Genomic Deletion of PFKFB3 Decreases In Vivo Tumorigenesis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Generation of Conditional Pfkfb3 Knockout Mice
2.2. Fibroblast Isolation and In Vitro Cre-Mediated Recombination
2.3. Western Blotting
2.4. F26BP Measurements
2.5. Glycolysis Assay
2.6. Immunohistochemistry
2.7. Tumor Model Generation
2.8. Tumor Growth Monitoring
2.9. Statistics and Sampling
3. Results
3.1. Generation of a Transgenic Pfkfb3 Inducible Knockout Mouse
3.2. Homozygous Pfkfb3 Deletion in Adult Mice Does Not Affect Organ Function In Vivo
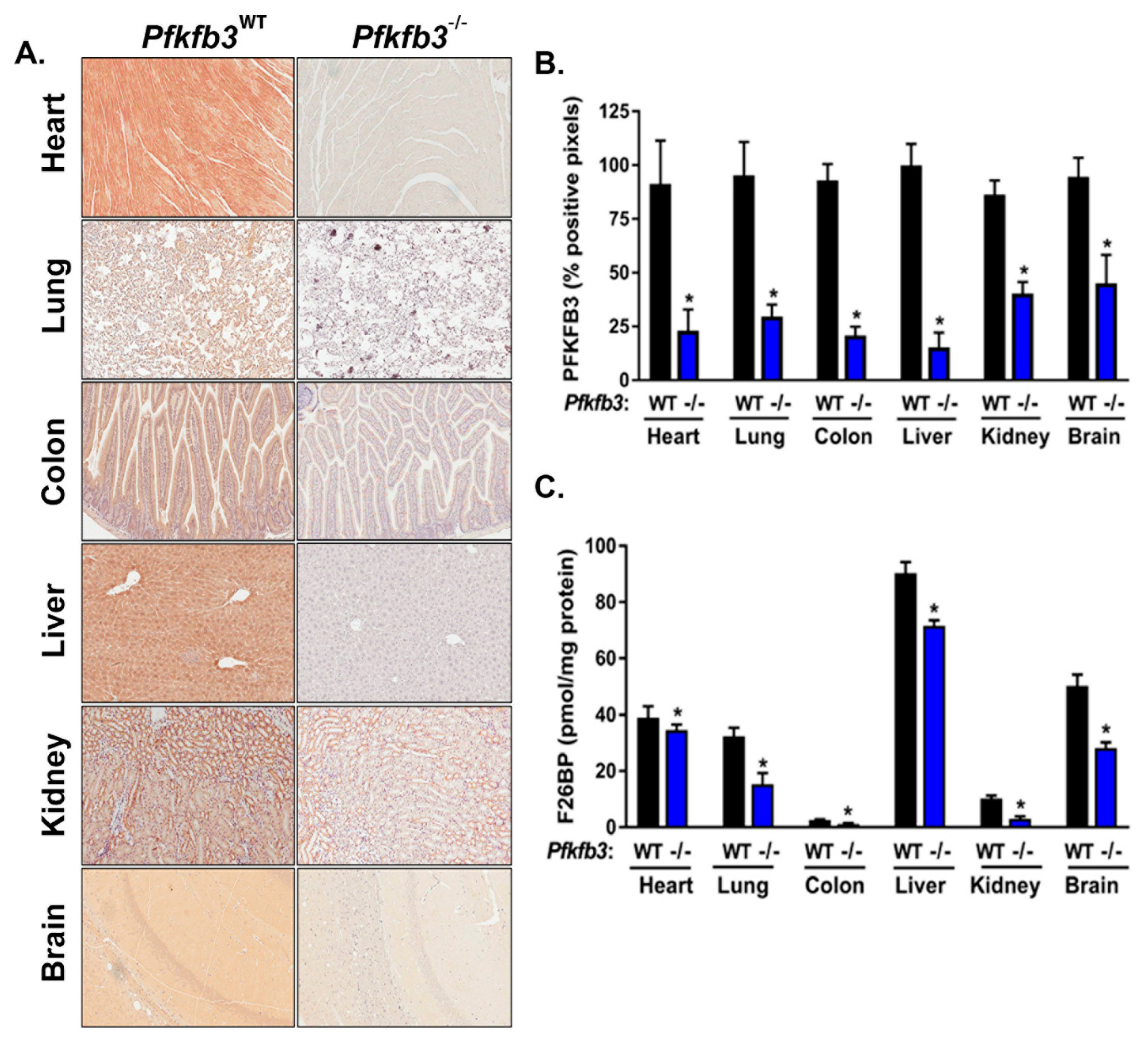
3.3. Cre-Mediated Recombination Effectively Decreases PFKFB3 Protein Expression and Activity in Diverse Organs In Vivo
3.4. Pfkfb3 Deletion Decreases Glucose Uptake and Growth of Spontaneous HER2-Driven Mammary Tumors
3.5. Pfkfb3 Genomic Deletion Causes Apoptosis and Cell Cycle Arrest in Spontaneous HER2-Driven Mammary Tumors
3.6. Pfkfb3 Genomic Deletion Decreases Tumorigenesis in K-Ras-Driven Lung Tumors In Vivo
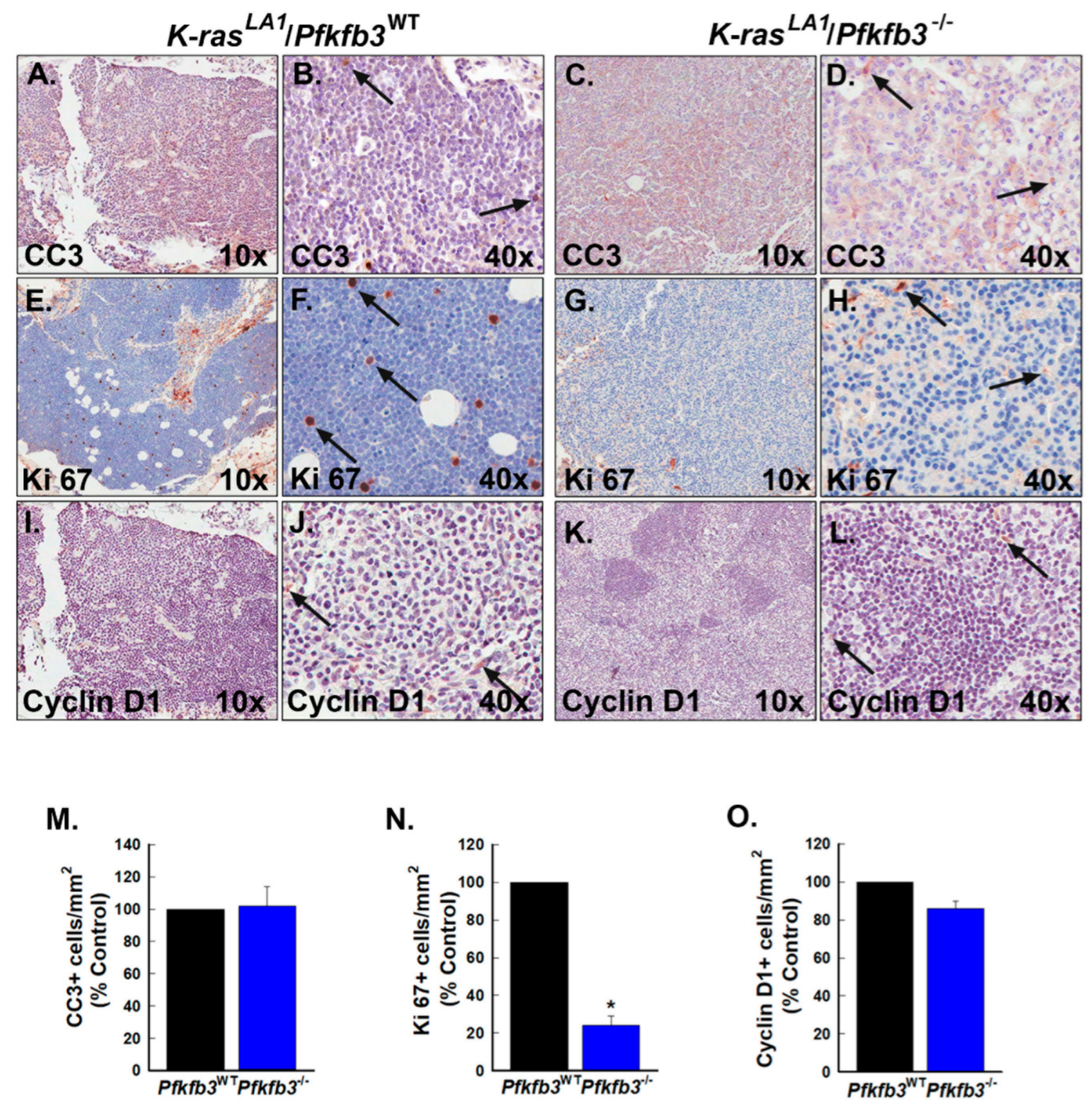
3.7. Pfkfb3 Deletion Decreases Cell Cycle Progression in K-Ras-Driven Lung Tumors
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeBerardinis, R.J.; Lum, J.J.; Hatzivassiliou, G.; Thompson, C.B. The biology of cancer: Metabolic reprogramming fuels cell growth and proliferation. Cell Metab. 2008, 7, 11–20. [Google Scholar] [CrossRef]
- Marshall, M.J.; Goldberg, D.M.; Neal, F.E.; Millar, D.R. Enzymes of glucose metabolism in carcinoma of the cervix and endometrium of the human uterus. Br. J. Cancer 1978, 37, 990–1001. [Google Scholar] [CrossRef]
- Weber, G. Enzymology of cancer cells (second of two parts). N. Engl. J. Med. 1977, 296, 541–551. [Google Scholar] [CrossRef]
- Okar, D.A.; Manzano, A.; Navarro-Sabate, A.; Riera, L.; Bartrons, R.; Lange, A.J. PFK-2/FBPase-2: Maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends Biochem. Sci. 2001, 26, 30–35. [Google Scholar] [CrossRef]
- Wu, C.; Khan, S.A.; Peng, L.J.; Lange, A.J. Roles for fructose-2,6-bisphosphate in the control of fuel metabolism: Beyond its allosteric effects on glycolytic and gluconeogenic enzymes. Adv. Enzym. Regul. 2006, 46, 72–88. [Google Scholar] [CrossRef]
- Van Schaftingen, E.; Hue, L.; Hers, H.G. Fructose 2,6-bisphosphate, the probably structure of the glucose- and glucagon-sensitive stimulator of phosphofructokinase. Biochem. J. 1980, 192, 897–901. [Google Scholar] [CrossRef]
