RNA-Independent Regulatory Functions of lncRNA in Complex Disease
Abstract
Simple Summary
Abstract
1. Introduction
2. The Multifaceted Regulatory Mechanism of lncRNAs
2.1. The Long Non-Coding DNA Locus as the Key Player in Complex Diseases
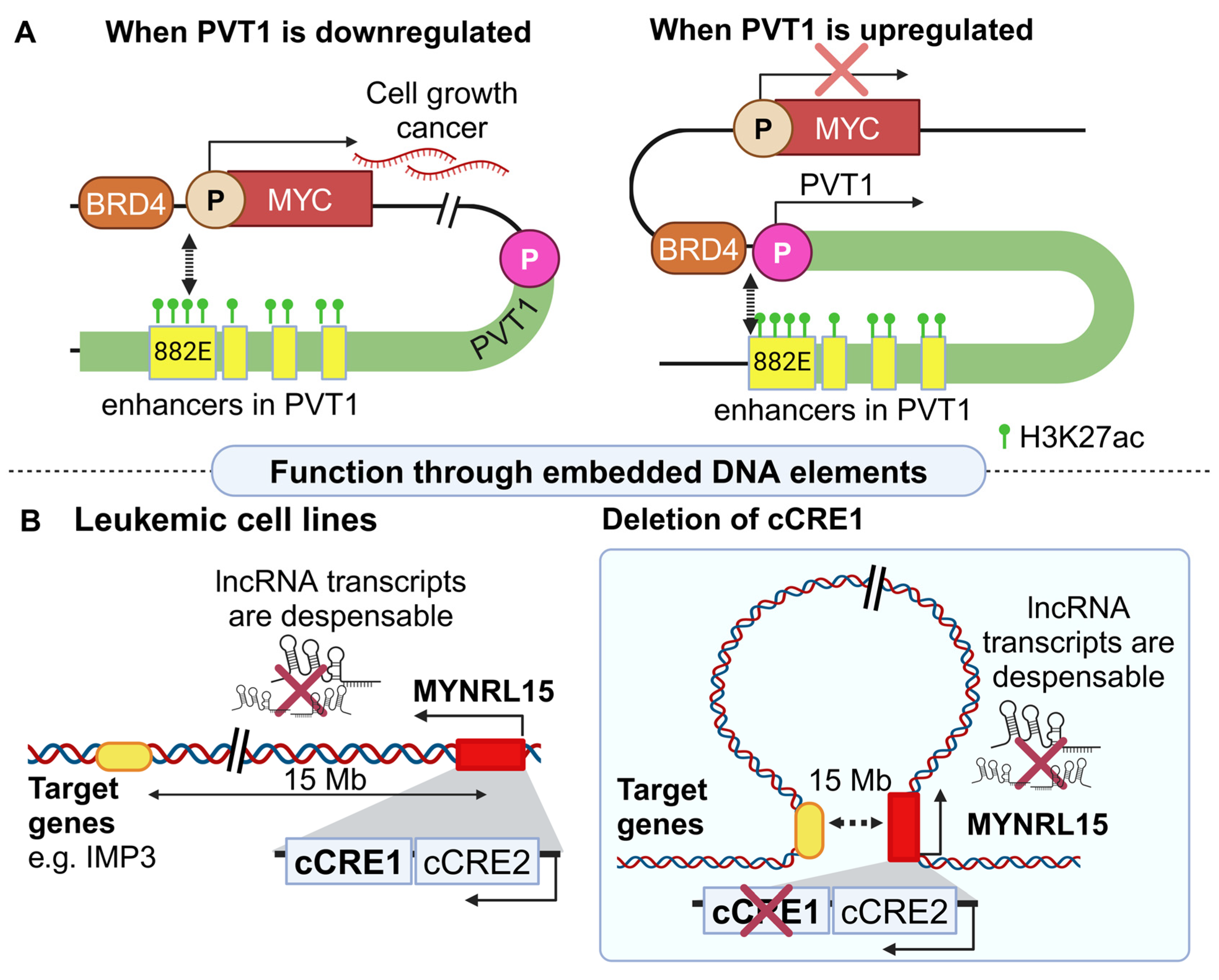
2.1.1. Regulatory Effects of lncRNA Loci in Cancer
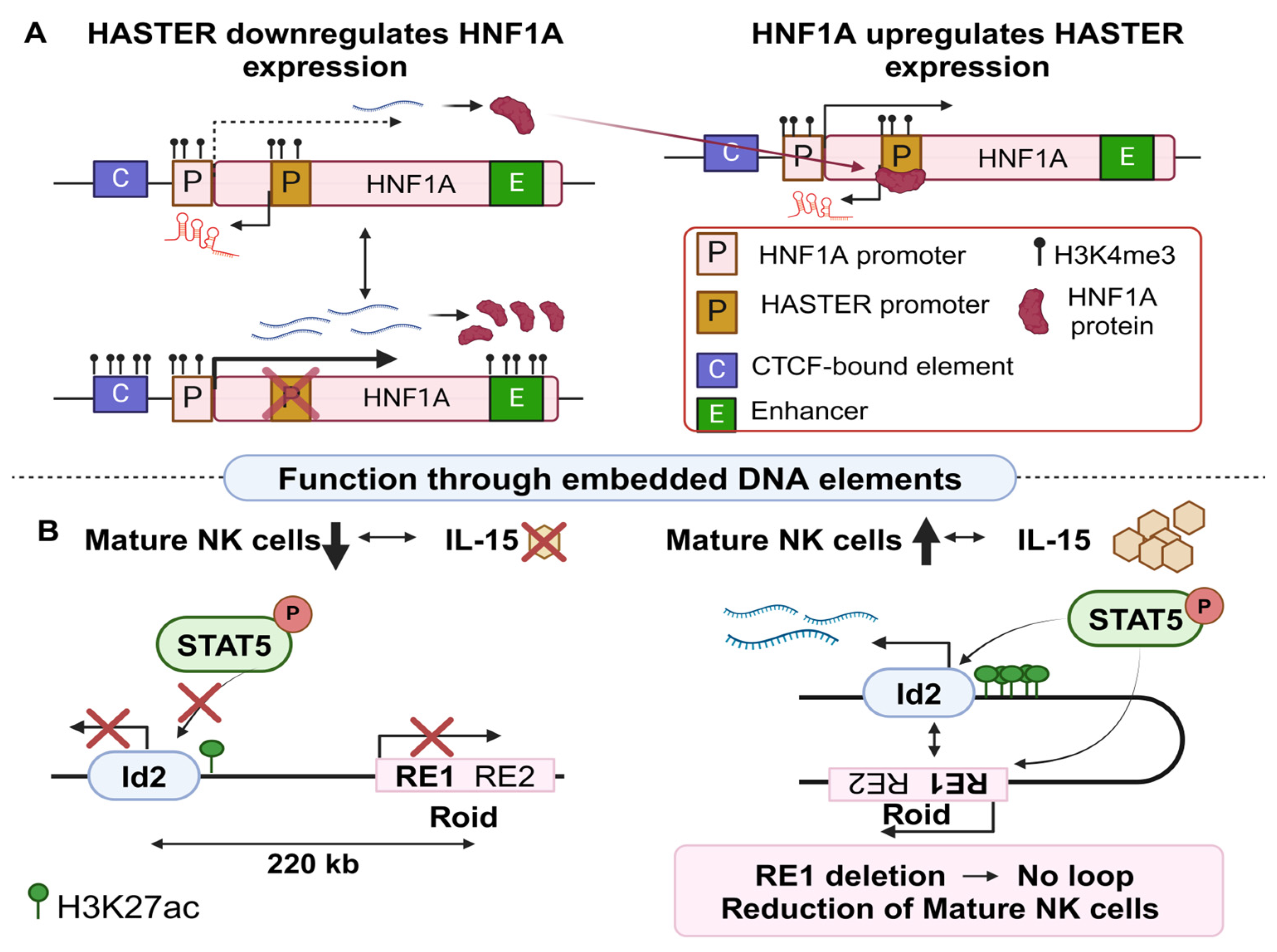
2.1.2. Regulatory Effects of lncRNA Loci in Other Complex Diseases
2.2. Nascent lncRNA Transcription as a Regulatory Mechanism
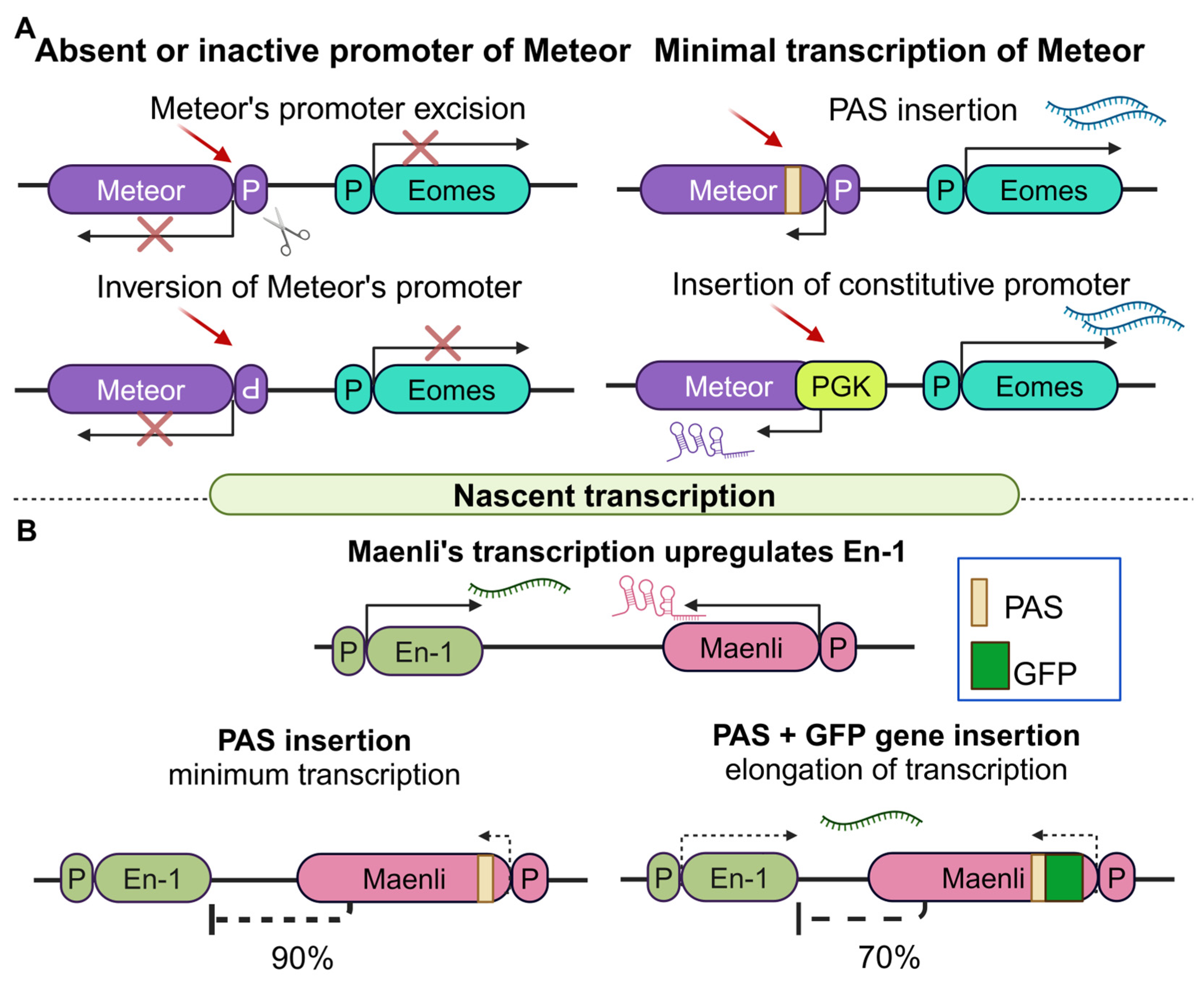
2.2.1. Regulatory Effects of Nascent lncRNA Transcription in Cancer
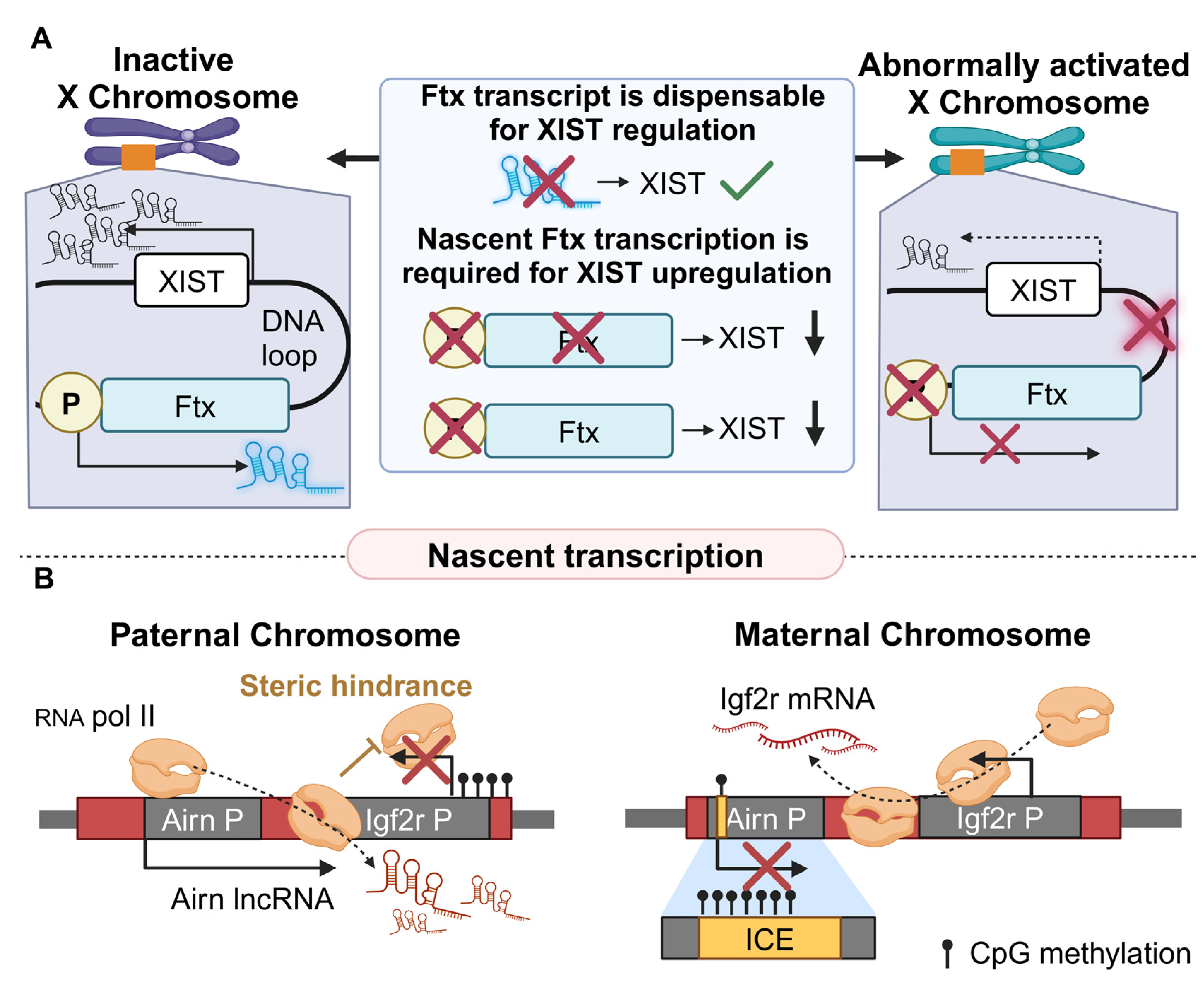
2.2.2. Nascent lncRNA Transcription in Other Diseases
2.3. Examples of lncRNA That Combine Diverse Mechanisms to Collectively Regulate Target Genes in Cancer
2.4. The Regulation of Heart Formation through the Contrasting Effects of Two lncRNA Loci on a Common Target Gene
3. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
List of Abbreviations
| 3C | chromatin conformation capture |
| 4C-Seq | chromosome conformation capture combined with high-throughput sequencing |
| Airn | antisense Igf2r RNA non-coding |
| as-lncRNA | antisense long non-coding RNA |
| ASO | antisense oligonucleotides |
| ATAC-seq | assay for transposase-accessible chromatin using sequencing |
| Bcll11b | B cell lymphoma/leukemia 11b |
| Blustr | bivalent locus upregulated by the splicing and transcription of an RNA |
| bp | base pair |
| BRD4 | bromodomain protein 4 |
| BVG-polyA | bovine growth hormone polyadenylation signal |
| CBSs | CTCF binding sites |
| cCREs | cis-regulatory elements |
| CEBPB CCAAT | enhancer binding protein beta |
| ChIP-seq | chromatin immunoprecipitation sequencing |
| ChIRP | chromatin isolation RNA purification |
| ciRNAs | circular RNAs |
| CMV | cytomegalovirus |
| CRISPRi | clustered regularly interspaced short palindromic repeats inhibition |
| CTCF | CCCTC-binding factor |
| DamID seq | DNA adenine methyltransferase identification sequencing |
| eCBS | exon CTCT binding site |
| En1 | engrailed-1 |
| ENCODE | Encyclopedia of DNA Elements |
| ESC | embryonic stem cells |
| FISH | fluorescence in situ hybridization |
| Ftx | Five prime to Xist |
| HAND2 | heart and neural crest derivatives expressed 2 |
| Haunt | HOXA upstream non-coding transcript |
| HGP | Human Genome Project |
| Hi-C | high-throughput chromosome conformation capture technique |
| Hi-ChIP | in situ Hi-C followed by chromatin immunoprecipitation |
| Hnd | Handsdown lncRNA |
| HNF1A | hepatic nuclear factor 1A |
| HS5-1 | hypersensitivity site 5-1 |
| ICE | imprinted control element |
| Igf2r | insulin-like growth factor 2 receptor |
| IL-15 | interleukin 15 |
| ILCs | innate lymphoid cells |
| Kb | kilo base |
| KD | knock-down |
| KI | knock-in |
| KO | knock-out |
| LNA | locked nucleic acids |
| lncRNA | long non-coding RNA |
| Maenli | master activator of engrailed-1 in the limb |
| meDIP | methylated DNA immunoprecipitation |
| mESC | mouse embryonic stem cell |
| Meteor | mesendoderm transcription enhancer organizing region |
| miRNAs | microRNAs |
| mRNAs | messenger RNAs |
| MYC | myelocytomatosis oncogene |
| MyoD | myoblast determination protein 1 |
| ncRNA | non-coding RNA |
| NGS | next-generation sequencing |
| NK | natural killers |
| ORF | open reading frame |
| P53RE | p53 response element |
| PAS | poly adenylation signal |
| PCA | principal component analysis |
| pCBS | proximal CBS |
| Pcdhα | protocadherin α |
| PDXs | patient-derived xenografts |
| piRNAs | PIWI-interacting RNAs |
| PRC | polycomb repressive complex |
| PVT1 | plasmacytoma variant translocation 1 |
| RA | retinoic acid |
| RE | regulatory element |
| RNA pol II | RNA polymerase II |
| rRNAs | ribosomal RNAs |
| Rroid | RNA demarcated regulatory region of Id2 |
| RUNX1 | runt-related transcription factor 1 |
| Rz | self-cleaving ribozyme |
| Sfmbt2 | Scm like with four Mbt domains 2 |
| shRNA | short hairpin RNA |
| snoRNAs | small nucleolus RNAs |
| snRNAs | small nuclear RNAs |
| STAT5 | signal transducer and activator of transcription 5 |
| TF | transcription factor |
| TG | target |
| tRNAs | transfer RNAs |
| TSS | transcription start site |
| UMI-4C | unique molecular identifier 4C |
| Uph | upperhand |
| WT | wild type |
| XCI | X chromosome inactivation |
| Xist | X-linked X-inactive-specific transcript |
References
- Birney, E. The International Human Genome Project. Hum. Mol. Genet. 2021, 30, R161–R163. [Google Scholar] [CrossRef] [PubMed]
- Consortium, E.P.; Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef] [PubMed]
- Jarroux, J.; Morillon, A.; Pinskaya, M. History, Discovery, and Classification of lncRNAs. Adv. Exp. Med. Biol. 2017, 1008, 1–46. [Google Scholar] [CrossRef] [PubMed]
- Wilkinson, M.E.; Charenton, C.; Nagai, K. RNA Splicing by the Spliceosome. Annu. Rev. Biochem. 2020, 89, 359–388. [Google Scholar] [CrossRef] [PubMed]
- Wajahat, M.; Bracken, C.P.; Orang, A. Emerging Functions for snoRNAs and snoRNA-Derived Fragments. Int. J. Mol. Sci. 2021, 22, 10193. [Google Scholar] [CrossRef] [PubMed]
- Tafrihi, M.; Hasheminasab, E. MiRNAs: Biology, Biogenesis, their Web-based Tools, and Databases. Microrna 2019, 8, 4–27. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.Y.; Cai, Z.R.; Liu, J.; Wang, D.S.; Ju, H.Q.; Xu, R.H. Circular RNA: Metabolism, functions and interactions with proteins. Mol. Cancer 2020, 19, 172. [Google Scholar] [CrossRef]
