Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Sets
2.2. Study Subject Characteristics
2.3. Data Preparation
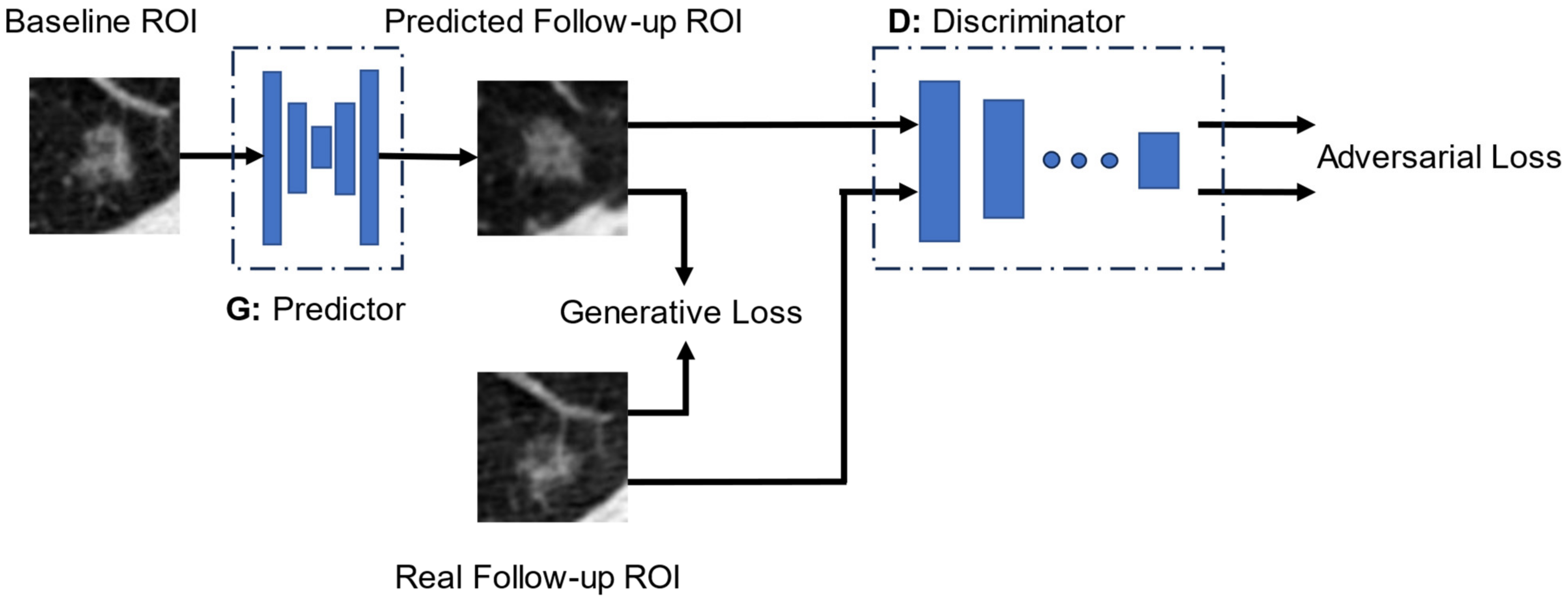
2.4. Growth Predictive Model Based on the Wasserstein Generative Adversarial Network
2.5. Generative Loss Function
2.6. Discriminator Loss Function
2.7. Performance Evaluation and Statistical Analysis
3. Results
Net Reclassification Improvement in Lung Cancer Risk Stratification
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A. Technical Information of LDCT Images
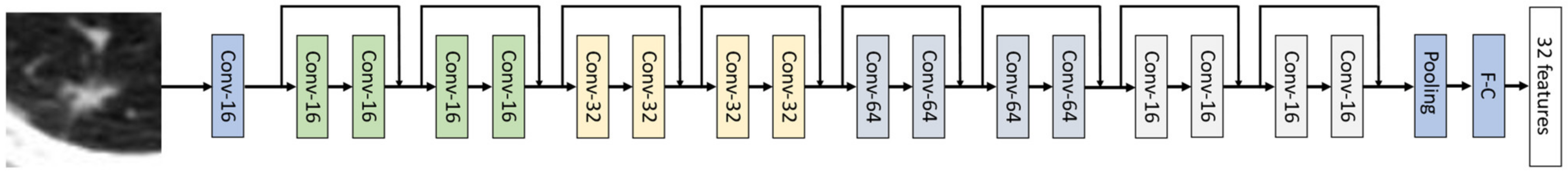
Appendix B. Network Structures of Predictor and Discriminator

Appendix C. Losses in the Generative Loss Function
- (1)
- loss measured the average absolute pixel-wise difference between the predicted and the real follow-up images:
- (2)
- Structural similarity index (SSIM) loss () quantified the degradation of structural information between two images (x and y) with three comparative measures: luminance l, contrast c, and structure s:
- (3)
- Learned perceptual loss () was a perceptual metric for assessing dissimilarities in feature spaces that encompass disparities in content and style discrepancies between images. We utilized a pre-trained modified Resnet-18 model (26) (described in Section Deep Residual Neural Network (ResNet-18) for Learned Perceptual Loss ()) to extract deep radiomics features for characterizing lung nodules. The was calculated by averaging the difference between the features extracted from the predicted follow-up image and the real image . The features were output from the last hidden layer of the Resnet-18 network.
