Discovering Potential in Non-Cancer Medications: A Promising Breakthrough for Multiple Myeloma Patients
Abstract
Simple Summary
Abstract
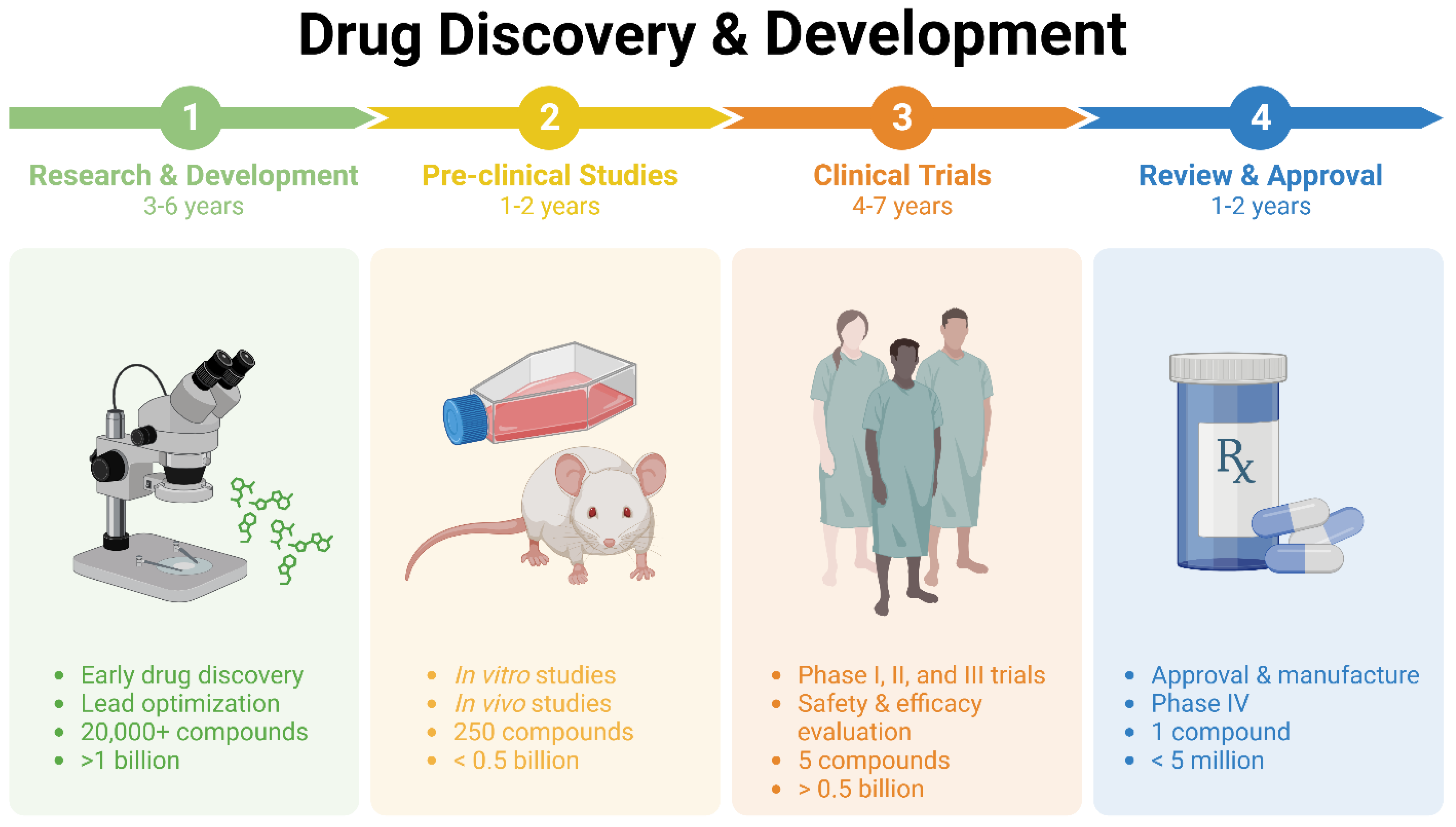
1. The Significance of Repurposing
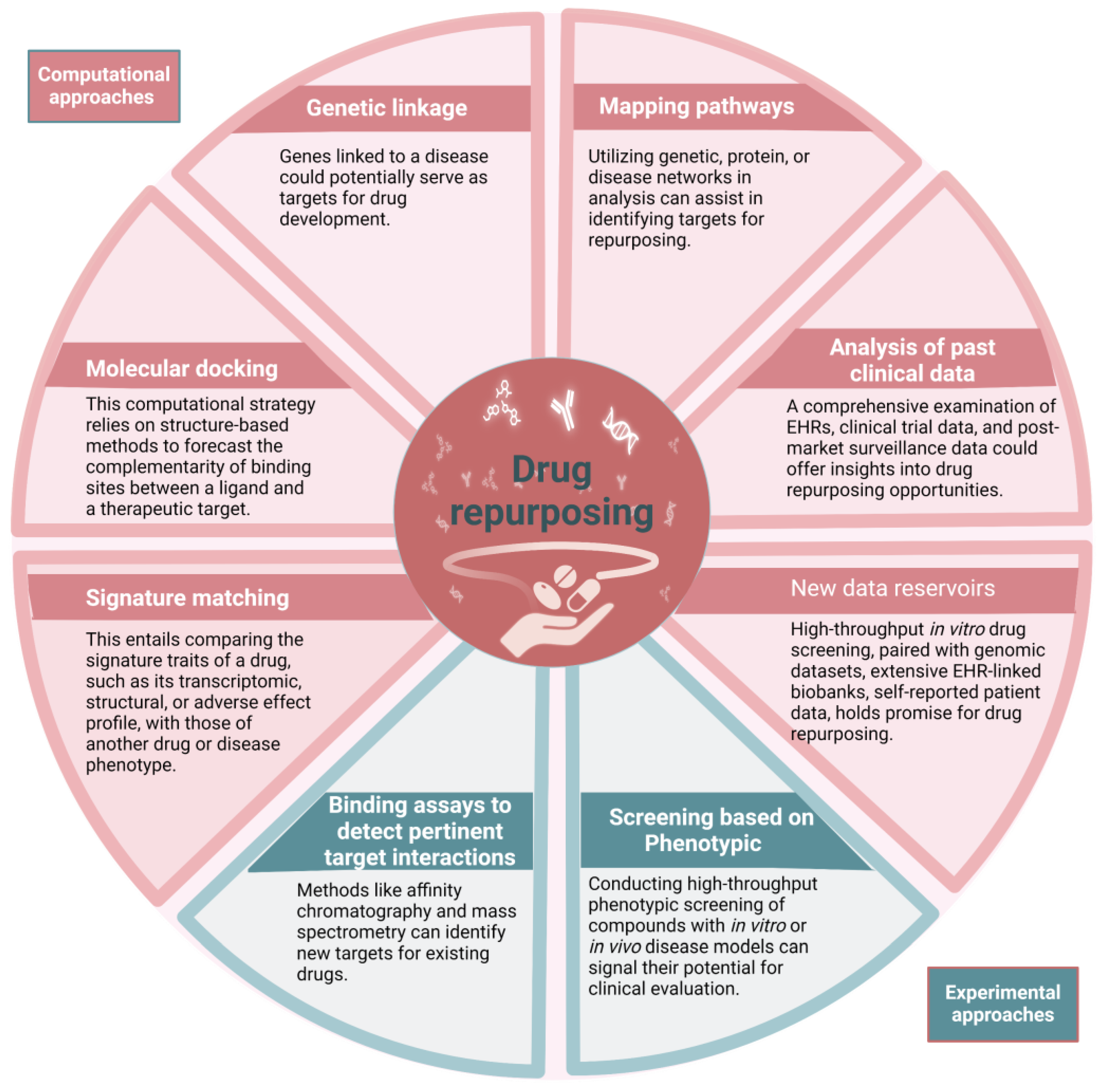
2. Pharmacological Repurposing Strategies and Tools
3. Medicines That Could Be Repurposed to Treat MM
3.1. Thalidomide
3.2. Statins
3.3. Celecoxib
3.4. Aspirin
3.5. Clarithromycin
3.6. Rapamycin
3.7. Valproic Acid
3.8. Nelfinavir
3.9. Metformin
3.10. Bisphosphonates
3.11. CuET
3.12. Albendazole
4. Conclusions and Future Prospectives
5. Practice Points
- Although there have been numerous clinical trials conducted to evaluate different approaches for treating cancer, the 5-year survival rate for individuals with MM in the US remains at a modest 55%.