- Yalcin, A.; Telang, S.; Clem, B.; Chesney, J. Regulation of glucose metabolism by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatases in cancer. Exp. Mol. Pathol. 2009, 86, 174–179. [Google Scholar] [CrossRef]
- Okar, D.A.; Lange, A.J. Fructose-2,6-bisphosphate and control of carbohydrate metabolism in eukaryotes. Biofactors 1999, 10, 1–14. [Google Scholar] [CrossRef]
- Sakakibara, R.; Kato, M.; Okamura, N.; Nakagawa, T.; Komada, Y.; Tominaga, N.; Shimojo, M.; Fukasawa, M. Characterization of a human placental fructose-6-phosphate, 2-kinase/fructose-2,6-bisphosphatase. J. Biochem. 1997, 122, 122–128. [Google Scholar] [CrossRef]
- Telang, S.; Yalcin, A.; Clem, A.L.; Bucala, R.; Lane, A.N.; Eaton, J.W.; Chesney, J. Ras transformation requires metabolic control by 6-phosphofructo-2-kinase. Oncogene 2006, 25, 7225–7234. [Google Scholar] [CrossRef]
- Minchenko, A.; Leshchinsky, I.; Opentanova, I.; Sang, N.; Srinivas, V.; Armstead, V.; Caro, J. Hypoxia-inducible factor-1-mediated expression of the 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase-3 (PFKFB3) gene. Its possible role in the Warburg effect. J. Biol. Chem. 2002, 277, 6183–6187. [Google Scholar] [PubMed]
- Obach, M.; Navarro-Sabate, A.; Caro, J.; Kong, X.; Duran, J.; Gomez, M.; Perales, J.C.; Ventura, F.; Rosa, J.L.; Bartrons, R. 6-Phosphofructo-2-kinase (pfkfb3) gene promoter contains hypoxia-inducible factor-1 binding sites necessary for transactivation in response to hypoxia. J. Biol. Chem. 2004, 279, 53562–53570. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Pan, H.; Liu, Z.; Xie, J.; Han, W. Roles of PFKFB3 in cancer. Signal Transduct. Target. Ther. 2017, 2, 17044. [Google Scholar] [CrossRef] [PubMed]
- Novellasdemunt, L.; Obach, M.; Millán-Ariño, L.; Manzano, A.; Ventura, F.; Rosa, J.L.; Jordan, A.; Navarro-Sabate, A.; Bartrons, R. Progestins activate 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3) in breast cancer cells. Biochem. J. 2012, 442, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Riera, L.; Manzano, A.; Navarro-Sabate, A.; Perales, J.C.; Bartrons, R. Insulin induces PFKFB3 gene expression in HT29 human colon adenocarcinoma cells. Biochim. Biophys. Acta 2002, 1589, 89–92. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Rodríguez-García, A.; Samsó, P.; Fontova, P.; Simon-Molas, H.; Manzano, A.; Castaño, E.; Rosa, J.L.; Martinez-Outshoorn, U.; Ventura, F.; Navarro-Sabaté, À.; et al. TGF-β1 targets Smad, p38 MAPK, and PI3K/Akt signaling pathways to induce PFKFB3 gene expression and glycolysis in glioblastoma cells. FEBS J. 2017, 284, 3437–3454. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-García, A.; Monsalve, E.; Novellasdemunt, L.; Navarro-Sabaté, A.; Manzano, A.; Rivero, S.; Castrillo, A.; Casado, M.; Laborda, J.; Bartrons, R.; et al. Cooperation of adenosine with macrophage Toll-4 receptor agonists leads to increased glycolytic flux through the enhanced expression of PFKFB3 gene. J. Biol. Chem. 2011, 286, 19247–19258. [Google Scholar] [CrossRef]
- Garcia-Cao, I.; Song, M.S.; Hobbs, R.M.; Laurent, G.; Giorgi, C.; de Boer, V.C.; Anastasiou, D.; Ito, K.; Sasaki, A.T.; Rameh, L.; et al. Systemic elevation of PTEN induces a tumor-suppressive metabolic state. Cell 2012, 149, 49–62. [Google Scholar] [CrossRef]
- Manes, N.P.; El-Maghrabi, M.R. The kinase activity of human brain 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase is regulated via inhibition by phosphoenolpyruvate. Arch. Biochem. Biophys. 2005, 438, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Qiao, P.; Sun, Y.; Ren, C.; Yu, Z. Positive regulation of PFKFB3 by PIM2 promotes glycolysis and paclitaxel resistance in breast cancer. Clin. Transl. Med. 2021, 11, e400. [Google Scholar] [CrossRef]
- Yalcin, A.; Clem, B.F.; Imbert-Fernandez, Y.; Ozcan, S.C.; Peker, S.; O’Neal, J.; Klarer, A.C.; Clem, A.L.; Telang, S.; Chesney, J. 6-Phosphofructo-2-kinase (PFKFB3) promotes cell cycle progression and suppresses apoptosis via Cdk1-mediated phosphorylation of p27. Cell Death Dis 2014, 5, e1337. [Google Scholar] [CrossRef]
- Chesney, J.; Telang, S.; Yalcin, A.; Clem, A.; Wallis, N.; Bucala, R. Targeted disruption of inducible 6-phosphofructo-2-kinase results in embryonic lethality. Biochem. Biophys. Res. Commun. 2005, 331, 139–146. [Google Scholar] [CrossRef] [PubMed]
- Leone, R.D.; Powell, J.D. Metabolism of immune cells in cancer. Nat. Rev. Cancer 2020, 20, 516–531. [Google Scholar] [CrossRef]
- Xu, Y.; Chen, Y.; Zhang, X.; Ma, J.; Liu, Y.; Cui, L.; Wang, F. Glycolysis in Innate Immune Cells Contributes to Autoimmunity. Front. Immunol. 2022, 13, 920029. [Google Scholar] [CrossRef]