- (4)
- Adversarial loss () was calculated from the discriminator’s classification result. Based on the principle that the predictor network should receive a reward when its synthesized image successfully deceives the discriminator (D), and be penalized otherwise, we defined the adversarial loss as
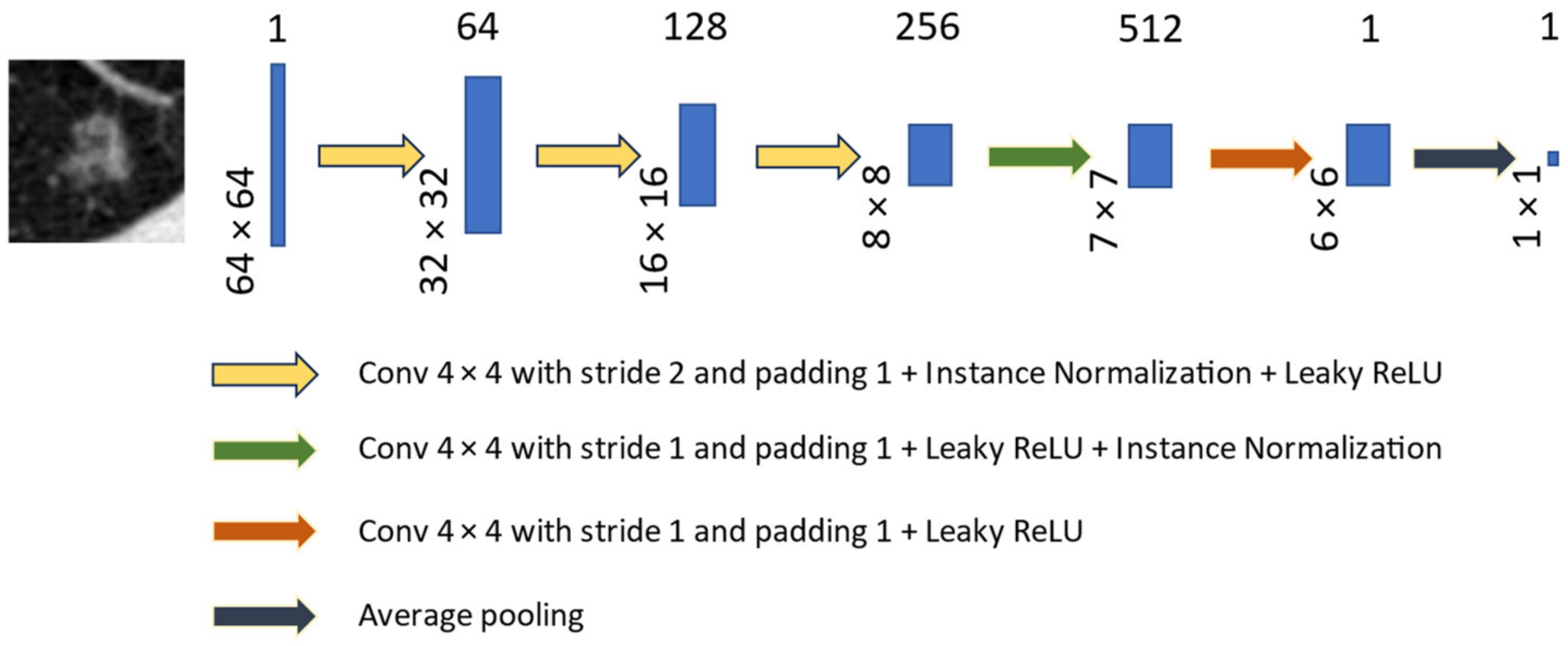
Deep Residual Neural Network (ResNet-18) for Learned Perceptual Loss ()
Appendix D. Net Reclassification Index (NRI)
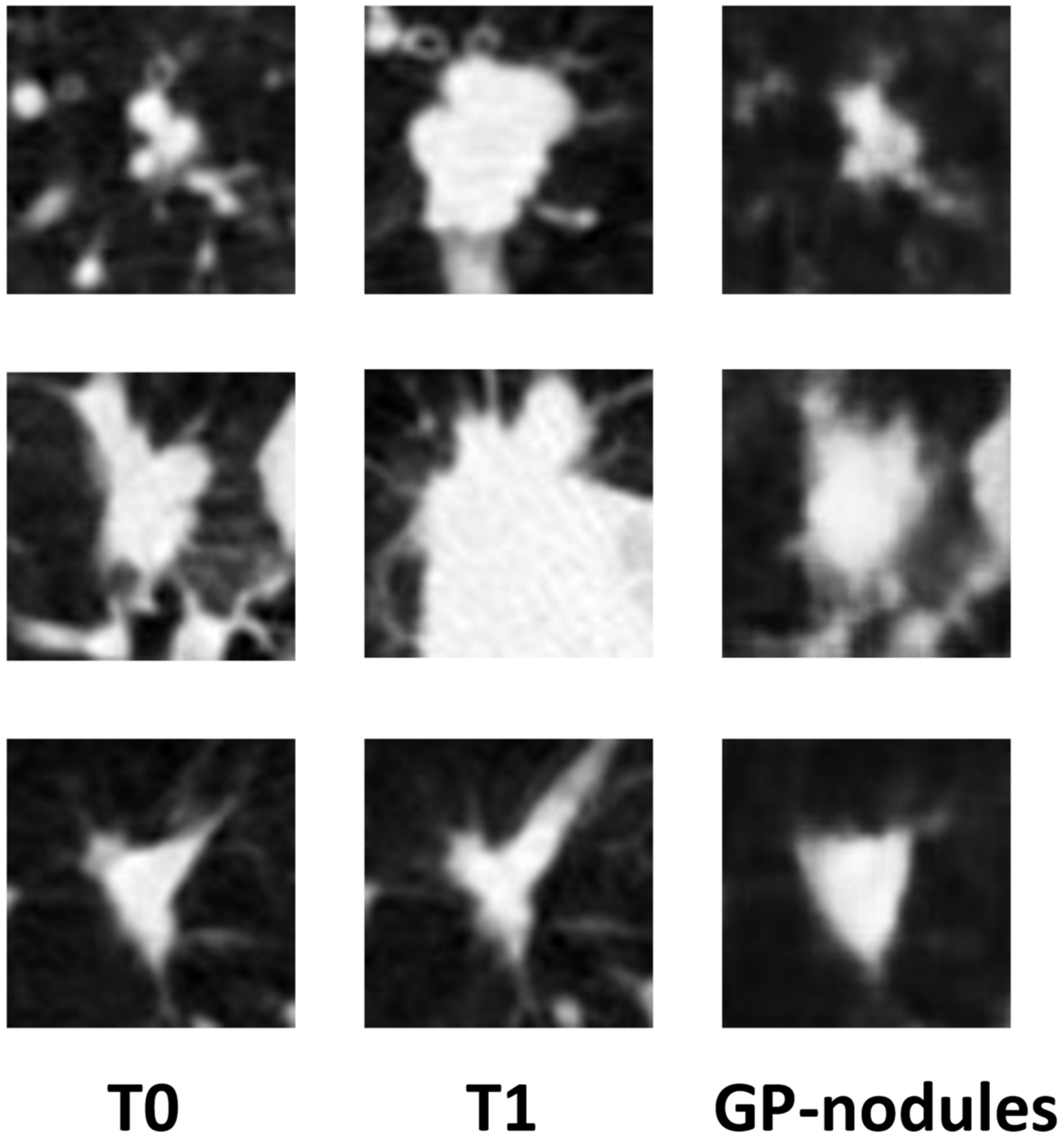
Appendix E. Examples of the Limited Capability of GP-WGAN in Predicting Nodule Growth Rates
References
- Cancer Stat Facts: Lung and Bronchus Cancer. Available online: https://seer.cancer.gov/statfacts/html/lungb.html (accessed on 1 September 2022).
- Howlader, N.; Noone, A.M.; Krapcho, M.; Miller, D.; Brest, A.; Yu, M.; Ruhl, J.; Tatalovich, Z.; Mariotto, A.; Lewis, D.R.; et al. SEER Cancer Statistics Review, 1975–2018. Available online: https://seer.cancer.gov/archive/csr/1975_2018/index.html (accessed on 1 September 2022).
- Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; Sicks, J.D. Reduced lung-cancer mortality with low-dose computed tomographic screening. N. Engl. J. Med. 2011, 365, 395–409. [Google Scholar] [CrossRef] [PubMed]
- McKee, B.J.; Regis, S.M.; McKee, A.B.; Flacke, S.; Wald, C. Performance of ACR Lung-RADS in a clinical CT lung screening program. J. Am. Coll. Radiol. 2016, 13, R25–R29. [Google Scholar] [CrossRef] [PubMed]
- Tao, G.; Zhu, L.; Chen, Q.; Yin, L.; Li, Y.; Yang, J.; Ni, B.; Zhang, Z.; Koo, C.W.; Patil, P.D. Prediction of future imagery of lung nodule as growth modeling with follow-up computed tomography scans using deep learning: A retrospective cohort study. Transl. Lung Cancer Res. 2022, 11, 250. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.J.; Sun, Y.L.; Li, D.C.; Ma, Z.X.; Jin, L. Prediction of pulmonary nodule growth: Current status and perspectives. J. Clin. Images Med. Case Rep. 2023, 4, 2393. [Google Scholar]
- Tabassum, S.; Rosli, N.B.; Binti Mazalan, M.S.A. Mathematical modeling of cancer growth process: A review. J. Phys. Conf. Ser. 2019, 1366, 012018. [Google Scholar] [CrossRef]
- Tan, M.; Ma, W.; Sun, Y.; Gao, P.; Huang, X.; Lu, J.; Chen, W.; Wu, Y.; Jin, L.; Tang, L. Prediction of the growth rate of early-stage lung adenocarcinoma by radiomics. Front. Oncol. 2021, 11, 658138. [Google Scholar] [CrossRef] [PubMed]
- Yang, R.; Hui, D.; Li, X.; Wang, K.; Li, C.; Li, Z. Prediction of single pulmonary nodule growth by CT radiomics and clinical features—A one-year follow-up study. Front. Oncol. 2022, 12, 1034817. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.; Deng, J.; She, Y.; Zhang, L.; Ren, Y.; Sun, W.; Su, H.; Dai, C.; Jiang, G.; Sun, X. Quantitative features can predict further growth of persistent pure ground-glass nodule. Quant. Imaging Med. Surg. 2019, 9, 283. [Google Scholar] [CrossRef]
- Krishnamurthy, S.; Narasimhan, G.; Rengasamy, U. Lung nodule growth measurement and prediction using auto cluster seed K-means morphological segmentation and shape variance analysis. Int. J. Biomed. Eng. Technol. 2017, 24, 53–71. [Google Scholar] [CrossRef]
- Esteva, A.; Robicquet, A.; Ramsundar, B.; Kuleshov, V.; DePristo, M.; Chou, K.; Cui, C.; Corrado, G.; Thrun, S.; Dean, J. A guide to deep learning in healthcare. Nat. Med. 2019, 25, 24–29. [Google Scholar] [CrossRef]
- Zhang, L.; Lu, L.; Wang, X.; Zhu, R.M.; Bagheri, M.; Summers, R.M.; Yao, J. Spatio-temporal convolutional LSTMs for tumor growth prediction by learning 4D longitudinal patient data. IEEE Trans. Med. Imaging 2019, 39, 1114–1126. [Google Scholar] [CrossRef] [PubMed]
- Rafael-Palou, X.; Aubanell, A.; Ceresa, M.; Ribas, V.; Piella, G.; Ballester, M.A.G. Prediction of lung nodule progression with an uncertainty-aware hierarchical probabilistic network. Diagnostics 2022, 12, 2639. [Google Scholar] [CrossRef] [PubMed]
- Sheng, J.; Li, Y.; Cao, G.; Hou, K. Modeling nodule growth via spatial transformation for follow-up prediction and diagnosis. In Proceedings of the 2021 International Joint Conference on Neural Networks (IJCNN), Shenzhen, China, 18–22 July 2021; pp. 1–7. [Google Scholar]
- Li, M.; Tang, H.; Chan, M.D.; Zhou, X.; Qian, X. DC-AL GAN: Pseudoprogression and true tumor progression of glioblastoma multiform image classification based on DCGAN and AlexNet. Med. Phys. 2020, 47, 1139–1150. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Ma, B.; Jiang, P.; Zeng, D.; Wang, X.; Li, S. Prediction of Alzheimer’s disease progression with multi-information generative adversarial network. IEEE J. Biomed. Health Inform. 2020, 25, 711–719. [Google Scholar] [CrossRef] [PubMed]