- Cai, A.; Hu, Y.; Zhou, Z.; Qi, Q.; Wu, Y.; Dong, P.; Chen, L.; Wang, F. PIWI-Interacting RNAs (piRNAs): Promising Applications as Emerging Biomarkers for Digestive System Cancer. Front. Mol. Biosci. 2022, 9, 848105. [Google Scholar] [CrossRef] [PubMed]
- Bridges, M.C.; Daulagala, A.C.; Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 2021, 220, e202009045. [Google Scholar] [CrossRef]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Kalvari, I.; Nawrocki, E.P.; Ontiveros-Palacios, N.; Argasinska, J.; Lamkiewicz, K.; Marz, M.; Griffiths-Jones, S.; Toffano-Nioche, C.; Gautheret, D.; Weinberg, Z.; et al. Rfam 14: Expanded coverage of metagenomic, viral and microRNA families. Nucleic Acids Res. 2021, 49, D192–D200. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Chen, M. Circular RNA Databases. Methods Mol. Biol. 2021, 2362, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. miRBase: From microRNA sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef] [PubMed]
- Cui, P.; Lin, Q.; Ding, F.; Xin, C.; Gong, W.; Zhang, L.; Geng, J.; Zhang, B.; Yu, X.; Yang, J.; et al. A comparison between ribo-minus RNA-sequencing and polyA-selected RNA-sequencing. Genomics 2010, 96, 259–265. [Google Scholar] [CrossRef] [PubMed]
- Mackowiak, S.D.; Zauber, H.; Bielow, C.; Thiel, D.; Kutz, K.; Calviello, L.; Mastrobuoni, G.; Rajewsky, N.; Kempa, S.; Selbach, M.; et al. Extensive identification and analysis of conserved small ORFs in animals. Genome Biol. 2015, 16, 179. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Hou, S.; Ye, Z.; Wang, W.; Hu, X.; Hang, Q. Long Non-Coding RNAs in Pancreatic Cancer: Biologic Functions, Mechanisms, and Clinical Significance. Cancers 2022, 14, 2115. [Google Scholar] [CrossRef] [PubMed]
- Begolli, R.; Sideris, N.; Giakountis, A. LncRNAs as Chromatin Regulators in Cancer: From Molecular Function to Clinical Potential. Cancers 2019, 11, 1524. [Google Scholar] [CrossRef]
- Alessio, E.; Bonadio, R.S.; Buson, L.; Chemello, F.; Cagnin, S. A Single Cell but Many Different Transcripts: A Journey into the World of Long Non-Coding RNAs. Int. J. Mol. Sci. 2020, 21, 302. [Google Scholar] [CrossRef] [PubMed]
- Brannan, C.I.; Dees, E.C.; Ingram, R.S.; Tilghman, S.M. The product of the H19 gene may function as an RNA. Mol. Cell. Biol. 1990, 10, 28–36. [Google Scholar] [CrossRef]
- Brockdorff, N.; Ashworth, A.; Kay, G.F.; McCabe, V.M.; Norris, D.P.; Cooper, P.J.; Swift, S.; Rastan, S. The product of the mouse Xist gene is a 15 kb inactive X-specific transcript containing no conserved ORF and located in the nucleus. Cell 1992, 71, 515–526. [Google Scholar] [CrossRef]
- Brockdorff, N.; Bowness, J.S.; Wei, G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020, 34, 733–744. [Google Scholar] [CrossRef]
- Aich, M.; Chakraborty, D. Role of lncRNAs in stem cell maintenance and differentiation. Curr. Top. Dev. Biol. 2020, 138, 73–112. [Google Scholar] [CrossRef] [PubMed]
- Constanty, F.; Shkumatava, A. lncRNAs in development and differentiation: From sequence motifs to functional characterization. Development 2021, 148, dev182741. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Safari, M.; Taheri, M.; Samadian, M. Expression of Linear and Circular lncRNAs in Alzheimer’s Disease. J. Mol. Neurosci. 2022, 72, 187–200. [Google Scholar] [CrossRef]
- Zhang, Y.; Wang, X.; Zhang, C.; Yi, H. The dysregulation of lncRNAs by epigenetic factors in human pathologies. Drug Discov. Today 2023, 28, 103664. [Google Scholar] [CrossRef]
- Ahmad, M.; Weiswald, L.B.; Poulain, L.; Denoyelle, C.; Meryet-Figuiere, M. Involvement of lncRNAs in cancer cells migration, invasion and metastasis: Cytoskeleton and ECM crosstalk. J. Exp. Clin. Cancer Res. 2023, 42, 173. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.; Zhou, Q.; Wang, C.Q.; Zhu, L.; Bi, C.; Zhang, S.; Wang, X.; Jin, H. LncRNAs regulate metabolism in cancer. Int. J. Biol. Sci. 2020, 16, 1194–1206. [Google Scholar] [CrossRef]
- Al-Noshokaty, T.M.; Mansour, A.; Abdelhamid, R.; Abdellatif, N.; Alaaeldien, A.; Reda, T.; Abdelmaksoud, N.M.; Doghish, A.S.; Abulsoud, A.I.; Elshaer, S.S. Role of long non-coding RNAs in pancreatic cancer pathogenesis and treatment resistance- A review. Pathol. Res. Pract. 2023, 245, 154438. [Google Scholar] [CrossRef]
- Elazazy, O.; Midan, H.M.; Shahin, R.K.; Elesawy, A.E.; Elballal, M.S.; Sallam, A.M.; Elbadry, A.M.M.; Elrebehy, M.A.; Bhnsawy, A.; Doghish, A.S. Long non-coding RNAs and rheumatoid arthritis: Pathogenesis and clinical implications. Pathol. Res. Pract. 2023, 246, 154512. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, P.; Lin, R.; Rong, L.; Xue, Y.; Fang, Y. LncRNA NALT interaction with NOTCH1 promoted cell proliferation in pediatric T cell acute lymphoblastic leukemia. Sci. Rep. 2015, 5, 13749. [Google Scholar] [CrossRef]
- Shi, Q.; Xue, C.; Zeng, Y.; Yuan, X.; Chu, Q.; Jiang, S.; Wang, J.; Zhang, Y.; Zhu, D.; Li, L. Notch signaling pathway in cancer: From mechanistic insights to targeted therapies. Signal Transduct. Target. Ther. 2024, 9, 128. [Google Scholar] [CrossRef] [PubMed]
- Lourenco, C.; Resetca, D.; Redel, C.; Lin, P.; MacDonald, A.S.; Ciaccio, R.; Kenney, T.M.G.; Wei, Y.; Andrews, D.W.; Sunnerhagen, M.; et al. MYC protein interactors in gene transcription and cancer. Nat. Rev. Cancer 2021, 21, 579–591. [Google Scholar] [CrossRef] [PubMed]
