Deep Residual Learning-Based Classification with Identification of Incorrect Predictions and Quantification of Cellularity and Nuclear Morphological Features in Digital Pathological Images of Common Astrocytic Tumors
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
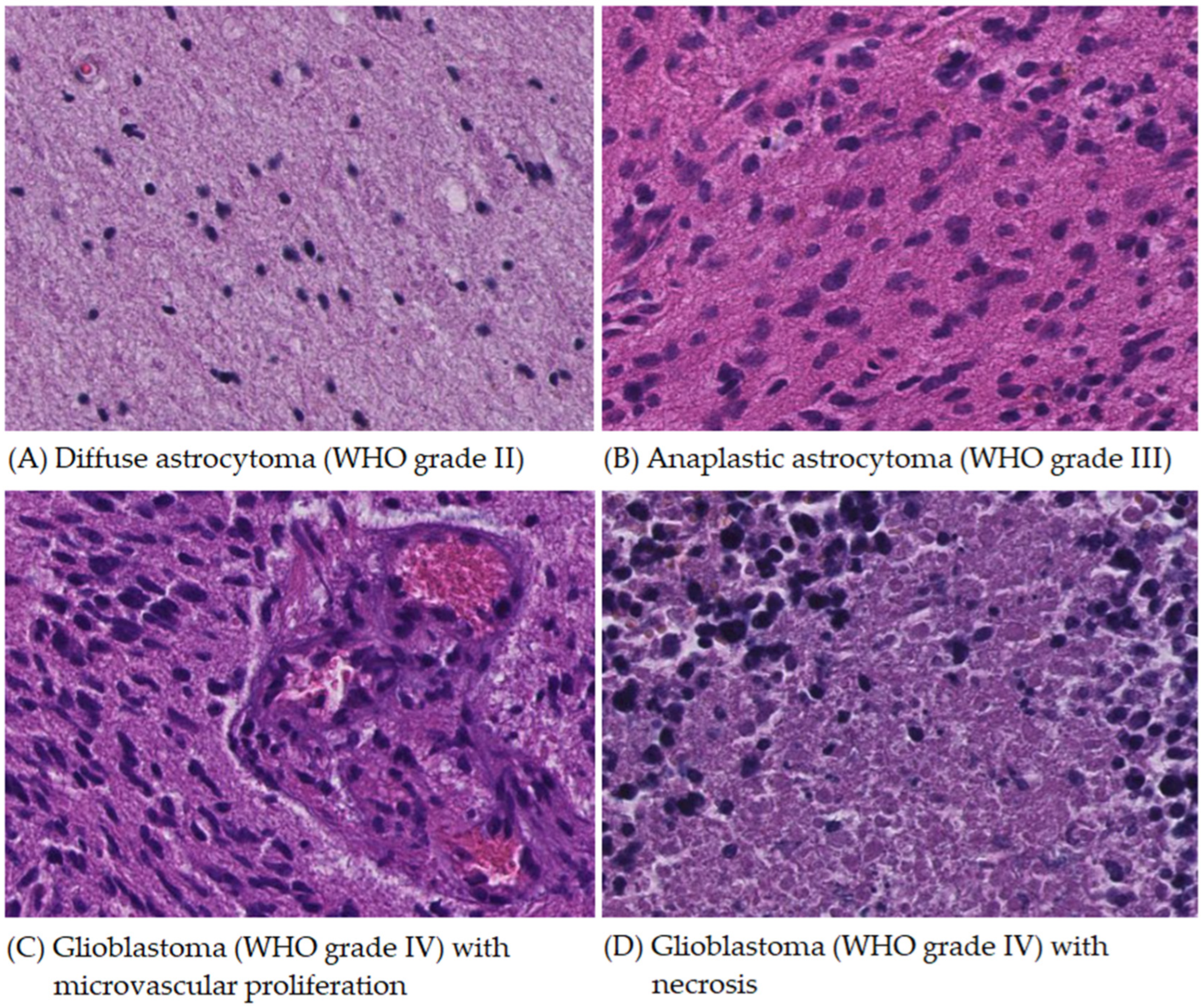
2.1. Cases
2.2. Application of Deep Residual Learning Model for Classification
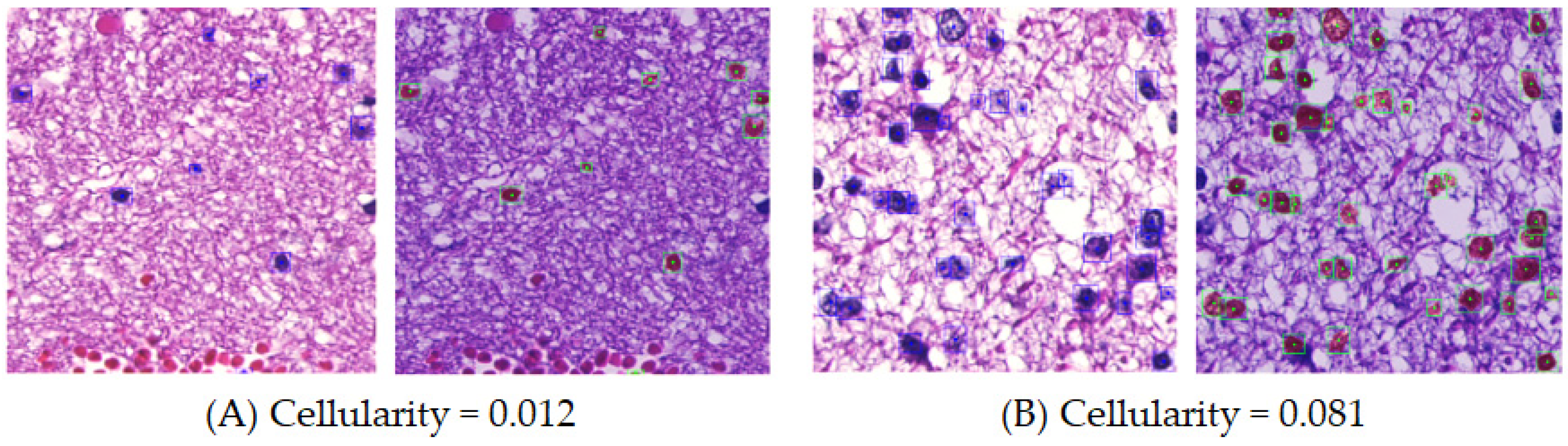
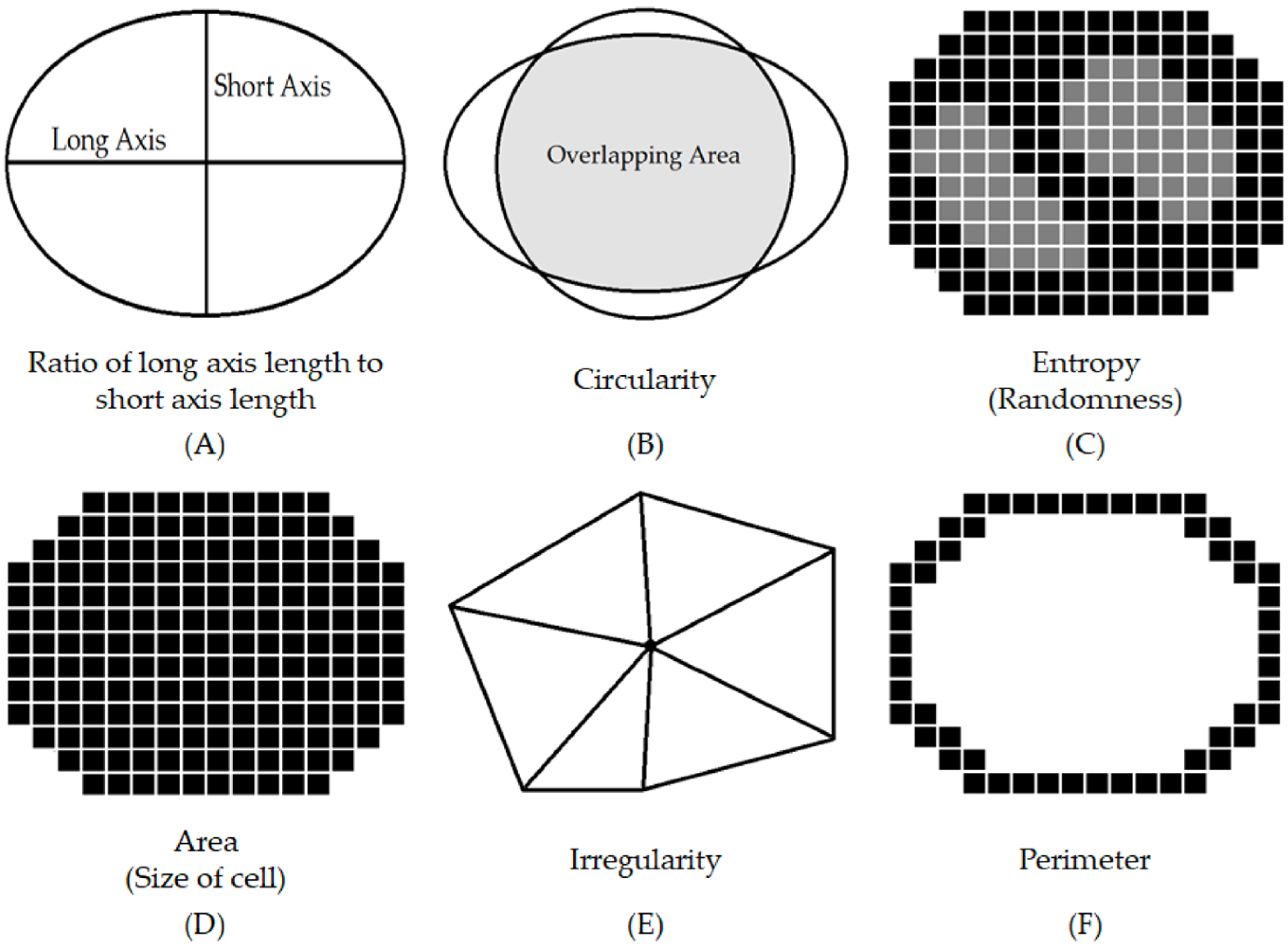
2.3. Quantification of Cellularity and Nuclear Morphological Features with Importance Weighting
3. Results
3.1. Data Summary
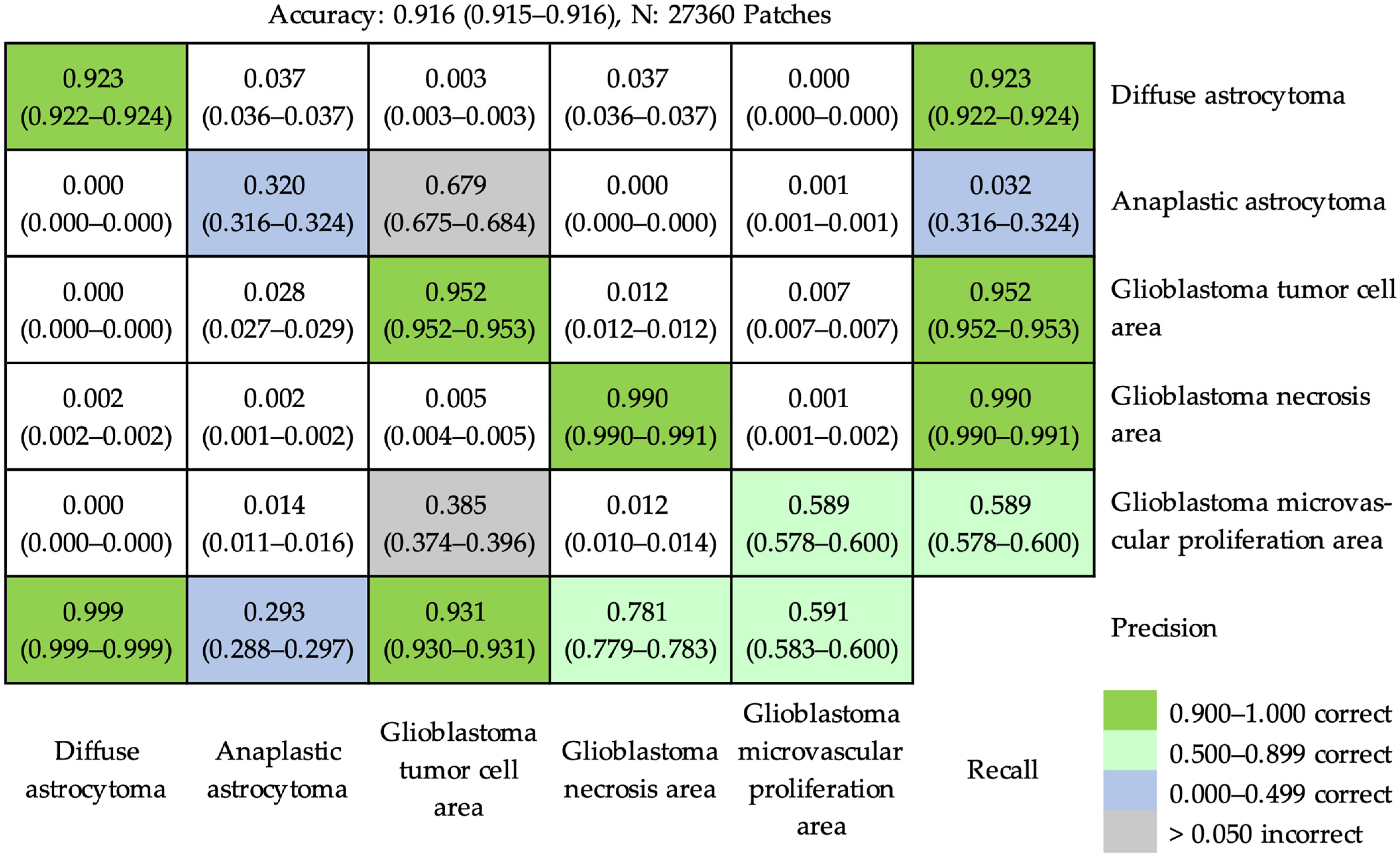
3.2. Outcomes of Application of Deep Residual Learning Model for Classification
3.2.1. At the Patch Level
3.2.2. At the Case Level
3.3. Outcomes of Quantification of Cellularity and Nuclear Morphological Features with Importance Weighting
3.3.1. Cellularity
3.3.2. Nuclear Morphological Features
3.3.3. Importance Weighting
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ostrom, Q.T.; Gittleman, H.; Stetson, L.; Virk, S.; Barnholtz-Sloan, J.S. Epidemiology of intracranial gliomas. In Progress in Neurological Surgery; S. Karger: Berlin, Germany, 2018; Volume 30, pp. 1–11. [Google Scholar] [CrossRef]
- Rasmussen, B.K.; Hansen, S.; Laursen, R.J.; Kosteljanetz, M.; Schultz, H.; Nørgård, B.M.; Guldberg, R.; Gradel, K.O. Epidemiology of glioma: Clinical characteristics, symptoms, and predictors of glioma patients grade I–IV in the Danish Neuro-Oncology Registry. J. Neurooncol. 2017, 135, 571–579. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Ohgaki, H.; Wiestier, O.D.; Cavenee, W.K.; Ellison, D.W.; Figarella-Branger, D.; Perry, A.; Reifenberger, G.; von Deimling, A. WHO Classification of Tumours of the Central Nervous System, 4th ed.; International Agency for Research on Cancer (IARC): Lyon, France, 2016; pp. 18–56. [Google Scholar]
- Kros, J.M.; Gorlia, T.; Kouwenhoven, M.C.; Zheng, P.-P.; Collins, V.P.; Figarella-Branger, D.; Giangaspero, F.; Giannini, C.; Mokhtari, K.; Mørk, S.J.; et al. Panel review of anaplastic oligodendroglioma from European Organization for Research and Treatment of Cancer Trial 26951: Assessment of consensus in diagnosis, influence of 1p/19q loss, and correlations with outcome. J. Neuropathol. Exp. Neurol. 2007, 66, 545–551. [Google Scholar] [CrossRef] [PubMed]