- Myeloma remains a challenging malignancy to treat due to the development of drug resistance, resulting in relapse for all patients.
- There is an ongoing demand for new medications. However, the process of finding a new treatment can often be quite time-consuming. Therefore, repurposing already approved non-cancer medication for MM can aid in the discovery of new effective drugs.
- The potential for repurposing approved drugs is promising, although a thorough analysis of these agents is necessary before they can be considered for clinical trials.
6. Research Agenda
- The potential of various non-anti-cancer drugs as an anti-myeloma treatment was discussed.
- Thalidomide stands out as an exemplary repurposed agent for treating MM.
- There is encouraging evidence that statins, rapamycin, clarithromycin, and leflunomide can inhibit MM.
- Extensive animal studies using the MM animal model, along with phase 1 clinical studies, are necessary to thoroughly investigate these agents as potential MM therapies.
Author Contributions
Funding
Conflicts of Interest
References
- Yang, J.; Liu, X.; Zhong, Q.-Z.; Yang, Y.; Wu, T.; Chen, S.-Y.; Chen, B.; Song, Y.-W.; Fang, H.; Wang, S.-L.; et al. Disparities in mortality risk after diagnosis of hematological malignancies in 185 countries: A global data analysis. Cancer Lett. 2024, 595, 216793. [Google Scholar] [CrossRef] [PubMed]
- Ashburn, T.T.; Thor, K.B. Drug repositioning: Identifying and developing new uses for existing drugs. Nat. Rev. Drug Discov. 2004, 3, 673–683. [Google Scholar] [CrossRef] [PubMed]
- Scannell, J.W.; Blanckley, A.; Boldon, H.; Warrington, B. Diagnosing the decline in pharmaceutical R&D efficiency. Nat. Rev. Drug Discov. 2012, 11, 191–200. [Google Scholar] [PubMed]
- Pammolli, F.; Magazzini, L.; Riccaboni, M. The productivity crisis in pharmaceutical R&D. Nat. Rev. Drug Discov. 2011, 10, 428–438. [Google Scholar] [PubMed]
- Waring, M.J.; Arrowsmith, J.; Leach, A.R.; Leeson, P.D.; Mandrell, S.; Owen, R.M.; Pairaudeau, G.; Pennie, W.D.; Pickett, S.D.; Wang, J.; et al. An analysis of the attrition of drug candidates from four major pharmaceutical companies. Nat. Rev. Drug Discov. 2015, 14, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Beachy, S.H.; Johnson, S.G.; Olson, S.; Berger, A.C. Drug Repurposing and Repositioning: Workshop Summary; National Academies Press: Washington, DC, USA, 2014. [Google Scholar]
- DiMasi, J.A.; Grabowski, H.G.; Hansen, R.W. Innovation in the pharmaceutical industry: New estimates of R&D costs. J. Health Econ. 2016, 47, 20–33. [Google Scholar] [PubMed]
- Paul, S.M.; Mytelka, D.S.; Dunwiddie, C.T.; Persinger, C.C.; Munos, B.H.; Lindborg, S.R.; Schacht, A.L. How to improve R&D productivity: The pharmaceutical industry’s grand challenge. Nat. Rev. Drug Discov. 2010, 9, 203–214. [Google Scholar] [PubMed]
- Papapetropoulos, A.; Szabo, C. Inventing new therapies without reinventing the wheel: The power of drug repurposing. Br. J. Pharmacol. 2018, 175, 165–167. [Google Scholar] [CrossRef]
- Hernandez, J.J.; Pryszlak, M.; Smith, L.; Yanchus, C.; Kurji, N.; Shahani, V.M.; Molinski, S.V. Giving drugs a second chance: Overcoming regulatory and financial hurdles in repurposing approved drugs as cancer therapeutics. Front. Oncol. 2017, 7, 273. [Google Scholar] [CrossRef] [PubMed]
- Nosengo, N. Can you teach old drugs new tricks? Nature 2016, 534, 314–316. [Google Scholar] [CrossRef] [PubMed]
- Ishida, J.; Konishi, M.; Ebner, N.; Springer, J. Repurposing of approved cardiovascular drugs. J. Transl. Med. 2016, 14, 269. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Lu, W.; Liu, C.; Fang, J.; Hou, Y.; Handy, D.E.; Wang, R.; Zhao, Y.; Yang, Y.; Huang, J.; et al. A genome-wide positioning systems network algorithm for in silico drug repurposing. Nat. Commun. 2019, 10, 3476. [Google Scholar] [CrossRef] [PubMed]
- Rudrapal, M.; Khairnar, S.J.; Jadhav, A.G. Drug repurposing (DR): An emerging approach in drug discovery. In Drug Repurposing-Hypothesis, Molecular Aspects and Therapeutic Applications; IntechOpe: London, UK, 2020; Volume 10. [Google Scholar]
- Rajkumar, S.V.; Kumar, S. Multiple myeloma current treatment algorithms. Blood Cancer J. 2020, 10, 94. [Google Scholar] [CrossRef] [PubMed]
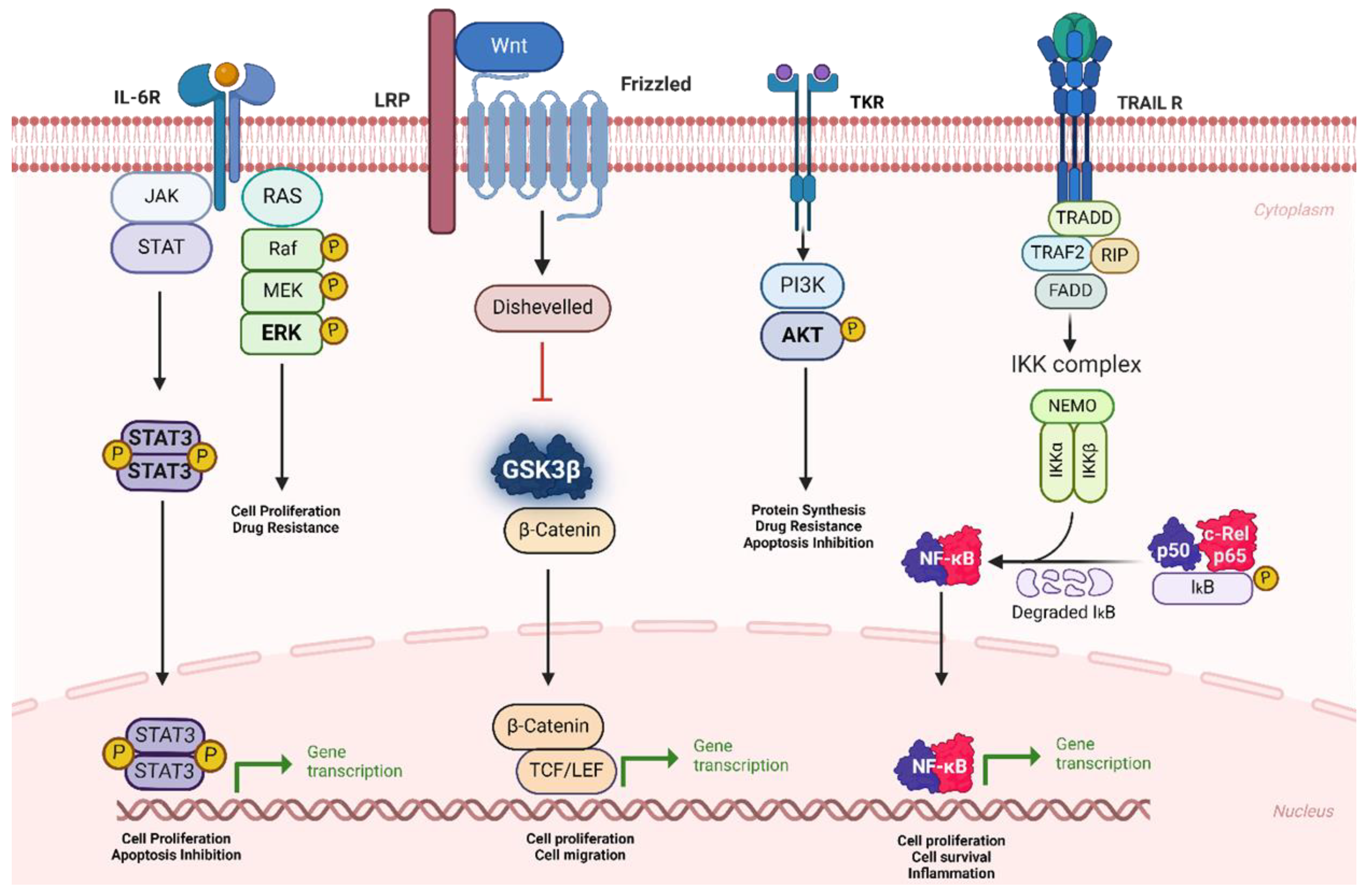
- Dehghanifard, A.; Kaviani, S.; Abroun, S.; Mehdizadeh, M.; Saiedi, S.; Maali, A.; Ghaffari, S.; Azad, M. Various signaling pathways in multiple myeloma cells and effects of treatment on these pathways. Clin. Lymphoma Myeloma Leuk. 2018, 18, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Al-Odat, O.S.; von Suskil, M.; Chitren, R.J.; Elbezanti, W.O.; Srivastava, S.K.; Budak-Alpddogan, T.; Jonnalagadda, S.C.; Aggarwal, B.B.; Pandey, M. Mcl-1 inhibition: Managing malignancy in multiple myeloma. Front. Pharmacol. 2021, 12, 699629. [Google Scholar] [CrossRef] [PubMed]
- Hurle, M.R.; Yang, L.; Xie, Q.; Rajpal, D.K.; Sanseau, P.; Agarwal, P. Computational drug repositioning: From data to therapeutics. Clin. Pharmacol. Ther. 2013, 93, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Keiser, M.J.; Setola, V.; Irwin, J.J.; Laggner, C.; Abbas, A.I.; Hufeisen, S.J.; Jensen, N.H.; Kuijer, M.B.; Matos, R.C.; Tran, T.B.; et al. Predicting new molecular targets for known drugs. Nature 2009, 462, 175–181. [Google Scholar] [CrossRef]
- Hieronymus, H.; Lamb, J.; Ross, K.N.; Peng, X.P.; Clement, C.; Rodina, A.; Nieto, M.; Du, J.; Stegmaier, K.; Raj, S.M.; et al. Gene expression signature-based chemical genomic prediction identifies a novel class of HSP90 pathway modulators. Cancer Cell 2006, 10, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Desai, R.J.; Handy, D.E.; Wang, R.; Schneeweiss, S.; Barabási, A.L.; Loscalzo, J. Network-based approach to prediction and population-based validation of in silico drug repurposing. Nat. Commun. 2018, 9, 2691. [Google Scholar] [CrossRef]
- Nam, Y.; Kim, M.; Chang, H.S.; Shin, H. Drug repurposing with network reinforcement. BMC Bioinform. 2019, 20, 383. [Google Scholar] [CrossRef]
- Zickenrott, S.; Angarica, V.E.; Upadhyaya, B.B.; Del Sol, A. Prediction of disease–gene–drug relationships following a differential network analysis. Cell Death Dis. 2016, 7, e2040. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Agarwal, P. Human disease-drug network based on genomic expression profiles. PLoS ONE 2009, 4, e6536. [Google Scholar] [CrossRef] [PubMed]