- Yi, X.; Walia, E.; Babyn, P. Generative adversarial network in medical imaging: A review. Med. Image Anal. 2019, 58, 101552. [Google Scholar] [CrossRef] [PubMed]
- Onishi, Y.; Teramoto, A.; Tsujimoto, M.; Tsukamoto, T.; Saito, K.; Toyama, H.; Imaizumi, K.; Fujita, H. Multiplanar analysis for pulmonary nodule classification in CT images using deep convolutional neural network and generative adversarial networks. Int. J. Comput. Assist. Radiol. Surg. 2020, 15, 173–178. [Google Scholar] [CrossRef] [PubMed]
- Baumgartner, C.F.; Koch, L.M.; Tezcan, K.C.; Ang, J.X.; Konukoglu, E. Visual feature attribution using wasserstein gans. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 8309–8319. [Google Scholar]
- Emami, H.; Dong, M.; Nejad-Davarani, S.P.; Glide-Hurst, C.K. Generating synthetic CTs from magnetic resonance images using generative adversarial networks. Med. Phys. 2018, 45, 3627–3636. [Google Scholar] [CrossRef] [PubMed]
- Vallières, M.; Freeman, C.R.; Skamene, S.R.; El Naqa, I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys. Med. Biol. 2015, 60, 5471–5496. [Google Scholar] [CrossRef] [PubMed]
- Ronneberger, O.; Fischer, P.; Brox, T. U-net: Convolutional networks for biomedical image segmentation. In Medical Image Computing and Computer-Assisted Intervention–MICCAI 2015: 18th International Conference, Munich, Germany, 5–9 October 2015; Proceedings, Part III 18; Springer International Publishing: New York, NY, USA, 2015; pp. 234–241. [Google Scholar]
- Wang, Z.; Bovik, A.C.; Sheikh, H.R.; Simoncelli, E.P. Image quality assessment: From error visibility to structural similarity. IEEE Trans. Image Process. 2004, 13, 600–612. [Google Scholar] [CrossRef]
- Zhang, R.; Isola, P.; Efros, A.A.; Shechtman, E.; Wang, O. The unreasonable effectiveness of deep features as a perceptual metric. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 586–595. [Google Scholar]
- Gulrajani, I.; Ahmed, F.; Arjovsky, M.; Dumoulin, V.; Courville, A.C. Improved training of wasserstein gans. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017. [Google Scholar]
- Wang, Y.; Zhou, C.; Ying, L.; Lee, E.; Chan, H.P.; Chughtai, A.; Hadjiiski, L.M.; Kazerooni, E.A. Leveraging Serial Low-Dose CT Scans in Radiomics-based Reinforcement Learning to Improve Early Diagnosis of Lung Cancer at Baseline Screening. Radiol. Cardiothorac. Imaging 2024, 6, e230196. [Google Scholar] [CrossRef]
- Winter, A.; Aberle, D.R.; Hsu, W. External validation and recalibration of the Brock model to predict probability of cancer in pulmonary nodules using NLST data. Thorax 2019, 74, 551–563. [Google Scholar] [CrossRef] [PubMed]
- Pinsky, P.F.; Gierada, D.S.; Black, W.; Munden, R.; Nath, H.; Aberle, D.; Kazerooni, E. Performance of Lung-RADS in the National Lung Screening Trial: A retrospective assessment. Ann. Intern. Med. 2015, 162, 485–491. [Google Scholar] [CrossRef] [PubMed]
- Bamber, D. The area above the ordinal dominance graph and the area below the receiver operating characteristic graph. J. Math. Psychol. 1975, 12, 387–415. [Google Scholar] [CrossRef]
- Swets, J.A. ROC analysis applied to the evaluation of medical imaging techniques. Investig. Radiol. 1979, 14, 109–121. [Google Scholar] [CrossRef] [PubMed]
- Mann, H.B.; Whitney, D.R. On a test of whether one of two random variables is stochastically larger than the other. Ann. Math. Stat. 1947, 18, 50–60. [Google Scholar] [CrossRef]