- Rumpf, S.; Sanal, N.; Marzano, M. Energy metabolic pathways in neuronal development and function. Oxf. Open Neurosci. 2023, 2, kvad00. [Google Scholar] [CrossRef] [PubMed]
- Yetkin-Arik, B.; Vogels, I.M.C.; Nowak-Sliwinska, P.; Weiss, A.; Houtkooper, R.H.; Van Noorden, C.J.F.; Klaassen, I.; Schlingemann, R.O. The role of glycolysis and mitochondrial respiration in the formation and functioning of endothelial tip cells during angiogenesis. Sci. Rep. 2019, 9, 12608. [Google Scholar] [CrossRef]
- Zheng, S.; Li, H.; Li, Y.; Chen, X.; Shen, J.; Chen, M.; Zhang, C.; Wu, J.; Sun, Q. The emerging role of glycolysis and immune evasion in gastric cancer. Cancer Cell Int. 2023, 23, 317. [Google Scholar] [CrossRef]
- Takata, N.; Miska, J.M.; Morgan, M.A.; Patel, P.; Billingham, L.K.; Joshi, N.; Schipma, M.J.; Dumar, Z.J.; Joshi, N.R.; Misharin, A.V.; et al. Lactate-dependent transcriptional regulation controls mammalian eye morphogenesis. Nat. Commun. 2023, 14, 4129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Duan, Z.; Li, Z.; Ge, F.; Wei, R.; Kong, L. The significance of glycolysis in tumor progression and its relationship with the tumor microenvironment. Front. Pharmacol. 2022, 13, 1091779. [Google Scholar] [CrossRef] [PubMed]
- Clem, B.; Telang, S.; Clem, A.; Yalcin, A.; Meier, J.; Simmons, A.; Rasku, M.A.; Arumugam, S.; Dean, W.L.; Eaton, J.; et al. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Mol. Cancer Ther. 2008, 7, 110–120. [Google Scholar] [CrossRef]
- Clem, B.F.; O’Neal, J.; Tapolsky, G.; Clem, A.L.; Imbert-Fernandez, Y.; Kerr, D.A.; Klarer, A.C.; Redman, R.; Miller, D.M.; Trent, J.O.; et al. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol. Cancer Ther. 2013, 12, 1461–1470. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, S.; McMahon, A.P. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: A tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol. 2002, 244, 305–318. [Google Scholar] [CrossRef]
- Van Schaftingen, E.; Lederer, B.; Bartrons, R.; Hers, H.G. A kinetic study of pyrophosphate: Fructose-6-phosphate phosphotransferase from potato tubers. Application to a microassay of fructose 2,6-bisphosphate. Eur. J. Biochem. 1982, 129, 191–195. [Google Scholar] [CrossRef] [PubMed]
- Chesney, J.; Clark, J.; Klarer, A.C.; Imbert-Fernandez, Y.; Lane, A.N.; Telang, S. Fructose-2,6-bisphosphate synthesis by 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 4 (PFKFB4) is required for the glycolytic response to hypoxia and tumor growth. Oncotarget 2014, 5, 6670–6686. [Google Scholar] [CrossRef] [PubMed]
