-21 regulates biological behavior through the PTEN/PI-3 K/Akt signaling pathway in human colorectal cancer cells. Int. J. Oncol. 2013, 42, 219–228. [Google Scholar] [CrossRef]
- Valeri, N.; Gasparini, P.; Braconi, C.; Paone, A.; Lovat, F.; Fabbri, M.; Sumani, K.M.; Alder, H.; Amadori, D.; Patel, T. MicroRNA-21 induces resistance to 5-fluorouracil by down-regulating human DNA MutS homolog 2 (hMSH2). Proc. Natl. Acad. Sci. USA 2010, 107, 21098–21103. [Google Scholar] [CrossRef] [PubMed]
- Mogilyansky, E.; Rigoutsos, I. The miR-17/92 cluster: A comprehensive update on its genomics, genetics, functions and increasingly important and numerous roles in health and disease. Cell Death Differ. 2013, 20, 1603–1614. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Zhou, H.; Xiao, H.; Liu, Z.; Tian, H.; Zhou, T. MicroRNA-92a functions as an oncogene in colorectal cancer by targeting PTEN. Dig. Dis. Sci. 2014, 59, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Nishida, N.; Nagahara, M.; Sato, T.; Mimori, K.; Sudo, T.; Tanaka, F.; Shibata, K.; Ishii, H.; Sugihara, K.; Doki, Y. Microarray analysis of colorectal cancer stromal tissue reveals upregulation of two oncogenic miRNA clusters. Clin. Cancer Res. 2012, 18, 3054–3070. [Google Scholar] [CrossRef] [PubMed]
- Zhou, T.; Zhang, G.; Liu, Z.; Xia, S.; Tian, H. Overexpression of miR-92a correlates with tumor metastasis and poor prognosis in patients with colorectal cancer. Int. J. Color. Dis. 2013, 28, 19–24. [Google Scholar] [CrossRef] [PubMed]
- Chang, P.-Y.; Chen, C.-C.; Chang, Y.-S.; Tsai, W.-S.; You, J.-F.; Lin, G.-P.; Chen, T.-W.; Chen, J.-S.; Chan, E.-C. MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget 2016, 7, 10663. [Google Scholar] [CrossRef]
- Kim, S.-A.; Kim, I.; Yoon, S.K.; Lee, E.K.; Kuh, H.-J. Indirect modulation of sensitivity to 5-fluorouracil by microRNA-96 in human colorectal cancer cells. Arch. Pharmacal Res. 2015, 38, 239–248. [Google Scholar] [CrossRef]
- Wang, Y.; Huang, J.-W.; Calses, P.; Kemp, C.J.; Taniguchi, T. MiR-96 Downregulates REV1 and RAD51 to Promote Cellular Sensitivity to Cisplatin and PARP Inhibition. Cancer Res. 2012, 72, 4037–4046. [Google Scholar] [CrossRef] [PubMed]
- Yuki, D.; Lin, Y.-M.; Fujii, Y.; Nakamura, Y.; Furukawa, Y. Isolation of LEM domain-containing 1, a novel testis-specific gene expressed in colorectal cancers. Oncol. Rep. 2004, 12, 275–280. [Google Scholar] [CrossRef]
- Slaby, O.; Svoboda, M.; Michalek, J.; Vyzula, R. MicroRNAs in colorectal cancer: Translation of molecular biology into clinical application. Mol. Cancer 2009, 8, 102. [Google Scholar] [CrossRef]
- He, Y.-q.; Sheng, J.-q.; Ling, X.-l.; Fu, L.; Jin, P.; Yen, L.; Rao, J. Estradiol regulates miR-135b and mismatch repair gene expressions via estrogen receptor-β in colorectal cells. Exp. Mol. Med. 2012, 44, 723–732. [Google Scholar] [CrossRef]
- Young, L.E.; Moore, A.E.; Sokol, L.; Meisner-Kober, N.; Dixon, D.A. The mRNA stability factor HuR inhibits microRNA-16 targeting of COX-2. Mol. Cancer Res. 2012, 10, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Chen, X.; Cheng, R.; Yang, F.; Yu, M.; Wang, C.; Cui, S.; Hong, Y.; Liang, H.; Liu, M.; et al. The Jun/miR-22/HuR regulatory axis contributes to tumourigenesis in colorectal cancer. Mol. Cancer 2018, 17, 11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Wang, J.; Li, N.; Liu, Z.; Chen, Z.; Li, Z.; Lai, Y.; Shen, L.; Gao, J. miR-34a increases the sensitivity of colorectal cancer cells to 5-fluorouracil in vitro and in vivo. Am. J. Cancer Res. 2018, 8, 280. [Google Scholar] [PubMed]