- van den Ben, M.J. Interobserver variation of the histopathological diagnosis in clinical trials on glioma: A clinician’s perspective. Acta Neuropathol. 2010, 120, 297–304. [Google Scholar] [CrossRef]
- Kim, I.; Kang, K.; Song, Y.; Kim, T.-J. Application of artificial intelligence in pathology: Trends and challenges. Diagnostics 2022, 12, 2794. [Google Scholar] [CrossRef] [PubMed]
- Pantanowitz, L.; Wu, U.; Seigh, L.; LoPresti, E.; Yeh, F.-C.; Salgia, P.; Michelow, P.; Hazelhurst, S.; Chen, W.-Y.; Hartman, D.; et al. Artificial intelligence–based screening for mycobacteria in whole-slide images of tissue samples. Am. J. Clin. Pathol. 2021, 156, 117–128. [Google Scholar] [CrossRef]
- Chuang, W.-Y.; Chang, S.-H.; Yu, W.-H.; Yang, C.-K.; Yeh, C.-J.; Ueng, S.-H.; Liu, Y.-J.; Chen, T.-D.; Chen, K.-H.; Hsieh, Y.-Y.; et al. Successful identification of nasopharyngeal carcinoma in nasopharyngeal biopsies using deep learning. Cancers 2020, 12, 507. [Google Scholar] [CrossRef]
- Chuang, W.-Y.; Chen, C.-C.; Yu, W.-H.; Yeh, C.-J.; Chang, S.-H.; Ueng, S.-H.; Wang, T.-H.; Hsueh, C.; Kuo, C.-F.; Yeh, C.-Y. Identification of nodal micrometastasis in colorectal cancer using deep learning on annotation-free whole-slide images. Mod. Pathol. 2021, 34, 1901–1911. [Google Scholar] [CrossRef]
- Li, L.; Han, D.; Yu, Y.; Li, J.; Liu, Y. Artificial intelligence-assisted interpretation of ki-67 expression and repeatability in breast cancer. Diagn. Pathol. 2022, 17, 20. [Google Scholar] [CrossRef]
- Bulten, W.; Balkenhol, M.; Belinga, J.-J.A.; Brilhante, A.; Çakır, A.; Egevad, L.; Eklund, M.; Farré, X.; Geronatsiou, K.; Molinié, V. Artificial intelligence assistance significantly improves Gleason grading of prostate biopsies by pathologists. Mod. Pathol. 2021, 34, 660–671. [Google Scholar] [CrossRef]
- Yang, C.-K.; Lee, C.-Y.; Wang, H.-S.; Huang, S.-C.; Liang, P.-I.; Chen, J.-S.; Kuo, C.-F.; Tu, K.-H.; Yeh, C.-Y.; Chen, T.-D. Glomerular disease classification and lesion identification by machine learning. Biomed. J. 2022, 45, 675–685. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Pang, J.; Wang, J.; Xiong, Y.; Li, X.; Sun, S.; Feng, W.; Liu, Z.; Shi, J.; Ouyang, W.; et al. Hybrid task cascade for instance segmentation. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), Long Beach, CA, USA, 15–20 June 2019. [Google Scholar] [CrossRef]
- Yu, W.-H.; Li, C.-H.; Wang, R.-C.; Yeh, C.-Y.; Chuang, S.-S. Machine learning based on morphological features enables classification of primary intestinal T-cell lymphomas. Cancers 2021, 13, 5463. [Google Scholar] [CrossRef] [PubMed]