- Dehingia, B.; Milewska, M.; Janowski, M.; Pekowska, A. CTCF shapes chromatin structure and gene expression in health and disease. EMBO Rep. 2022, 23, e55146. [Google Scholar] [CrossRef] [PubMed]
- Xiang, J.F.; Yin, Q.F.; Chen, T.; Zhang, Y.; Zhang, X.O.; Wu, Z.; Zhang, S.; Wang, H.B.; Ge, J.; Lu, X.; et al. Human colorectal cancer-specific CCAT1-L lncRNA regulates long-range chromatin interactions at the MYC locus. Cell Res. 2014, 24, 513–531. [Google Scholar] [CrossRef] [PubMed]
- Paralkar, V.R.; Taborda, C.C.; Huang, P.; Yao, Y.; Kossenkov, A.V.; Prasad, R.; Luan, J.; Davies, J.O.; Hughes, J.R.; Hardison, R.C.; et al. Unlinking an lncRNA from Its Associated cis Element. Mol. Cell 2016, 62, 104–110. [Google Scholar] [CrossRef] [PubMed]
- Cho, S.W.; Xu, J.; Sun, R.; Mumbach, M.R.; Carter, A.C.; Chen, Y.G.; Yost, K.E.; Kim, J.; He, J.; Nevins, S.A.; et al. Promoter of lncRNA Gene PVT1 Is a Tumor-Suppressor DNA Boundary Element. Cell 2018, 173, 1398–1412.e22. [Google Scholar] [CrossRef] [PubMed]
- Ng, M.; Verboon, L.; Issa, H.; Bhayadia, R.; Vermunt, M.W.; Winkler, R.; Schuler, L.; Alejo, O.; Schuschel, K.; Regenyi, E.; et al. Myeloid leukemia vulnerabilities embedded in long noncoding RNA locus MYNRL15. iScience 2023, 26, 107844. [Google Scholar] [CrossRef] [PubMed]
- Mowel, W.K.; McCright, S.J.; Kotzin, J.J.; Collet, M.A.; Uyar, A.; Chen, X.; DeLaney, A.; Spencer, S.P.; Virtue, A.T.; Yang, E.; et al. Group 1 Innate Lymphoid Cell Lineage Identity Is Determined by a cis-Regulatory Element Marked by a Long Non-coding RNA. Immunity 2017, 47, 435–449.e8. [Google Scholar] [CrossRef] [PubMed]
- Beucher, A.; Miguel-Escalada, I.; Balboa, D.; De Vas, M.G.; Maestro, M.A.; Garcia-Hurtado, J.; Bernal, A.; Gonzalez-Franco, R.; Vargiu, P.; Heyn, H.; et al. The HASTER lncRNA promoter is a cis-acting transcriptional stabilizer of HNF1A. Nat. Cell Biol. 2022, 24, 1528–1540. [Google Scholar] [CrossRef]
- Engreitz, J.M.; Haines, J.E.; Perez, E.M.; Munson, G.; Chen, J.; Kane, M.; McDonel, P.E.; Guttman, M.; Lander, E.S. Local regulation of gene expression by lncRNA promoters, transcription and splicing. Nature 2016, 539, 452–455. [Google Scholar] [CrossRef]
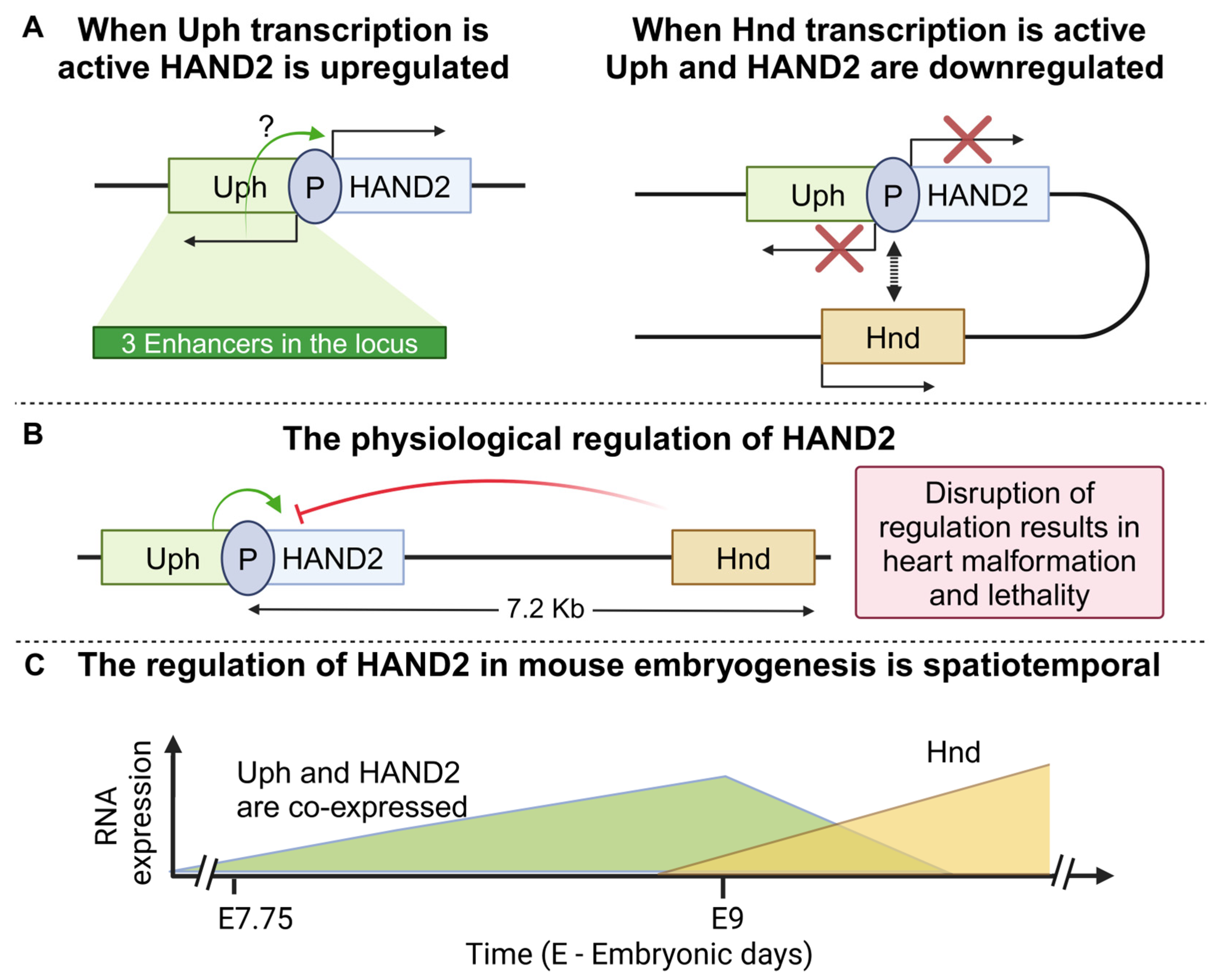
- Anderson, K.M.; Anderson, D.M.; McAnally, J.R.; Shelton, J.M.; Bassel-Duby, R.; Olson, E.N. Transcription of the non-coding RNA upperhand controls Hand2 expression and heart development. Nature 2016, 539, 433–436. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Zhang, J.; Liu, Y.; Fan, X.; Ai, S.; Luo, Y.; Li, X.; Jin, H.; Luo, S.; Zheng, H.; et al. The lncRNA Hand2os1/Uph locus orchestrates heart development through regulation of precise expression of Hand2. Development 2019, 146, dev176198. [Google Scholar] [CrossRef] [PubMed]
- Nunez-Martinez, H.N.; Recillas-Targa, F. Emerging Functions of lncRNA Loci beyond the Transcript Itself. Int. J. Mol. Sci. 2022, 23, 6258. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef] [PubMed]
- Abbastabar, M.; Kheyrollah, M.; Azizian, K.; Bagherlou, N.; Tehrani, S.S.; Maniati, M.; Karimian, A. Multiple functions of p27 in cell cycle, apoptosis, epigenetic modification and transcriptional regulation for the control of cell growth: A double-edged sword protein. DNA Repair 2018, 69, 63–72. [Google Scholar] [CrossRef] [PubMed]
- Razavipour, S.F.; Harikumar, K.B.; Slingerland, J.M. p27 as a Transcriptional Regulator: New Roles in Development and Cancer. Cancer Res. 2020, 80, 3451–3458. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Han, X.; Yuan, J.; Geng, T.; Chen, S.; Hu, X.; Cui, I.H.; Cui, H. Biallelic insertion of a transcriptional terminator via the CRISPR/Cas9 system efficiently silences expression of protein-coding and non-coding RNA genes. J. Biol. Chem. 2017, 292, 5624–5633. [Google Scholar] [CrossRef]
- Davies, J.O.; Telenius, J.M.; McGowan, S.J.; Roberts, N.A.; Taylor, S.; Higgs, D.R.; Hughes, J.R. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 2016, 13, 74–80. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Wang, X.; Zhu, C.; Wang, K. lncRNA PVT1: A novel oncogene in multiple cancers. Cell Mol. Biol. Lett. 2022, 27, 84. [Google Scholar] [CrossRef]
- Sheng, X.F.; Hong, L.L.; Fan, L.; Zhang, Y.; Chen, K.L.; Mu, J.; Shen, S.Y.; Zhuang, H.F. Circular RNA PVT1 Regulates Cell Proliferation, Migration, and Apoptosis by Stabilizing c-Myc and Downstream Target CXCR4 Expression in Acute Myeloid Leukemia. Turk. J. Haematol. 2023, 40, 82–91. [Google Scholar] [CrossRef]
- Tseng, Y.Y.; Moriarity, B.S.; Gong, W.; Akiyama, R.; Tiwari, A.; Kawakami, H.; Ronning, P.; Reuland, B.; Guenther, K.; Beadnell, T.C.; et al. PVT1 dependence in cancer with MYC copy-number increase. Nature 2014, 512, 82–86. [Google Scholar] [CrossRef]
- Tabury, K.; Monavarian, M.; Listik, E.; Shelton, A.K.; Choi, A.S.; Quintens, R.; Arend, R.C.; Hempel, N.; Miller, C.R.; Gyorrfy, B.; et al. PVT1 is a stress-responsive lncRNA that drives ovarian cancer metastasis and chemoresistance. Life Sci. Alliance 2022, 5, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shigeyasu, K.; Toden, S.; Ozawa, T.; Matsuyama, T.; Nagasaka, T.; Ishikawa, T.; Sahoo, D.; Ghosh, P.; Uetake, H.; Fujiwara, T.; et al. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol. Cancer 2020, 19, 155. [Google Scholar] [CrossRef]