- Leening, M.J.G.; Vedder, M.M.; Witteman, J.C.M.; Pencina, M.J.; Steyerberg, E.W. Net reclassification improvement: Computation, interpretation, and controversies: A literature review and clinician’s guide. Ann. Intern. Med. 2014, 160, 122–131. [Google Scholar] [CrossRef] [PubMed]
- DeLong, E.R.; DeLong, D.M.; Clarke-Pearson, D.L. Comparing the areas under two or more correlated receiver operating characteristic curves: A nonparametric approach. Biometrics 1988, 44, 837–845. [Google Scholar] [CrossRef]
- Hochberg, Y. A sharper Bonferroni procedure for multiple tests of significance. Biometrika 1988, 75, 800–802. [Google Scholar] [CrossRef]
- Huang, P.; Lin, C.T.; Li, Y.; Tammemagi, M.C.; Brock, M.V.; Atkar-Khattra, S.; Xu, Y.; Hu, P.; Mayo, J.R.; Schmidt, H. Prediction of lung cancer risk at follow-up screening with low-dose CT: A training and validation study of a deep learning method. Lancet Digit. Health 2019, 1, e353–e362. [Google Scholar] [CrossRef]
- Ardila, D.; Kiraly, A.P.; Bharadwaj, S.; Choi, B.; Reicher, J.J.; Peng, L.; Tse, D.; Etemadi, M.; Ye, W.; Corrado, G. End-to-end lung cancer screening with three-dimensional deep learning on low-dose chest computed tomography. Nat. Med. 2019, 25, 954–961. [Google Scholar] [CrossRef]
- Chung, K.; Mets, O.M.; Gerke, P.K.; Jacobs, C.; den Harder, A.M.; Scholten, E.T.; Prokop, M.; de Jong, P.A.; van Ginneken, B.; Schaefer-Prokop, C.M. Brock malignancy risk calculator for pulmonary nodules: Validation outside a lung cancer screening population. Thorax 2018, 73, 857–863. [Google Scholar] [CrossRef] [PubMed]
- Baldwin, D.R.; Gustafson, J.; Pickup, L.; Arteta, C.; Novotny, P.; Declerck, J.; Kadir, T.; Figueiras, C.; Sterba, A.; Exell, A. External validation of a convolutional neural network artificial intelligence tool to predict malignancy in pulmonary nodules. Thorax 2020, 75, 306–312. [Google Scholar] [CrossRef] [PubMed]
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Dorfman, D.D.; Berbaum, K.S.; Metz, C.E. Receiver operating characteristic rating analysis. Generalization to the population of readers and patients with the jackknife method. Investig. Radiol. 1992, 27, 723–731. [Google Scholar] [CrossRef]
- Abu-Srhan, A.; Abushariah, M.A.M.; Al-Kadi, O.S. The effect of loss function on conditional generative adversarial networks. J. King Saud. Univ. -Comput. Inf. Sci. 2022, 34, 6977–6988. [Google Scholar] [CrossRef]
- Isola, P.; Zhu, J.-Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Hammernik, K.; Knoll, F.; Sodickson, D.K.; Pock, T. L2 or not L2: Impact of loss function design for deep learning MRI reconstruction. In Proceedings of the ISMRM 25th Annual Meeting & Exhibition, Honolulu, HI, USA, 22–27 April 2017; p. 0687. [Google Scholar]


| Characteristic | Dataset (n = 1226) | Training/Validation Set (n = 776) | Test Set (n = 450) | ||||
|---|---|---|---|---|---|---|---|
| Positive (n = 218) | Negative (n = 1008) | Positive (n = 165) | Negative (n = 611) | Positive (n = 53) | Negative (n = 397) | ||
| Age (y), mean ± SD | 63.50 ± 5.09 | 61.89 ± 5.15 | 63.54 ± 4.97 | 61.55 ± 5.14 | 63.38 ± 5.45 | 62.40 ± 5.11 | |
| Gender | Female | 94 (43.12%) | 419 (41.57%) | 71 (43.03%) | 248 (40.59%) | 23 (43.40%) | 171 (43.07%) |
| Male | 124 (56.88%) | 589 (58.43%) | 94 (56.97%) | 363 (59.41%) | 30 (56.60%) | 226 (56.93%) | |
| Race | White | 205 (94.04%) | 939 (93.7%) | 157 (95.15%) | 566 (92.64%) | 48 (90.57%) | 373 (93.95%) |
| Other | 13 (5.96%) | 69 (6.85%) | 8 (4.85%) | 45 (7.36%) | 5 (9.43%) | 24 (6.05%) | |