- Chuang, W.-Y.; Yu, W.-H.; Lee, Y.-C.; Zhang, Q.-Y.; Chang, H.; Shih, L.-Y.; Yeh, C.-J.; Lin, S.M.-T.; Chang, S.-H.; Ueng, S.-H.; et al. Deep learning-based nuclear morphometry reveals an independent prognostic factor in mantle cell lymphoma. Am. J. Pathol. 2022, 192, 1763–1778. [Google Scholar] [CrossRef] [PubMed]
- Pei, L.; Jones, K.A.; Shboul, Z.A.; Chen, J.Y.; Iftekharuddin, K.M. Deep neural network analysis of pathology images with integrated molecular data for enhanced glioma classification and grading. Front. Oncol. 2021, 11, 668694. [Google Scholar] [CrossRef] [PubMed]
- Reza, S.M.S.; Iftekharuddin, K.M. Glioma grading using cell nuclei morphologic features in digital pathology images. In Proceedings of the SPIE Conference on Medical Imaging, San Diego, CA, USA, 24 March 2016. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016. [Google Scholar] [CrossRef]
- Su, F.; Cheng, Y.; Chang, L.; Wang, L.; Huang, G.; Yuan, P.; Zhang, C.; Ma, Y. Annotation-free glioma grading from pathological images using ensemble deep learning. Heliyon 2023, 9, e14654. [Google Scholar] [CrossRef]
- Rathore, S.; Niazi, T.; Iftikhar, M.A.; Chaddad, A. Glioma grading via analysis of digital pathology images using machine learning. Cancers 2020, 12, 578. [Google Scholar] [CrossRef] [PubMed]
- Jose, L.; Liu, S.; Russo, C.; Cong, C.; Song, Y.; Rodriguez, M.; Ieva, A.D. Artificial intelligence–assisted classification of gliomas using whole slide images. Arch. Pathol. Lab. Med. 2023, 147, 916–924. [Google Scholar] [CrossRef] [PubMed]
- Im, S.; Hyeon, J.; Rha, E.; Lee, J.; Choi, H.; Jung, Y.; Kim, T. Classification of diffuse glioma subtype from clinical-grade pathological images using deep transfer learning. Sensors 2021, 21, 3500. [Google Scholar] [CrossRef] [PubMed]
- Efron, B.; DiCiccio, T. Bootstrap confidence intervals. Stat. Sci. 1996, 11, 189–228. [Google Scholar]
- Kim, H.-Y. Statistical notes for clinical researchers: The independent samples t-test. Restor. Dent. Endod. 2019, 44, e26. [Google Scholar] [CrossRef]
- Doane, D.P.; Seward, L.E. Measuring skewness: A forgotten statistic? J. Stat. Educ. 2011, 9, 1–18. [Google Scholar] [CrossRef]
- Groeneveld, R.A.; Meeden, G. Measuring skewness and kurtosis. J. R. Stat. Soc. Ser. D Stat. 1984, 33, 391–399. [Google Scholar] [CrossRef]
