Cadherin Expression Profiles Define Glioblastoma Differentiation and Patient Prognosis
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Patients and Materials
2.2. Neuropathological Evaluation
2.3. Immunohistochemical Staining
2.4. Data Mining from Publicly Available Datasets
2.5. Statistics
3. Results
3.1. Clinical and Neuropathological Characteristics of Glioblastoma Series
3.2. Cadherins’ Expression Associated with Clinical, Imaging and Neuropathological GBM Features
3.3. Cadherins’ Co-Expression Profiles Improve the Identification of GBM Subgroups
3.4. Cadherins Expression Profiles Define GBM Patients’ Prognosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miller, P.W.; Clarke, D.N.; Weis, W.I.; Lowe, C.J.; Nelson, W.J. The evolutionary origin of epithelial cell-cell adhesion mechanisms. Curr. Top. Membr. 2013, 72, 267–311. [Google Scholar] [CrossRef]
- Larue, L.; Antos, C.; Butz, S.; Huber, O.; Delmas, V.; Dominis, M.; Kemler, R. A role for cadherins in tissue formation. Development 1996, 122, 3185–3194. [Google Scholar] [CrossRef]
- Saito, M.; Tucker, D.K.; Kohlhorst, D.; Niessen, C.M.; Kowalczyk, A.P. Classical and desmosomal cadherins at a glance. J. Cell Sci. 2012, 125, 2547–2552. [Google Scholar] [CrossRef]
- Inuzuka, H.; Redies, C.; Takeichi, M. Differential expression of R- and N-cadherin in neural and mesodermal tissues during early chicken development. Development 1991, 113, 959–967. [Google Scholar] [CrossRef]
- Shimoyama, Y.; Tsujimoto, G.; Kitajima, M.; Natori, M. Identification of three human type-II classic cadherins and frequent heterophilic interactions between different subclasses of type-II classic cadherins. Biochem. J. 2000, 349, 159–167. [Google Scholar] [CrossRef]
- Cavallaro, U.; Christofori, G. Cell adhesion and signalling by cadherins and Ig-CAMs in cancer. Nat. Rev. Cancer 2004, 4, 118–132. [Google Scholar] [CrossRef]
- Niessen, C.M.; Leckband, D.; Yap, A.S. Tissue organization by cadherin adhesion molecules: Dynamic molecular and cellular mechanisms of morphogenetic regulation. Physiol. Rev. 2011, 91, 691–731. [Google Scholar] [CrossRef]
- Ratheesh, A.; Yap, A.S. A bigger picture: Classical cadherins and the dynamic actin cytoskeleton. Nat. Rev. Mol. Cell Biol. 2012, 13, 673–679. [Google Scholar] [CrossRef]
- Wheelock, M.J.; Johnson, K.R. Cadherin-mediated cellular signaling. Curr. Opin. Cell Biol. 2003, 15, 509–514. [Google Scholar] [CrossRef]