- RezaSoltani, M.; Forouzesh, F.; Salehi, Z.; Zabihi, M.R.; Rejali, L.; Nazemalhosseini-Mojarad, E. Identification of LncPVT1 and CircPVT1 as prognostic biomarkers in human colorectal polyps. Sci. Rep. 2023, 13, 13113. [Google Scholar] [CrossRef] [PubMed]
- Venkatraman, S.; Balasubramanian, B.; Thuwajit, C.; Meller, J.; Tohtong, R.; Chutipongtanate, S. Targeting MYC at the intersection between cancer metabolism and oncoimmunology. Front. Immunol. 2024, 15, 1324045. [Google Scholar] [CrossRef] [PubMed]
- Bendixen, L.; Jensen, T.I.; Bak, R.O. CRISPR-Cas-mediated transcriptional modulation: The therapeutic promises of CRISPRa and CRISPRi. Mol. Ther. 2023, 31, 1920–1937. [Google Scholar] [CrossRef]
- Liu, G.; Lin, Q.; Jin, S.; Gao, C. The CRISPR-Cas toolbox and gene editing technologies. Mol. Cell 2022, 82, 333–347. [Google Scholar] [CrossRef] [PubMed]
- Rosspopoff, O.; Trono, D. Take a walk on the KRAB side. Trends Genet. 2023, 39, 844–857. [Google Scholar] [CrossRef]
- Gagnon, K.T.; Corey, D.R. Guidelines for Experiments Using Antisense Oligonucleotides and Double-Stranded RNAs. Nucleic Acid. Ther. 2019, 29, 116–122. [Google Scholar] [CrossRef]
- Crooke, S.T.; Liang, X.H.; Baker, B.F.; Crooke, R.M. Antisense technology: A review. J. Biol. Chem. 2021, 296, 100416. [Google Scholar] [CrossRef]
- Marchese, F.P.; Huarte, M. A “Counter-Enhancer” in Tumor Suppression. Cell 2018, 173, 1318–1319. [Google Scholar] [CrossRef] [PubMed]
- Mumbach, M.R.; Rubin, A.J.; Flynn, R.A.; Dai, C.; Khavari, P.A.; Greenleaf, W.J.; Chang, H.Y. HiChIP: Efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 2016, 13, 919–922. [Google Scholar] [CrossRef] [PubMed]
- Krijger, P.H.L.; Geeven, G.; Bianchi, V.; Hilvering, C.R.E.; de Laat, W. 4C-seq from beginning to end: A detailed protocol for sample preparation and data analysis. Methods 2020, 170, 17–32. [Google Scholar] [CrossRef] [PubMed]
- Kotekar, A.; Singh, A.K.; Devaiah, B.N. BRD4 and MYC: Power couple in transcription and disease. FEBS J. 2023, 290, 4820–4842. [Google Scholar] [CrossRef] [PubMed]
- Katz, L.S.; Brill, G.; Wang, P.; Lambertini, L.; Zhang, P.; Haldeman, J.M.; Liu, H.; Newgard, C.B.; Stewart, A.F.; Garcia-Ocana, A.; et al. Transcriptional activation of the Myc gene by glucose in beta-cells requires a ChREBP-dependent 3-D chromatin interaction between the Myc and Pvt1 genes. Mol. Metab. 2024, 79, 101848. [Google Scholar] [CrossRef] [PubMed]
- Taiana, E.; Favasuli, V.; Ronchetti, D.; Morelli, E.; Tassone, P.; Viglietto, G.; Munshi, N.C.; Neri, A.; Amodio, N. In Vitro Silencing of lncRNAs Using LNA GapmeRs. Methods Mol. Biol. 2021, 2348, 157–166. [Google Scholar] [CrossRef] [PubMed]
- Invrea, F.; Rovito, R.; Torchiaro, E.; Petti, C.; Isella, C.; Medico, E. Patient-derived xenografts (PDXs) as model systems for human cancer. Curr. Opin. Biotechnol. 2020, 63, 151–156. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Xu, R.; Yan, S.; Li, P.; Jia, C.; Sun, H.; Sheng, K.; Wang, Y.; Zhang, Q.; Guo, J.; et al. Hi-C, a chromatin 3D structure technique advancing the functional genomics of immune cells. Front. Genet. 2024, 15, 1377238. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Yin, C.; Chen, S.J.; Wen, L.Z.; Ding, K.; Lei, S.J.; Liu, J.P.; Wang, J.; Chen, K.X.; Jiang, H.L.; et al. The HNF1alpha-regulated lncRNA HNF1A-AS1 reverses the malignancy of hepatocellular carcinoma by enhancing the phosphatase activity of SHP-1. Mol. Cancer 2018, 17, 63. [Google Scholar] [CrossRef]
- Fujino, S.; Miyoshi, N.; Ito, A.; Yasui, M.; Matsuda, C.; Ohue, M.; Uemura, M.; Mizushima, T.; Doki, Y.; Eguchi, H. HNF1A regulates colorectal cancer progression and drug resistance as a downstream of POU5F1. Sci. Rep. 2021, 11, 10363. [Google Scholar] [CrossRef]
- Maruyama, R.; Yokota, T. Knocking Down Long Noncoding RNAs Using Antisense Oligonucleotide Gapmers. Methods Mol. Biol. 2020, 2176, 49–56. [Google Scholar] [CrossRef]
- Barral, A.; Dejardin, J. The chromatin signatures of enhancers and their dynamic regulation. Nucleus 2023, 14, 2160551. [Google Scholar] [CrossRef]
- Schwartzman, O.; Mukamel, Z.; Oded-Elkayam, N.; Olivares-Chauvet, P.; Lubling, Y.; Landan, G.; Izraeli, S.; Tanay, A. UMI-4C for quantitative and targeted chromosomal contact profiling. Nat. Methods 2016, 13, 685–691. [Google Scholar] [CrossRef]
- Nabekura, T.; Shibuya, A. Type 1 innate lymphoid cells: Soldiers at the front line of immunity. Biomed. J. 2021, 44, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Abel, A.M.; Yang, C.; Thakar, M.S.; Malarkannan, S. Natural Killer Cells: Development, Maturation, and Clinical Utilization. Front. Immunol. 2018, 9, 1869. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.; Cherrier, D.E.; Chea, S.; Vosshenrich, C.; Serafini, N.; Petit, M.; Liu, P.; Golub, R.; Di Santo, J.P. An Id2(RFP)-Reporter Mouse Redefines Innate Lymphoid Cell Precursor Potentials. Immunity 2019, 50, 1054–1068.e3. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhao, X.Y. Transcription Factors Associated With IL-15 Cytokine Signaling During NK Cell Development. Front. Immunol. 2021, 12, 610789. [Google Scholar] [CrossRef] [PubMed]
- Nakato, R.; Sakata, T. Methods for ChIP-seq analysis: A practical workflow and advanced applications. Methods 2021, 187, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Ando-Kuri, M.; Rivera, I.S.M.; Rowley, M.J.; Corces, V.G. Analysis of Chromatin Interactions Mediated by Specific Architectural Proteins in Drosophila Cells. Methods Mol. Biol. 2018, 1766, 239–256. [Google Scholar] [CrossRef]