- Peyvandipour, A.; Saberian, N.; Shafi, A.; Donato, M.; Draghici, S. A novel computational approach for drug repurposing using systems biology. Bioinformatics 2018, 34, 2817–2825. [Google Scholar] [CrossRef] [PubMed]
- Naylor, D.M.; Kauppi, D.; Schonfeld, J. Therapeutic drug repurposing, repositioning and rescue. Drug Discov. 2015, 57, 1–16. [Google Scholar]
- Chatr-Aryamontri, A.; Breitkreutz, B.-J.; Heinicke, S.; Boucher, L.; Winter, A.; Stark, C.; Nixon, J.; Ramage, L.; Kolas, N.; O’Donnell, L.; et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2012, 41, D816–D823. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P. STRING v10: Protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.Y.; Pandey, R.; Nguyen, T.M. HAPPI-2: A comprehensive and high-quality map of human annotated and predicted protein interactions. BMC Genom. 2017, 18, 182. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef] [PubMed]
- Croft, D.; O’Kelly, G.; Wu, G.; Haw, R.; Gillespie, M.; Matthews, L.; Caudy, M.; Garapati, P.; Gopinath, G.; Jassal, B.; et al. Reactome: A database of reactions, pathways and biological processes. Nucleic Acids Res. 2011, 39, D691–D697. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jensen, M.; Zenklusen, J. A Practical Guide to the Cancer Genome Atlas (TCGA). Statistical Genomics. Methods Protoc. 2016, 1418, 111–141. [Google Scholar]
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Tissue-based map of the human proteome. Science 2015, 347, 1260419. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Oksvold, P.; Fagerberg, L.; Lundberg, E.; Jonasson, K.; Forsberg, M.; Zwahlen, M.; Kampf, C.; Wester, K.; Hober, S.; et al. Towards a knowledge-based human protein atlas. Nat. Biotechnol. 2010, 28, 1248–1250. [Google Scholar] [CrossRef]
- Uhlén, M.; Björling, E.; Agaton, C.; Szigyarto, C.A.-K.; Amini, B.; Andersen, E.; Andersson, A.-C.; Angelidou, P.; Asplund, A.; Asplund, C.; et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol. Cell. Proteom. 2005, 4, 1920–1932. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J.; Crawford, E.D.; Peck, D.; Modell, J.W.; Blat, I.C.; Wrobel, M.J.; Lerner, J.; Brunet, J.-P.; Subramanian, A.; Ross, K.N.; et al. The Connectivity Map: Using gene-expression signatures to connect small molecules, genes, and disease. Science 2006, 313, 1929–1935. [Google Scholar] [CrossRef] [PubMed]
- Lamb, J. The Connectivity Map: A new tool for biomedical research. Nat. Rev. Cancer 2007, 7, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Borate, B.; Baxevanis, A.D. Searching Online Mendelian Inheritance in Man (OMIM) for information on genetic loci involved in human disease. Curr. Protoc. Bioinform. 2009, 27, 1.2.1–1.2.13. [Google Scholar] [CrossRef] [PubMed]
- Goh, K.-I.; Cusick, M.E.; Valle, D.; Childs, B.; Vidal, M.; Barabási, A.-L. The human disease network. Proc. Natl. Acad. Sci. USA 2007, 104, 8685–8690. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wu, S.-J.; Yang, S.-Y.; Peng, J.-W.; Wang, S.-N.; Wang, F.-Y.; Song, Y.-X.; Qi, T.; Li, Y.-X.; Li, Y.-Y. DNetDB: The human disease network database based on dysfunctional regulation mechanism. BMC Syst. Biol. 2016, 10, 36. [Google Scholar] [CrossRef] [PubMed]
- Law, V.; Knox, C.; Djoumbou, Y.; Jewison, T.; Guo, A.C.; Liu, Y.; Maciejewski, A.; Arndt, D.; Wilson, M.; Neveu, V.; et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014, 42, D1091–D1097. [Google Scholar] [CrossRef] [PubMed]
- Andersson, M.L.; Böttiger, Y.; Bastholm-Rahmner, P.; Ovesjö, M.-L.; Veg, A.; Eiermann, B. Evaluation of usage patterns and user perception of the drug–drug interaction database SFINX. Int. J. Med. Inform. 2015, 84, 327–333. [Google Scholar] [CrossRef] [PubMed]
- Kale, V.P.; Habib, H.; Chitren, R.; Patel, M.; Pramanik, K.C.; Jonnalagadda, S.C.; Challagundla, K.; Pandey, M.K. Old drugs, new uses: Drug repurposing in hematological malignancies. Semin. Cancer Biol. 2021, 68, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Letunic, I.; Jensen, L.J.; Bork, P. The SIDER database of drugs and side effects. Nucleic Acids Res. 2016, 44, D1075–D1079. [Google Scholar] [CrossRef] [PubMed]
- Siramshetty, V.B.; Nickel, J.; Omieczynski, C.; Gohlke, B.-O.; Drwal, M.N.; Preissner, R. WITHDRAWN—A resource for withdrawn and discontinued drugs. Nucleic Acids Res. 2016, 44, D1080–D1086. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M.; Szklarczyk, D.; Franceschini, A.; Von Mering, C.; Jensen, L.J.; Bork, P. STITCH 3: Zooming in on protein–chemical interactions. Nucleic Acids Res. 2012, 40, D876–D880. [Google Scholar] [CrossRef] [PubMed]
- Shameer, K.; Glicksberg, B.S.; Hodos, R.; Johnson, K.W.; Badgeley, M.A.; Readhead, B.; Tomlinson, M.S.; O’Connor, T.; Miotto, R.; Kidd, B.A.; et al. Systematic analyses of drugs and disease indications in RepurposeDB reveal pharmacological, biological and epidemiological factors influencing drug repositioning. Brief. Bioinform. 2018, 19, 656–678. [Google Scholar] [CrossRef] [PubMed]
- Garnett, M.J.; Edelman, E.J.; Heidorn, S.J.; Greenman, C.D.; Dastur, A.; Lau, K.W.; Greninger, P.; Thompson, I.R.; Luo, X.; Soares, J.; et al. Systematic identification of genomic markers of drug sensitivity in cancer cells. Nature 2012, 483, 570–575. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.K.B.; Tiong, K.H.; Chang, J.K.; Liew, C.S.; Abdul Rahman, Z.A.; Tan, A.C.; Khang, T.F.; Cheong, S.C. DeSigN: Connecting gene expression with therapeutics for drug repurposing and development. BMC Genom. 2017, 18, 934. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.; Mehta, J.; Desikan, R.; Ayers, D.; Roberson, P.; Eddlemon, P.; Munshi, N.; Anaissie, E.; Wilson, C.; Dhodapkar, M.; et al. Antitumor activity of thalidomide in refractory multiple myeloma. N. Engl. J. Med. 1999, 341, 1565–1571. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, S.V.; Blood, E.; Vesole, D.; Fonseca, R.; Greipp, P.R. Phase III clinical trial of thalidomide plus dexamethasone compared with dexamethasone alone in newly diagnosed multiple myeloma: A clinical trial coordinated by the Eastern Cooperative Oncology Group. J. Clin. Oncol. 2006, 24, 431–436. [Google Scholar] [CrossRef] [PubMed]
- Afzal, A.; Fiala, M.A.; Gage, B.F.; Wildes, T.M.; Sanfilippo, K. Statins reduce mortality in multiple myeloma: A population-based US study. Clin. Lymphoma Myeloma Leuk. 2020, 20, e937–e943. [Google Scholar] [CrossRef] [PubMed]
