Isoform-Level Transcriptome Analysis of Peripheral Blood Mononuclear Cells from Breast Cancer Patients Identifies a Disease-Associated RASGEF1A Isoform
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. The Study Population and Clinicopathological Characteristics
2.2. Processing of Blood Samples
2.3. Sequencing of RNA Isolated from PBMCs
2.4. RNA-Seq Data Alignment and Identification of Differentially Expressed Isoforms
2.5. Preparation of cDNA and Isoform-Specific RT-qPCR Analysis
3. Results
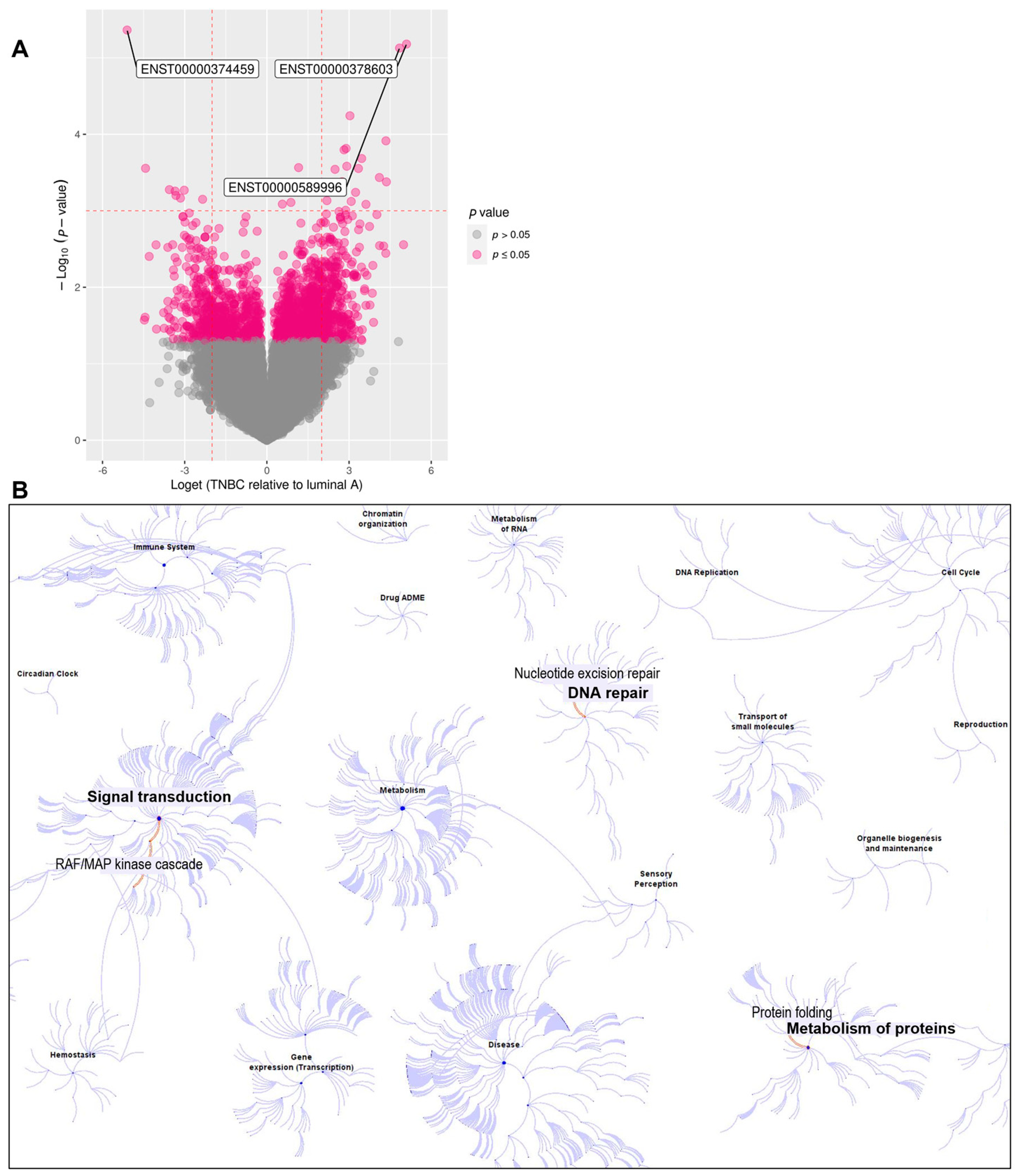
3.1. RNA Sequencing Identified Differences in Isoform Expression between Luminal A and TNBC Patients
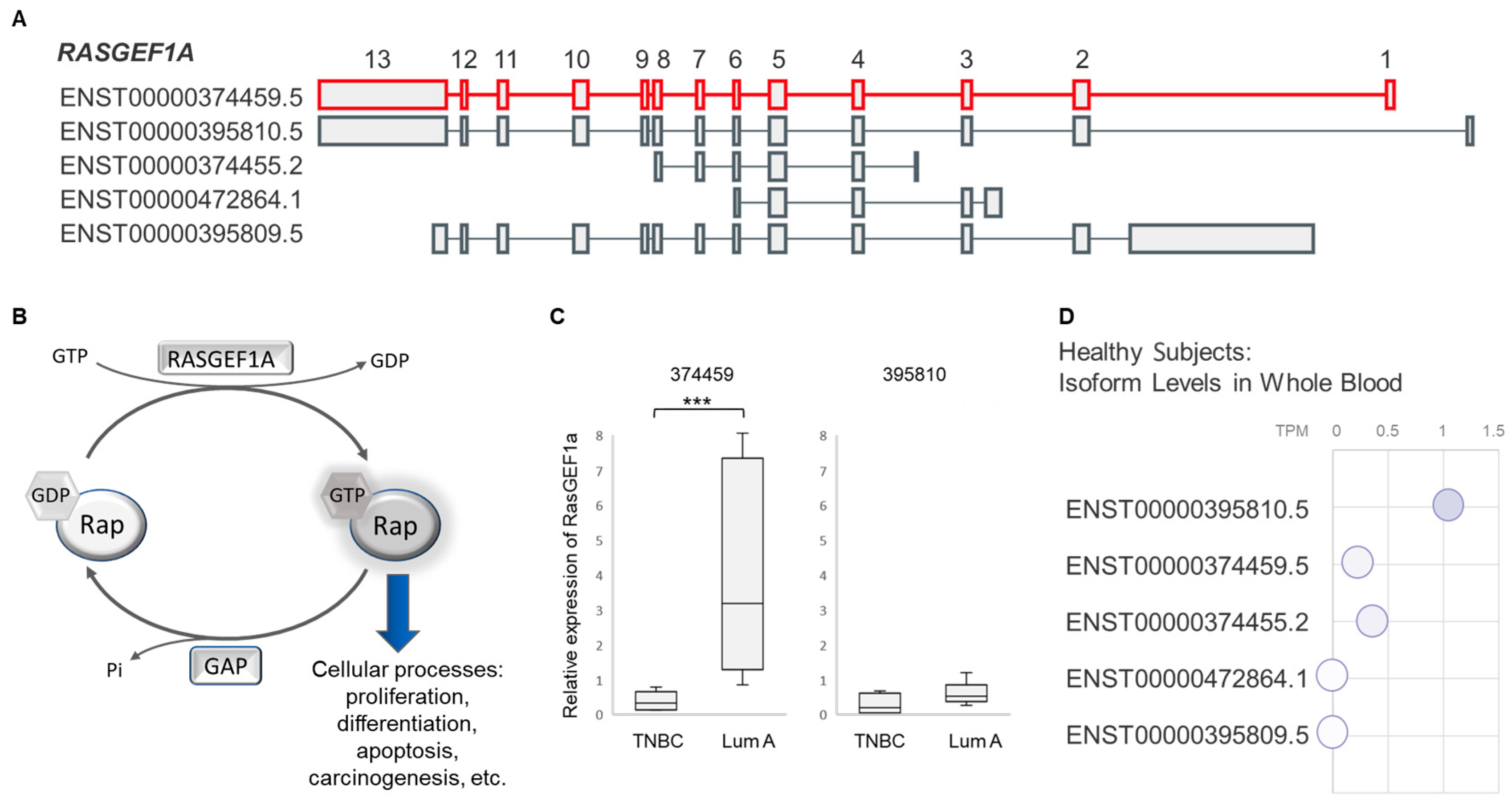
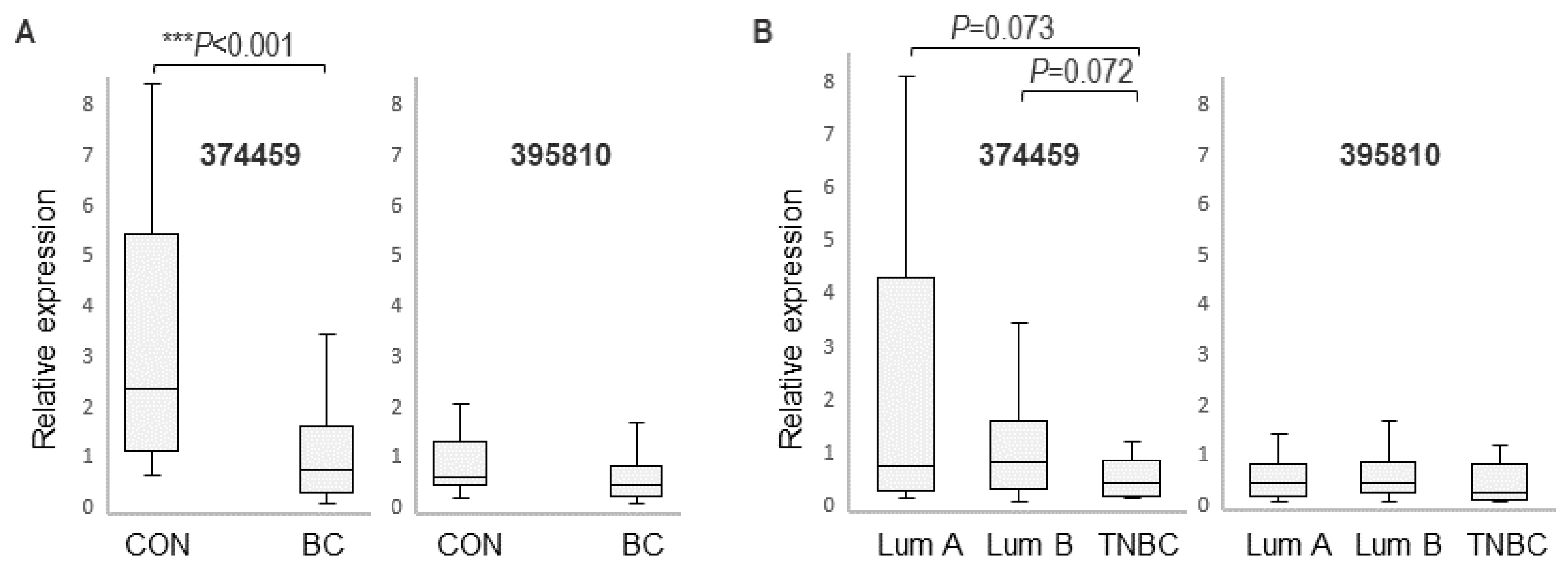
3.2. Expression of RASGEF1A Isoforms in a Larger Cohort Comprising BC Patients and Healthy Female Controls
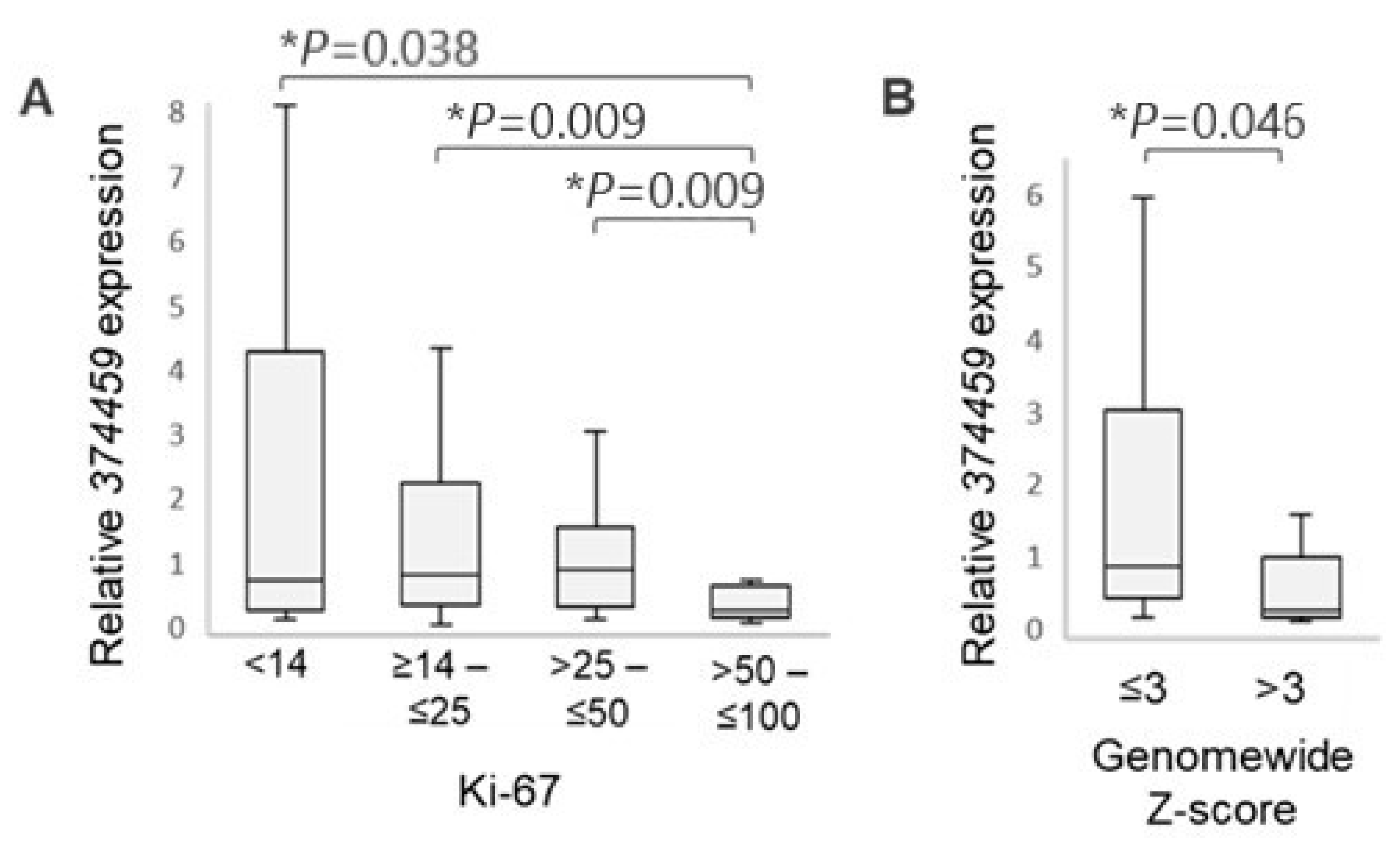
3.3. RASGEF1A Isoform Expression and Clinicopathological Characteristics of BC Patients
3.3.1. Association with Ki-67 Proliferation Index
3.3.2. Association with Circulating Tumor DNA (ctDNA) Content
3.3.3. Other Clinicopathological Characteristics
4. Discussion
4.1. The Advantages of Blood Analyses over Standard Methods for Cancer Detection
4.2. RASGEF1A Function
4.3. RASGEF1A 374459 Isoform and Cancer Proliferation and Shedding
4.4. Advantages of Isoform-Level Bioinformatics Analysis of RNA-Seq Data
4.5. Other Dysregulated Isoforms Identified in Our Study
4.6. Limitations and Future Perspectives
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wilkinson, L.; Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 2021, 95, 20211033. [Google Scholar] [CrossRef] [PubMed]
- Čelešnik, H.; Potočnik, U. Peripheral Blood Transcriptome in Breast Cancer Patients as a Source of Less Invasive Immune Biomarkers for Personalized Medicine, and Implications for Triple Negative Breast Cancer. Cancers 2022, 14, 591. [Google Scholar] [CrossRef] [PubMed]
- Skok, K.; Gradišnik, L.; Čelešnik, H.; Milojević, M.; Potočnik, U.; Jezernik, G.; Gorenjak, M.; Sobočan, M.; Takač, I.; Kavalar, R.; et al. MFUM-BrTNBC-1, a Newly Established Patient-Derived Triple-Negative Breast Cancer Cell Line: Molecular Characterisation, Genetic Stability, and Comprehensive Comparison with Commercial Breast Cancer Cell Lines. Cells 2021, 11, 117. [Google Scholar] [CrossRef] [PubMed]
- Szymiczek, A.; Lone, A.; Akbari, M.R. Molecular intrinsic versus clinical subtyping in breast cancer: A comprehensive review. Clin. Genet. 2021, 99, 613–637. [Google Scholar] [CrossRef]
- Park, S.; Koo, J.S.; Kim, M.S.; Park, H.S.; Lee, J.S.; Lee, J.S.; Kim, S.I.; Park, B.-W. Characteristics and outcomes according to molecular subtypes of breast cancer as classified by a panel of four biomarkers using immunohistochemistry. Breast 2012, 21, 50–57. [Google Scholar] [CrossRef]
- Prat, A.; Adamo, B.; Cheang, M.C.U.; Anders, C.K.; Carey, L.A.; Perou, C.M. Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 2013, 18, 123–133. [Google Scholar] [CrossRef]
- Rajtak, A.; Ostrowska-Leśko, M.; Żak, K.; Tarkowski, R.; Kotarski, J.; Okła, K. Integration of local and systemic immunity in ovarian cancer: Implications for immunotherapy. Front. Immunol. 2022, 13, 1018256. [Google Scholar] [CrossRef]