- Yamakuchi, M.; Ferlito, M.; Lowenstein, C.J. miR-34a repression of SIRT1 regulates apoptosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13421–13426. [Google Scholar] [CrossRef] [PubMed]
- Chang, H.-Y.; Ye, S.-P.; Pan, S.-L.; Kuo, T.-T.; Liu, B.C.; Chen, Y.-L.; Huang, T.-C. Overexpression of miR-194 reverses HMGA2-driven signatures in colorectal cancer. Theranostics 2017, 7, 3889. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Shen, Z.-l.; Gao, Z.-d.; Zhao, G.; Wang, C.-y.; Yang, Y.; Zhang, J.-z.; Yan, Y.-c.; Shen, C.; Jiang, K.-w. MiR-194, commonly repressed in colorectal cancer, suppresses tumor growth by regulating the MAP4K4/c-Jun/MDM2 signaling pathway. Cell Cycle 2015, 14, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.-H.; Ren, L.-L.; Zheng, P.; Zheng, H.-M.; Yu, Y.-N.; Wang, J.-L.; Lin, Y.-W.; Chen, Y.-X.; Ge, Z.-Z.; Chen, X.-Y. miR-194 as a predictor for adenoma recurrence in patients with advanced colorectal adenoma after polypectomy. Cancer Prev. Res. 2014, 7, 607–616. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.-J.; Ren, L.-L.; Wang, Z.-H.; Sun, T.-T.; Yu, Y.-N.; Wang, Y.-C.; Yan, T.-T.; Zou, W.; He, J.; Zhang, Y. MiR-194 deregulation contributes to colorectal carcinogenesis via targeting AKT2 pathway. Theranostics 2014, 4, 1193. [Google Scholar] [CrossRef]
- Meng, W.-J.; Yang, L.; Ma, Q.; Zhang, H.; Adell, G.; Arbman, G.; Wang, Z.-Q.; Li, Y.; Zhou, Z.-G.; Sun, X.-F. MicroRNA expression profile reveals miR-17-92 and miR-143-145 cluster in synchronous colorectal cancer. Medicine 2015, 94, e1297. [Google Scholar] [CrossRef] [PubMed]
- Gomes, S.E.; Simões, A.E.; Pereira, D.M.; Castro, R.E.; Rodrigues, C.M.; Borralho, P.M. miR-143 or miR-145 overexpression increases cetuximab-mediated antibody-dependent cellular cytotoxicity in human colon cancer cells. Oncotarget 2016, 7, 9368. [Google Scholar] [CrossRef]
- Al-Haidari, A.; Algaber, A.; Madhi, R.; Syk, I.; Thorlacius, H. MiR-155-5p controls colon cancer cell migration via post-transcriptional regulation of Human Antigen R (HuR). Cancer Lett. 2018, 421, 145–151. [Google Scholar] [CrossRef]
- Abdelmohsen, K.; Srikantan, S.; Kuwano, Y.; Gorospe, M. miR-519 reduces cell proliferation by lowering RNA-binding protein HuR levels. Proc. Natl. Acad. Sci. USA 2008, 105, 20297–20302. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Kim, M.M.; Srikantan, S.; Mercken, E.M.; Brennan, S.E.; Wilson, G.M.; de Cabo, R.; Gorospe, M. miR-519 suppresses tumor growth by reducing HuR levels. Cell Cycle 2010, 9, 1354–1359. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Zhang, M.; Sun, W.; Dong, C. Upregulation of miR-324-5p Inhibits Proliferation and Invasion of Colorectal Cancer Cells by Targeting ELAVL1. Oncol. Res. 2019, 27, 515–524. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Tian, Z.; Ye, H.; Sun, X.; Zhang, H.; Sun, Y.; Mao, Y.; Yang, Z.; Li, M. Emerging functions of circular RNA in the regulation of adipocyte metabolism and obesity. Cell Death Discov. 2022, 8, 268. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Zheng, L.; Xu, Y.; Liang, Y.; Li, D. Role of microRNA-34b-5p in cancer and injury: How does it work? Cancer Cell Int. 2022, 22, 381. [Google Scholar] [CrossRef] [PubMed]