- Muller, W.J.; Sinn, E.; Pattengale, P.K.; Wallace, R.; Leder, P. Single-step induction of mammary adenocarcinoma in transgenic mice bearing the activated c-neu oncogene. Cell 1988, 54, 105–115. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.; Mercer, K.; Greenbaum, D.; Bronson, R.T.; Crowley, D.; Tuveson, D.A.; Jacks, T. Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 2001, 410, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Taetle, R.; Rosen, F.; Abramson, I.; Venditti, J.; Howell, S. Use of nude mouse xenografts as preclinical drug screens: In vivo activity of established chemotherapeutic agents against melanoma and ovarian carcinoma xenografts. Cancer Treat. Rep. 1987, 71, 297–304. [Google Scholar] [PubMed]
- Bartrons, R.; Simon-Molas, H.; Rodríguez-García, A.; Castaño, E.; Navarro-Sabaté, À.; Manzano, A.; Martinez-Outschoorn, U.E. Fructose 2,6-Bisphosphate in Cancer Cell Metabolism. Front. Oncol. 2018, 8, 331. [Google Scholar] [CrossRef] [PubMed]
- O’Neal, J.; Clem, A.; Reynolds, L.; Dougherty, S.; Imbert-Fernandez, Y.; Telang, S.; Chesney, J.; Clem, B.F. Inhibition of 6-phosphofructo-2-kinase (PFKFB3) suppresses glucose metabolism and the growth of HER2+ breast cancer. Breast Cancer Res. Treat. 2016, 160, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Murakami, T.; Kawasaki, M.; Takahashi, M. The cell cycle associated change of the Ki-67 reactive nuclear antigen expression. J. Cell. Physiol. 1987, 133, 579–584. [Google Scholar] [CrossRef]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A Molecular Connection for Cell Cycle Control, Adhesion and Invasion in Tumor and Stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef] [PubMed]
- Marcucci, F.; Rumio, C. On the Role of Glycolysis in Early Tumorigenesis—Permissive and Executioner Effects. Cells 2023, 12, 1124. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Lu, X.; Huang, L.; Wang, W.; Jiang, G.; Dean, K.C.; Clem, B.; Telang, S.; Jenson, A.B.; Cuatrecasas, M.; et al. Different thresholds of ZEB1 are required for Ras-mediated tumour initiation and metastasis. Nat. Commun. 2014, 5, 5660. [Google Scholar] [CrossRef] [PubMed]