- Han, J.; Zhang, Z.; Wang, K. 3C and 3C-based techniques: The powerful tools for spatial genome organization deciphering. Mol. Cytogenet. 2018, 11, 21. [Google Scholar] [CrossRef]
- Chen, X.; Shen, Y.; Draper, W.; Buenrostro, J.D.; Litzenburger, U.; Cho, S.W.; Satpathy, A.T.; Carter, A.C.; Ghosh, R.P.; East-Seletsky, A.; et al. ATAC-see reveals the accessible genome by transposase-mediated imaging and sequencing. Nat. Methods 2016, 13, 1013–1020. [Google Scholar] [CrossRef] [PubMed]
- Kara, B.; Uyguner, O.; Maras Genc, H.; Islek, E.E.; Kasap, M.; Toksoy, G.; Akpinar, G.; Uyur Yalcin, E.; Anik, Y.; Ustek, D. BEND4 as a Candidate Gene for an Infection-Induced Acute Encephalopathy Characterized by a Cyst and Calcification of the Pons and Cerebellar Atrophy. Mol. Syndromol. 2022, 13, 12–22. [Google Scholar] [CrossRef] [PubMed]
- Canzio, D.; Nwakeze, C.L.; Horta, A.; Rajkumar, S.M.; Coffey, E.L.; Duffy, E.E.; Duffie, R.; Monahan, K.; O’Keeffe, S.; Simon, M.D.; et al. Antisense lncRNA Transcription Mediates DNA Demethylation to Drive Stochastic Protocadherin alpha Promoter Choice. Cell 2019, 177, 639–653.e15. [Google Scholar] [CrossRef] [PubMed]
- Furlan, G.; Gutierrez Hernandez, N.; Huret, C.; Galupa, R.; van Bemmel, J.G.; Romito, A.; Heard, E.; Morey, C.; Rougeulle, C. The Ftx Noncoding Locus Controls X Chromosome Inactivation Independently of Its RNA Products. Mol. Cell 2018, 70, 462–472.e8. [Google Scholar] [CrossRef] [PubMed]
- Latos, P.A.; Pauler, F.M.; Koerner, M.V.; Senergin, H.B.; Hudson, Q.J.; Stocsits, R.R.; Allhoff, W.; Stricker, S.H.; Klement, R.M.; Warczok, K.E.; et al. Airn transcriptional overlap, but not its lncRNA products, induces imprinted Igf2r silencing. Science 2012, 338, 1469–1472. [Google Scholar] [CrossRef] [PubMed]
- Gil, N.; Perry, R.B.; Mukamel, Z.; Tuck, A.; Buhler, M.; Ulitsky, I. Complex regulation of Eomes levels mediated through distinct functional features of the Meteor long non-coding RNA locus. Cell Rep. 2023, 42, 112569. [Google Scholar] [CrossRef] [PubMed]
- Allou, L.; Balzano, S.; Magg, A.; Quinodoz, M.; Royer-Bertrand, B.; Schopflin, R.; Chan, W.L.; Speck-Martins, C.E.; Carvalho, D.R.; Farage, L.; et al. Non-coding deletions identify Maenli lncRNA as a limb-specific En1 regulator. Nature 2021, 592, 93–98. [Google Scholar] [CrossRef]
- Ritter, N.; Ali, T.; Kopitchinski, N.; Schuster, P.; Beisaw, A.; Hendrix, D.A.; Schulz, M.H.; Muller-McNicoll, M.; Dimmeler, S.; Grote, P. The lncRNA Locus Handsdown Regulates Cardiac Gene Programs and Is Essential for Early Mouse Development. Dev. Cell 2019, 50, 644–657.e8. [Google Scholar] [CrossRef]
- Guo, Y.; Xu, Q.; Canzio, D.; Shou, J.; Li, J.; Gorkin, D.U.; Jung, I.; Wu, H.; Zhai, Y.; Tang, Y.; et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell 2015, 162, 900–910. [Google Scholar] [CrossRef]
- Dai, Z.; Li, R.; Hou, Y.; Li, Q.; Zhao, K.; Li, T.; Li, M.J.; Wu, X. Inducible CRISPRa screen identifies putative enhancers. J. Genet. Genom. 2021, 48, 917–927. [Google Scholar] [CrossRef]
- Hsu, H.K.; Weng, Y.I.; Hsu, P.Y.; Huang, T.H.; Huang, Y.W. Detection of DNA Methylation by MeDIP and MBDCap Assays: An Overview of Techniques. Methods Mol. Biol. 2020, 2102, 225–234. [Google Scholar] [CrossRef]
- Robin, J.D. C-HiC: A High-Resolution Method for Unbiased Chromatin Conformation Capture Targeting Small Locus. Methods Mol. Biol. 2021, 2157, 85–102. [Google Scholar] [CrossRef]
- Vega-Benedetti, A.F.; Loi, E.; Moi, L.; Blois, S.; Fadda, A.; Antonelli, M.; Arcella, A.; Badiali, M.; Giangaspero, F.; Morra, I.; et al. Clustered protocadherins methylation alterations in cancer. Clin. Epigenetics 2019, 11, 100. [Google Scholar] [CrossRef]
- Dallosso, A.R.; Hancock, A.L.; Szemes, M.; Moorwood, K.; Chilukamarri, L.; Tsai, H.H.; Sarkar, A.; Barasch, J.; Vuononvirta, R.; Jones, C.; et al. Frequent long-range epigenetic silencing of protocadherin gene clusters on chromosome 5q31 in Wilms’ tumor. PLoS Genet. 2009, 5, e1000745. [Google Scholar] [CrossRef]
- Patrat, C.; Ouimette, J.F.; Rougeulle, C. X chromosome inactivation in human development. Development 2020, 147, dev183095. [Google Scholar] [CrossRef] [PubMed]
- Loda, A.; Heard, E. Xist RNA in action: Past, present, and future. PLoS Genet. 2019, 15, e1008333. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Li, Y.; Guo, L.; Zhang, C.; Tang, S. Long non-coding RNA FTX predicts a poor prognosis of human cancers: A meta-analysis. Biosci. Rep. 2021, 41, BSR20203995. [Google Scholar] [CrossRef] [PubMed]
- Liang, Y.; Lu, H. Long noncoding RNA FTX is associated with prognosis of glioma patients. J. Gene Med. 2020, 22, e3237. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Qu, T.; Li, Y.; Ma, J.; Yu, H. Biological role of long non-coding RNA FTX in cancer progression. Biomed. Pharmacother. 2022, 153, 113446. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, J.; Chen, Z.; Wu, G.; Zhou, Z.; Wu, T.; Wang, W.; Luo, Y.; Liu, T. Prognostic significance of long non-coding RNA five prime to XIST in various cancers. BMC Cancer 2022, 22, 61. [Google Scholar] [CrossRef]
- Huret, C.; Ferraye, L.; David, A.; Mohamed, M.; Valentin, N.; Charlotte, F.; Savignac, M.; Goodhardt, M.; Guery, J.C.; Rougeulle, C.; et al. Altered X-chromosome inactivation predisposes to autoimmunity. Sci. Adv. 2024, 10, eadn6537. [Google Scholar] [CrossRef] [PubMed]