- Xu, L.; Zou, C.; Zhang, S.; Chu, T.S.M.; Zhang, Y.; Chen, W.; Zhao, C.; Yang, L.; Xu, Z.; Dong, S.; et al. Reshaping the systemic tumor immune environment (STIE) and tumor immune microenvironment (TIME) to enhance immunotherapy efficacy in solid tumors. J. Hematol. Oncol. 2022, 15, 87. [Google Scholar] [CrossRef]
- Kalantari, S.; Kazemi, B.; Roudi, R.; Zali, H.; D’Angelo, A.; Mohamadkhani, A.; Madjd, Z.; Pourshams, A. RNA-sequencing for transcriptional profiling of whole blood in early stage and metastatic pancreatic cancer patients. Cell Biol. Int. 2023, 47, 238–249. [Google Scholar] [CrossRef]
- Kumar, S.; Das, A. Peripheral blood mononuclear cell derived biomarker detection using eXplainable Artificial Intelligence (XAI) provides better diagnosis of breast cancer. Comput. Biol. Chem. 2023, 104, 107867. [Google Scholar]
- Twine, N.C.; A Stover, J.; Marshall, B.; Dukart, G.; Hidalgo, M.; Stadler, W.; Logan, T.; Dutcher, J.; Hudes, G.; Dorner, A.J.; et al. Disease-associated expression profiles in peripheral blood mononuclear cells from patients with advanced renal cell carcinoma. Cancer Res. 2003, 63, 6069–6075. [Google Scholar] [PubMed]
- Yang, Y.; Zhang, T.; Xiao, R.; Hao, X.; Zhang, H.; Qu, H.; Xie, B.; Wang, T.; Fang, X. Platform-independent approach for cancer detection from gene expression profiles of peripheral blood cells. Brief. Bioinform. 2020, 21, 1006–1015. [Google Scholar] [CrossRef]
- de Fraipont, F.; Gazzeri, S.; William, C.C.; Eymin, B. Circular RNAs and RNA Splice Variants as Biomarkers for Prognosis and Therapeutic Response in the Liquid Biopsies of Lung Cancer Patients. Front. Genet. 2019, 10, 390. [Google Scholar] [CrossRef] [PubMed]
- Stricker, T.P.; Brown, C.D.; Bandlamudi, C.; McNerney, M.; Kittler, R.; Montoya, V.; Peterson, A.; Grossman, R.; White, K.P. Robust stratification of breast cancer subtypes using differential patterns of transcript isoform expression. PLoS Genet. 2017, 13, e1006589. [Google Scholar] [CrossRef]
- Weber, R.; Ghoshdastider, U.; Spies, D.; Duré, C.; Valdivia-Francia, F.; Forny, M.; Ormiston, M.; Renz, P.F.; Taborsky, D.; Yigit, M.; et al. Monitoring the 5′UTR landscape reveals isoform switches to drive translational efficiencies in cancer. Oncogene 2023, 42, 638–650. [Google Scholar] [CrossRef]
- Vitting-Seerup, K.; Sandelin, A. The Landscape of Isoform Switches in Human Cancers. Mol. Cancer Res. 2017, 15, 1206–1220. [Google Scholar] [CrossRef]
- Zhang, Y.; Qian, J.; Gu, C.; Yang, Y. Alternative splicing and cancer: A systematic review. Signal Transduct. Target. Ther. 2021, 6, 78. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Pal, S.; Bi, Y.; Tchou, J.; Davuluri, R.V. Isoform level expression profiles provide better cancer signatures than gene level expression profiles. Genome Med. 2013, 5, 33. [Google Scholar] [CrossRef]
- Erdem, M.; Ozgul, I.; Dioken, D.N.; Gurcuoglu, I.; Ergun, S.G.; Cetin-Atalay, R.; Can, T.; Erson-Bensan, A.E. Identification of an mRNA isoform switch for HNRNPA1 in breast cancers. Sci. Rep. 2021, 11, 24444. [Google Scholar] [CrossRef]