- Kim, T.K. Understanding one-way ANOVA using conceptual figures. Korean J. Anesthesiol. 2017, 70, 22–26. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Guestrin, C. XGBoost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016. [Google Scholar] [CrossRef]
- Ciga, O.; Xu, T.; Nofech-Mozes, S.; Noy, S.; Lu, F.-I.; Martel, A.L. Overcoming the limitations of patch-based learning to detect cancer in whole slide images. Sci. Rep. 2021, 11, 8894. [Google Scholar] [CrossRef]
- Li, W.; Nguyen, V.D.; Liao, H.; Wilder, M.; Cheng, K.; Luo, J. Patch transformer for multi-tagging whole slide histopathology images. In Proceedings of the 22nd International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI), Shenzhen, China, 13–17 October 2019. [Google Scholar]
- Hans, P.; Wouter, B.; Jeroen, V.D.L.; Geert, L. Detection of prostate cancer in whole-slide images through end-to-end training with image-level labels. IEEE Trans. Med. Imaging 2021, 40, 1817–1826. [Google Scholar]
- Cheung, E.Y.W.; Wu, R.W.K.; Li, A.S.M.; Chu, E.S.M. AI deployment on GBM diagnosis: A novel approach to analyze histopathological images using image feature-based analysis. Cancers 2023, 15, 5063. [Google Scholar] [CrossRef]
- Nahm, F.S. Receiver operating characteristic curve: Overview and practical use for clinicians. Korean J. Anesthesiol. 2022, 75, 25–36. [Google Scholar] [CrossRef]
- Unal, I. Defining an optimal cut-point value in ROC analysis: An alternative approach. Comput. Math. Methods Med. 2017, 2017, 3762651. [Google Scholar] [CrossRef]
- Boateng, E.Y.; Abaye, D.A. A review of the logistic regression model with emphasis on medical research. J. Data Anal. Inf. Process. 2019, 7, 190–207. [Google Scholar] [CrossRef]
- Lee, W.; Lam, S.-K.; Zhang, Y.; Yang, R.; Cai, J. Review of methodological workflow, interpretation and limitations of nomogram application in cancer study. Radiat. Med. Prot. 2022, 3, 200–207. [Google Scholar] [CrossRef]

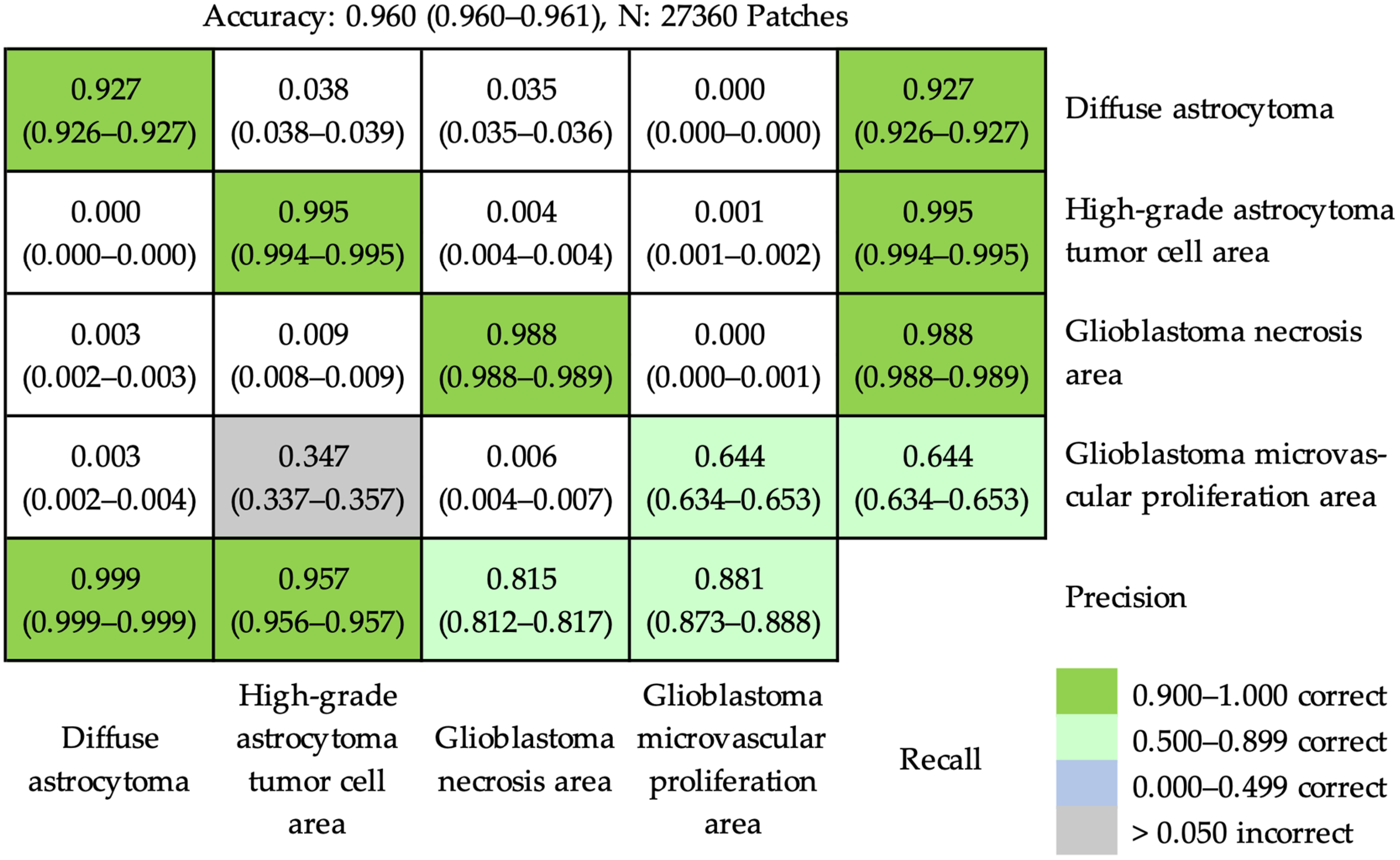
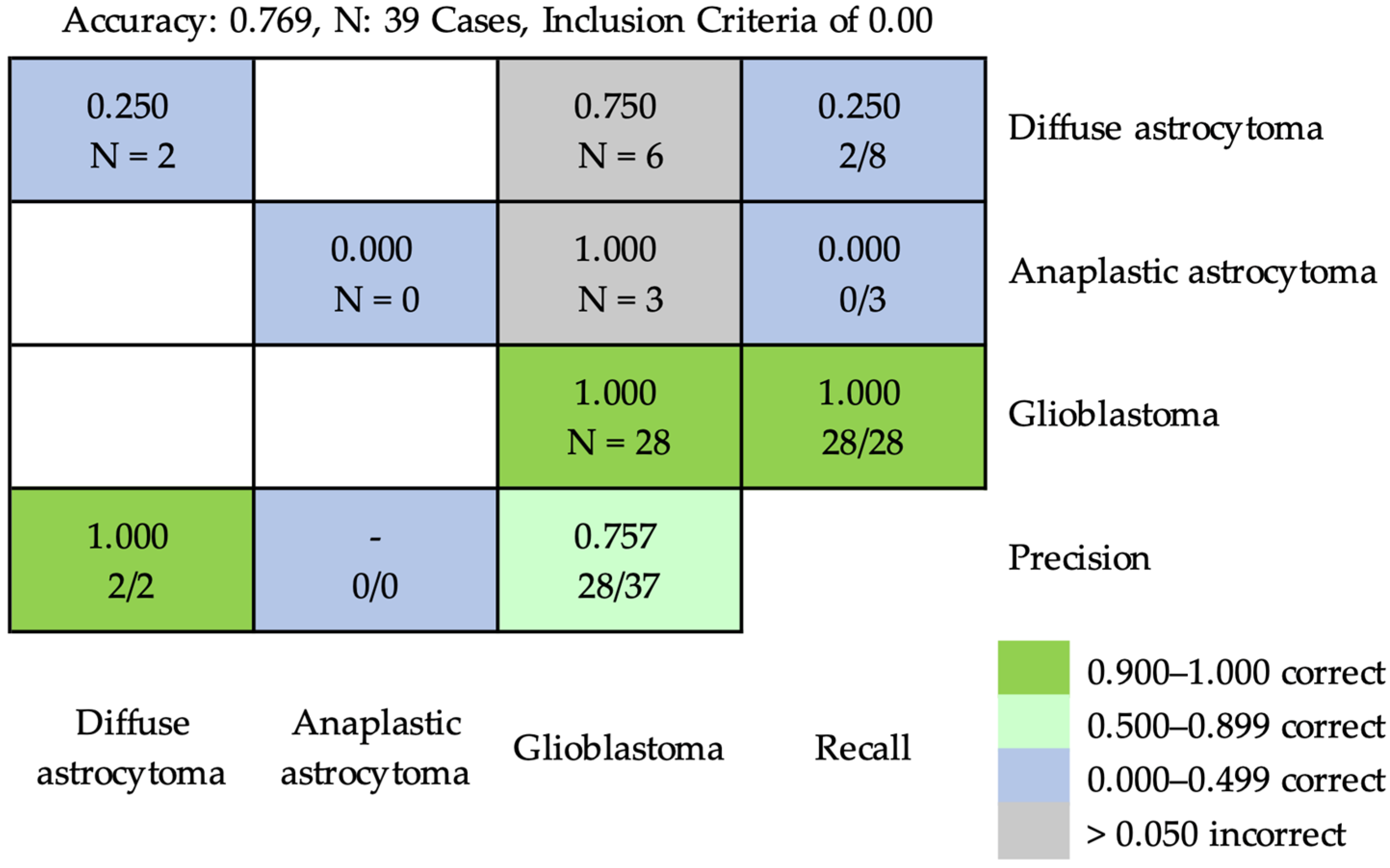


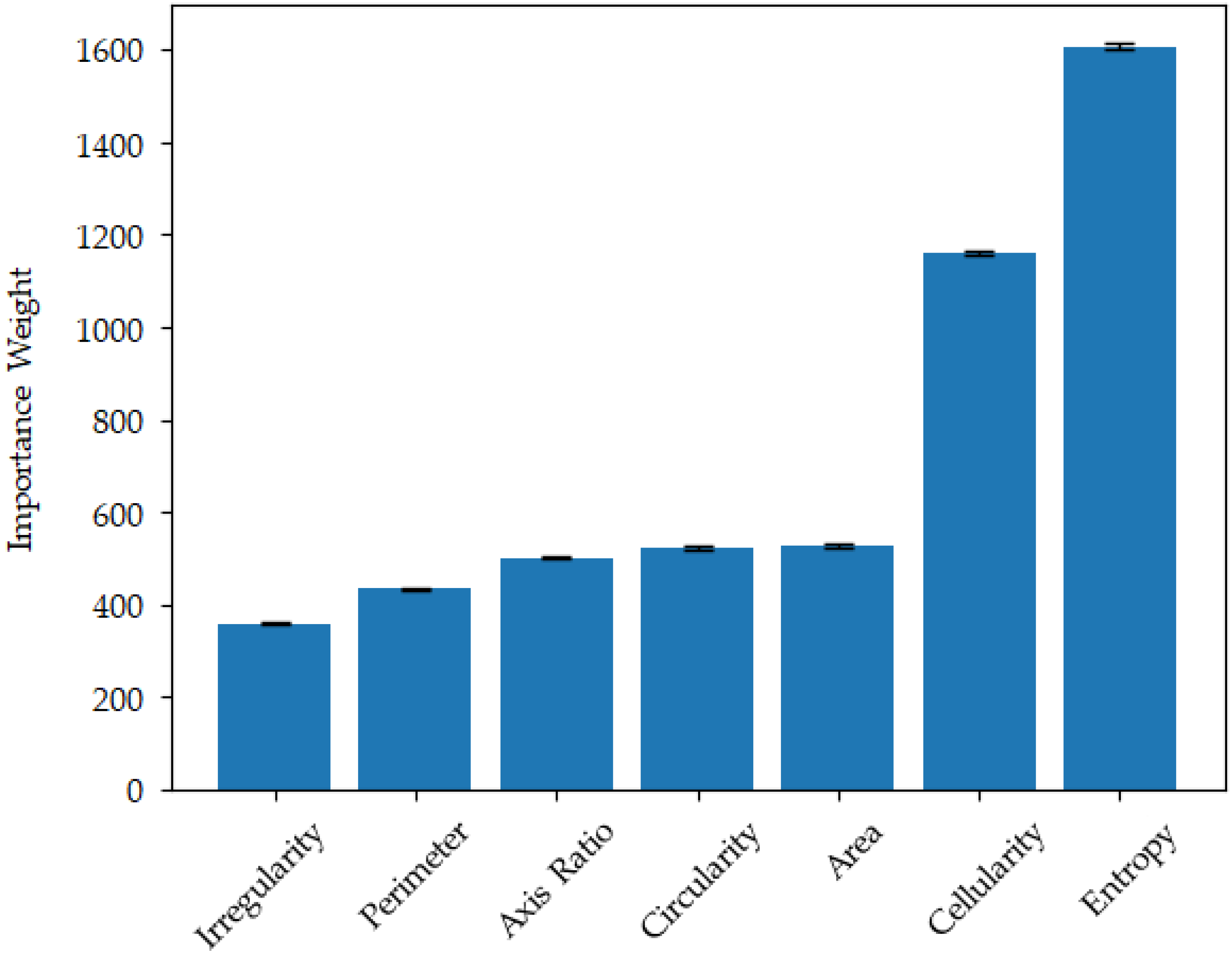
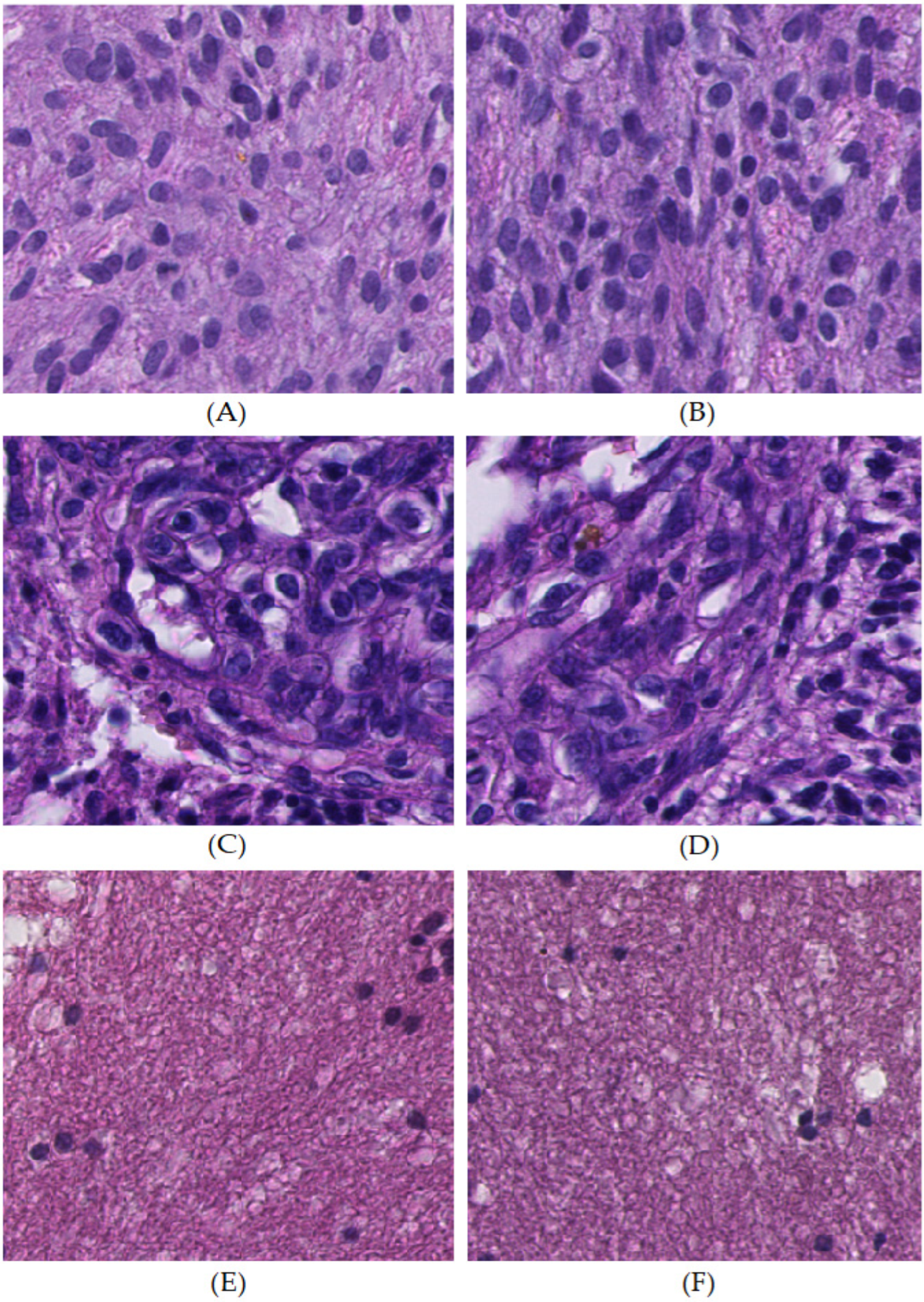
| Category | Training | Validation | Testing | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Cases | ROIs | Patches | Cases | ROIs | Patches | Cases | ROIs | Patches | |
| Diffuse astrocytoma | 4 | 729 | 15,176 | 3 | 212 | 1877 | 8 | 833 | 12,316 |
| Anaplastic astrocytoma | 6 | 1487 | 3551 | 2 | 177 | 401 | 3 | 256 | 1014 |