- Vestweber, D. Cadherins in tissue architecture and disease. J. Mol. Med. 2015, 93, 5–11. [Google Scholar] [CrossRef]
- Birchmeier, W.; Behrens, J. Cadherin expression in carcinomas: Role in the formation of cell junctions and the prevention of invasiveness. Biochim. Biophys. Acta 1994, 1198, 11–26. [Google Scholar] [CrossRef] [PubMed]
- Bracke, M.E.; Van Roy, F.M.; Mareel, M.M. The E-cadherin/catenin complex in invasion and metastasis. Curr. Top. Microbiol. Immunol. 1996, 213 Pt 1, 123–161. [Google Scholar] [CrossRef]
- Frixen, U.H.; Behrens, J.; Sachs, M.; Eberle, G.; Voss, B.; Warda, A.; Lochner, D.; Birchmeier, W. E-cadherin-mediated cell-cell adhesion prevents invasiveness of human carcinoma cells. J. Cell Biol. 1991, 113, 173–185. [Google Scholar] [CrossRef]
- Paredes, J.; Figueiredo, J.; Albergaria, A.; Oliveira, P.; Carvalho, J.; Ribeiro, A.S.; Caldeira, J.; Costa, A.M.; Simoes-Correia, J.; Oliveira, M.J.; et al. Epithelial E- and P-cadherins: Role and clinical significance in cancer. Biochim. Biophys. Acta 2012, 1826, 297–311. [Google Scholar] [CrossRef] [PubMed]
- Hazan, R.B.; Qiao, R.; Keren, R.; Badano, I.; Suyama, K. Cadherin switch in tumor progression. Ann. N. Y. Acad. Sci. 2004, 1014, 155–163. [Google Scholar] [CrossRef]
- Li, H.; Leung, T.C.; Hoffman, S.; Balsamo, J.; Lilien, J. Coordinate regulation of cadherin and integrin function by the chondroitin sulfate proteoglycan neurocan. J. Cell Biol. 2000, 149, 1275–1288. [Google Scholar] [CrossRef]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Tomita, K.; van Bokhoven, A.; van Leenders, G.J.; Ruijter, E.T.; Jansen, C.F.; Bussemakers, M.J.; Schalken, J.A. Cadherin switching in human prostate cancer progression. Cancer Res. 2000, 60, 3650–3654. [Google Scholar]
- Hu, Q.P.; Kuang, J.Y.; Yang, Q.K.; Bian, X.W.; Yu, S.C. Beyond a tumor suppressor: Soluble E-cadherin promotes the progression of cancer. Int. J. Cancer 2016, 138, 2804–2812. [Google Scholar] [CrossRef]
- Lammens, T.; Swerts, K.; Derycke, L.; De Craemer, A.; De Brouwer, S.; De Preter, K.; Van Roy, N.; Vandesompele, J.; Speleman, F.; Philippe, J.; et al. N-cadherin in neuroblastoma disease: Expression and clinical significance. PLoS ONE 2012, 7, e31206. [Google Scholar] [CrossRef]
- Padmanaban, V.; Krol, I.; Suhail, Y.; Szczerba, B.M.; Aceto, N.; Bader, J.S.; Ewald, A.J. E-cadherin is required for metastasis in multiple models of breast cancer. Nature 2019, 573, 439–444. [Google Scholar] [CrossRef]