- Brown, R.L.; Reinke, L.M.; Damerow, M.S.; Perez, D.; Chodosh, L.A.; Yang, J.; Cheng, C. CD44 splice isoform switching in human and mouse epithelium is essential for epithelial-mesenchymal transition and breast cancer progression. J. Clin. Investig. 2011, 121, 1064–1074. [Google Scholar] [CrossRef]
- Lombardi, G.; Falaschi, E.; Di Cristofano, C.; Naccarato, A.G.; Sensi, E.; Aretini, P.; Roncella, M.; Bevilacqua, G.; Caligo, M.A. Identification of novel alternatively spliced BRCA1-associated RING domain (BARD1) messenger RNAs in human peripheral blood lymphocytes and in sporadic breast cancer tissues. Genes Chromosomes Cancer 2007, 46, 791–795. [Google Scholar] [CrossRef]
- Okumura, N.; Yoshida, H.; Kitagishi, Y.; Nishimura, Y.; Matsuda, S. Alternative splicings on p53, BRCA1 and PTEN genes involved in breast cancer. Biochem. Biophys. Res. Commun. 2011, 413, 395–399. [Google Scholar] [CrossRef] [PubMed]
- Shapiro, I.M.; Cheng, A.W.; Flytzanis, N.C.; Balsamo, M.; Condeelis, J.S.; Oktay, M.H.; Burge, C.B.; Gertler, F.B. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS Genet. 2011, 7, e1002218. [Google Scholar] [CrossRef] [PubMed]
- Zhao, W.; Hoadley, K.A.; Parker, J.S.; Perou, C.M. Identification of mRNA isoform switching in breast cancer. BMC Genom. 2016, 17, 181. [Google Scholar] [CrossRef] [PubMed]
- Fackenthal, J.D.; Yoshimatsu, T.; Zhang, B.; de Garibay, G.R.; Colombo, M.; De Vecchi, G.; Ayoub, S.C.; Lal, K.; I Olopade, O.; Vega, A.; et al. Naturally occurring BRCA2 alternative mRNA splicing events in clinically relevant samples. J. Med. Genet. 2016, 53, 548–558. [Google Scholar] [CrossRef] [PubMed]
- Sanz, D.J.; Acedo, A.; Infante, M.; Durán, M.; Pérez-Cabornero, L.; Esteban-Cardeñosa, E.; Lastra, E.; Pagani, F.; Miner, C.; Velasco, E.A. A High Proportion of DNA Variants of BRCA1 and BRCA2 Is Associated with Aberrant Splicing in Breast/Ovarian Cancer Patients. Clin. Cancer Res. 2010, 16, 1957–1967. [Google Scholar] [CrossRef] [PubMed]
- Lend, A.K.; Kazantseva, A.; Kivil, A.; Valvere, V.; Palm, K. Diagnostic significance of alternative splice variants of REST and DOPEY1 in the peripheral blood of patients with breast cancer. Tumour Biol. 2015, 36, 2473–2480. [Google Scholar] [CrossRef]
- Dumeaux, V.; Fjukstad, B.; Fjosne, H.E.; Frantzen, J.-O.; Holmen, M.M.; Rodegerdts, E.; Schlichting, E.; Børresen-Dale, A.-L.; Bongo, L.A.; Lund, E.; et al. Interactions between the tumor and the blood systemic response of breast cancer patients. PLoS Comput. Biol. 2017, 13, e1005680. [Google Scholar] [CrossRef]
- Nicolini, A.; Ferrari, P.; Silvestri, R.; Gemignani, F. The breast cancer tumor microenvironment and precision medicine: Immunogenicity and conditions favoring response to immunotherapy. J. Natl. Cancer Cent. 2024, 4, 14–24. [Google Scholar] [CrossRef]
- Yao, J.; Li, S.; Wang, X. Identification of Breast Cancer Immune Subtypes by Analyzing Bulk Tumor and Single Cell Transcriptomes. Front. Cell Dev. Biol. 2021, 9, 781848. [Google Scholar]