- Zhao, C.; Popel, A.S. Computational model of microRNA control of HIF-VEGF pathway: Insights into the pathophysiology of ischemic vascular disease and cancer. PLoS Comput. Biol. 2015, 11, e1004612. [Google Scholar] [CrossRef] [PubMed]
- Slabáková, E.; Culig, Z.; Remšík, J.; Souček, K. Alternative mechanisms of miR-34a regulation in cancer. Cell Death Dis. 2017, 8, e3100. [Google Scholar] [CrossRef]
- Iyengar, B.R.; Pillai, B.; Venkatesh, K.; Gadgil, C.J. Systematic comparison of the response properties of protein and RNA mediated gene regulatory motifs. Mol. BioSystems 2017, 13, 1235–1245. [Google Scholar] [CrossRef]
- Takahashi, M.; Sung, B.; Shen, Y.; Hur, K.; Link, A.; Boland, C.R.; Aggarwal, B.B.; Goel, A. Boswellic acid exerts antitumor effects in colorectal cancer cells by modulating expression of the let-7 and miR-200 microRNA family. Carcinogenesis 2012, 33, 2441–2449. [Google Scholar] [CrossRef]
- Saridaki, Z.; Weidhaas, J.B.; Lenz, H.-J.; Laurent-Puig, P.; Jacobs, B.; De Schutter, J.; De Roock, W.; Salzman, D.W.; Zhang, W.; Yang, D. A let-7 microRNA-binding site polymorphism in KRAS predicts improved outcome in patients with metastatic colorectal cancer treated with salvage cetuximab/panitumumab monotherapy. Clin. Cancer Res. 2014, 20, 4499–4510. [Google Scholar] [CrossRef]
- Cappuzzo, F.; Sacconi, A.; Landi, L.; Ludovini, V.; Biagioni, F.; D’Incecco, A.; Capodanno, A.; Salvini, J.; Corgna, E.; Cupini, S. MicroRNA signature in metastatic colorectal cancer patients treated with anti-EGFR monoclonal antibodies. Clin. Color. Cancer 2014, 13, 37–45.e34. [Google Scholar] [CrossRef]
- Eslamizadeh, S.; Heidari, M.; Agah, S.; Faghihloo, E.; Ghazi, H.; Mirzaei, A.; Akbari, A. The role of microRNA signature as diagnostic biomarkers in different clinical stages of colorectal cancer. Cell J. 2018, 20, 220. [Google Scholar]
- Lasda, E.; Parker, R. Circular RNAs: Diversity of form and function. RNA 2014, 20, 1829–1842. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 2020, 21, 475–490. [Google Scholar] [CrossRef] [PubMed]
- Baumann, K. CircRNAs in lifespan. Nat. Rev. Mol. Cell Biol. 2020, 21, 420. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Chinnaiyan, A.M.; Conn, S.J. Circular RNA in cancer. Nat. Rev. Cancer 2024, 24, 597–613. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, X.O.; Chen, T.; Xiang, J.F.; Yin, Q.F.; Xing, Y.H.; Zhu, S.; Yang, L.; Chen, L.L. Circular intronic long noncoding RNAs. Mol. Cell 2013, 51, 792–806. [Google Scholar] [CrossRef] [PubMed]
- Rybak-Wolf, A.; Stottmeister, C.; Glazar, P.; Jens, M.; Pino, N.; Giusti, S.; Hanan, M.; Behm, M.; Bartok, O.; Ashwal-Fluss, R.; et al. Circular RNAs in the Mammalian Brain Are Highly Abundant, Conserved, and Dynamically Expressed. Mol. Cell 2015, 58, 870–885. [Google Scholar] [CrossRef]
- Patop, I.L.; Wust, S.; Kadener, S. Past, present, and future of circRNAs. EMBO J. 2019, 38, e100836. [Google Scholar] [CrossRef] [PubMed]