- Rodriguez, F.J.; Lewis-Tuffin, L.J.; Anastasiadis, P.Z. E-cadherin’s dark side: Possible role in tumor progression. Biochim. Biophys. Acta 2012, 1826, 23–31. [Google Scholar] [CrossRef]
- Ribeiro, A.S.; Sousa, B.; Carreto, L.; Mendes, N.; Nobre, A.R.; Ricardo, S.; Albergaria, A.; Cameselle-Teijeiro, J.F.; Gerhard, R.; Soderberg, O.; et al. P-cadherin functional role is dependent on E-cadherin cellular context: A proof of concept using the breast cancer model. J. Pathol. 2013, 229, 705–718. [Google Scholar] [CrossRef]
- Kroger, C.; Afeyan, A.; Mraz, J.; Eaton, E.N.; Reinhardt, F.; Khodor, Y.L.; Thiru, P.; Bierie, B.; Ye, X.; Burge, C.B.; et al. Acquisition of a hybrid E/M state is essential for tumorigenicity of basal breast cancer cells. Proc. Natl. Acad. Sci. USA 2019, 116, 7353–7362. [Google Scholar] [CrossRef]
- Brown, M.S.; Muller, K.E.; Pattabiraman, D.R. Quantifying the Epithelial-to-Mesenchymal Transition (EMT) from Bench to Bedside. Cancers 2022, 14, 1138. [Google Scholar] [CrossRef]
- Appolloni, I.; Barilari, M.; Caviglia, S.; Gambini, E.; Reisoli, E.; Malatesta, P. A cadherin switch underlies malignancy in high-grade gliomas. Oncogene 2015, 34, 1991–2002. [Google Scholar] [CrossRef]
- Asano, K.; Duntsch, C.D.; Zhou, Q.; Weimar, J.D.; Bordelon, D.; Robertson, J.H.; Pourmotabbed, T. Correlation of N-cadherin expression in high grade gliomas with tissue invasion. J. Neurooncol. 2004, 70, 3–15. [Google Scholar] [CrossRef]
- Noh, M.G.; Oh, S.J.; Ahn, E.J.; Kim, Y.J.; Jung, T.Y.; Jung, S.; Kim, K.K.; Lee, J.H.; Lee, K.H.; Moon, K.S. Prognostic significance of E-cadherin and N-cadherin expression in Gliomas. BMC Cancer 2017, 17, 583. [Google Scholar] [CrossRef]
- Noronha, C.; Ribeiro, A.S.; Taipa, R.; Castro, D.S.; Reis, J.; Faria, C.; Paredes, J. Cadherin Expression and EMT: A Focus on Gliomas. Biomedicines 2021, 9, 1328. [Google Scholar] [CrossRef]
- Alexander, B.M.; Cloughesy, T.F. Adult Glioblastoma. J. Clin. Oncol. 2017, 35, 2402–2409. [Google Scholar] [CrossRef]
- Weller, M. Challenges in the diagnoses and treatment of CNS tumors. Neurooncol. Pract. 2019, 6, 329. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Gittleman, H.; Patil, N.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2012–2016. Neuro Oncol. 2019, 21 (Suppl. S5), v1–v100. [Google Scholar] [CrossRef]