| Glioblastoma tumor cell area | 2207 | 7382 | 515 | 913 | 1521 | 11,653 | |||
| Glioblastoma necrosis area | 29 | 657 | 2357 | 12 | 1072 | 2088 | 28 | 460 | 2145 |
| Glioblastoma microvascular proliferation area | 423 | 465 | 114 | 126 | 257 | 232 | |||
| Total | 39 | 5503 | 28,931 | 17 | 2090 | 5405 | 39 | 3327 | 27,360 |
| Testing Case No. | Diagnosis | Characteristic Morphological Features | Prediction | Inclusion Criterion of 0.00 | Inclusion Criterion of 0.02 | Inclusion Criterion of 0.05 | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Count | Ratio | Ratio | Classification | Ratio | Classification | Ratio | Classification | |||
| Case 13 | Diffuse astrocytoma | Diffuse astrocytoma | 5025 | 0.998 | 0.998 | Glioblastoma | 0.998 | Diffuse astrocytoma | 0.998 | Diffuse astrocytoma |
| Anaplastic astrocytoma | 3 | 0.001 | 0.001 | |||||||
| Glioblastoma tumor cell area | ||||||||||
| Glioblastoma necrosis area | 9 | 0.002 | 0.002 | |||||||
| Glioblastoma microvascular proliferation area | ||||||||||
| Case 14 | Diffuse astrocytoma | Diffuse astrocytoma | 216 | 0.722 | 0.722 | Glioblastoma | 0.722 | Glioblastoma | 0.722 | Glioblastoma |
| Anaplastic astrocytoma | ||||||||||
| Glioblastoma tumor cell area | 22 | 0.074 | 0.074 | 0.074 | 0.074 | |||||
| Glioblastoma necrosis area | 60 | 0.201 | 0.201 | 0.201 | 0.201 | |||||
| Glioblastoma microvascular proliferation area | 1 | 0.003 | 0.003 | |||||||
| Case 15 | Anaplastic astrocytoma | Diffuse astrocytoma | Glioblastoma | Glioblastoma | Glioblastoma | |||||
| Anaplastic astrocytoma | ||||||||||
| Glioblastoma tumor cell area | 146 | 1.000 | 1.000 | 1.000 | 1.000 | |||||
| Glioblastoma necrosis area | ||||||||||
| Glioblastoma microvascular proliferation area | ||||||||||
| Case 17 | Diffuse astrocytoma | Diffuse astrocytoma | 5280 | 0.998 | 0.998 | Glioblastoma | 0.998 | Diffuse astrocytoma | 0.998 | Diffuse astrocytoma |
| Anaplastic astrocytoma | ||||||||||
| Glioblastoma tumor cell area | ||||||||||
| Glioblastoma necrosis area | 9 | 0.002 | 0.002 | |||||||
| Glioblastoma microvascular proliferation area | ||||||||||
| Case 22 | Diffuse astrocytoma | Diffuse astrocytoma | 530 | 0.994 | 0.994 | Glioblastoma | 0.994 | Diffuse astrocytoma | 0.994 | Diffuse astrocytoma |
| Anaplastic astrocytoma | ||||||||||
| Glioblastoma tumor cell area | ||||||||||
| Glioblastoma necrosis area | 1 | 0.002 | 0.002 | |||||||