- Loizides, S.; Constantinidou, A. Triple negative breast cancer: Immunogenicity, tumor microenvironment, and immunotherapy. Front. Genet. 2022, 13, 1095839. [Google Scholar] [CrossRef] [PubMed]
- Murazawa, C.; Hashimoto, N.; Kuraishi, K.; Motoyama, M.; Hashimoto, S.-I.; Ikeuchi, M.; Norimura, S.; Matsunaga, T.; Teramoto, K.; Haba, R.; et al. Status and prognostic value of immunological biomarkers of breast cancer. Oncol. Let. 2023, 25, 164. [Google Scholar] [CrossRef] [PubMed]
- Belic, J.; Koch, M.; Ulz, P.; Auer, M.; Gerhalter, T.; Mohan, S.; Fischereder, K.; Petru, E.; Bauernhofer, T.; Geigl, J.B.; et al. Rapid Identification of Plasma DNA Samples with Increased ctDNA Levels by a Modified FAST-SeqS Approach. Clin. Chem. 2015, 61, 838–849. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-optimal probabilistic RNA-seq quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef]
- Gorenjak, M.; Zupin, M.; Jezernik, G.; Skok, P.; Potočnik, U. Omics data integration identifies ELOVL7 and MMD gene regions as novel loci for adalimumab response in patients with Crohn’s disease. Sci. Rep. 2021, 11, 5449. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Fabregat, A.; Sidiropoulos, K.; Viteri, G.; Marin-Garcia, P.; Ping, P.; Stein, L.; D’eustachio, P.; Hermjakob, H. Reactome diagram viewer: Data structures and strategies to boost performance. Bioinformatics 2018, 34, 1208–1214. [Google Scholar] [CrossRef]
- The UniProt Consortium. UniProt: The Universal Protein Knowledgebase in 2023. Nucleic Acids Res. 2023, 51, D523–D531. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 2023, 51, D638–D646. [Google Scholar] [CrossRef] [PubMed]
- Bahar, M.E.; Kim, H.J.; Kim, D.R. Targeting the RAS/RAF/MAPK pathway for cancer therapy: From mechanism to clinical studies. Signal Transduct. Target. Ther. 2023, 8, 455. [Google Scholar] [CrossRef] [PubMed]
- Andrade, W.A.; Silva, A.M.; Alves, V.S.; Salgado, A.P.C.; Melo, M.B.; Andrade, H.M.; Dall’Orto, F.V.; A Garcia, S.; Silveira, T.N.; Gazzinelli, R.T. Early endosome localization and activity of RasGEF1b, a toll-like receptor-inducible Ras guanine-nucleotide exchange factor. Genes Immun. 2010, 11, 447–457. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Fernandes, H.B.; de Oliveira, I.M.; Postler, T.S.; Lima, S.Q.; Santos, C.A.C.; Oliveira, M.S.; Leão, F.B.; Ghosh, S.; Souza, M.C.; Andrade, W.; et al. Transcriptomic analysis reveals that RasGEF1b deletion alters basal and LPS-induced expression of genes involved in chemotaxis and cytokine responses in macrophages. Sci. Rep. 2023, 13, 19614. [Google Scholar] [CrossRef]
- Leão, F.B.; Vaughn, L.S.; Bhatt, D.; Liao, W.; Maloney, D.; Carvalho, B.C.; Oliveira, L.; Ghosh, S.; Silva, A.M. Toll-like Receptor (TLR)-induced Rasgef1b expression in macrophages is regulated by NF-κB through its proximal promoter. Int. J. Biochem. Cell Biol. 2020, 127, 105840. [Google Scholar] [CrossRef]
- Sequera, C.; Manzano, S.; Guerrero, C.; Porras, A. How Rap and its GEFs control liver physiology and cancer development. C3G alterations in human hepatocarcinoma. Hepatic Oncol. 2018, 5, HEP05. [Google Scholar] [CrossRef]