- Weller, M.; Tabatabai, G.; Kastner, B.; Felsberg, J.; Steinbach, J.P.; Wick, A.; Schnell, O.; Hau, P.; Herrlinger, U.; Sabel, M.C.; et al. MGMT Promoter Methylation Is a Strong Prognostic Biomarker for Benefit from Dose-Intensified Temozolomide Rechallenge in Progressive Glioblastoma: The DIRECTOR Trial. Clin. Cancer Res. 2015, 21, 2057–2064. [Google Scholar] [CrossRef]
- Verhaak, R.G.; Hoadley, K.A.; Purdom, E.; Wang, V.; Qi, Y.; Wilkerson, M.D.; Miller, C.R.; Ding, L.; Golub, T.; Mesirov, J.P.; et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 2010, 17, 98–110. [Google Scholar] [CrossRef]
- Nagaishi, M.; Paulus, W.; Brokinkel, B.; Vital, A.; Tanaka, Y.; Nakazato, Y.; Giangaspero, F.; Ohgaki, H. Transcriptional factors for epithelial-mesenchymal transition are associated with mesenchymal differentiation in gliosarcoma. Brain Pathol. 2012, 22, 670–676. [Google Scholar] [CrossRef]
- Wood, M.D.; Reis, G.F.; Reuss, D.E.; Phillips, J.J. Protein Analysis of Glioblastoma Primary and Posttreatment Pairs Suggests a Mesenchymal Shift at Recurrence. J. Neuropathol. Exp. Neurol. 2016, 75, 925–935. [Google Scholar] [CrossRef]
- Asano, K.; Kubo, O.; Tajika, Y.; Huang, M.C.; Takakura, K.; Ebina, K.; Suzuki, S. Expression and role of cadherins in astrocytic tumors. Brain Tumor Pathol. 1997, 14, 27–33. [Google Scholar] [CrossRef]
- Asano, K.; Kubo, O.; Tajika, Y.; Takakura, K.; Suzuki, S. Expression of cadherin and CSF dissemination in malignant astrocytic tumors. Neurosurg. Rev. 2000, 23, 39–44. [Google Scholar] [CrossRef]
- Camand, E.; Peglion, F.; Osmani, N.; Sanson, M.; Etienne-Manneville, S. N-cadherin expression level modulates integrin-mediated polarity and strongly impacts on the speed and directionality of glial cell migration. J. Cell Sci. 2012, 125, 844–857. [Google Scholar] [CrossRef]
- Utsuki, S.; Oka, H.; Miyajima, Y.; Kijima, C.; Yasui, Y.; Fujii, K. Adult cerebellar glioblastoma cases have different characteristics from supratentorial glioblastoma. Brain Tumor Pathol. 2012, 29, 87–95. [Google Scholar] [CrossRef]
- Wu, W.; Tian, Y.; Wan, H.; Ma, J.; Song, Y.; Wang, Y.; Zhang, L. Expression of beta-catenin and E- and N-cadherin in human brainstem gliomas and clinicopathological correlations. Int. J. Neurosci. 2013, 123, 318–323. [Google Scholar] [CrossRef]