- Arizaca-Maquera, K.A.; Fernández-Álvarez, A.J. Circular RNAs: Molecules with past and future. Actual. Biol. 2022, 44, 1–12. [Google Scholar] [CrossRef]
- Wan, L.; Zhang, L.; Fan, K.; Cheng, Z.X.; Sun, Q.C.; Wang, J.J. Circular RNA-ITCH Suppresses Lung Cancer Proliferation via Inhibiting the Wnt/beta-Catenin Pathway. Biomed. Res. Int. 2016, 2016, 1579490. [Google Scholar] [CrossRef]
- Li, X.; Wang, J.; Zhang, C.; Lin, C.; Zhang, J.; Zhang, W.; Zhang, W.; Lu, Y.; Zheng, L.; Li, X. Circular RNA circITGA7 inhibits colorectal cancer growth and metastasis by modulating the Ras pathway and upregulating transcription of its host gene ITGA7. J. Pathol. 2018, 246, 166–179. [Google Scholar] [CrossRef]
- Li, Z.; Huang, C.; Bao, C.; Chen, L.; Lin, M.; Wang, X.; Zhong, G.; Yu, B.; Hu, W.; Dai, L.; et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 2015, 22, 256–264. [Google Scholar] [CrossRef]
- Chen, N.; Zhao, G.; Yan, X.; Lv, Z.; Yin, H.; Zhang, S.; Song, W.; Li, X.; Li, L.; Du, Z.; et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 2018, 19, 218. [Google Scholar] [CrossRef]
- Li, H.; Yang, F.; Hu, A.; Wang, X.; Fang, E.; Chen, Y.; Li, D.; Song, H.; Wang, J.; Guo, Y.; et al. Therapeutic targeting of circ-CUX1/EWSR1/MAZ axis inhibits glycolysis and neuroblastoma progression. EMBO Mol. Med. 2019, 11, e10835. [Google Scholar] [CrossRef] [PubMed]
- Conn, V.M.; Hugouvieux, V.; Nayak, A.; Conos, S.A.; Capovilla, G.; Cildir, G.; Jourdain, A.; Tergaonkar, V.; Schmid, M.; Zubieta, C.; et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 2017, 3, 17053. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Zhang, J.; Tian, Y.; Gao, Y.; Dong, X.; Chen, W.; Yuan, X.; Yin, W.; Xu, J.; Chen, K.; et al. CircRNA inhibits DNA damage repair by interacting with host gene. Mol. Cancer 2020, 19, 128. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Gao, Y.; Liao, J.; Miao, M.; Chen, K.; Xi, F.; Wei, W.; Wang, H.; Wang, Y.; Xu, X.; et al. Genome-wide profiling of circular RNAs, alternative splicing, and R-loops in stem-differentiating xylem of Populus trichocarpa. J. Integr. Plant Biol. 2021, 63, 1294–1308. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Klungland, A. Modifications and interactions at the R-loop. DNA Repair 2020, 96, 102958. [Google Scholar] [CrossRef]
- Garcia-Muse, T.; Aguilera, A. R Loops: From Physiological to Pathological Roles. Cell 2019, 179, 604–618. [Google Scholar] [CrossRef]
- Yu, F.X.; Guan, K.L. The Hippo pathway: Regulators and regulations. Genes. Dev. 2013, 27, 355–371. [Google Scholar] [CrossRef] [PubMed]
- Wu, N.; Yuan, Z.; Du, K.Y.; Fang, L.; Lyu, J.; Zhang, C.; He, A.; Eshaghi, E.; Zeng, K.; Ma, J.; et al. Translation of yes-associated protein (YAP) was antagonized by its circular RNA via suppressing the assembly of the translation initiation machinery. Cell Death Differ. 2019, 26, 2758–2773. [Google Scholar] [CrossRef] [PubMed]
- Abdelmohsen, K.; Panda, A.C.; Munk, R.; Grammatikakis, I.; Dudekula, D.B.; De, S.; Kim, J.; Noh, J.H.; Kim, K.M.; Martindale, J.L.; et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 2017, 14, 361–369. [Google Scholar] [CrossRef]
- Haupt, Y.; Maya, R.; Kazaz, A.; Oren, M. Mdm2 promotes the rapid degradation of p53. Nature 1997, 387, 296–299. [Google Scholar] [CrossRef]