- Chandel, N.S. Metabolism of Proliferating Cells. Cold Spring Harb. Perspect. Biol. 2021, 13, a040618. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Han, S.; Chen, J.; Sun, J.; Sun, X. PFKFB3 knockdown attenuates Amyloid β-Induced microglial activation and retinal pigment epithelium disorders in mice. Int. Immunopharmacol. 2023, 115, 109691. [Google Scholar] [CrossRef]
- Ganeshan, K.; Chawla, A. Metabolic regulation of immune responses. Annu. Rev. Immunol. 2014, 32, 609–634. [Google Scholar] [CrossRef] [PubMed]
- Soto-Heredero, G.; Gómez de las Heras, M.M.; Gabandé-Rodríguez, E.; Oller, J.; Mittelbrunn, M. Glycolysis—A key player in the inflammatory response. FEBS J. 2020, 287, 3350–3369. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef] [PubMed]
- Calvo, M.N.; Bartrons, R.; Castaño, E.; Perales, J.C.; Navarro-Sabaté, A.; Manzano, A. PFKFB3 gene silencing decreases glycolysis, induces cell-cycle delay and inhibits anchorage-independent growth in HeLa cells. FEBS Lett. 2006, 580, 3308–3314. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Zhang, X.; Wang, L.; Yang, Q.; Ma, Q.; Xu, J.; Wang, J.; Kovacs, L.; Ayon, R.J.; Liu, Z.; et al. PFKFB3-mediated endothelial glycolysis promotes pulmonary hypertension. Proc. Natl. Acad. Sci. USA 2019, 116, 13394–13403. [Google Scholar] [CrossRef]
- Li, J.; Zhang, S.; Liao, D.; Zhang, Q.; Chen, C.; Yang, X.; Jiang, D.; Pang, J. Overexpression of PFKFB3 promotes cell glycolysis and proliferation in renal cell carcinoma. BMC Cancer 2022, 22, 83. [Google Scholar] [CrossRef] [PubMed]
- De Bock, K.; Georgiadou, M.; Schoors, S.; Kuchnio, A.; Wong, B.W.; Cantelmo, A.R.; Quaegebeur, A.; Ghesquiere, B.; Cauwenberghs, S.; Eelen, G.; et al. Role of PFKFB3-driven glycolysis in vessel sprouting. Cell 2013, 154, 651–663. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Chen, Y.; Zhu, Y. The molecular basis of targeting PFKFB3 as a therapeutic strategy against cancer. Oncotarget 2017, 8, 62793–62802. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.; Ghosh, S.; Kumar, S. Tumor glycolysis, an essential sweet tooth of tumor cells. Semin. Cancer Biol. 2022, 86, 1216–1230. [Google Scholar] [CrossRef] [PubMed]
- Gregoriou, M.; Cornish-Bowden, A.; Trayer, I.P. Isotope-exchange evidence for allosteric regulation of hexokinase II by glucose 6-phosphate and for an obligatory addition of substrates. Biochem. Soc. Trans. 1981, 9, 62–63. [Google Scholar] [CrossRef] [PubMed]