| Ethnicity | Hispanic/Latino | 0 (0.00%) | 14 (1.39%) | 0 (0.00%) | 9 (1.47%) | 0 (0.00%) | 5 (1.26%) |
| Other | 218 (100%) | 994 (98.61%) | 165 (100%) | 602 (98.53%) | 53 (100%) | 392 (98.74%) | |
| Smoking | Current | 120 (55.05%) | 494 (49.01%) | 92 (55.76%) | 297 (48.61%) | 28 (52.83%) | 197 (49.62%) |
| Former | 98 (44.95%) | 514 (50.99%) | 73 (44.24%) | 314 (51.39%) | 25 (47.17%) | 200 (50.38%) | |
| Smoked, mean ± SD | Packs/yr. | 65.10 ± 26.11 | 56.16 ± 25.06 | 65.07 ± 26.01 | 56.13 ± 25.37 | 65.17 ± 26.44 | 56.21 ± 24.58 |
| Avg/day | 30.00 ± 11.93 | 28.48 ± 1201 | 30.05 ± 11.89 | 28.49 ± 11.63 | 39.85 ± 12.06 | 28.47 ± 12.47 | |
| Years | 43.86 ± 7.00 | 39.93 ± 7.23 | 43.83 ± 6.89 | 39.79 ± 7.23 | 43.96 ± 7.31 | 40.14 ± 7.23 | |
| Family cancer History, N (%) | Positive | 50 (22.94%) | 227 (22.52%) | 39 (23.64%) | 133 (21.77%) | 11 (20.75%) | 94 (23.68%) |
| Medical history | COPD | 16 (7.34%) | 46 (4.56%) | 10 (6.06%) | 28 (4.58%) | 6 (11.32%) | 18 (4.53%) |
| Emphysema | 28 (12.84%) | 79 (7.74%) | 19 (11.52%) | 45 (7.36%) | 9 (16.98%) | 34 (8.56%) | |
| TNM stage | Stage IA | 115 (52.7%) | 81 (49.1%) | 34 (64.1%) | |||
| Stage IB | 26 (11.9%) | 19 (11.5%) | 7 (13.2%) | ||||
| Stage IIA | 13 (6.0%) | 11 (6.7%) | 2 (3.8%) | ||||
| Stage IIB | 9 (4.1%) | 9 (5.5%) | 0 (0.0%) | ||||
| Stage IIIA | 20 (9.2%) | 17 (10.3%) | 3 (5.7%) | ||||
| Stage IIIB | 5 (2.3%) | 4 (2.4%) | 1 (1.9%) | ||||
| Stage IV | 24 (11.0%) | 19 (11.5%) | 5 (9.4%) | ||||
| Other * | 6 (2.8%) | 5 (3.0%) | 1 (1.9%) | ||||
| Histopathological subtype ** | Bronchioloalveolar carcinoma | 40 (18.3%) | 30 (18.2%) | 10 (18.9%) | |||
| Adenocarcinoma | 92 (42.2%) | 63 (38.2%) | 29 (54.7%) | ||||
| Squamous cell carcinoma | 42 (19.3%) | 35 (21.2%) | 7 (13.2%) | ||||
| Large cell carcinoma | 4 (1.8%) | 4 (2.4%) | 0 (0.0%) | ||||
| Non-small cell, other | 21 (9.6%) | 17 (10.3%) | 4 (7.5%) | ||||
| Small cell carcinoma | 16 (7.3%) | 14 (8.5%) | 2 (3.8%) | ||||
| Carcinoid | 2 (0.9%) | 1 (0.6%) | 1 (1.9%) | ||||
| Margins | Spiculated | 60 (27.5%) | 80 (8.0%) | 44 (26.6%) | 42 (6.9%) | 16 (30.2%) | 38 (9.6%) |
| Smooth | 76 (34.9%) | 720 (71.4%) | 60 (36.4%) | 444 (72.7%) | 16 (30.2%) | 276 (69.5%) | |
| Other † | 82 (37.6%) | 208 (20.6%) | 61 (37.0%) | 125 (20.4%) | 21 (39.6%) | 83 (20.9%) | |
| Internal characteristics | Soft Tissue | 148 (67.9%) | 773 (76.7%) | 112 (67.9%) | 475 (77.7%) | 36 (67.9%) | 298 (75.1%) |
| Ground glass | 42 (19.3%) | 130 (12.9%) | 33 (20.0%) | 74 (12.1%) | 9 (17.0%) | 56 (14.1%) | |
| Mixed | 19 (8.7%) | 55 (5.4%) | 14 (8.5%) | 23 (3.8%) | 5 (9.4%) | 22 (5.5%) | |
| Other †† | 9 (4.1%) | 50 (5.0%) | 6 (3.6%) | 39 (6.4%) | 3 (5.7%) | 21 (5.3%) | |
| Model\Criteria | All Nodule | Solid Nodule | Spiculated Nodule | Diameter 6 to 14 mm | ||
|---|---|---|---|---|---|---|
| n (Benign + Malignant) | 397 + 53 | 298 + 36 | 38 + 16 | 267 + 38 | ||
| LCRP + GP-nodule | AUC | 0.827 ± 0.028 | 0.828 ± 0.037 | 0.850 ± 0.055 | 0.782 ± 0.041 | |
| 95% CI | (0.772, 0.883) | (0.762, 0.895) | (0.743, 0.958) | (0.702, 0.862) | ||
| LCRP + real follow-up | AUC | 0.862 ± 0.028 | 0.864 ± 0.034 | 0.922 ± 0.037 | 0.826 ± 0.039 | |
| 95% CI | (0.806, 0.917) | (0.797, 0.931) | (0.848, 0.995) | (0.750, 0.902) | ||
| p-value | LCRP + GP-nodule | 0.071 | 0.091 | 0.150 | 0.077 | |
| LCRP + real baseline | 0.024 | 0.020 | 0.018 | 0.018 | ||
| Brock model | <0.001 | 0.002 | <0.001 | 0.006 | ||
| LCRP + real baseline | AUC | 0.805 ± 0.031 | 0.793 ± 0.039 | 0.793 ± 0.067 | 0.749 ± 0.045 | |