- Hosoi, Y.; Soma, M.; Shiura, H.; Sado, T.; Hasuwa, H.; Abe, K.; Kohda, T.; Ishino, F.; Kobayashi, S. Female mice lacking Ftx lncRNA exhibit impaired X-chromosome inactivation and a microphthalmia-like phenotype. Nat. Commun. 2018, 9, 3829. [Google Scholar] [CrossRef] [PubMed]
- Schmit, T.; Klomp, M.; Khan, M.N. An Overview of Flow Cytometry: Its Principles and Applications in Allergic Disease Research. Methods Mol. Biol. 2021, 2223, 169–182. [Google Scholar] [CrossRef] [PubMed]
- Marcho, C.; Bevilacqua, A.; Tremblay, K.D.; Mager, J. Tissue-specific regulation of Igf2r/Airn imprinting during gastrulation. Epigenetics Chromatin 2015, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, F.C.; Charalambous, M. Genomic imprinting, growth and maternal-fetal interactions. J. Exp. Biol. 2018, 221, jeb164517. [Google Scholar] [CrossRef] [PubMed]
- Hosen, M.R.; Militello, G.; Weirick, T.; Ponomareva, Y.; Dassanayaka, S.; Moore, J.B.t.; Doring, C.; Wysoczynski, M.; Jones, S.P.; Dimmeler, S.; et al. Airn Regulates Igf2bp2 Translation in Cardiomyocytes. Circ. Res. 2018, 122, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Torrente, Y.; Bella, P.; Tripodi, L.; Villa, C.; Farini, A. Role of Insulin-Like Growth Factor Receptor 2 across Muscle Homeostasis: Implications for Treating Muscular Dystrophy. Cells 2020, 9, 441. [Google Scholar] [CrossRef] [PubMed]
- Alberini, C.M. IGF2 in memory, neurodevelopmental disorders, and neurodegenerative diseases. Trends Neurosci. 2023, 46, 488–502. [Google Scholar] [CrossRef]
- Zhong, Y.; Ren, X.; Cao, X.; Xu, Y.; Song, Y.; Zhou, Y.; Mao, F.; Shen, S.; Wang, Z.; Sun, Q. Insulin-like growth factor 2 receptor is a key immune-related gene that is correlated with a poor prognosis in patients with triple-negative breast cancer: A bioinformatics analysis. Front. Oncol. 2022, 12, 871786. [Google Scholar] [CrossRef]
- Bartolomei, M.S.; Oakey, R.J.; Wutz, A. Genomic imprinting: An epigenetic regulatory system. PLoS Genet. 2020, 16, e1008970. [Google Scholar] [CrossRef]
- Andergassen, D.; Muckenhuber, M.; Bammer, P.C.; Kulinski, T.M.; Theussl, H.C.; Shimizu, T.; Penninger, J.M.; Pauler, F.M.; Hudson, Q.J. The Airn lncRNA does not require any DNA elements within its locus to silence distant imprinted genes. PLoS Genet. 2019, 15, e1008268. [Google Scholar] [CrossRef] [PubMed]
- Braceros, A.K.; Schertzer, M.D.; Omer, A.; Trotman, J.B.; Davis, E.S.; Dowen, J.M.; Phanstiel, D.H.; Aiden, E.L.; Calabrese, J.M. Proximity-dependent recruitment of Polycomb repressive complexes by the lncRNA Airn. Cell Rep. 2023, 42, 112803. [Google Scholar] [CrossRef] [PubMed]
- Probst, S.; Arnold, S.J. Eomesodermin-At Dawn of Cell Fate Decisions During Early Embryogenesis. Curr. Top. Dev. Biol. 2017, 122, 93–115. [Google Scholar] [CrossRef] [PubMed]
- Costello, I.; Pimeisl, I.M.; Drager, S.; Bikoff, E.K.; Robertson, E.J.; Arnold, S.J. The T-box transcription factor Eomesodermin acts upstream of Mesp1 to specify cardiac mesoderm during mouse gastrulation. Nat. Cell Biol. 2011, 13, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Latifi, B.; Muller, S.; Luptak, A.; Chen, I.A. Self-cleaving ribozymes: Substrate specificity and synthetic biology applications. RSC Chem. Biol. 2021, 2, 1370–1383. [Google Scholar] [CrossRef]
- Huettl, R.E.; Luxenhofer, G.; Bianchi, E.; Haupt, C.; Joshi, R.; Prochiantz, A.; Huber, A.B. Engrailed 1 mediates correct formation of limb innervation through two distinct mechanisms. PLoS ONE 2015, 10, e0118505. [Google Scholar] [CrossRef] [PubMed]
- Gwak, J.; Shin, J.Y.; Lee, K.; Hong, S.K.; Oh, S.; Goh, S.H.; Kim, W.S.; Ju, B.G. SFMBT2 (Scm-like with four mbt domains 2) negatively regulates cell migration and invasion in prostate cancer cells. Oncotarget 2016, 7, 48250–48264. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.; Na, W.; Maeng, J.H.; Wu, H.; Ju, B.G. Regulation of DU145 prostate cancer cell growth by Scm-like with four mbt domains 2. J. Biosci. 2013, 38, 105–112. [Google Scholar] [CrossRef] [PubMed]
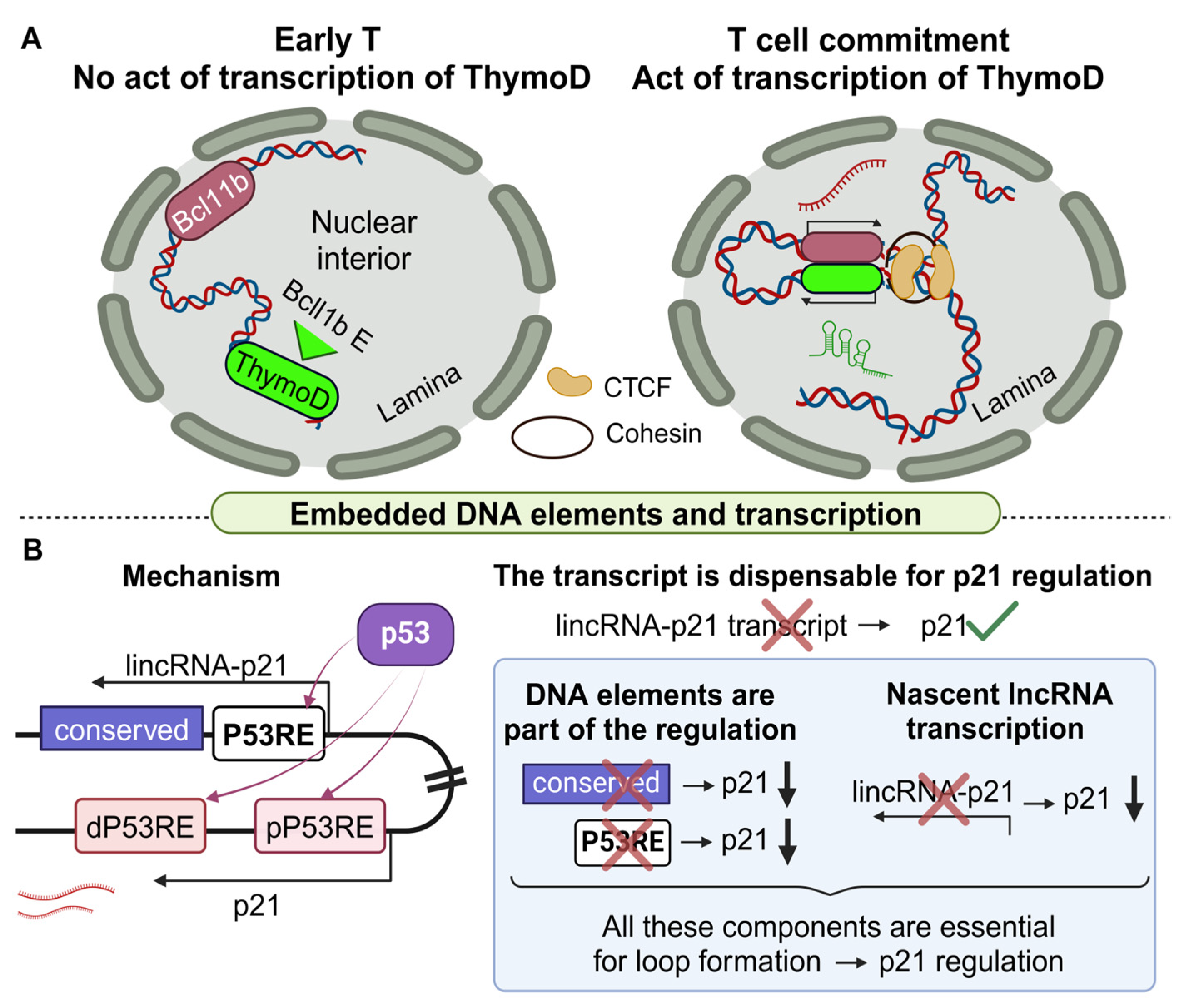
- Isoda, T.; Moore, A.J.; He, Z.; Chandra, V.; Aida, M.; Denholtz, M.; Piet van Hamburg, J.; Fisch, K.M.; Chang, A.N.; Fahl, S.P.; et al. Non-coding Transcription Instructs Chromatin Folding and Compartmentalization to Dictate Enhancer-Promoter Communication and T Cell Fate. Cell 2017, 171, 103–119.e18. [Google Scholar] [CrossRef] [PubMed]
- Winkler, L.; Jimenez, M.; Zimmer, J.T.; Williams, A.; Simon, M.D.; Dimitrova, N. Functional elements of the cis-regulatory lincRNA-p21. Cell Rep. 2022, 39, 110687. [Google Scholar] [CrossRef]
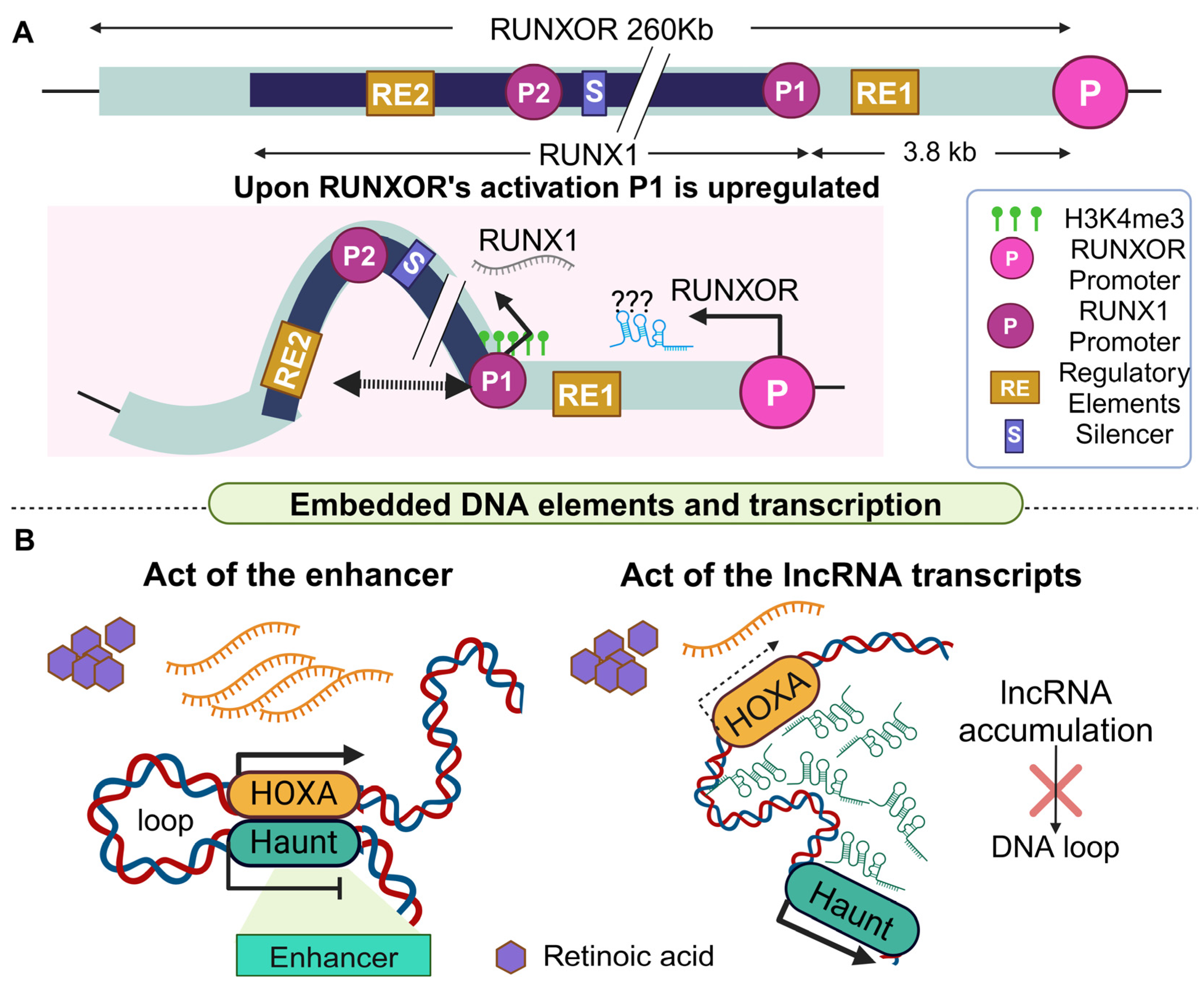
- Nie, Y.; Zhou, L.; Wang, H.; Chen, N.; Jia, L.; Wang, C.; Wang, Y.; Chen, J.; Wen, X.; Niu, C.; et al. Profiling the epigenetic interplay of lncRNA RUNXOR and oncogenic RUNX1 in breast cancer cells by gene in situ cis-activation. Am. J. Cancer Res. 2019, 9, 1635–1649. [Google Scholar] [PubMed]
- Yin, Y.; Yan, P.; Lu, J.; Song, G.; Zhu, Y.; Li, Z.; Zhao, Y.; Shen, B.; Huang, X.; Zhu, H.; et al. Opposing Roles for the lncRNA Haunt and Its Genomic Locus in Regulating HOXA Gene Activation during Embryonic Stem Cell Differentiation. Cell Stem Cell 2015, 16, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Trujillo-Ochoa, J.L.; Kazemian, M.; Afzali, B. The role of transcription factors in shaping regulatory T cell identity. Nat. Rev. Immunol. 2023, 23, 842–856. [Google Scholar] [CrossRef] [PubMed]