- Brånvall, E.; Ekberg, S.; Eloranta, S.; Wästerlid, T.; Birmann, B.M.; Smedby, K.E. Statin use is associated with improved survival in multiple myeloma: A Swedish population-based study of 4315 patients. Am. J. Hematol. 2020, 95, 652–661. [Google Scholar] [CrossRef] [PubMed]
- Sanfilippo, K.M.; Keller, J.; Gage, B.F.; Luo, S.; Wang, T.-F.; Moskowitz, G.; Gumbel, J.; Blue, B.; O’Brian, K.; Carson, K.R. Statins are associated with reduced mortality in multiple myeloma. J. Clin. Oncol. 2016, 34, 4008–4014. [Google Scholar] [CrossRef] [PubMed]
- Epstein, M.M.; Divine, G.; Chao, C.R.; Wells, K.E.; Feigelson, H.S.; Scholes, D.; Roblin, D.; Ulcickas Yood, M.; Engel, L.S.; Taylor, A.; et al. Statin use and risk of multiple myeloma: An analysis from the cancer research network. Int. J. Cancer 2017, 141, 480–487. [Google Scholar] [CrossRef] [PubMed]
- Bernard, M.; Bancos, S.; Sime, P.; Phipps, R. Targeting cyclooxygenase-2 in hematological malignancies: Rationale and promise. Curr. Pharm. Des. 2008, 14, 2051–2060. [Google Scholar] [CrossRef] [PubMed]
- Tołoczko-Iwaniuk, N.; Dziemiańczyk-Pakieła, D.; Nowaszewska, B.K.; Celińska-Janowicz, K.; Miltyk, W. Celecoxib in cancer therapy and prevention—Review. Curr. Drug Targets 2019, 20, 302–315. [Google Scholar] [CrossRef] [PubMed]
- Kardosh, A.; Soriano, N.; Liu, Y.-T.; Uddin, J.; Petasis, N.A.; Hofman, F.M.; Chen, T.C.; Schönthal, A.H. Multitarget inhibition of drug-resistant multiple myeloma cell lines by dimethyl-celecoxib (DMC), a non–COX-2 inhibitory analog of celecoxib. Blood 2005, 106, 4330–4338. [Google Scholar] [CrossRef]
- Marinac, C.R.; Colditz, G.A.; Rosner, B.; Ghobrial, I.M.; Birmann, B.M. Aspirin Use and Survival in Multiple Myeloma Patients. Blood 2018, 132, 3250. [Google Scholar] [CrossRef]
- Birmann, B.M.; Giovannucci, E.L.; Rosner, B.A.; Colditz, G.A. Regular aspirin use and risk of multiple myeloma: A prospective analysis in the health professionals follow-up study and nurses’ health study. Cancer Prev. Res. 2014, 7, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Holien, T.; Olsen, O.E.; Misund, K.; Hella, H.; Waage, A.; Rø, T.B.; Sundan, A. Lymphoma and myeloma cells are highly sensitive to growth arrest and apoptosis induced by artesunate. Eur. J. Haematol. 2013, 91, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Papanikolaou, X.; Johnson, S.; Garg, T.; Tian, E.; Tytarenko, R.; Zhang, Q.; Stein, C.; Barlogie, B.; Epstein, J.; Heuck, C. Artesunate overcomes drug resistance in multiple myeloma by inducing mitochondrial stress and non-caspase apoptosis. Oncotarget 2014, 5, 4118–4128. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xue, F.; Cheng, Z.; Yang, X.; Wang, S.; Geng, F.; Pan, L. Effect of artesunate on inhibiting proliferation and inducing apoptosis of SP2/0 myeloma cells through affecting NFκB p65. Int. J. Hematol. 2009, 90, 513–521. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.-F.; Dong, K.; Dong, J.-F.; Wang, Y.; Gao, W. Effects of Artesunate Combined with Arsenious Acid on Proliferation and Apoptosis of Multiple Myeloma Cells via PI3K/AKT Signaling Pathway. Zhongguo Shi Yan Xue Ye Xue Za Zhi 2021, 29, 1819–1824. [Google Scholar] [PubMed]
- Baumann, P.; Mandl-Weber, S.; Volkl, A.; Adam, C.; Bumeder, I.; Oduncu, F.; Schmidmaier, R. Dihydroorotate dehydrogenase inhibitor A771726 (leflunomide) induces apoptosis and diminishes proliferation of multiple myeloma cells. Mol. Cancer Ther. 2009, 8, 366–375. [Google Scholar] [CrossRef]
- Rosenzweig, M.; Palmer, J.; Tsai, N.-C.; Synold, T.; Wu, X.; Tao, S.; Hammond, S.N.; Buettner, R.; Duarte, L.; Htut, M.; et al. Repurposing leflunomide for relapsed/refractory multiple myeloma: A phase 1 study. Leuk. Lymphoma 2020, 61, 1669–1677. [Google Scholar] [CrossRef] [PubMed]
- Van Nuffel, A.M.; Sukhatme, V.; Pantziarka, P.; Meheus, L.; Sukhatme, V.P.; Bouche, G. Repurposing Drugs in Oncology (ReDO)—Clarithromycin as an anti-cancer agent. Ecancermedicalscience 2015, 9, 513. [Google Scholar] [CrossRef] [PubMed]
- Mark, T.M.; Bowman, I.A.; Rossi, A.C.; Shah, M.; Rodriguez, M.; Quinn, R.; Pearse, R.N.; Zafar, F.; Pekle, K.; Jayabalan, D.; et al. Thalidomide, clarithromycin, lenalidomide and dexamethasone therapy in newly diagnosed, symptomatic multiple myeloma. Leuk. Lymphoma 2014, 55, 2842–2849. [Google Scholar] [CrossRef] [PubMed]
- Strömberg, T.; Dimberg, A.; Hammarberg, A.; Carlson, K.; Osterborg, A.; Nilsson, K.; Jernberg-Wiklund, H. Rapamycin sensitizes multiple myeloma cells to apoptosis induced by dexamethasone. Blood 2004, 103, 3138–3147. [Google Scholar] [CrossRef] [PubMed]
- Gera, J.; Lichtenstein, A. The mammalian target of rapamycin pathway as a therapeutic target in multiple myeloma. Leuk. Lymphoma 2011, 52, 1857–1866. [Google Scholar] [CrossRef]
- Raje, N.; Kumar, S.; Hideshima, T.; Ishitsuka, K.; Chauhan, D.; Mitsiades, C.; Podar, K.; Le Gouill, S.; Richardson, P.; Munshi, N.C.; et al. Combination of the mTOR inhibitor rapamycin and CC-5013 has synergistic activity in multiple myeloma. Blood 2004, 104, 4188–4193. [Google Scholar] [CrossRef] [PubMed]
- Yee, A.J.; Hari, P.; Marcheselli, R.; Mahindra, A.K.; Cirstea, D.D.; Scullen, T.A.; Burke, J.N.; Rodig, S.J.; Hideshima, T.; Laubach, J.P.; et al. Outcomes in patients with relapsed or refractory multiple myeloma in a phase I study of everolimus in combination with lenalidomide. Br. J. Haematol. 2014, 166, 401–409. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Wada, T.; Shimizu, R.; Izumi, T.; Akutsu, M.; Mitsunaga, K.; Noborio-Hatano, K.; Nobuyoshi, M.; Ozawa, K.; Kano, Y.; et al. Histone deacetylases are critical targets of bortezomib-induced cytotoxicity in multiple myeloma. Blood J. Am. Soc. Hematol. 2010, 116, 406–417. [Google Scholar] [CrossRef] [PubMed]
- Bono, C.; Karlin, L.; Harel, S.; Mouly, E.; Labaume, S.; Galicier, L.; Apcher, S.; Sauvageon, H.; Fermand, J.-P.; Bories, J.-C.; et al. The human immunodeficiency virus-1 protease inhibitor nelfinavir impairs proteasome activity and inhibits the proliferation of multiple myeloma cells in vitro and in vivo. Haematologica 2012, 97, 1101–1109. [Google Scholar] [CrossRef] [PubMed]
- Kraus, M.; Bader, J.; Overkleeft, H.; Driessen, C. Nelfinavir augments proteasome inhibition by bortezomib in myeloma cells and overcomes bortezomib and carfilzomib resistance. Blood Cancer J. 2013, 3, e103. [Google Scholar] [CrossRef] [PubMed]