| Glioblastoma microvascular proliferation area | 2 | 0.004 | 0.004 | |||||||
| Case 25 | Diffuse astrocytoma | Diffuse astrocytoma | 1 | 0.002 | 0.002 | Glioblastoma | Glioblastoma | Anaplastic astrocytoma | ||
| Anaplastic astrocytoma | 449 | 0.943 | 0.943 | 0.943 | 0.943 | |||||
| Glioblastoma tumor cell area | 16 | 0.034 | 0.034 | 0.034 | ||||||
| Glioblastoma necrosis area | 8 | 0.017 | 0.017 | |||||||
| Glioblastoma microvascular proliferation area | 2 | 0.004 | 0.004 | |||||||
| Case 27 | Anaplastic astrocytoma | Diffuse astrocytoma | Glioblastoma | Glioblastoma | Glioblastoma | |||||
| Anaplastic astrocytoma | 28 | 0.113 | 0.113 | 0.113 | 0.113 | |||||
| Glioblastoma tumor cell area | 220 | 0.887 | 0.887 | 0.887 | 0.887 | |||||
| Glioblastoma necrosis area | ||||||||||
| Glioblastoma microvascular proliferation area | ||||||||||
| Case 29 | Anaplastic astrocytoma | Diffuse astrocytoma | Glioblastoma | Glioblastoma | Glioblastoma | |||||
| Anaplastic astrocytoma | 296 | 0.477 | 0.477 | 0.477 | 0.477 | |||||
| Glioblastoma tumor cell area | 323 | 0.521 | 0.521 | 0.521 | 0.521 | |||||
| Glioblastoma necrosis area | ||||||||||
| Glioblastoma microvascular proliferation area | 1 | 0.002 | 0.002 | |||||||
| Case 38 | Diffuse astrocytoma | Diffuse astrocytoma | 6 | 0.016 | 0.016 | Glioblastoma | Glioblastoma | Glioblastoma | ||
| Anaplastic astrocytoma | ||||||||||
| Glioblastoma tumor cell area | ||||||||||
| Glioblastoma necrosis area | 367 | 0.984 | 0.984 | 0.984 | 0.984 | |||||
| Glioblastoma microvascular proliferation area | ||||||||||
| Category | ROIs | Cellularity |
|---|---|---|
| Diffuse astrocytoma | 1774 | 0.052 ± 0.018 |
| Anaplastic astrocytoma | 1915 | 0.180 ± 0.063 |
| Glioblastoma tumor cell area | 4238 | 0.195 ± 0.051 |
| Glioblastoma necrosis area | 2184 | 0.003 ± 0.008 |
| Glioblastoma microvascular proliferation area | 794 | 0.122 ± 0.052 |
| Diffuse Astrocytoma | Anaplastic Astrocytoma | Glioblastoma Tumor Cell Area | Glioblastoma Necrosis Area | Glioblastoma Microvascular Proliferation Area | |
|---|---|---|---|---|---|
| Diffuse astrocytoma | <0.001 * | <0.001 * | <0.001 * | <0.001 * | |
| Anaplastic astrocytoma | <0.001 * | <0.001 * | <0.001 * | ||
| Glioblastoma tumor cell area | <0.001 * | <0.001 * | |||
| Glioblastoma necrosis area | <0.001 * | ||||
| Glioblastoma microvascular proliferation area |