- Permatasari, H.K.; Nakahata, S.; Ichikawa, T.; Fauzi, Y.R.; Kiyonari, H.; Shide, K.; Kameda, T.; Shimoda, K.; Ono, M.; Taki, T.; et al. Oncogenic isoform switch of tumor suppressor BCL11B in adult T-cell leukemia/lymphoma. Exp. Hematol. 2022, 111, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Zhang, J.A.; Dose, M.; Kueh, H.Y.; Mosadeghi, R.; Gounari, F.; Rothenberg, E.V. A far downstream enhancer for murine Bcl11b controls its T-cell specific expression. Blood 2013, 122, 902–911. [Google Scholar] [CrossRef] [PubMed]
- Isoda, T.; Morio, T.; Takagi, M. Noncoding RNA transcription at enhancers and genome folding in cancer. Cancer Sci. 2019, 110, 2328–2336. [Google Scholar] [CrossRef] [PubMed]
- Mi, J.X.; Zhang, Y.N.; Lai, Z.; Li, W.; Zhou, L.; Zhong, F. Principal Component Analysis based on Nuclear norm Minimization. Neural Netw. 2019, 118, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Shamloo, B.; Usluer, S. p21 in Cancer Research. Cancers 2019, 11, 1178. [Google Scholar] [CrossRef]
- Groff, A.F.; Sanchez-Gomez, D.B.; Soruco, M.M.L.; Gerhardinger, C.; Barutcu, A.R.; Li, E.; Elcavage, L.; Plana, O.; Sanchez, L.V.; Lee, J.C.; et al. In Vivo Characterization of Linc-p21 Reveals Functional cis-Regulatory DNA Elements. Cell Rep. 2016, 16, 2178–2186. [Google Scholar] [CrossRef]
- Felletti, M.; Stifel, J.; Wurmthaler, L.A.; Geiger, S.; Hartig, J.S. Twister ribozymes as highly versatile expression platforms for artificial riboswitches. Nat. Commun. 2016, 7, 12834. [Google Scholar] [CrossRef]
- Rose, J.C.; Popp, N.A.; Richardson, C.D.; Stephany, J.J.; Mathieu, J.; Wei, C.T.; Corn, J.E.; Maly, D.J.; Fowler, D.M. Suppression of unwanted CRISPR-Cas9 editing by co-administration of catalytically inactivating truncated guide RNAs. Nat. Commun. 2020, 11, 2697. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Chen, F.; Chen, Q.; Wan, X.; Shi, M.; Chen, A.K.; Ma, Z.; Li, G.; Wang, M.; Ying, Y.; et al. MyoD is a 3D genome structure organizer for muscle cell identity. Nat. Commun. 2022, 13, 205. [Google Scholar] [CrossRef]
- Zhigulev, A.; Sahlen, P. Targeted Chromosome Conformation Capture (HiCap). Methods Mol. Biol. 2022, 2532, 75–94. [Google Scholar] [CrossRef] [PubMed]
- Allen, M.A.; Andrysik, Z.; Dengler, V.L.; Mellert, H.S.; Guarnieri, A.; Freeman, J.A.; Sullivan, K.D.; Galbraith, M.D.; Luo, X.; Kraus, W.L.; et al. Global analysis of p53-regulated transcription identifies its direct targets and unexpected regulatory mechanisms. Elife 2014, 3, e02200. [Google Scholar] [CrossRef] [PubMed]
- Al-Harbi, S.; Aljurf, M.; Mohty, M.; Almohareb, F.; Ahmed, S.O.A. An update on the molecular pathogenesis and potential therapeutic targeting of AML with t(8;21)(q22;q22.1);RUNX1-RUNX1T1. Blood Adv. 2020, 4, 229–238. [Google Scholar] [CrossRef] [PubMed]
- Ariffin, N.S. RUNX1 as a Novel Molecular Target for Breast Cancer. Clin. Breast Cancer 2022, 22, 499–506. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.C. RUNX1 and cancer. Biochim. Biophys. Acta Rev. Cancer 2022, 1877, 188715. [Google Scholar] [CrossRef]
- Li, J.; Liang, Q.; Zhou, H.; Zhou, M.; Huang, H. Correction: Profiling the impact of the promoters on CRISPR-Cas12a system in human cells. Cell Mol. Biol. Lett. 2023, 28, 82. [Google Scholar] [CrossRef]
- Wang, H.; Li, W.; Guo, R.; Sun, J.; Cui, J.; Wang, G.; Hoffman, A.R.; Hu, J.F. An intragenic long noncoding RNA interacts epigenetically with the RUNX1 promoter and enhancer chromatin DNA in hematopoietic malignancies. Int. J. Cancer 2014, 135, 2783–2794. [Google Scholar] [CrossRef]
- Oh, J.H.; Kim, C.Y.; Lee, J.Y.; Kim, M.H. Retinoic acid and CTCF play key roles in inducing the collinear expression of the Hoxa cluster. Acta Biochim. Biophys. Sin. 2018, 50, 555–559. [Google Scholar] [CrossRef]
- Abuhantash, M.; Collins, E.M.; Thompson, A. Role of the HOXA cluster in HSC emergence and blood cancer. Biochem. Soc. Trans. 2021, 49, 1817–1827. [Google Scholar] [CrossRef] [PubMed]
- Alexopoulou, A.N.; Couchman, J.R.; Whiteford, J.R. The CMV early enhancer/chicken beta actin (CAG) promoter can be used to drive transgene expression during the differentiation of murine embryonic stem cells into vascular progenitors. BMC Cell Biol. 2008, 9, 2. [Google Scholar] [CrossRef] [PubMed]
- Alfeghaly, C.; Behm-Ansmant, I.; Maenner, S. Study of Genome-Wide Occupancy of Long Non-Coding RNAs Using Chromatin Isolation by RNA Purification (ChIRP). Methods Mol. Biol. 2021, 2300, 107–117. [Google Scholar] [CrossRef] [PubMed]
- Videira, R.F.; Koop, A.M.C.; Ottaviani, L.; Poels, E.M.; Kocken, J.M.M.; Dos Remedios, C.; Mendes-Ferreira, P.; Van De Kolk, K.W.; Du Marchie Sarvaas, G.J.; Lourenco, A.; et al. The adult heart requires baseline expression of the transcription factor Hand2 to withstand right ventricular pressure overload. Cardiovasc. Res. 2022, 118, 2688–2702. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xu, Y.J.; Li, R.G.; Wang, Z.S.; Zhang, M.; Qu, X.K.; Qiao, Q.; Li, X.M.; Di, R.M.; Qiu, X.B.; et al. HAND2 loss-of-function mutation causes familial dilated cardiomyopathy. Eur. J. Med. Genet. 2019, 62, 103540. [Google Scholar] [CrossRef] [PubMed]
- Yanagisawa, H.; Clouthier, D.E.; Richardson, J.A.; Charite, J.; Olson, E.N. Targeted deletion of a branchial arch-specific enhancer reveals a role of dHAND in craniofacial development. Development 2003, 130, 1069–1078. [Google Scholar] [CrossRef] [PubMed]