- Alodhaibi, I.; Ailawadhi, S.; Burbano, G.P.; O’Brien, P.J.; Buadi, F.K.; Hayman, S.; Kumar, S.K.; Gonsalves, W.I. An Open-Label Phase I Study of Metformin and Nelfinavir in Combination with Bortezomib in Patients With Relapsed and Refractory Multiple Myeloma. Clin. Lymphoma Myeloma Leuk. 2024, 24, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Koltai, T. Nelfinavir and other protease inhibitors in cancer: Mechanisms involved in anticancer activity. F1000Research 2015, 4, 9. [Google Scholar] [CrossRef] [PubMed]
- Dalva-Aydemir, S.; Bajpai, R.; Martinez, M.; Adekola, K.U.; Kandela, I.; Wei, C.; Singhal, S.; Koblinski, J.E.; Raje, N.S.; Rosen, S.T.; et al. Targeting the metabolic plasticity of multiple myeloma with FDA-approved ritonavir and metformin. Clin. Cancer Res. 2015, 21, 1161–1171. [Google Scholar] [CrossRef] [PubMed]
- Mishra, R.K.; Wei, C.; Hresko, R.C.; Bajpai, R.; Heitmeier, M.; Matulis, S.M.; Nooka, A.K.; Rosen, S.T.; Hruz, P.W.; Schiltz, G.E.; et al. In silico modeling-based identification of glucose transporter 4 (GLUT4)-selective inhibitors for cancer therapy. J. Biol. Chem. 2015, 290, 14441–14453. [Google Scholar] [CrossRef]
- Jagannathan, S.; Abdel-Malek, M.; Malek, E.; Vad, N.; Latif, T.; Anderson, K.; Driscoll, J. Pharmacologic screens reveal metformin that suppresses GRP78-dependent autophagy to enhance the anti-myeloma effect of bortezomib. Leukemia 2015, 29, 2184–2191. [Google Scholar] [CrossRef] [PubMed]
- Mansour, A.; Wakkach, A.; Blin-Wakkach, C. Emerging roles of osteoclasts in the modulation of bone microenvironment and immune suppression in multiple myeloma. Front. Immunol. 2017, 8, 954. [Google Scholar] [CrossRef]
- Tai, Y.-T.; Cho, S.-F.; Anderson, K.C. Osteoclast immunosuppressive effects in multiple myeloma: Role of programmed cell death ligand 1. Front. Immunol. 2018, 9, 1822. [Google Scholar] [CrossRef] [PubMed]
- Chroma, K.; Skrott, Z.; Gursky, J.; Bacovsky, J.; Moudry, P.; Buchtova, T.; Mistrik, M.; Bartek, J. A drug repurposing strategy for overcoming human multiple myeloma resistance to standard-of-care treatment. Cell Death Dis. 2022, 13, 203. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.; Liang, L.; Wang, H.; Luo, S.; Hu, L.; Wang, Y.; Shen, X.; Xiao, L.; Zhang, Y.; Peng, H.; et al. Albendazole inhibits NF-κB signaling pathway to overcome tumor stemness and bortezomib resistance in multiple myeloma. Cancer Lett. 2021, 520, 307–320. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Witzig, T.; Rajkumar, S.V. Thalidomide as an anti-cancer agent. J. Cell. Mol. Med. 2002, 6, 160–174. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.T. Thalidomide embryopathy: A model for the study of congenital incomitant horizontal strabismus. Trans. Am. Ophthalmol. Soc. 1991, 89, 623–674. [Google Scholar] [PubMed]
- Grover, J.; Vats, V.; Gopalakrishna, R.; Ramam, M. Thalidomide: A re-look. Natl. Med. J. India 2000, 13, 132–141. [Google Scholar] [PubMed]
- Perri, A.J., III; Hsu, S. A review of thalidomide’s history and current dermatological applications. Dermatol. Online J. 2003, 9, 5. [Google Scholar] [CrossRef] [PubMed]
- Gordon, J.; Goggin, P. Thalidomide and its derivatives: Emerging from the wilderness. Postgrad. Med. J. 2003, 79, 127–132. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.; McMenamin, M.; Mulcahy, F.; Bergin, C. Thalidomide Therapy for the Treatment of Hypertrophic Herpes Simplex Virus—Related Genitalis in HIV-Infected Individuals. Clin. Infect. Dis. 2007, 44, e96–e99. [Google Scholar] [CrossRef] [PubMed]
- Holland, S. Cytokine therapy of mycobacterial infections. Adv. Intern. Med. 2000, 45, 431–452. [Google Scholar] [PubMed]
- Gupta, S.C.; Sung, B.; Prasad, S.; Webb, L.J.; Aggarwal, B.B. Cancer drug discovery by repurposing: Teaching new tricks to old dogs. Trends Pharmacol. Sci. 2013, 34, 508–517. [Google Scholar] [CrossRef]
- Locksley, R.M.; Killeen, N.; Lenardo, M.J. The TNF and TNF receptor superfamilies: Integrating mammalian biology. Cell 2001, 104, 487–501. [Google Scholar] [CrossRef] [PubMed]
- Hideshima, T.; Chauhan, D.; Richardson, P.; Mitsiades, C.; Mitsiades, N.; Hayashi, T.; Munshi, N.; Dang, L.; Castro, A.; Palombella, V.; et al. NF-κB as a therapeutic target in multiple myeloma. J. Biol. Chem. 2002, 277, 16639–16647. [Google Scholar] [CrossRef] [PubMed]
- Singhal, S.; Mehta, J. Thalidomide in cancer. Biomed. Pharmacother. 2002, 56, 4–12. [Google Scholar] [CrossRef] [PubMed]
- Kumar, V.; Chhibber, S. Thalidomide: An old drug with new action. J. Chemother. 2011, 23, 326–334. [Google Scholar] [CrossRef] [PubMed]
- Solèr, R.A.; Howard, M.; Brink, N.S.; Gibb, D.; Tedder, R.S.; Nadal, D. Regression of AIDS-related Kaposi’s sarcoma during therapy with thalidomide. Clin. Infect. Dis. 1996, 23, 501–503. [Google Scholar] [CrossRef] [PubMed]
- Tunio, M.A.; Hashmi, A.; Qayyum, A.; Naimatullah, N.; Masood, R. Low-dose thalidomide in patients with metastatic renal cell carcinoma. Brain 2012, 3, 3.75. [Google Scholar]
- Franks, M.E.; Macpherson, G.R.; Figg, W.D. Thalidomide. Lancet 2004, 363, 1802–1811. [Google Scholar] [CrossRef] [PubMed]
- Figg, W.D.; Hussain, M.H.; Gulley, J.L.; Arlen, P.M.; Aragon-Ching, J.B.; Petrylak, D.P.; Higano, C.S.; Steinberg, S.M.; Chatta, G.S.; Parnes, H.; et al. A double-blind randomized crossover study of oral thalidomide versus placebo for androgen dependent prostate cancer treated with intermittent androgen ablation. J. Urol. 2009, 181, 1104–1113. [Google Scholar] [CrossRef][Green Version]
- Ghobrial, I.M.; Rajkumar, S.V. Management of thalidomide toxicity. J. Support. Oncol. 2003, 1, 194–205. [Google Scholar] [PubMed]
- Fadul, C.E.; Kingman, L.S.; Meyer, L.P.; Cole, B.F.; Eskey, C.J.; Rhodes, C.H.; Roberts, D.W.; Newton, H.B.; Pipas, J.M. A phase II study of thalidomide and irinotecan for treatment of glioblastoma multiforme. J. Neuro-Oncol. 2008, 90, 229–235. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Penas-Prado, M.; Hess, K.R.; Fisch, M.J.; Lagrone, L.W.; Groves, M.D.; Levin, V.A.; De Groot, J.F.; Puduvalli, V.K.; Colman, H.; Volas-Redd, G.; et al. Randomized phase II adjuvant factorial study of dose-dense temozolomide alone and in combination with isotretinoin, celecoxib, and/or thalidomide for glioblastoma. Neuro-Oncology 2015, 17, 266–273. [Google Scholar] [CrossRef] [PubMed]
- Chapman-Shimshoni, D.; Yuklea, M.; Radnay, J.; Shapiro, H.; Lishner, M. Simvastatin induces apoptosis of B-CLL cells by activation of mitochondrial caspase 9. Exp. Hematol. 2003, 31, 779–783. [Google Scholar] [CrossRef] [PubMed]