| Attributes | Moments (Mean ± SD) | Diffuse Astrocytoma | Anaplastic Astrocytoma | Glioblastoma Tumor Cell Area | Glioblastoma Necrosis Area | Glioblastoma Microvascular Proliferation Area | F-Test | |
|---|---|---|---|---|---|---|---|---|
| N = 15 | N = 11 | N = 69 | F-Statistics | p Value | ||||
| Axis Ratio | Mean | 1.437 ± 0.103 | 1.540 ± 0.118 | 1.570 ± 0.123 | 1.504 ± 0.193 | 1.734 ± 0.144 | 20.377 | <0.001 * |
| Variance | 0.139 ± 0.066 | 0.192 ± 0.092 | 0.194 ± 0.101 | 0.169 ± 0.195 | 0.359 ± 0.182 | 12.489 | <0.001 * | |
| Skewness | 2.351 ± 0.285 | 2.113 ± 0.409 | 1.844 ± 0.332 | 1.704 ± 0.903 | 1.839 ± 0.614 | 10.38 | <0.001 * | |
| Kurtosis | 10.106 ± 2.979 | 9.264 ± 4.545 | 6.201 ± 2.959 | 5.264 ± 6.070 | 5.368 ± 3.936 | 21.733 | <0.001 * | |
| Circularity | Mean | 0.672 ± 0.037 | 0.634 ± 0.040 | 0.622 ± 0.038 | 0.641 ± 0.053 | 0.574 ± 0.035 | 25.248 | <0.001 * |
| Variance | 0.014 ± 0.003 | 0.015 ± 0.003 | 0.015 ± 0.003 | 0.013 ± 0.007 | 0.018 ± 0.003 | 10.478 | <0.001 * | |
| Skewness | −0.642 ± 0.153 | −0.422 ± 0.153 | −0.330 ± 0.163 | −0.423 ± 0.365 | −0.250 ± 0.223 | 11.425 | <0.001 * | |
| Kurtosis | 0.140 ± 0.308 | −0.235 ± 0.274 | −0.341 ± 0.248 | −0.282 ± 0.657 | −0.525 ± 0.307 | 11.417 | <0.001 * | |
| Entropy | Mean | 4.921 ± 0.198 | 4.745 ± 0.165 | 4.808 ± 0.187 | 4.925 ± 0.202 | 4.711 ± 0.227 | 7.833 | <0.001 * |
| Variance | 0.127 ± 0.035 | 0.131 ± 0.034 | 0.135 ± 0.035 | 0.138 ± 0.072 | 0.192 ± 0.059 | 9.655 | <0.001 * | |
| Skewness | −0.401 ± 0.187 | −0.398 ± 0.229 | −0.408 ± 0.290 | −0.474 ± 0.453 | −0.517 ± 0.348 | 0.806 | 0.546 | |
| Kurtosis | 0.993 ± 0.568 | 0.885 ± 0.568 | 0.900 ± 0.692 | 0.766 ± 1.745 | 0.866 ± 0.941 | 0.323 | 0.899 | |
| Area (μm2) | Mean | 21.311 ± 3.466 | 25.958 ± 4.901 | 30.546 ± 6.183 | 13.676 ± 3.536 | 25.345 ± 6.750 | 53.112 | <0.001 * |
| Variance | 83.678 ± 40.508 | 164.005 ± 78.404 | 231.174 ± 126.803 | 55.408 ± 37.289 | 185.253 ± 181.007 | 13.109 | <0.001 * | |
| Skewness | 1.460 ± 0.377 | 1.588 ± 0.355 | 1.471 ± 0.511 | 1.575 ± 1.037 | 1.362 ± 0.567 | 1.639 | 0.151 | |
| Kurtosis | 6.493 ± 7.097 | 4.493 ± 2.177 | 4.257 ± 5.202 | 6.362 ± 14.965 | 3.129 ± 3.774 | 1.153 | 0.334 | |
| Irregularity | Mean | 3.339 ± 0.948 | 5.342 ± 1.327 | 6.783 ± 1.907 | 2.648 ± 1.115 | 7.784 ± 2.161 | 59.537 | <0.001 * |
| Variance | 17.540 ± 9.779 | 40.655 ± 15.941 | 61.880 ± 32.027 | 9.060 ± 9.498 | 90.134 ± 115.829 | 9.488 | <0.001 * | |