- Yaman, E.; Gasper, R.; Koerner, C.; Wittinghofer, A.; Tazebay, U.H. RasGEF1A and RasGEF1B are guanine nucleotide exchange factors that discriminate between Rap GTP-binding proteins and mediate Rap2-specific nucleotide exchange. FEBS J. 2009, 276, 4607–4616. [Google Scholar] [CrossRef]
- GTEx Portal. 7/21/2023. Available online: www.gtexportal.org (accessed on 13 September 2024).
- Hashmi, A.A.; Hashmi, K.A.; Irfan, M.; Khan, S.M.; Edhi, M.M.; Ali, J.P.; Hashmi, S.K.; Asif, H.; Faridi, N.; Khan, A. Ki67 index in intrinsic breast cancer subtypes and its association with prognostic parameters. BMC Res. Notes 2019, 12, 605. [Google Scholar] [CrossRef]
- Cheang, M.C.U.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 Index, HER2 Status, and Prognosis of Patients with Luminal B Breast Cancer. JNCI J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef]
- de Azambuja, E.; Cardoso, F.; de Castro, G.; Colozza, M.; Mano, M.S.; Durbecq, V.; Sotiriou, C.; Larsimont, D.; Piccart-Gebhart, M.J.; Paesmans, M. Ki-67 as prognostic marker in early breast cancer: A meta-analysis of published studies involving 12 155 patients. Br. J. Cancer 2007, 96, 1504–1513. [Google Scholar] [CrossRef]
- Goldhirsch, A.; Winer, E.P.; Coates, A.S.; Gelber, R.D.; Piccart-Gebhart, M.; Thürlimann, B.; Senn, H.-J. Personalizing the treatment of women with early breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann. Oncol. 2013, 24, 2206–2223. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, R.; Osako, T.; Okumura, Y.; Hayashi, M.; Toyozumi, Y.; Arima, N. Ki-67 as a prognostic marker according to breast cancer subtype and a predictor of recurrence time in primary breast cancer. Exp. Ther. Med. 2010, 1, 747–754. [Google Scholar] [CrossRef] [PubMed]
- Spyratos, F.; Ferrero-Poüs, M.; Trassard, M.; Hacène, K.; Phillips, E.; Tubiana-Hulin, M.; Le Doussal, V. Correlation between MIB-1 and other proliferation markers: Clinical implications of the MIB-1 cutoff value. Cancer 2002, 94, 2151–2159. [Google Scholar] [CrossRef]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Belic, J.; Koch, M.; Ulz, P.; Auer, M.; Gerhalter, T.; Mohan, S.; Fischereder, K.; Petru, E.; Bauernhofer, T.; Geigl, J.B.; et al. mFast-SeqS as a Monitoring and Pre-screening Tool for Tumor-Specific Aneuploidy in Plasma DNA. Adv. Exp. Med. Biol. 2016, 924, 147–155. [Google Scholar]
- Mendelaar, P.A.J.; Robbrecht, D.G.J.; Rijnders, M.; de Wit, R.; de Weerd, V.; Deger, T.; Westgeest, H.M.; Aarts, M.J.B.; Voortman, J.; Martens, J.W.M.; et al. Genome-wide aneuploidy detected by mFast-SeqS in circulating cell-free DNA is associated with poor response to pembrolizumab in patients with advanced urothelial cancer. Mol. Oncol. 2022, 16, 2086–2097. [Google Scholar] [CrossRef] [PubMed]
- Čelešnik, H.; Potočnik, U. Blood-Based mRNA Tests as Emerging Diagnostic Tools for Personalised Medicine in Breast Cancer. Cancers 2023, 15, 1087. [Google Scholar] [CrossRef]
- Chen, S.; Liu, M.; Liang, B.; Ge, S.; Peng, J.; Huang, H.; Xu, Y.; Tang, X.; Deng, L. Identification of human peripheral blood monocyte gene markers for early screening of solid tumors. PLoS ONE 2020, 15, e0230905. [Google Scholar] [CrossRef]