- Osuka, S.; Zhu, D.; Zhang, Z.; Li, C.; Stackhouse, C.T.; Sampetrean, O.; Olson, J.J.; Gillespie, G.Y.; Saya, H.; Willey, C.D.; et al. N-cadherin upregulation mediates adaptive radioresistance in glioblastoma. J. Clin. Investig. 2021, 131, e136098. [Google Scholar] [CrossRef]
- Utsuki, S.; Sato, Y.; Oka, H.; Tsuchiya, B.; Suzuki, S.; Fujii, K. Relationship between the expression of E-, N-cadherins and beta-catenin and tumor grade in astrocytomas. J. Neurooncol. 2002, 57, 187–192. [Google Scholar] [CrossRef]
- Bar, J.K.; Zub, L.; Lis-Nawara, A.; Noga, L.; Jelen, M.; Paradowski, B. Expression and Interactions between Cell Adhesion Molecules CD44v6 and E-Cadherin in Human Gliomas. Adv. Clin. Exp. Med. 2014, 23, 827–834. [Google Scholar] [CrossRef]
- Motta, F.J.; Valera, E.T.; Lucio-Eterovic, A.K.; Queiroz, R.G.; Neder, L.; Scrideli, C.A.; Machado, H.R.; Carlotti-Junior, C.G.; Marie, S.K.; Tone, L.G. Differential expression of E-cadherin gene in human neuroepithelial tumors. Genet. Mol. Res. 2008, 7, 295–304. [Google Scholar] [CrossRef]
- Martins, E.P.; Goncalves, C.S.; Pojo, M.; Carvalho, R.; Ribeiro, A.S.; Miranda-Goncalves, V.; Taipa, R.; Pardal, F.; Pinto, A.A.; Custodia, C.; et al. Cadherin-3 is a novel oncogenic biomarker with prognostic value in glioblastoma. Mol. Oncol. 2022, 16, 2611–2631. [Google Scholar] [CrossRef]
- Brennan, C.W.; Verhaak, R.G.; McKenna, A.; Campos, B.; Noushmehr, H.; Salama, S.R.; Zheng, S.; Chakravarty, D.; Sanborn, J.Z.; Berman, S.H.; et al. The somatic genomic landscape of glioblastoma. Cell 2013, 155, 462–477. [Google Scholar] [CrossRef]
- Stupp, R.; Mason, W.P.; van den Bent, M.J.; Weller, M.; Fisher, B.; Taphoorn, M.J.; Belanger, K.; Brandes, A.A.; Marosi, C.; Bogdahn, U.; et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 2005, 352, 987–996. [Google Scholar] [CrossRef]
- Xie, J.; Feng, Y.; Lin, T.; Huang, X.Y.; Gan, R.H.; Zhao, Y.; Su, B.H.; Ding, L.C.; She, L.; Chen, J.; et al. CDH4 suppresses the progression of salivary adenoid cystic carcinoma via E-cadherin co-expression. Oncotarget 2016, 7, 82961–82971. [Google Scholar] [CrossRef]
- Ma, C.; Zhao, J.Z.; Lin, R.T.; Zhou, L.; Chen, Y.N.; Yu, L.J.; Shi, T.Y.; Wang, M.; Liu, M.M.; Liu, Y.R.; et al. Combined overexpression of cadherin 6, cadherin 11 and cluster of differentiation 44 is associated with lymph node metastasis and poor prognosis in oral squamous cell carcinoma. Oncol. Lett. 2018, 15, 9498–9506. [Google Scholar] [CrossRef]