- Du, W.W.; Fang, L.; Yang, W.; Wu, N.; Awan, F.M.; Yang, Z.; Yang, B.B. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 2017, 24, 357–370. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D. The evolution of post-translational modifications. Curr. Opin. Genet. Dev. 2022, 76, 101956. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, D.H.; Wu, N.; Xiao, J.H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.B.; Jensen, T.I.; Clausen, B.H.; Bramsen, J.B.; Finsen, B.; Damgaard, C.K.; Kjems, J. Natural RNA circles function as efficient microRNA sponges. Nature 2013, 495, 384–388. [Google Scholar] [CrossRef]
- Li, F.; Zhang, L.; Li, W.; Deng, J.; Zheng, J.; An, M.; Lu, J.; Zhou, Y. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/beta-catenin pathway. Oncotarget 2015, 6, 6001–6013. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Du, Y.; Yang, X.; Mo, Y.; Fan, C.; Xiong, F.; Ren, D.; Ye, X.; Li, C.; Wang, Y.; et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol. Cancer 2018, 17, 79. [Google Scholar] [CrossRef]
- Yao, B.; Zhang, Q.; Yang, Z.; An, F.; Nie, H.; Wang, H.; Yang, C.; Sun, J.; Chen, K.; Zhou, J.; et al. CircEZH2/miR-133b/IGF2BP2 aggravates colorectal cancer progression via enhancing the stability of m(6)A-modified CREB1 mRNA. Mol. Cancer 2022, 21, 140. [Google Scholar] [CrossRef]
- Mehta, S.L.; Chokkalla, A.K.; Bathula, S.; Arruri, V.; Chelluboina, B.; Vemuganti, R. CDR1as regulates alpha-synuclein-mediated ischemic brain damage by controlling miR-7 availability. Mol. Ther. Nucleic Acids 2023, 31, 57–67. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; Zeng, X.; Shan, R.; Wen, W.; Li, J.; Tan, J.; Li, L.; Wan, R. The Emerging Picture of the Roles of CircRNA-CDR1as in Cancer. Front. Cell Dev. Biol. 2020, 8, 590478. [Google Scholar] [CrossRef] [PubMed]
- Tan, P.; Sun, H.; Xu, M.; Liu, X.; Qin, J.; Nie, J.; Qin, X.; Wang, S.; Pan, Y. Circular RNA circ0104103 inhibits colorectal cancer progression through interactions with HuR and miR-373-5p. Cancer Sci. 2023, 114, 1396–1409. [Google Scholar] [CrossRef] [PubMed]
- Pellecchia, S.; Quintavalle, C.; Pallante, P. The control of tumor progression by circular RNAs: Novel prognostic and therapeutic insights resulting from the analysis of the circAGO2/human antigen R complex. Transl. Cancer Res. 2019, 8, S211–S215. [Google Scholar] [CrossRef] [PubMed]
- Du, W.W.; Yang, W.; Liu, E.; Yang, Z.; Dhaliwal, P.; Yang, B.B. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016, 44, 2846–2858. [Google Scholar] [CrossRef] [PubMed]
- Yang, T.; Li, Y.; Zhao, F.; Zhou, L.; Jia, R. Circular RNA Foxo3: A Promising Cancer-Associated Biomarker. Front. Genet. 2021, 12, 652995. [Google Scholar] [CrossRef] [PubMed]
- Panda, A.C.; Grammatikakis, I.; Munk, R.; Gorospe, M.; Abdelmohsen, K. Emerging roles and context of circular RNAs. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef]
- Singh, D.; Kesharwani, P.; Alhakamy, N.A.; Siddique, H.R. Accentuating CircRNA-miRNA-Transcription Factors Axis: A Conundrum in Cancer Research. Front. Pharmacol. 2021, 12, 784801. [Google Scholar] [CrossRef]
- Zhang, Y.; Luo, J.; Yang, W.; Ye, W.C. CircRNAs in colorectal cancer: Potential biomarkers and therapeutic targets. Cell Death Dis. 2023, 14, 353. [Google Scholar] [CrossRef] [PubMed]