- Holden, M.; Holden, L.; Olsen, K.S.; Lund, E. Local in Time Statistics for detecting weak gene expression signals in blood—illustrated for prediction of metastases in breast cancer in the NOWAC Post-genome Cohort. Adv. Genom. Genet. 2017, 7, 11–28. [Google Scholar] [CrossRef]
- Holsbø, E.; Olsen, K.S. Metastatic Breast Cancer and Pre-Diagnostic Blood Gene Expression Profiles-The Norwegian Women and Cancer (NOWAC) Post-Genome Cohort. Front. Oncol. 2020, 10, 575461. [Google Scholar] [CrossRef]
- Nøst, T.H.; Holden, M.; Dønnem, T.; Bøvelstad, H.; Rylander, C.; Lund, E.; Sandanger, T.M. Transcriptomic signals in blood prior to lung cancer focusing on time to diagnosis and metastasis. Sci. Rep. 2021, 11, 7406. [Google Scholar] [CrossRef] [PubMed]
- Weedon-Fekjær, H.; Lindqvist, B.H.; Vatten, L.J.; O Aalen, O.; Tretli, S. Breast cancer tumor growth estimated through mammography screening data. Breast Cancer Res. 2008, 10, R41. [Google Scholar] [CrossRef] [PubMed]
- Russo, C.; Wyld, L.; Aubreu, M.D.C.; Bury, C.S.; Heaton, C.; Cole, L.M.; Francese, S. Non-invasive screening of breast cancer from fingertip smears—A proof of concept study. Sci. Rep. 2023, 13, 1868. [Google Scholar] [CrossRef] [PubMed]
- Thigpen, D.; Kappler, A.; Brem, R. The Role of Ultrasound in Screening Dense Breasts—A Review of the Literature and Practical Solutions for Implementation. Diagnostics 2018, 8, 20. [Google Scholar] [CrossRef] [PubMed]
- Ming, W.; Xie, H.; Hu, Z.; Chen, Y.; Zhu, Y.; Bai, Y.; Liu, H.; Sun, X.; Liu, Y.; Gu, W. Two Distinct Subtypes Revealed in Blood Transcriptome of Breast Cancer Patients With an Unsupervised Analysis. Front. Oncol. 2019, 9, 985. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, E.; Sugimoto, M.; Kawaguchi, K.; Pu, F.; Uozumi, R.; Yamaguchi, A.; Nishie, M.; Tsuda, M.; Kotake, T.; Morita, S.; et al. Gene expression profile of peripheral blood mononuclear cells may contribute to the identification and immunological classification of breast cancer patients. Breast Cancer 2019, 26, 282–289. [Google Scholar] [CrossRef]
- Cho, K.J.; Liang, J.R.; Crespo, P.; Aran, V. Editorial: Ras and Other GTPases in Cancer: From Basic to Applied Research. Front. Mol. Biosci. 2021, 8, 804818. [Google Scholar] [CrossRef]
- Yaman, E. Functional Identification of RASGEF1 Family of Exchange Factors as Activators of RAP2, and as Interacting Partners of CCDC124. Ph.D. Thesis, Department of Molecular Biology and Genetics and The Institute of Engineering and Science of Bilkent University, Ankara, Türkiye, 2009. [Google Scholar]
- Bokoch, G.M. Biology of the Rap proteins, members of the ras superfamily of GTP-binding proteins. Biochem. J. 1993, 289, 17–24. [Google Scholar] [CrossRef]
- Paganini, S.; Guidetti, G.F.; Catricalà, S.; Trionfini, P.; Panelli, S.; Balduini, C.; Torti, M. Identification and biochemical characterization of Rap2C, a new member of the Rap family of small GTP-binding proteins. Biochimie 2006, 88, 285–295. [Google Scholar] [CrossRef]
- The Human Protein Atlas. 7/21/2023. Available online: www.proteinatlas.org (accessed on 13 September 2024).