- Ribeiro, A.S.; Paredes, J. P-Cadherin Linking Breast Cancer Stem Cells and Invasion: A Promising Marker to Identify an “Intermediate/Metastable” EMT State. Front. Oncol. 2014, 4, 371. [Google Scholar] [CrossRef] [PubMed]
- Lewis-Tuffin, L.J.; Rodriguez, F.; Giannini, C.; Scheithauer, B.; Necela, B.M.; Sarkaria, J.N.; Anastasiadis, P.Z. Misregulated E-cadherin expression associated with an aggressive brain tumor phenotype. PLoS ONE 2010, 5, e13665. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, M.; Aladowicz, E.; Lanfrancone, L.; Goding, C.R. Tbx3 represses E-cadherin expression and enhances melanoma invasiveness. Cancer Res. 2008, 68, 7872–7881. [Google Scholar] [CrossRef] [PubMed]
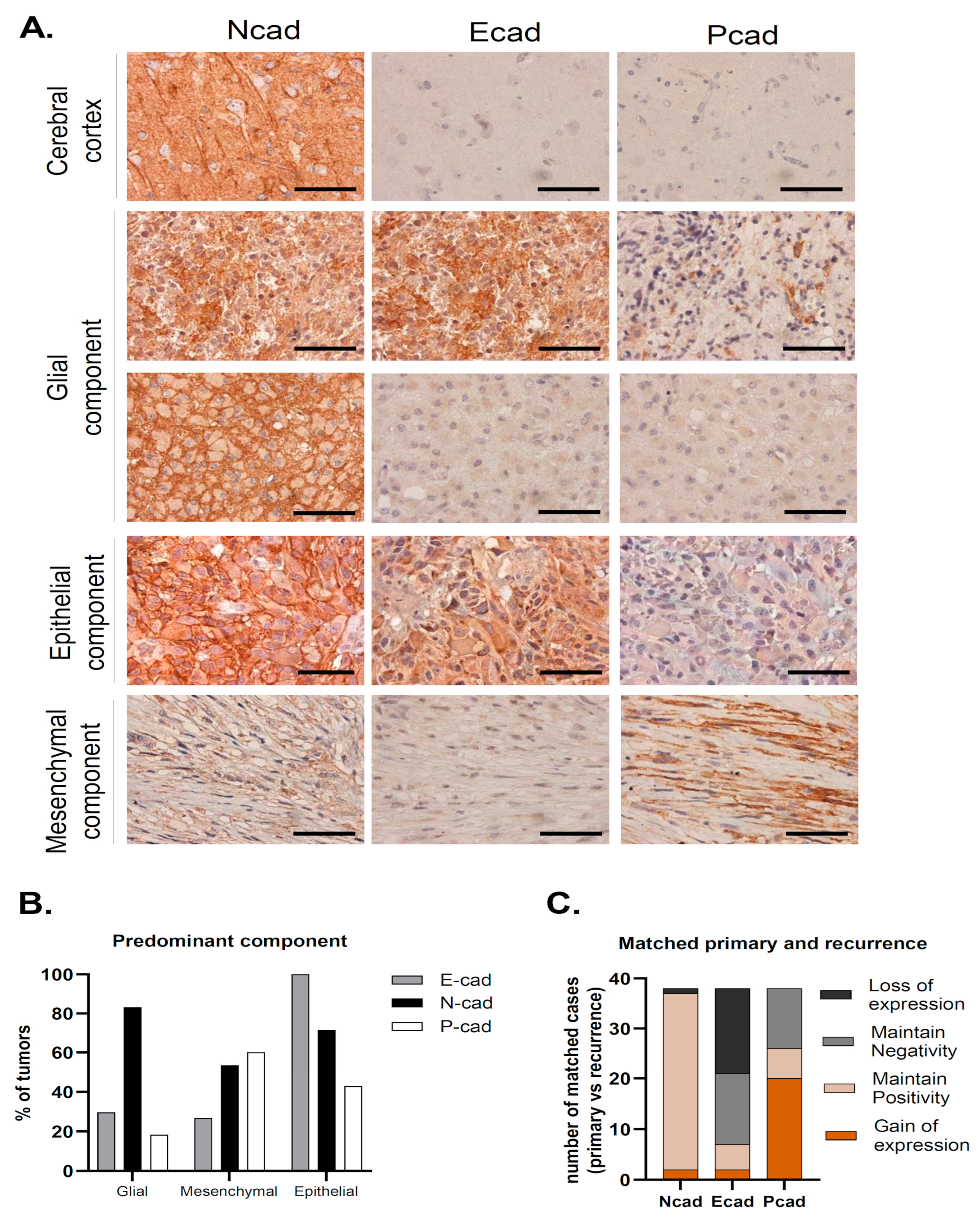
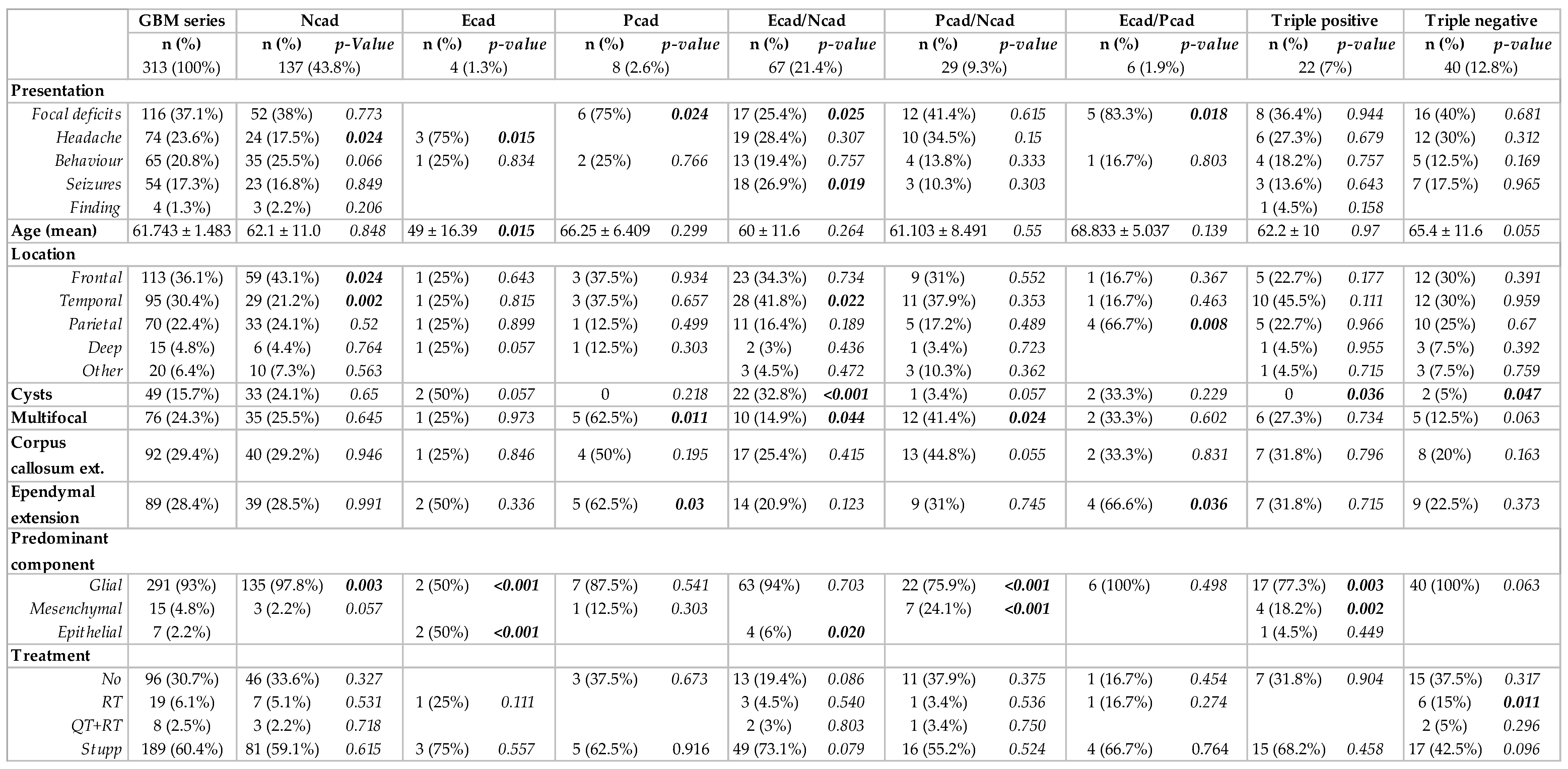
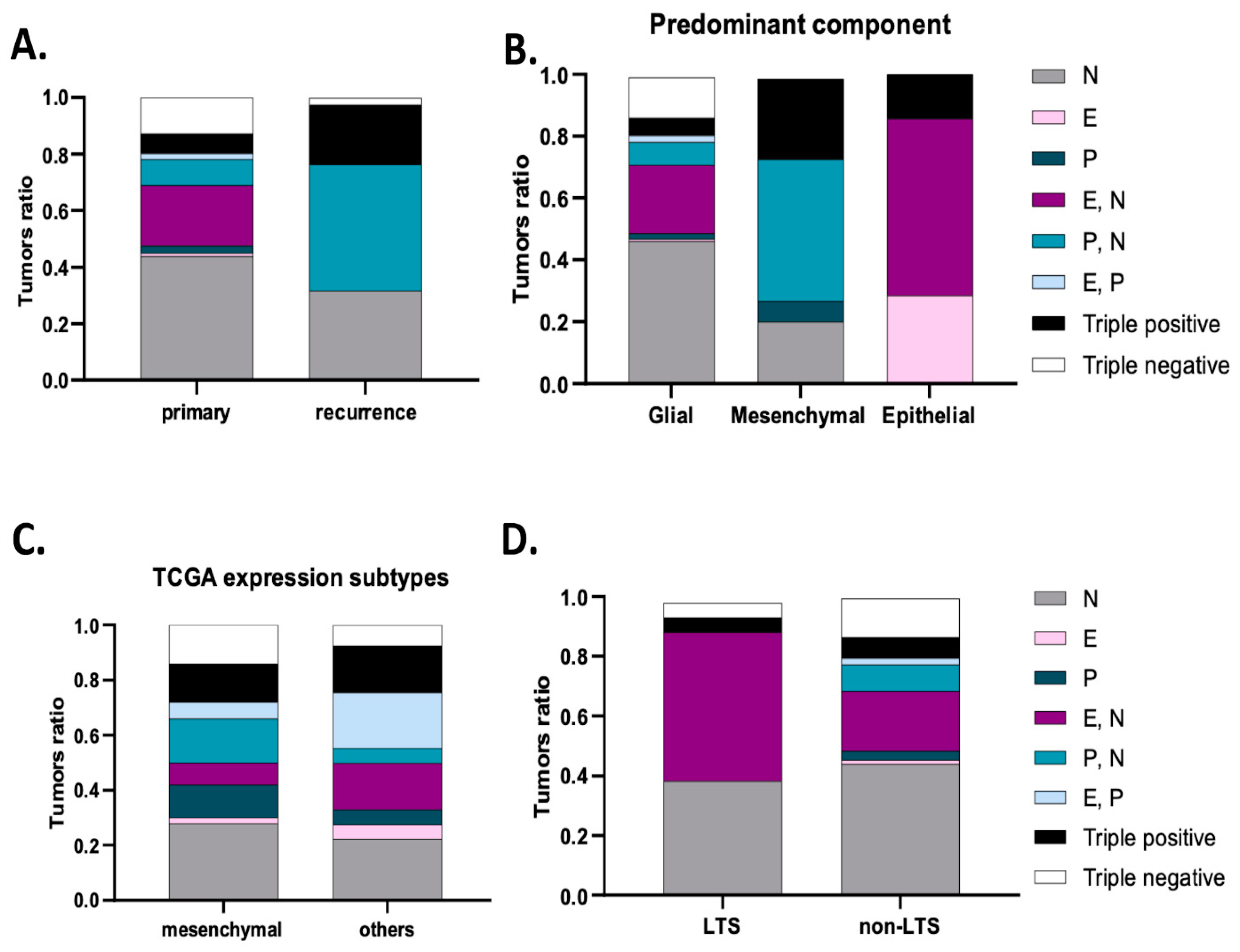

| GBM Series | N-Cadherin Positive | E-Cadherin Positive | P-Cadherin Positive | ||||
|---|---|---|---|---|---|---|---|
| n (%) | n (%) | p-Value | n (%) | p-Value | n (%) | p-Value | |
| 313 (100%) | 255 (81.5%) | 97 (31%) | 65 (20.8%) | ||||
| Papillary structures | 17 (5.4%) | 15 (5.9%) | 0.46 | 17 (17.5%) | <0.001 | 6 (9.2%) | 0.126 |
| Whorls | 8 (2.6%) | 5 (2%) | 0.162 | 4 (4.1%) | 0.239 | 6 (9.2%) | <0.001 |
| Glomeruloid vessels | 47 (15%) | 39 (15.3%) | 0.773 | 20 (20.6%) | 0.063 | 9 (13.8%) | 0.767 |
| Myxoid stroma | 18 (5.7%) | 16 (6.3%) | 0.404 | 9 (9.3%) | 0.072 | 4 (6.2%) | 0.875 |
| Inflammation | 80 (25.6%) | 69 (27%) | 0.502 | 39 (40.2%) | 0.001 | 33 (50.8%) | <0.001 |
| Calcifications | 10 (3.2%) | 10 (3.9%) | 0.125 | 3 (3.1%) | 0.945 | 0 | 0.1 |
| Oligodendroglial component | 36 (11.7%) | 32 (12.5%) | 0.643 | 9 (9.3%) | 0.299 | 0 | 0.014 |
| Gemistocytes | 49 (15.7%) | 49 (19.5%) | <0.001 | 13 (13.4%) | 0.424 | 5 (7.8%) | 0.058 |
| Glial component | 312 (99.7%) | 254 (99.6%) | 0.633 | 96 (99%) | 0.135 | 65 (100%) | 0.608 |
| Mesenchymal component | 63 (20.1%) | 44 (17.3%) | 0.75 | 25 (25.8%) | 0.005 | 43 (66.2%) | <0.001 |
| Epithelial component | 41 (13.1%) | 36 (14.1%) | 0.263 | 39 (40.2%) | <0.001 | 15 (23%) | 0.007 |
| Predominant component | |||||||
| Glial | 291 (93%) | 242 (94.9%) | 0.981 | 86 (88.7%) | <0.001 | 53 (81.5%) | 0.001 |
| Mesenchymal | 15 (4.8%) | 8 (3.1%) | 0.561 | 4 (4.1%) | 0.376 | 9 (13.8%) | <0.001 |
| Epithelial | 7 (2.2%) | 5 (2%) | 0.489 | 7 (7.2%) | <0.001 | 3 (4.6%) | 0.145 |
| Tumor recurrence | 38 | 37 (97.4%) | 8 (20.5%) | 25 (65.8%) | |||