- Pradelli, D.; Soranna, D.; Zambon, A.; Catapano, A.; Mancia, G.; La Vecchia, C.; Corrao, G. Statins use and the risk of all and subtype hematological malignancies: A meta-analysis of observational studies. Cancer Med. 2015, 4, 770–780. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Jia, W.; Jin, Y.; Zhen, S. Statin use is associated with reduced risk of haematological malignancies: Evidence from a meta-analysis. PLoS ONE 2014, 9, e87019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Liu, B. Statin use and the risk of multiple myeloma: A PRISMA-compliant meta-analysis. Ann. Hematol. 2020, 99, 1805–1812. [Google Scholar] [CrossRef] [PubMed]
- Longo, J.; Smirnov, P.; Li, Z.; Branchard, E.; van Leeuwen, J.E.; Licht, J.D.; Haibe-Kains, B.; Andrews, D.W.; Keats, J.J.; Pugh, T.J.; et al. The mevalonate pathway is an actionable vulnerability of t (4; 14)-positive multiple myeloma. Leukemia 2021, 35, 796–808. [Google Scholar] [CrossRef] [PubMed]
- Juarez, D.; Buono, R.; Matulis, S.M.; Gupta, V.A.; Duong, M.R.; Yudiono, J.; Paul, M.; Mallya, S.; Diep, G.; Hsin, P.; et al. Statin-induced mitochondrial priming sensitizes multiple myeloma cells to BCL2 and MCL1 inhibitors. Cancer Res. Commun. 2023, 3, 2497–2509. [Google Scholar] [CrossRef]
- Cetin, M.; Buyukberber, S.; Demir, M.; Sari, I.; Sari, I.; Deniz, K.; Eser, B.; Altuntas, F.; Camcı, C.; Öztürk, A.; et al. Overexpression of cyclooxygenase-2 in multiple myeloma: Association with reduced survival. Am. J. Hematol. 2005, 80, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Ladetto, M.; Vallet, S.; Trojan, A.; Dell’Aquila, M.; Monitillo, L.; Rosato, R.; Santo, L.; Drandi, D.; Bertola, A.; Falco, P.; et al. Cyclooxygenase-2 (COX-2) is frequently expressed in multiple myeloma and is an independent predictor of poor outcome. Blood 2005, 105, 4784–4791. [Google Scholar] [CrossRef] [PubMed]
- Trojan, A.; Tinguely, M.; Vallet, S.; Seifert, B.; Jenni, B.; Zippelius, A.; Witzens-Harig, M.; Mechtersheimer, G.; Ho, A.; Goldschmidt, H.; et al. Clinical significance of cyclooxygenase-2 (COX-2) in multiple myeloma. Swiss Med. Wkly. 2006, 136, 400–403. [Google Scholar] [PubMed]
- Jendrossek, V. Targeting apoptosis pathways by Celecoxib in cancer. Cancer Lett. 2013, 332, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Steinbach, G.; Lynch, P.M.; Phillips, R.K.; Wallace, M.H.; Hawk, E.; Gordon, G.B.; Wakabayashi, N.; Saunders, B.; Shen, Y.; Fujimura, T.; et al. The effect of celecoxib, a cyclooxygenase-2 inhibitor, in familial adenomatous polyposis. N. Engl. J. Med. 2000, 342, 1946–1952. [Google Scholar] [CrossRef] [PubMed]
- Scilimati, A. Patient Bone Marrow Aspiration to Explore the Cyclooxygenases (COXs) Involvement in Multiple Myeloma. J. Cancer Res. Therap. Oncol. 2021, 9, 1–19. [Google Scholar]
- Roy, P.; Sarkar, U.A.; Basak, S. The NF-κB activating pathways in multiple myeloma. Biomedicines 2018, 6, 59. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Hong, J.; Huang, H.; Fu, D.; Wu, S.; Wang, Q.; Ye, Y.; Liu, Y. High expression of phosphorylated extracellular signal-regulated kinase (ERK1/2) is associated with poor prognosis in newly diagnosed patients with multiple myeloma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 2636–2643. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Liu, H.; Xiong, C.; Liu, J.; Sun, T.; Ren, Z.; Li, Y.; Geng, J.; Li, X. Aspirin exerts anti-tumor effect through inhibiting Blimp1 and activating ATF4/CHOP pathway in multiple myeloma. Biomed. Pharmacother. 2020, 125, 110005. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Mikasa, K.; Hamada, K.; Konishi, M.; Maeda, K.; Yoshimoto, E.; Ueda, K.; Majima, T.; Sawaki, M.; Kita, E.; et al. Effect of clarithromycin treatment of natural killer cell activity in patients with advanced non-small cell lung cancer. Gan Kagaku Ryoho. Cancer Chemother. 1998, 25, 2259–2266. [Google Scholar] [CrossRef]
- Musto, P.; Falcone, A.; Sanpaolo, G.; Bodenizza, C.; Carotenuto, M.; Carella, A.M. Inefficacy of clarithromycin in advanced multiple myeloma: A definitive report. Haematologica 2002, 87, 658–659. [Google Scholar] [PubMed]
- Durie, B. Clarithromycin (Biaxin) as primary treatment for myeloma. Blood 1997, 90, 579. [Google Scholar]
- Stewart, A.; Trudel, S.; Al-Berouti, B.; Sutton, D.; Meharchand, J.; Shustik, C. Lack of response to short-term use of clarithromycin (BIAXIN) in multiple myeloma. Blood J. Am. Soc. Hematol. 1999, 93, 4441–4442. [Google Scholar]
- Moreau, P.; Huynh, A.; Facon, T.; Bouilly, I.; Sotto, J.; Legros, L.; Milpied, N.; Attal, M.; Bataille, R.; Harousseau, J.; et al. Lack of efficacy of clarithromycin in advanced multiple myeloma. Leukemia 1999, 13, 490–491. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.; Ranaghan, L.; Morrison, J.; Group, N.I.R.H. Phase II trial of clarithromycin and pamidronate therapy in myeloma. Med. Oncol. 2001, 18, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Coleman, M.; Leonard, J.; Lyons, L.; Pekle, K.; Nahum, K.; Pearse, R.; Niesvizky, R.; Michaeli, J. BLT-D (clarithromycin [Biaxin], low-dose thalidomide, and dexamethasone) for the treatment of myeloma and Waldenström’s macroglobulinemia. Leuk. Lymphoma 2002, 43, 1777–1782. [Google Scholar] [CrossRef] [PubMed]
- Morris, T.; Kettle, P.; Drake, M.; Jones, F.; Hull, D.; Boyd, K.; Morrison, A.; Clarke, P.; O’Reilly, P.; Quinn, J. Clarithromycin with low dose dexamethasone and thalidomide is effective therapy in relapsed/refractory myeloma. Br. J. Haematol. 2008, 143, 349–354. [Google Scholar] [CrossRef] [PubMed]
- Puig, N.; Hernández, M.T.; Rosiñol, L.; González, E.; de Arriba, F.; Oriol, A.; González-Calle, V.; Escalante, F.; de la Rubia, J.; Gironella, M.; et al. Lenalidomide and dexamethasone with or without clarithromycin in patients with multiple myeloma ineligible for autologous transplant: A randomized trial. Blood Cancer J. 2021, 11, 101. [Google Scholar] [CrossRef] [PubMed]
- Klein, B.; Zhang, X.-G.; Lu, Z.-Y.; Bataille, R. Interleukin-6 in human multiple myeloma. Blood 1995, 85, 863–872. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Slifer, T.; Araujo, F.; Remington, J. Effect of clarithromycin and azithromycin on production of cytokines by human monocytes. Int. J. Antimicrob. Agents 1999, 11, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Moriya, S.; Komatsu, S.; Yamasaki, K.; Kawai, Y.; Kokuba, H.; Hirota, A.; Che, X.F.; Inazu, M.; Gotoh, A.; Hiramoto, M. Targeting the integrated networks of aggresome formation, proteasome, and autophagy potentiates ER stress-mediated cell death in multiple myeloma cells. Int. J. Oncol. 2015, 46, 474–486. [Google Scholar] [CrossRef] [PubMed]