- Shao, Q.; Huang, Y.; Zhang, C.; Gao, X.; Gao, S. Emerging landscape of circHIPK3 and its role in cancer and other diseases (Review). Mol. Med. Rep. 2021, 23. [Google Scholar] [CrossRef]
- Tang, X.; Ren, H.; Guo, M.; Qian, J.; Yang, Y.; Gu, C. Review on circular RNAs and new insights into their roles in cancer. Comput. Struct. Biotechnol. J. 2021, 19, 910–928. [Google Scholar] [CrossRef]
- Mao, Y.; Miao, J.; Xi, L.; Tong, H.; Shen, X.; Li, Q.; Yu, C. circSKA3 promotes colorectal cancer metastases through miR-1238 and methylation. Mol. Cell Biochem. 2024, 479, 941–950. [Google Scholar] [CrossRef]
- Ji, H.; Kim, T.W.; Lee, W.J.; Jeong, S.D.; Cho, Y.B.; Kim, H.H. Two circPPFIA1s negatively regulate liver metastasis of colon cancer via miR-155-5p/CDX1 and HuR/RAB36. Mol. Cancer 2022, 21, 197. [Google Scholar] [CrossRef]
- Yuan, M.; Zhang, X.; Yue, F.; Zhang, F.; Jiang, S.; Zhou, X.; Lv, J.; Zhang, Z.; Sun, Y.; Chen, Z.; et al. CircNOLC1 Promotes Colorectal Cancer Liver Metastasis by Interacting with AZGP1 and Sponging miR-212-5p to Regulate Reprogramming of the Oxidative Pentose Phosphate Pathway. Adv. Sci. 2023, 10, 2205229. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Lin, C.; Liu, Y. Molecular mechanism of miR-34b-5p and RNA binding protein HuR binding to lncRNA OIP5-AS1 in colon cancer cells. Cancer Gene Ther. 2022, 29, 612–624. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Sun, D.L.; Yang, S.P.; Song, J.; Wu, M.Y.; Niu, W.W.; Song, M.; Zhang, X.L. Long noncoding RNA TNFRSF10A-AS1 promotes colorectal cancer through upregulation of HuR. World J. Gastroenterol. 2022, 28, 2184–2200. [Google Scholar] [CrossRef] [PubMed]
- Fontemaggi, G.; Turco, C.; Esposito, G.; Di Agostino, S. New Molecular Mechanisms and Clinical Impact of circRNAs in Human Cancer. Cancers 2021, 13, 3154. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Wu, Y.; Luo, X.; Jin, D.; Zhou, W.; Ju, Z.; Wang, D.; Meng, Q.; Wang, H.; Fu, X.; et al. Circular RNA circRHOBTB3 represses metastasis by regulating the HuR-mediated mRNA stability of PTBP1 in colorectal cancer. Theranostics 2021, 11, 7507–7526. [Google Scholar] [CrossRef] [PubMed]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Wang, D.; Miao, Y.R.; Wu, X.; Luo, H.; Cao, W.; Yang, W.; Yang, J.; Guo, A.Y.; Gong, J. lncRNASNP v3: An updated database for functional variants in long non-coding RNAs. Nucleic Acids Res. 2023, 51, D192–D198. [Google Scholar] [CrossRef]
- Dueva, R.; Akopyan, K.; Pederiva, C.; Trevisan, D.; Dhanjal, S.; Lindqvist, A.; Farnebo, M. Neutralization of the Positive Charges on Histone Tails by RNA Promotes an Open Chromatin Structure. Cell Chem. Biol. 2019, 26, 1436–1449.e1435. [Google Scholar] [CrossRef]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef]
- Yang, F.; Deng, X.; Ma, W.; Berletch, J.B.; Rabaia, N.; Wei, G.; Moore, J.M.; Filippova, G.N.; Xu, J.; Liu, Y.; et al. The lncRNA Firre anchors the inactive X chromosome to the nucleolus by binding CTCF and maintains H3K27me3 methylation. Genome Biol. 2015, 16, 52. [Google Scholar] [CrossRef]