| Progression-Free Survival | Overall Survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95.0% CI | p-Value | HR | 95.0% CI | p-Value | ||||
| Inferior | Superior | Inferior | Superior | ||||||
| Surgery | Surgery | 1 | 1 | ||||||
| Biopsy | 2.120 | 1.626 | 2.763 | <0.001 | 2.329 | 1.842 | 2.945 | <0.001 | |
| Treatment | Yes | 1 | 1 | ||||||
| No | 13.244 | 8.492 | 20.665 | <0.001 | 4.730 | 3.547 | 6.308 | <0.001 | |
| KPS | 100 | 1 | 1 | ||||||
| 50 | 0.505 | 0.056 | 4.550 | 0.543 | 1.772 | 1.183 | 2.654 | 0.006 | |
| 60 | 0.580 | 0.078 | 4.318 | 0.595 | 1.411 | 0.866 | 2.298 | 0.167 | |
| 70 | 0.587 | 0.080 | 4.295 | 0.600 | 2.575 | 1.223 | 5.421 | 0.013 | |
| 80 | 0.430 | 0.059 | 3.105 | 0.403 | 2.596 | 1.035 | 6.508 | 0.042 | |
| 90 | 0.391 | 0.054 | 2.826 | 0.352 | 1.599 | 1.022 | 2.503 | 0.040 | |
| Age | < 65 yo | 1 | 1 | ||||||
| > 65 yo | 1.258 | 0.974 | 1.625 | 0.079 | 1.400 | 1.113 | 1.762 | 0.004 | |
| Ncad | Negative | 1 | 1 | ||||||
| Positive | 0.725 | 0.522 | 1.007 | 0.055 | 0.627 | 0.468 | 0.840 | 0.002 | |
| Ecad | Negative | 1 | 1 | ||||||
| Positive | 0.716 | 0.546 | 0.938 | 0.016 | 0.779 | 0.608 | 0.998 | 0.048 | |
| Pcad | Negative | 1 | 1 | ||||||
| Positive | 1.367 | 1.003 | 1.863 | 0.048 | 1.364 | 1.029 | 1.806 | 0.031 | |
| Cadherins co-expression | Ecad/Ncad | 1 | 1 | ||||||
| Ncad | 1.367 | 0.984 | 1.900 | 0.063 | 1.316 | 0.969 | 1.787 | 0.079 | |
| Ecad | 1.507 | 0.471 | 4.826 | 0.490 | 2.550 | 0.923 | 7.041 | 0.071 | |
| Pcad | 1.864 | 0.884 | 3.933 | 0.102 | 2.611 | 1.239 | 5.502 | 0.012 | |
| Pcad/Ncad | 1.910 | 1.186 | 3.076 | 0.008 | 1.619 | 1.033 | 2.536 | 0.036 | |
| Ecad/Pcad | 2.203 | 1.068 | 5.868 | 0.035 | 2.728 | 1.169 | 6.366 | 0.020 | |
| Triple positive | 1.194 | 0.683 | 2.087 | 0.533 | 1.427 | 0.875 | 2.328 | 0.154 | |
| Triple negative | 1.712 | 1.084 | 2.704 | 0.021 | 1.794 | 1.196 | 2.690 | 0.005 | |
| Progression-Free Survival | Overall Survival | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| HR | 95.0% CI | p-Value | HR | 95.0% CI | p-Value | ||||
| Inferior | Superior | Inferior | Superior | ||||||
| Mesenchymal Component | yes | 1.000 | 1.000 | ||||||
| no | 1.060 | 0.670 | 1.660 | 0.811 | 0.810 | 0.530 | 1.250 | 0.345 | |
| Epithelial Component | yes | 1.000 | 1.000 | ||||||
| no | 0.930 | 0.580 | 1.470 | 0.743 | 0.720 | 0.450 | 1.140 | 0.160 | |
| Cadherins co-expression | Ecad/N-cad | 1.000 | 1.000 | ||||||
| Ncad | 0.990 | 0.690 | 1.440 | 0.979 | 1.300 | 0.900 | 1.870 | 0.162 | |