- Chiang, T.W.; le Sage, C.; Larrieu, D.; Demir, M.; Jackson, S.P. CRISPR-Cas9(D10A) nickase-based genotypic and phenotypic screening to enhance genome editing. Sci. Rep. 2016, 6, 24356. [Google Scholar] [CrossRef] [PubMed]
- Omachi, K.; Miner, J.H. Comparative analysis of dCas9-VP64 variants and multiplexed guide RNAs mediating CRISPR activation. PLoS ONE 2022, 17, e0270008. [Google Scholar] [CrossRef] [PubMed]
- da Costa-Nunes, J.A.; Noordermeer, D. TADs: Dynamic structures to create stable regulatory functions. Curr. Opin. Struct. Biol. 2023, 81, 102622. [Google Scholar] [CrossRef]
- George, M.R.; Duan, Q.; Nagle, A.; Kathiriya, I.S.; Huang, Y.; Rao, K.; Haldar, S.M.; Bruneau, B.G. Minimal in vivo requirements for developmentally regulated cardiac long intergenic non-coding RNAs. Development 2019, 146, dev185314. [Google Scholar] [CrossRef]
- Lange, M.; Begolli, R.; Giakountis, A. Non-Coding Variants in Cancer: Mechanistic Insights and Clinical Potential for Personalized Medicine. Noncoding RNA 2021, 7, 47. [Google Scholar] [CrossRef] [PubMed]

| lncRNA (L) | Target Gene (TG) | Distance ** | Effect on TG | Mechanism of Function | Physiological Processes | Dysregulated Phenotype of TG | Ref. |
|---|---|---|---|---|---|---|---|
| LockD | Cdkn1b (p27) | +4 Kb | Down | Promoter-like element, chromatin interaction | Cell cycle progression | Cancer | [35] |
| PVT1 | MYC | +58 Kb | Down | Chromatin conformation, promoter competition for enhancer | Cell proliferation | Cancer | [36] |
| MYNRL15 | IMP3 WDR61 COMMD4 SNUPN | +12–15 Mb | Down | CTCF-mediated long-range looping (12–15 Mb radius) | RNA splicing, DNA replication | Anti- leukemic effect | [37] |
| Rroid | Id2 | −220 Kb | Up | Epigenetic modifications, long distance interactions | NK cell lineage | Cancer | [38] |
| HASTER | HNF1A | +2 Kb * | Down | Epigenetic modifications, insulator like element, stability of HNF1A | Hepatic cell differentiation | Cancer | [39] |
| Bendr | Bend4 | ND, + TSS | Up | Enhancer-like element in the Bendr promoter/5’ proximal region | Promotion of germ cell differentiation | Acute encephalopathy | [40] |
| Upperhand (Uph, Hand2os1) | HAND2 | −123 bp (antisense) | Up | Super-enhancer—promoter interactions | Heart development | Heart morphogenesis leading to lethality | [41,42] |
| lncRNA (L) | Target Gene (TG) | Distance ** | Effect on TG Expression | Mechanism of Function | Physiological Processes | Dysregulated Phenotype of TG | Ref. |
|---|---|---|---|---|---|---|---|
| Pcdhα | Pcdha cluster | Antisense overlapping | Up | Nascent lncRNA transcription, target demethylation, CTCF – cohesin looping, | Cell surface identity signals of neurons | Possible cancer association Wilms tumor | [83] |
| Ftx | Xist | +195 Kb * | Up | Loop formation, nascent transcription | X chromosome inactivation (XCI) | Cancer | [84] |
| Airn | Igf2r | −30 Kb | Down | Perturbation of RNA pol II from the target gene promoter | Genomic imprinting, development | Cancer | [85] |
| Meteor | Eomes | +70 Kb | Up | Minimal transcriptional initiation | Cardiac mesoderm and neuronal differentiation programs | CD4+ T cell retainment in inflamation | [86] |
| Maenli | En1 | −251 Kb | Up | Transcription elongation, epigenetic modifications | Dorsal–ventral patterning in the limb | Brain abnormalities and limb malformation | [87] |
| Blustr | Sfmbt2 | +5 Kb | Up | Transcription elongation, epigenetic modifications and 5’ splicing site of intron 1 | ND | ND | [40] |
| Handsdown | HAND2 | +7.2 Kb | Down | CTCF independent chromatin interactions (yet not fully understood) | Heart development | Heart hyperplasia, lethality | [88] |
| lncRNA (L) | Target Gene (TG) | Distance of L from TG | Regulation of the TG | Mechanism of Function | Physiological Processes | Dysregulated Phenotype | Ref. |
|---|---|---|---|---|---|---|---|
| ThymoD | Bcl11b | +850 Kb * | Up | Demethylation, CTCF binding, cohesin, loop, chromatin reposition | T-lineage determination | Lymphoma and leukemia | [119] |
| lincRNA p21 | Cdk1a (p21) | +12 Kb | Up | Nascent lncRNA transcription, p53 elements | Apoptosis, cell cycle control, hypoxia | Increased cell proliferation | [120] |
| RUNXOR | RUNX1 | −3.8 Kb | Up (RUNX1c) | Enhancer and promoter interaction in the target gene | Cell development, hematopoiesis | Cancer (breast cancer and leukemia) | [121] |
| Haunt | HOXA cluster | −40 Kb | DNA locus: Up lncRNA transcript: down | Nascent transcription, enhancer promoter interaction | Development | Cancer | [122] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kafida, M.; Karela, M.; Giakountis, A. RNA-Independent Regulatory Functions of lncRNA in Complex Disease. Cancers 2024, 16, 2728. https://doi.org/10.3390/cancers16152728
Kafida M, Karela M, Giakountis A. RNA-Independent Regulatory Functions of lncRNA in Complex Disease. Cancers. 2024; 16(15):2728. https://doi.org/10.3390/cancers16152728
Chicago/Turabian StyleKafida, Michaela, Maria Karela, and Antonis Giakountis. 2024. "RNA-Independent Regulatory Functions of lncRNA in Complex Disease" Cancers 16, no. 15: 2728. https://doi.org/10.3390/cancers16152728
APA StyleKafida, M., Karela, M., & Giakountis, A. (2024). RNA-Independent Regulatory Functions of lncRNA in Complex Disease. Cancers, 16(15), 2728. https://doi.org/10.3390/cancers16152728