- Saldaña-Meyer, R.; Rodriguez-Hernaez, J.; Escobar, T.; Nishana, M.; Jácome-López, K.; Nora, E.P.; Bruneau, B.G.; Tsirigos, A.; Furlan-Magaril, M.; Skok, J.; et al. RNA Interactions Are Essential for CTCF-Mediated Genome Organization. Mol. Cell 2019, 76, 412–422.e415. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; Lu, J.Y.; Liu, L.; Yin, Y.; Chen, C.; Han, X.; Wu, B.; Xu, R.; Liu, W.; Yan, P.; et al. Divergent lncRNAs Regulate Gene Expression and Lineage Differentiation in Pluripotent Cells. Cell Stem Cell 2016, 18, 637–652. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Mattick, J.S.; Amaral, P.P.; Carninci, P.; Carpenter, S.; Chang, H.Y.; Chen, L.L.; Chen, R.; Dean, C.; Dinger, M.E.; Fitzgerald, K.A.; et al. Long non-coding RNAs: Definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 2023, 24, 430–447. [Google Scholar] [CrossRef] [PubMed]
- West, J.A.; Mito, M.; Kurosaka, S.; Takumi, T.; Tanegashima, C.; Chujo, T.; Yanaka, K.; Kingston, R.E.; Hirose, T.; Bond, C.; et al. Structural, super-resolution microscopy analysis of paraspeckle nuclear body organization. J. Cell Biol. 2016, 214, 817–830. [Google Scholar] [CrossRef]
- Johnsson, P.; Ziegenhain, C.; Hartmanis, L.; Hendriks, G.J.; Hagemann-Jensen, M.; Reinius, B.; Sandberg, R. Transcriptional kinetics and molecular functions of long noncoding RNAs. Nat. Genet. 2022, 54, 306–317. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharjee, R.; Prabhakar, N.; Kumar, L.; Bhattacharjee, A.; Kar, S.; Malik, S.; Kumar, D.; Ruokolainen, J.; Negi, A.; Jha, N.K.; et al. Crosstalk between long noncoding RNA and microRNA in Cancer. Cell. Oncol. 2023, 46, 885–908. [Google Scholar] [CrossRef] [PubMed]
- Lulli, M.; Napoli, C.; Landini, I.; Mini, E.; Lapucci, A. Role of Non-Coding RNAs in Colorectal Cancer: Focus on Long Non-Coding RNAs. Int. J. Mol. Sci. 2022, 23, 13431. [Google Scholar] [CrossRef] [PubMed]
- Entezari, M.; Taheriazam, A.; Orouei, S.; Fallah, S.; Sanaei, A.; Hejazi, E.S.; Kakavand, A.; Rezaei, S.; Heidari, H.; Behroozaghdam, M.; et al. LncRNA-miRNA axis in tumor progression and therapy response: An emphasis on molecular interactions and therapeutic interventions. Biomed. Pharmacother. 2022, 154, 113609. [Google Scholar] [CrossRef]
- Ma, B.; Wang, S.; Wu, W.; Shan, P.; Chen, Y.; Meng, J.; Xing, L.; Yun, J.; Hao, L.; Wang, X.; et al. Mechanisms of circRNA/lncRNA-miRNA interactions and applications in disease and drug research. Biomed. Pharmacother. 2023, 162, 114672. [Google Scholar] [CrossRef]
- Yoon, J.H.; Abdelmohsen, K.; Gorospe, M. Functional interactions among microRNAs and long noncoding RNAs. Semin. Cell Dev. Biol. 2014, 34, 9–14. [Google Scholar] [CrossRef] [PubMed]
- Forrest, M.E.; Saiakhova, A.; Beard, L.; Buchner, D.A.; Scacheri, P.C.; LaFramboise, T.; Markowitz, S.; Khalil, A.M. Colon Cancer-Upregulated Long Non-Coding RNA lincDUSP Regulates Cell Cycle Genes and Potentiates Resistance to Apoptosis. Sci. Rep. 2018, 8, 7324. [Google Scholar] [CrossRef]
- Shakhpazyan, N.K.; Mikhaleva, L.M.; Bedzhanyan, A.L.; Sadykhov, N.K.; Midiber, K.Y.; Konyukova, A.K.; Kontorschikov, A.S.; Maslenkina, K.S.; Orekhov, A.N. Long Non-Coding RNAs in Colorectal Cancer: Navigating the Intersections of Immunity, Intercellular Communication, and Therapeutic Potential. Biomedicines 2023, 11, 2411. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Shen, X. Long noncoding RNAs: Functions and mechanisms in colon cancer. Mol. Cancer 2020, 19, 167. [Google Scholar] [CrossRef]