| Ecad | 2.200 | 0.650 | 7.430 | 0.205 | 4.240 | 1.290 | 13.930 | 0.017 | |
| Pcad | 1.330 | 0.600 | 2.980 | 0.482 | 2.330 | 1.050 | 5.130 | 0.036 | |
| Pcad/Ncad | 1.340 | 0.770 | 2.360 | 0.303 | 1.440 | 0.850 | 2.460 | 0.178 | |
| Ecad/Pcad | 2.960 | 1.180 | 7.400 | 0.020 | 3.350 | 1.330 | 8.440 | 0.010 | |
| Triple positive | 1.190 | 0.640 | 2.210 | 0.577 | 2.110 | 1.190 | 3.760 | 0.011 | |
| Triple negative | 1.090 | 0.660 | 1.820 | 0.728 | 1.490 | 0.910 | 2.450 | 0.113 | |
| Surgery | Surgery | 1.000 | 1.000 | ||||||
| Biopsy | 1.760 | 1.290 | 2.390 | <0.001 | 1.550 | 1.170 | 2.060 | 0.003 | |
| Treatment | yes | 1.000 | 1.000 | ||||||
| no | 15.000 | 9.020 | 24.930 | <0.001 | 4.570 | 3.300 | 6.330 | <0.0001 | |
| KPS | 100 | 1.000 | 1.000 | ||||||
| 50 | 0.150 | 0.020 | 1.390 | 0.094 | 0.700 | 0.080 | 6.170 | 0.752 | |
| 60 | 0.250 | 0.030 | 1.900 | 0.179 | 2.400 | 0.320 | 18.040 | 0.396 | |
| 70 | 0.240 | 0.030 | 1.790 | 0.162 | 2.190 | 0.300 | 16.220 | 0.442 | |
| 80 | 0.210 | 0.030 | 1.550 | 0.125 | 1.520 | 0.210 | 11.150 | 0.681 | |
| 90 | 0.200 | 0.030 | 1.510 | 0.118 | 1.310 | 0.180 | 9.660 | 0.790 | |
| Age | <65 yo | 1.000 | 1.000 | ||||||
| >65 yo | 1.210 | 0.920 | 1.600 | 0.176 | 1.270 | 0.980 | 1.660 | 0.072 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Noronha, C.; Ribeiro, A.S.; Carvalho, R.; Mendes, N.; Reis, J.; Faria, C.C.; Taipa, R.; Paredes, J. Cadherin Expression Profiles Define Glioblastoma Differentiation and Patient Prognosis. Cancers 2024, 16, 2298. https://doi.org/10.3390/cancers16132298
Noronha C, Ribeiro AS, Carvalho R, Mendes N, Reis J, Faria CC, Taipa R, Paredes J. Cadherin Expression Profiles Define Glioblastoma Differentiation and Patient Prognosis. Cancers. 2024; 16(13):2298. https://doi.org/10.3390/cancers16132298
Chicago/Turabian StyleNoronha, Carolina, Ana Sofia Ribeiro, Rita Carvalho, Nuno Mendes, Joaquim Reis, Claudia C. Faria, Ricardo Taipa, and Joana Paredes. 2024. "Cadherin Expression Profiles Define Glioblastoma Differentiation and Patient Prognosis" Cancers 16, no. 13: 2298. https://doi.org/10.3390/cancers16132298
APA StyleNoronha, C., Ribeiro, A. S., Carvalho, R., Mendes, N., Reis, J., Faria, C. C., Taipa, R., & Paredes, J. (2024). Cadherin Expression Profiles Define Glioblastoma Differentiation and Patient Prognosis. Cancers, 16(13), 2298. https://doi.org/10.3390/cancers16132298