- Gregoriou, M.; Trayer, I.P.; Cornish-Bowden, A. Isotope-exchange evidence that glucose 6-phosphate inhibits rat-muscle hexokinase II at an allosteric site. Eur. J. Biochem. 1983, 134, 283–288. [Google Scholar] [CrossRef] [PubMed]
- Lazo, P.A.; Bosca, L. Mitochondrial membrane-bound hexokinase of ascites tumor cells. Functional implications of lysine residues studied by modification with imidoesters. Functional implications of lysine residues studied by modification with imidoesters. Hoppe Seylers Z. Physiol. Chem. 1982, 363, 635–641. [Google Scholar] [CrossRef] [PubMed]
- Yao, X.; He, Z.; Qin, C.; Zhang, P.; Sui, C.; Deng, X.; Fang, Y.; Li, G.; Shi, J. Inhibition of PFKFB3 in HER2-positive gastric cancer improves sensitivity to trastuzumab by inducing tumour vessel normalisation. Br. J. Cancer 2022, 127, 811–823. [Google Scholar] [CrossRef]
- Hu, K.-F.; Shu, C.-W.; Lee, C.-H.; Tseng, C.-J.; Chou, Y.-H.; Liu, P.-F. Comparative clinical significance and biological roles of PFKFB family members in oral squamous cell carcinoma. Cancer Cell Int. 2023, 23, 257. [Google Scholar] [CrossRef]
- Kotowski, K.; Rosik, J.; Machaj, F.; Supplitt, S.; Wiczew, D.; Jabłońska, K.; Wiechec, E.; Ghavami, S.; Dzięgiel, P. Role of PFKFB3 and PFKFB4 in Cancer: Genetic Basis, Impact on Disease Development/Progression, and Potential as Therapeutic Targets. Cancers 2021, 13, 909. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Imbert-Fernandez, Y.; Chang, S.M.; Lanceta, L.; Sanders, N.M.; Chesney, J.; Clem, B.F.; Telang, S. Genomic Deletion of PFKFB3 Decreases In Vivo Tumorigenesis. Cancers 2024, 16, 2330. https://doi.org/10.3390/cancers16132330
Imbert-Fernandez Y, Chang SM, Lanceta L, Sanders NM, Chesney J, Clem BF, Telang S. Genomic Deletion of PFKFB3 Decreases In Vivo Tumorigenesis. Cancers. 2024; 16(13):2330. https://doi.org/10.3390/cancers16132330
Chicago/Turabian StyleImbert-Fernandez, Yoannis, Simone M. Chang, Lilibeth Lanceta, Nicole M. Sanders, Jason Chesney, Brian F. Clem, and Sucheta Telang. 2024. "Genomic Deletion of PFKFB3 Decreases In Vivo Tumorigenesis" Cancers 16, no. 13: 2330. https://doi.org/10.3390/cancers16132330
APA StyleImbert-Fernandez, Y., Chang, S. M., Lanceta, L., Sanders, N. M., Chesney, J., Clem, B. F., & Telang, S. (2024). Genomic Deletion of PFKFB3 Decreases In Vivo Tumorigenesis. Cancers, 16(13), 2330. https://doi.org/10.3390/cancers16132330