- Takemori, N.; Ooi, H.-K.; Imai, G.; Hoshino, K.; Saio, M. Possible mechanisms of action of clarithromycin and its clinical application as a repurposing drug for treating multiple myeloma. Ecancermedicalscience 2020, 14, 1088. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, S.; Baker, H.; Vézina, C. Rapamycin (AY-22, 989), a new antifungal antibiotic II. Fermentation, isolation and characterization. J. Antibiot. 1975, 28, 727–732. [Google Scholar] [CrossRef] [PubMed]
- Saunders, R.N.; Metcalfe, M.S.; Nicholson, M.L. Rapamycin in transplantation: A review of the evidence. Kidney Int. 2001, 59, 3–16. [Google Scholar] [CrossRef] [PubMed]
- Laplante, M.; Sabatini, D.M. mTOR signaling at a glance. J. Cell Sci. 2009, 122, 3589–3594. [Google Scholar] [CrossRef]
- Ichiyama, T.; Okada, K.; Lipton, J.M.; Matsubara, T.; Hayashi, T.; Furukawa, S. Sodium valproate inhibits production of TNF-α and IL-6 and activation of NF-κB. Brain Res. 2000, 857, 246–251. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Hao, C.-L.; Zhang, Z.-H.; Wang, L.-H.; Yan, L.-N.; Zhang, R.-J.; Lin, L.; Yang, Y. Valproic acid increased autophagic flux in human multiple myeloma cells in vitro. Biomed. Pharmacother. 2020, 127, 110167. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, Z.; Zhao, R.; Li, H.; Wang, T.; Yan, L.; Gu, C.; Zhao, L.; Hao, C. Valproic acid activates autophagy in multiple myeloma cell lines RPMI8226 and U266. Zhonghua Xue Ye Xue Za Zhi Zhonghua Xueyexue Zazhi 2016, 37, 478–483. [Google Scholar]
- Kitazoe, K.-I.; Abe, M.; Hiasa, M.; Oda, A.; Amou, H.; Harada, T.; Nakano, A.; Takeuchi, K.; Hashimoto, T.; Ozaki, S.; et al. Valproic acid exerts anti-tumor as well as anti-angiogenic effects on myeloma. Int. J. Hematol. 2009, 89, 45–57. [Google Scholar] [CrossRef]
- Yang, Y.; Ikezoe, T.; Takeuchi, T.; Adachi, Y.; Ohtsuki, Y.; Takeuchi, S.; Koeffler, H.P.; Taguchi, H. HIV-1 protease inhibitor induces growth arrest and apoptosis of human prostate cancer LNCaP cells in vitro and in vivo in conjunction with blockade of androgen receptor STAT3 and AKT signaling. Cancer Sci. 2005, 96, 425–433. [Google Scholar] [CrossRef] [PubMed]
- Kawabata, S.; Gills, J.; Mercado-Matos, J.; Lopiccolo, J.; Wilson, W.; Hollander, M.; Dennis, P. Synergistic effects of nelfinavir and bortezomib on proteotoxic death of NSCLC and multiple myeloma cells. Cell Death Dis. 2012, 3, e353. [Google Scholar] [CrossRef] [PubMed]
- Kalender, A.; Selvaraj, A.; Kim, S.Y.; Gulati, P.; Brûlé, S.; Viollet, B.; Kemp, B.E.; Bardeesy, N.; Dennis, P.; Schlager, J.J.; et al. Metformin, independent of AMPK, inhibits mTORC1 in a rag GTPase-dependent manner. Cell Metab. 2010, 11, 390–401. [Google Scholar] [CrossRef] [PubMed]
- Del Barco, S.; Vazquez-Martin, A.; Cufí, S.; Oliveras-Ferraros, C.; Bosch-Barrera, J.; Joven, J.; Martin-Castillo, B.; Menendez, J.A. Metformin: Multi-faceted protection against cancer. Oncotarget 2011, 2, 896–917. [Google Scholar] [CrossRef] [PubMed]
- Greaves, D.; Calle, Y. Epithelial Mesenchymal Transition (EMT) and Associated Invasive Adhesions in Solid and Haematological Tumours. Cells 2022, 11, 649. [Google Scholar] [CrossRef] [PubMed]
- Noto, H.; Goto, A.; Tsujimoto, T.; Noda, M. Cancer risk in diabetic patients treated with metformin: A systematic review and meta-analysis. PLoS ONE 2012, 7, e33411. [Google Scholar] [CrossRef] [PubMed]
- Bodmer, M.; Meier, C.; Krähenbühl, S.; Jick, S.S.; Meier, C.R. Long-term metformin use is associated with decreased risk of breast cancer. Diabetes Care 2010, 33, 1304–1308. [Google Scholar] [CrossRef] [PubMed]
- Machado-Neto, J.A.; Fenerich, B.A.; Scopim-Ribeiro, R.; Eide, C.A.; Coelho-Silva, J.L.; Dechandt, C.R.P.; Fernandes, J.C.; Rodrigues Alves, A.P.N.; Scheucher, P.S.; Simões, B.P.; et al. Metformin exerts multitarget antileukemia activity in JAK2V617F-positive myeloproliferative neoplasms. Cell Death Dis. 2018, 9, 311. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.K.; Dingli, D. Metformin inhibits IL-6 signaling by decreasing IL-6R expression on multiple myeloma cells. Leukemia 2019, 33, 2695–2709. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.-H.; Luo, S.; O’Brian, K.K.; Thomas, T.S.; Colditz, G.A.; Carlsson, N.P.; Carson, K.R. Association between metformin use and progression of monoclonal gammopathy of undetermined significance to multiple myeloma in US veterans with diabetes mellitus: A population-based retrospective cohort study. Lancet Haematol. 2015, 2, e30–e36. [Google Scholar] [CrossRef] [PubMed]
- Boursi, B.; Mamtani, R.; Yang, Y.-X.; Weiss, B.M. Impact of metformin on the progression of MGUS to multiple myeloma. Leuk. Lymphoma 2017, 58, 1265–1267. [Google Scholar] [CrossRef] [PubMed]
- LeGrand, J.; Park, E.S.; Wang, H.; Gupta, S.; Owens, J.D., Jr.; Nelson, P.J.; DuBois, W.; Bair, T.; Janz, S.; Mushinski, J.F. Global gene expression profiling in mouse plasma cell tumor precursor and bystander cells reveals potential intervention targets for plasma cell neoplasia. Blood J. Am. Soc. Hematol. 2012, 119, 1018–1028. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, E.; Lv, N.; Ma, L.; Yao, S.; Yan, M.; Zi, F.; Deng, G.; Liu, X.; He, J. Metformin and FTY720 synergistically induce apoptosis in multiple myeloma cells. Cell. Physiol. Biochem. 2018, 48, 785–800. [Google Scholar] [CrossRef] [PubMed]
- Berenson, J.R. Antitumor effects of bisphosphonates: From the laboratory to the clinic. Curr. Opin. Support. Palliat. Care 2011, 5, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, R.S.; Roberson, P.K.; Manolagas, S.C. Giant osteoclast formation and long-term oral bisphosphonate therapy. N. Engl. J. Med. 2009, 360, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Hou, Y.; Weng, X.; Pang, W.; Hou, L.; Liang, Y.; Wang, Y.; Du, L.; Wu, T.; Yao, M.; et al. Diethyldithiocarbamate-copper complex (CuET) inhibits colorectal cancer progression via miR-16-5p and 15b-5p/ALDH1A3/PKM2 axis-mediated aerobic glycolysis pathway. Oncogenesis 2021, 10, 4. [Google Scholar] [CrossRef] [PubMed]