- Guo, X.-X.; An, S.; Yang, Y.; Liu, Y.; Hao, Q.; Xu, T.-R. Rap-Interacting Proteins are Key Players in the Rap Symphony Orchestra. Cell. Physiol. Biochem. 2016, 39, 137–156. [Google Scholar] [CrossRef]
- Qu, D.; Huang, H.; DI, J.; Gao, K.; Lu, Z.; Zheng, J. Structure, functional regulation and signaling properties of Rap2B. Oncol. Lett. 2016, 11, 2339–2346. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Duan, H.B.; Yang, Y.S. Knockdown of Rap2B Inhibits the Proliferation and Invasion in Hepatocellular Carcinoma Cells. Oncol. Res. 2017, 25, 19–27. [Google Scholar] [CrossRef]
- Zhang, X.; He, Y.; Lee, K.-H.; Dubois, W.; Li, Z.; Wu, X.; Kovalchuk, A.; Zhang, W.; Huang, J. Rap2b, a novel p53 target, regulates p53-mediated pro-survival function. Cell Cycle 2013, 12, 1279–1291. [Google Scholar] [CrossRef]
- Sciacchitano, S.; Sacconi, A.; De Vitis, C.; Blandino, G.; Piaggio, G.; Salvati, V.; Napoli, C.; Marchetti, P.; Taurelli, B.S.; Coluzzi, F.; et al. H-Ras gene takes part to the host immune response to COVID-19. Cell Death Discov. 2021, 7, 158. [Google Scholar] [CrossRef] [PubMed]
- Ura, K.; Obama, K.; Satoh, S.; Sakai, Y.; Nakamura, Y.; Furukawa, Y. Enhanced RASGEF1A Expression Is Involved in the Growth and Migration of Intrahepatic Cholangiocarcinoma. Clin. Cancer Res. 2006, 12, 6611–6616. [Google Scholar] [CrossRef][Green Version]
- Bates, J.P.; Derakhshandeh, R.; Jones, L.; Webb, T.J. Mechanisms of immune evasion in breast cancer. BMC Cancer 2018, 18, 556. [Google Scholar] [CrossRef] [PubMed]
- Goldhirsch, A.; Wood, W.C.; Coates, A.S.; Gelber, R.D.; Thürlimann, B.; Senn, H.-J.; Panel members. Strategies for subtypes—Dealing with the diversity of breast cancer: Highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2011. Ann. Oncol. 2011, 22, 1736–1747. [Google Scholar] [CrossRef] [PubMed]
- Cailleux, F.; Agostinetto, E.; Lambertini, M.; Rothé, F.; Wu, H.-T.; Balcioglu, M.; Kalashnikova, E.; Vincent, D.; Viglietti, G.; Gombos, A.; et al. Circulating Tumor DNA After Neoadjuvant Chemotherapy in Breast Cancer Is Associated with Disease Relapse. JCO Precis. Oncol. 2022, 6, e2200148. [Google Scholar] [CrossRef]
- Magbanua, M.J.M.; Swigart, L.B.; Ahmed, Z.; Sayaman, R.W.; Renner, D.; Kalashnikova, E.; Hirst, G.L.; Yau, C.; Wolf, D.M.; Li, W.; et al. Clinical significance and biology of circulating tumor DNA in high-risk early-stage HER2-negative breast cancer receiving neoadjuvant chemotherapy. Cancer Cell 2023, 41, 1091–1102.e4. [Google Scholar] [CrossRef]
- Gong, J.; Wang, J.; Tian, Y.; Zhang, J.; Liang, W.; Li, Z.; Yu, J.; Tang, B.; He, S. Expression of tubulin folding cofactor B in mouse hepatic ischemia-reperfusion injury. Biomed. Rep. 2017, 6, 525–531. [Google Scholar] [CrossRef]
- Carranza, G.; Castaño, R.; Fanarraga, M.L.; Villegas, J.C.; Gonçalves, J.; Soares, H.; Avila, J.; Marenchino, M.; Campos-Olivas, R.; Montoya, G.; et al. Autoinhibition of TBCB regulates EB1-mediated microtubule dynamics. Cell. Mol. Life Sci. 2013, 70, 357–371. [Google Scholar] [CrossRef] [PubMed]
- Kortazar, D.; Fanarraga, M.; Carranza, G.; Bellido, J.; Villegas, J.; Avila, J.; Zabala, J. Role of cofactors B (TBCB) and E (TBCE) in tubulin heterodimer dissociation. Exp. Cell Res. 2007, 313, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Schoumacher, M.; Goldman, R.D.; Louvard, D.; Vignjevic, D.M. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 2010, 189, 541–556. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Wang, W.; Li, Q.; Guo, T.; Yang, S.; Shi, J.; Yuan, W.; Chu, Y. High Expression of Microtubule-associated Protein TBCB Predicts Adverse Outcome and Immunosuppression in Acute Myeloid Leukemia. J. Cancer 2023, 14, 1707–1724. [Google Scholar] [CrossRef] [PubMed]
- Tan, H.; Liao, H.; Zhao, L.; Lu, Y.; Jiang, S.; Tao, D.; Liu, Y.; Ma, Y. HILI destabilizes microtubules by suppressing phosphorylation and Gigaxonin-mediated degradation of TBCB. Sci. Rep. 2017, 7, 46376. [Google Scholar] [CrossRef]
- Hamel, E.; Sackett, D.L.; Vourloumis, D.; Nicolaou, K.C. The Coral-Derived Natural Products Eleutherobin and Sarcodictyins A and B: Effects on the Assembly of Purified Tubulin with and without Microtubule-Associated Proteins and Binding at the Polymer Taxoid Site. Biochemistry 1999, 38, 5490–5498. [Google Scholar] [CrossRef]
- Gilson, P.; Drouot, G.; Witz, A.; Merlin, J.-L.; Becuwe, P.; Harlé, A. Emerging Roles of DDB2 in Cancer. Int. J. Mol. Sci. 2019, 20, 5168. [Google Scholar] [CrossRef]
- Chen, H.H.; Fan, P.; Chang, S.-W.; Tsao, Y.-P.; Huang, H.-P.; Chen, S.-L. NRIP/DCAF6 stabilizes the androgen receptor protein by displacing DDB2 from the CUL4A-DDB1 E3 ligase complex in prostate cancer. Oncotarget 2017, 8, 21501–21515. [Google Scholar] [CrossRef]
- Stoyanova, T.; Roy, N.; Bhattacharjee, S.; Kopanja, D.; Valli, T.; Bagchi, S.; Raychaudhuri, P. p21 Cooperates with DDB2 Protein in Suppression of Ultraviolet Ray-induced Skin Malignancies. J. Biol. Chem. 2012, 287, 3019–3028. [Google Scholar] [CrossRef]
- Bommi, P.V.; Ravindran, S.; Raychaudhuri, P.; Bagchi, S. DDB2 regulates Epithelial-to-Mesenchymal Transition (EMT) in Oral/Head and Neck Squamous Cell Carcinoma. Oncotarget 2018, 9, 34708–34718. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Sun, L.; Feng, X.; Wang, Z.; Yuan, Y.; Xing, C. The Differential Expression of Core Genes in Nucleotide Excision Repair Pathway Indicates Colorectal Carcinogenesis and Prognosis. BioMed Res. Int. 2018, 2018, 9651320. [Google Scholar] [CrossRef] [PubMed]