| Skewness | 4.279 ± 0.728 | 3.691 ± 0.865 | 3.200 ± 0.663 | 2.710 ± 1.650 | 2.747 ± 1.243 | 7.622 | <0.001 * | |
| Kurtosis | 33.584 ± 11.747 | 26.882 ± 14.452 | 18.175 ± 9.300 | 14.466 ± 17.311 | 13.132 ± 15.371 | 9.002 | <0.001 * | |
| Perimeter (μm) | Mean | 17.887 ± 1.390 | 19.942 ± 1.760 | 21.745 ± 2.075 | 14.327 ± 1.827 | 20.254 ± 2.39 | 79.245 | <0.001 * |
| Variance | 14.608 ± 5.064 | 24.255 ± 5.941 | 30.381 ± 8.932 | 13.589 ± 7.075 | 30.875 ± 13.953 | 24.395 | <0.001 * | |
| Skewness | 0.824 ± 0.205 | 1.014 ± 0.313 | 0.833 ± 0.300 | 0.827 ± 0.509 | 0.846 ± 0.405 | 1.413 | 0.221 | |
| Kurtosis | 2.137 ± 1.224 | 1.732 ± 0.952 | 1.190 ± 1.089 | 1.392 ± 3.147 | 1.062 ± 1.275 | 1.35 | 0.245 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, Y.-C.; Lin, S.-Z.; Wu, J.-R.; Yu, W.-H.; Harn, H.-J.; Tsai, W.-C.; Liu, C.-A.; Kuo, K.-L.; Yeh, C.-Y.; Tsai, S.-T. Deep Residual Learning-Based Classification with Identification of Incorrect Predictions and Quantification of Cellularity and Nuclear Morphological Features in Digital Pathological Images of Common Astrocytic Tumors. Cancers 2024, 16, 2449. https://doi.org/10.3390/cancers16132449
Chen Y-C, Lin S-Z, Wu J-R, Yu W-H, Harn H-J, Tsai W-C, Liu C-A, Kuo K-L, Yeh C-Y, Tsai S-T. Deep Residual Learning-Based Classification with Identification of Incorrect Predictions and Quantification of Cellularity and Nuclear Morphological Features in Digital Pathological Images of Common Astrocytic Tumors. Cancers. 2024; 16(13):2449. https://doi.org/10.3390/cancers16132449
Chicago/Turabian StyleChen, Yen-Chang, Shinn-Zong Lin, Jia-Ru Wu, Wei-Hsiang Yu, Horng-Jyh Harn, Wen-Chiuan Tsai, Ching-Ann Liu, Ken-Leiang Kuo, Chao-Yuan Yeh, and Sheng-Tzung Tsai. 2024. "Deep Residual Learning-Based Classification with Identification of Incorrect Predictions and Quantification of Cellularity and Nuclear Morphological Features in Digital Pathological Images of Common Astrocytic Tumors" Cancers 16, no. 13: 2449. https://doi.org/10.3390/cancers16132449
APA StyleChen, Y.-C., Lin, S.-Z., Wu, J.-R., Yu, W.-H., Harn, H.-J., Tsai, W.-C., Liu, C.-A., Kuo, K.-L., Yeh, C.-Y., & Tsai, S.-T. (2024). Deep Residual Learning-Based Classification with Identification of Incorrect Predictions and Quantification of Cellularity and Nuclear Morphological Features in Digital Pathological Images of Common Astrocytic Tumors. Cancers, 16(13), 2449. https://doi.org/10.3390/cancers16132449