| 95% CI | (0.744, 0.866) | (0.716, 0.870) | (0.661, 0.924) | (0.660, 0.838) | ||
| p-value | LCRP + GP-nodule | 0.099 | 0.058 | 0.105 | 0.083 | |
| Brock model | 0.146 | 0.249 | 0.007 | 0.201 | ||
| Brock model + real baseline | AUC | 0.754 ± 0.035 | 0.768 ± 0.040 | 0.595 ± 0.080 | 0.704 ± 0.042 | |
| 95% CI | (0.686, 0.823) | (0.690, 0.846) | (0.439, 0.751) | (0.621, 0.787) | ||
| p-value | LCRP + GP-nodule | 0.043 | 0.045 | <0.001 | 0.048 | |
| Risk groups (Number of subjects, %) | Reclassified by LCRP model + GP-nodules | |||||
| Stratify real nodules | Low | Medium | High | |||
| Cancer (N = 53) | (N = 3, 5.66%) | (N = 21, 39.62%) | (N = 29, 54.72%) | |||
| Negative (N = 397) | (N = 164, 41.31%) | (N = 189, 47.61%) | (N = 44, 11.08%) | |||
| Lung-RADS at baseline | Cancer (N = 53) | Low (N = 12, 22.64%) | 2 | 5 + | 5 + | event NRI = 0.15 p = 0.08 |
| Medium (N = 15, 28.30%) | 0 − | 11 | 4 + | |||
| High (N = 26, 49.06%) | 1 − | 5 − | 20 | |||
| NRI = 0.38, p < 0.001 | Negative (N = 397) | Low (N = 144, 36.27%) | 89 | 46 + | 9 + | nonevent NRI = 0.24 p < 0.001 |
| Medium (N = 136, 34.26%) | 67 − | 67 | 2 + | |||
| High (N = 117, 29.47%) | 8 − | 76 − | 33 | |||
| Brock at baseline | Cancer (N = 53) | low (N = 5, 9.43%) | 1 | 4 + | 0 + | event NRI = 0.19 p = 0.02 |
| Medium (N = 27, 50.95%) | 2 − | 14 | 11 + | |||
| High (N = 21, 39.62%) | 0 − | 3 − | 18 | |||
| NRI = 0.20, p = 0.03 | Negative (N = 397) | Low (N = 162, 40.81%) | 105 | 57 + | 0 + | nonevent NRI = 0.01 p = 0.75 |
| Medium (N = 183, 46.10%) | 54 − | 110 | 19 + | |||
| High (N = 52, 13.10%) | 5 − | 22 − | 25 | |||
| LCRP at baseline | Cancer (N = 53) | Low (N = 5, 9.43%) | 3 | 2 + | 0 | event NRI = 0.23 p < 0.001 |
| Medium (N = 29, 54.72%) | 0 − | 19 | 10 + | |||
| High (N = 19, 35.85%) | 0 − | 0 − | 19 | |||
| NRI = 0.20, p = 0.004 | Negative (N = 397) | Low (N = 158, 39.80%) | 136 | 22 + | 0 + | nonevent NRI = −0.03 p = 0.16 |
| Medium (N = 213, 53.65%) | 28 − | 165 | 20 + | |||
| High (N = 26, 6.55%) | 0 − | 2 − | 24 | |||
| LCRP at 1-year follow-up | Cancer (N = 53) | Low (N = 3, 5.66%) | 2 | 1 + | 0 + | event NRI = 0.04 p = 0.60 |
| Medium (N = 23, 43.40%) | 1 − | 14 | 8 + | |||
| High (N = 27, 50.94%) | 0 − | 6 − | 21 | |||
| NRI = 0.04, p = 0.62 | Negative (N = 397) | Low (N = 155, 39.05%) | 123 | 32 + | 0 + | nonevent NRI = −0.003 p = 0.91 |
| Medium (N = 208, 52.39%) | 41 − | 149 | 18 + | |||
| High (N = 34, 8.56%) | 0 − | 8 − | 26 | |||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.; Zhou, C.; Ying, L.; Chan, H.-P.; Lee, E.; Chughtai, A.; Hadjiiski, L.M.; Kazerooni, E.A. Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model. Cancers 2024, 16, 2229. https://doi.org/10.3390/cancers16122229
Wang Y, Zhou C, Ying L, Chan H-P, Lee E, Chughtai A, Hadjiiski LM, Kazerooni EA. Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model. Cancers. 2024; 16(12):2229. https://doi.org/10.3390/cancers16122229
Chicago/Turabian StyleWang, Yifan, Chuan Zhou, Lei Ying, Heang-Ping Chan, Elizabeth Lee, Aamer Chughtai, Lubomir M. Hadjiiski, and Ella A. Kazerooni. 2024. "Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model" Cancers 16, no. 12: 2229. https://doi.org/10.3390/cancers16122229
APA StyleWang, Y., Zhou, C., Ying, L., Chan, H.-P., Lee, E., Chughtai, A., Hadjiiski, L. M., & Kazerooni, E. A. (2024). Enhancing Early Lung Cancer Diagnosis: Predicting Lung Nodule Progression in Follow-Up Low-Dose CT Scan with Deep Generative Model. Cancers, 16(12), 2229. https://doi.org/10.3390/cancers16122229