- Kattan, Z.; Marchal, S.; Brunner, E.; Ramacci, C.; Leroux, A.; Merlin, J.L.; Domenjoud, L.; Dauça, M.; Becuwe, P. Damaged DNA Binding Protein 2 Plays a Role in Breast Cancer Cell Growth. PLoS ONE 2008, 3, e2002. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Han, C.; Zhao, R.; Liu, X.; Srivastava, A.; Gong, L.; Mao, H.; Qu, M.; Zhao, W.; Yu, J.; Wang, Q.-E. DDB2 suppresses tumorigenicity by limiting the cancer stem cell population in ovarian cancer. Mol. Cancer Res. 2014, 12, 784–794. [Google Scholar] [CrossRef] [PubMed]
- Bogaert, A.; Fijalkowska, D.; Staes, A.; Van de Steene, T.; Vuylsteke, M.; Stadler, C.; Eyckerman, S.; Spirohn, K.; Hao, T.; A Calderwood, M.; et al. N-terminal proteoforms may engage in different protein complexes. Life Sci. Alliance 2023, 6, e202301972. [Google Scholar] [CrossRef]
- Bogaert, A.; Fernandez, E.; Gevaert, K. N-Terminal Proteoforms in Human Disease. Trends Biochem. Sci. 2020, 45, 308–320. [Google Scholar] [CrossRef]
- Nakahara, K.; Shoun, H. N-Terminal Processing and Amino Acid Sequence of Two Isoforms of Nitric Oxide Reductase Cytochrome P450nor from Fusarium oxysporum1. J. Biochem. 1996, 120, 1082–1087. [Google Scholar] [CrossRef]
- Müntener, K.; Willimann, A.; Zwicky, R.; Svoboda, B.; Mach, L.; Baici, A. Folding Competence of N-terminally Truncated Forms of Human Procathepsin B. J. Biol. Chem. 2005, 280, 11973–11980. [Google Scholar] [CrossRef]
- Ree, R.; Varland, S.; Arnesen, T. Spotlight on protein N-terminal acetylation. Exp. Mol. Med. 2018, 50, 1–13. [Google Scholar] [CrossRef]
| Study Cohort: Female Breast Cancer Patients (n = 156) | |||||
|---|---|---|---|---|---|
| BC subtype | Luminal A | Luminal B | HER2(+) | TNBC | |
| 45 (28.85%) | 90 (57.69%) | 5 (3.21%) | 16 (10.26%) | ||
| Histological type | ILC | IDC | IDC + DCIS | * Other | |
| 18 (11.54%) | 57 (36.54%) | 73 (46.79%) | 8 (5.13%) | ||
| Localization/containment | Localized/contained | Locally advanced | Metastatic | Not known | |
| 124 (79.49%) | 17 (10.90%) | 8 (5.13%) | 7 (4.49%) | ||
| Grade | 1 | 2 | 3 | Not known | |
| 36 (23.08%) | 73 (46.79%) | 40 (25.64%) | 7 (4.49%) | ||
| Tumor size | T1, ≤2 cm | T2, >2 to ≤5 cm | T3, >5 mm | Not known | |
| 95 (60.90%) | 43 (27.56%) | 3 (1.92%) | 15 (9.61%) | ||
| Lymph nodes | Negative | Micrometastasis ≤ 2 mm | Macrometastasis > 2 mm | Not known | |
| 98 (62.82%) | 7 (4.49%) | 23 (14.74%) | 28 (17.95%) | ||
| Ki-67 index | <14% | ≥14 to ≤25% | >25 to ≤50% | >50 to ≤100% | Not known |
| 44 (28.21%) | 56 (35.90%) | 41 (26.28%) | 13 (8.33%) | 2 (1.28%) | |
| Genome-wide z-score | ≤3% | >3% | Not determin. | ||
| 30 (19.23%) | 11 (7.05%) | 115 (73.72%) | |||
| Gene | GO Term ID | GO Term Name | GO Category | GO Term Description |
|---|---|---|---|---|
| RASGEF1A | GO:0005085 | guanyl-nucleotide exchange factor activity | Molecular Function | Stimulates the exchange of GDP to GTP on a signaling GTPase |
| RASGEF1A | GO:0007265 | Ras protein signal transduction | Biological Process | Involved in the transmission of signals through Ras proteins |
| DDB2 | GO:0003684 | Damaged DNA binding | Molecular Function | The ability to bind to DNA that has been damaged |
| DDB2 | GO:0006281 | DNA repair | Biological Process | Cellular processes of restoring DNA after damage |
| TBCB | GO:0043014 | Alpha-tubulin binding | Molecular Function | Binding to the microtubule constituent protein alpha-tubulin |
| TBCB | GO:0007021 | Tubulin complex assembly | Biological Process | Assembly of alpha- and beta-tubulin to form a tubulin heterodimer |
| TBCB | GO:0007023 | Post-chaperonin tubulin folding pathway | Biological Process | Completion of folding of alpha- and beta-tubulin after chaperonin-mediated partial folding |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Čelešnik, H.; Gorenjak, M.; Krušič, M.; Crnobrnja, B.; Sobočan, M.; Takač, I.; Arko, D.; Potočnik, U. Isoform-Level Transcriptome Analysis of Peripheral Blood Mononuclear Cells from Breast Cancer Patients Identifies a Disease-Associated RASGEF1A Isoform. Cancers 2024, 16, 3171. https://doi.org/10.3390/cancers16183171
Čelešnik H, Gorenjak M, Krušič M, Crnobrnja B, Sobočan M, Takač I, Arko D, Potočnik U. Isoform-Level Transcriptome Analysis of Peripheral Blood Mononuclear Cells from Breast Cancer Patients Identifies a Disease-Associated RASGEF1A Isoform. Cancers. 2024; 16(18):3171. https://doi.org/10.3390/cancers16183171
Chicago/Turabian StyleČelešnik, Helena, Mario Gorenjak, Martina Krušič, Bojana Crnobrnja, Monika Sobočan, Iztok Takač, Darja Arko, and Uroš Potočnik. 2024. "Isoform-Level Transcriptome Analysis of Peripheral Blood Mononuclear Cells from Breast Cancer Patients Identifies a Disease-Associated RASGEF1A Isoform" Cancers 16, no. 18: 3171. https://doi.org/10.3390/cancers16183171
APA StyleČelešnik, H., Gorenjak, M., Krušič, M., Crnobrnja, B., Sobočan, M., Takač, I., Arko, D., & Potočnik, U. (2024). Isoform-Level Transcriptome Analysis of Peripheral Blood Mononuclear Cells from Breast Cancer Patients Identifies a Disease-Associated RASGEF1A Isoform. Cancers, 16(18), 3171. https://doi.org/10.3390/cancers16183171