- Kasagi, Y.; Oki, E.; Ando, K.; Ito, S.; Iguchi, T.; Sugiyama, M.; Nakashima, Y.; Ohgaki, K.; Saeki, H.; Mimori, K.; et al. The Expression of CCAT2, a Novel Long Noncoding RNA Transcript, and rs6983267 Single-Nucleotide Polymorphism Genotypes in Colorectal Cancers. Oncology 2017, 92, 48–54. [Google Scholar] [CrossRef]
- Ling, H.; Spizzo, R.; Atlasi, Y.; Nicoloso, M.; Shimizu, M.; Redis, R.S.; Nishida, N.; Gafà, R.; Song, J.; Guo, Z.; et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. 2013, 23, 1446–1461. [Google Scholar] [CrossRef]
- Rajtmajerová, M.; Trailin, A.; Liška, V.; Hemminki, K.; Ambrozkiewicz, F. Long Non-Coding RNA and microRNA Interplay in Colorectal Cancer and Their Effect on the Tumor Microenvironment. Cancers 2022, 14, 5450. [Google Scholar] [CrossRef]
- Alshahrani, S.H.; Al-Hadeithi, Z.S.M.; Almalki, S.G.; Malviya, J.; Hjazi, A.; Mustafa, Y.F.; Alawady, A.H.R.; Alsaalamy, A.H.; Joshi, S.K.; Alkhafaji, A.T. LncRNA-miRNA interaction is involved in colorectal cancer pathogenesis by modulating diverse signaling pathways. Pathol. Res. Pract. 2023, 251, 154898. [Google Scholar] [CrossRef] [PubMed]
- Srikantan, S.; Tominaga, K.; Gorospe, M. Functional interplay between RNA-binding protein HuR and microRNAs. Curr. Protein Pept. Sci. 2012, 13, 372–379. [Google Scholar] [CrossRef] [PubMed]
- Höck, J.; Weinmann, L.; Ender, C.; Rüdel, S.; Kremmer, E.; Raabe, M.; Urlaub, H.; Meister, G. Proteomic and functional analysis of Argonaute-containing mRNA–protein complexes in human cells. EMBO Rep. 2007, 8, 1052–1060. [Google Scholar] [CrossRef] [PubMed]
- Kedde, M.; Agami, R. Interplay between microRNAs and RNA-binding proteins determines developmental processes. Cell Cycle 2008, 7, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Merino, E.J.; Wilkinson, K.A.; Coughlan, J.L.; Weeks, K.M. RNA structure analysis at single nucleotide resolution by selective 2 ‘-hydroxyl acylation and primer extension (SHAPE). J. Am. Chem. Soc. 2005, 127, 4223–4231. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, C.B.; Shomron, N.; Sandberg, R.; Hornstein, E.; Kitzman, J.; Burge, C.B. Determinants of targeting by endogenous and exogenous microRNAs and siRNAs. RNA 2007, 13, 1894–1910. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yadav, V.; Singh, T.; Sharma, D.; Garg, V.K.; Chakraborty, P.; Ghatak, S.; Satapathy, S.R. Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis. Cancers 2024, 16, 3183. https://doi.org/10.3390/cancers16183183
Yadav V, Singh T, Sharma D, Garg VK, Chakraborty P, Ghatak S, Satapathy SR. Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis. Cancers. 2024; 16(18):3183. https://doi.org/10.3390/cancers16183183
Chicago/Turabian StyleYadav, Vikas, Tejveer Singh, Deepika Sharma, Vivek Kumar Garg, Payel Chakraborty, Souvik Ghatak, and Shakti Ranjan Satapathy. 2024. "Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis" Cancers 16, no. 18: 3183. https://doi.org/10.3390/cancers16183183
APA StyleYadav, V., Singh, T., Sharma, D., Garg, V. K., Chakraborty, P., Ghatak, S., & Satapathy, S. R. (2024). Unraveling the Regulatory Role of HuR/microRNA Axis in Colorectal Cancer Tumorigenesis. Cancers, 16(18), 3183. https://doi.org/10.3390/cancers16183183