- Suh, J.J.; Pettinati, H.M.; Kampman, K.M.; O’Brien, C.P. The status of disulfiram: A half of a century later. J. Clin. Psychopharmacol. 2006, 26, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Kannappan, V.; Ali, M.; Small, B.; Rajendran, G.; Elzhenni, S.; Taj, H.; Wang, W.; Dou, Q.P. Recent advances in repurposing disulfiram and disulfiram derivatives as copper-dependent anticancer agents. Front. Mol. Biosci. 2021, 8, 741316. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Wang, H.; Chen, P.; Shen, X.; Zhang, B.; Liu, J.; Peng, H.; Xiao, X. Identification and characterization of multiple myeloma stem cell-like cells. Cancers 2021, 13, 3523. [Google Scholar] [CrossRef] [PubMed]
- Jin, N.; Zhu, X.; Cheng, F.; Zhang, L. Disulfiram/copper targets stem cell-like ALDH+ population of multiple myeloma by inhibition of ALDH1A1 and Hedgehog pathway. J. Cell. Biochem. 2018, 119, 6882–6893. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.-Y.; Jung, B.-K.; Hong, S.-J. Albendazole and mebendazole as anti-parasitic and anti-cancer agents: An update. Korean J. Parasitol. 2021, 59, 189–225. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-J.; Lee, Y.-C.; Huang, C.-H.; Shi, Y.-J.; Chen, Y.-J.; Pei, S.-N.; Chou, Y.-W.; Chang, L.-S. Non-mitotic effect of albendazole triggers apoptosis of human leukemia cells via SIRT3/ROS/p38 MAPK/TTP axis-mediated TNF-α upregulation. Biochem. Pharmacol. 2019, 162, 154–168. [Google Scholar] [CrossRef] [PubMed]

| Purpose | Resource | Refs. |
|---|---|---|
| Human pathways and protein–protein interaction (PPI) | BiGRID, STRING, HAPPI, KEGG, Reactome | [27,28,29,30,31] |
| Molecular classification of more than 20,000 main cancers matched normal tissue from 33 types of cancer | Cancer Genome Atlas | [32] |
| Protein expression in cancer, matched normal tissues, and human cancer cell lines | The Human Protein Atlas | [33,34,35] |
| Drug sensitivity, gene expression, and genotype for human cancer cell lines | Cancer Cell Line Encyclopedia | [36] |
| Data of genome-wide transcription expression from cultured human cancer cells with many small compounds | Connectivity Map 02 (CMap) | [37,38] |
| Disease-specific gene curation and analysis | OMIM, GEO | [39,40] |
| Disease–disease connectivity; connectivity of two genes elaborated within the same disease | The human disease network | [40] |
| Disease similarities as seen through the lens of gene regulatory mechanisms; comprehension of disease etiology and pathophysiology | Human Disease Network Database (DNetDB) | [41] |
| Drug–drug interaction; comprehensive drug-target information on tens of thousands of drugs and targets | DrugBank | [42] |
| Drug–drug interaction | SFINX | [43] |
| Database of more than 270 non-cancer drugs for potential repurposing for anti-cancer therapy | Repurposing Drugs in Oncology (ReDO) | [44] |
| Database of drugs and adverse drug reactions (ADRs) | Side Effect Resource (SIDER) | [45] |
| Withdrawn or discontinued drugs | WITHDRAWN | [46,47] |
| An inventory of main and secondary uses for repurposed pharmaceuticals | RepurposeDB | [48] |
| Chemical (including drugs)–protein interaction network | STITCH | [47] |
| Data on the sensitivity of hundreds of compounds and over a thousand cancer cell lines | Genomics of Drug Sensitivity in Cancer (GDSC) | [49] |
| Gene expression pattern-based prediction of drug effectiveness against cancer | DeSigN | [50] |
| Drug Name | Old-Indication | New-Indication | Mechanism of Action | Clinical Trials Status | Refs. |
|---|---|---|---|---|---|
| Thalidomide | Sedative, anti-nausea | MM | Inhibits IKK (also NF-κB); inhibits TNF; inhibits IL-1, IL-6, IL-12, VEGF | Approved in combination with dexamethasone | [51,52] |
| Statins | High Cholesterol | MM | HMG-CoA reductase inhibitors, upregulation of PUMA and NOXA | Smouldering MM, phase II | [53,54,55,56] |
| Celecoxib | Anti- inflammatory | MM and drug-resistant MM | Inhibits COX-2, inhibits Mcl-1, Bcl-2, survivin, Akt | Not for MM, approved for FAP | [57,58,59] |
| Aspirin | Anti- inflammatory | MM | Inhibits COX-1 and COX-2, suppresses cytokines and NF-κB, inhibits EKR and Blimp1, activates ATF4/CHOP | Preclinical | [60,61] |
| Artesunate | Malaria | MM and drug-resistant MM | Decreased expression of MYC and Bcl-2, triggers cleavage of caspase-3 | Preclinical | [62,63,64,65] |
| Leflunomide | Rheumatism | MM | DHODH inhibitor, cyclin D2 and pRb inhibition | Phase II | [66,67] |
| Clarithromycin | Antibiotic | MM and drug-resistant MM | Inhibits IL-6 and MGFs | Phase III | [68,69] |
| Rapamycin | Fungal infections | MM | Antagonist of mTOR | Phase I | [70,71,72,73] |
| Valproic acid | Seizures, migraine, and epilepsy | MM | Blocks HDAC, inhibits NF-κB and cytokines | Preclinical | [74] |
| Nelfinavir | HIV Infection | MM and drug-resistant MM | Inhibits 26S proteasome- disrupts Akt and STAT3, ERK1/2 | Phase I | [75,76,77,78] |
| Metformin | Diabetes mellitus type 2 | MM | Activates AMPK (suppresses mTORC1, activates p53), inhibits EMT, regulates cell cycle proteins (ERK1/2, JAK2/STAT), IL-6 suppression | Smoldering Myeloma and Monoclonal gammopathy of undetermined significance phase II, MM phase I | [77,79,80,81] |
| Bisphosphonates | Osteoporosis | MM | HMG-CoA pathway suppression, osteoclast apoptosis | Preclinical | [82,83] |
| CuET | Alcohol-abuse drug disulfiram (DSF) | Drug-resistant MM | ALDH inhibition | Preclinical | [84] |
| Albendazole | Parasitic infections | Drug-resistant MM | Microtubule system interference, p65/NF-κB pathway inhibition | Preclinical | [85] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Al-Odat, O.S.; Nelson, E.; Budak-Alpdogan, T.; Jonnalagadda, S.C.; Desai, D.; Pandey, M.K. Discovering Potential in Non-Cancer Medications: A Promising Breakthrough for Multiple Myeloma Patients. Cancers 2024, 16, 2381. https://doi.org/10.3390/cancers16132381
Al-Odat OS, Nelson E, Budak-Alpdogan T, Jonnalagadda SC, Desai D, Pandey MK. Discovering Potential in Non-Cancer Medications: A Promising Breakthrough for Multiple Myeloma Patients. Cancers. 2024; 16(13):2381. https://doi.org/10.3390/cancers16132381
Chicago/Turabian StyleAl-Odat, Omar S., Emily Nelson, Tulin Budak-Alpdogan, Subash C. Jonnalagadda, Dhimant Desai, and Manoj K. Pandey. 2024. "Discovering Potential in Non-Cancer Medications: A Promising Breakthrough for Multiple Myeloma Patients" Cancers 16, no. 13: 2381. https://doi.org/10.3390/cancers16132381
APA StyleAl-Odat, O. S., Nelson, E., Budak-Alpdogan, T., Jonnalagadda, S. C., Desai, D., & Pandey, M. K. (2024). Discovering Potential in Non-Cancer Medications: A Promising Breakthrough for Multiple Myeloma Patients. Cancers, 16(13), 2381. https://doi.org/10.3390/cancers16132381







